Abstract
In a screen for mutants that display synthetic lethal interaction with hpr1Δ, a hyperrecombination mutant of Saccharomyces cerevisiae, we have isolated a novel cold-sensitive allele of the acetyl coenzyme A (CoA) carboxylase gene, acc1cs, encoding the rate-limiting enzyme of fatty acid synthesis. The synthetic lethal phenotype of the acc1cs hpr1Δ double mutant was only partially complemented by exogenous fatty acids. hpr1Δ was also synthetically lethal with a previously isolated, temperature-sensitive allele of ACC1, mtr7 (mRNA transport), indicating that the lethality of the acc1cs hpr1Δ double mutant was not allele specific. The basis for the interaction between conditional acc1 alleles and hpr1Δ was investigated in more detail. In the hpr1Δ mutant background, acetyl-CoA carboxylase enzyme activity was reduced about 15-fold and steady-state levels of biotinylated Acc1p and ACC1 mRNA were reduced 2-fold. The reduced Acc1p activity in hpr1Δ cells, however, did not result in an altered lipid or fatty acid composition of the mutant membranes but rendered cells hypersensitive to soraphen A, an inhibitor of Acc1p. Similar to mtr7, hpr1Δ and acc1cs mutant cells displayed a defect in nuclear export of polyadenylated RNA. Oversized transcripts were detected in hpr1Δ, and rRNA processing was disturbed, but pre-mRNA splicing appeared wild type. Surprisingly, the transport defect of hpr1Δ and acc1cs mutant cells was accompanied by an altered ring-shaped structure of the nucleolus. These observations suggest that the basis for the synthetic lethal interaction between hpr1Δ and acc1 may lie in a functional overlap of the two mutations in nuclear poly(A)+ RNA production and export that results in an altered structure of the nucleolus.
The hpr1Δ mutant of Saccharomyces cerevisiae was isolated in a screen for mutations that confer an increased mitotic recombination (1, 2). The hpr1Δ null mutant is temperature sensitive for growth at 37°C and displays a 700-fold-elevated rate of mitotic intrachromatid recombination. Hpr1p has two regions of homology to topoisomerase I, Top1p (3, 40), and hpr1Δ mutants display synthetic lethality with mutations in all three DNA topoisomerase genes, TOP1, TOP2, and TOP3 (3). A fourth synthetic lethal interaction has been found between hpr1Δ and a mutant carrying a deletion of one copy of the histone H3-H4 genes (10).
In hpr1Δ null mutants, transcription of many physiologically unrelated genes is affected (44) and the temperature-sensitive growth phenotype of hpr1Δ mutants is suppressed by mutations in components of the general transcription machinery (12, 27, 41). The Hpr1 protein has been found to be in a distinct RNA polymerase II complex (8) and has been suggested to have a functional role in transcription elongation (9).
To better understand the in vivo function of Hpr1p, a screen was initiated to isolate additional mutants that exhibit synthetic lethal interaction with hpr1Δ. This screen yielded a novel cold-sensitive allele of the acetyl coenzyme A (CoA) carboxylase gene, acc1-200cs, hereafter referred to as acc1cs (14).
The acetyl-CoA carboxylase gene, ACC1, encodes a biotin-containing enzyme that synthesizes malonyl-CoA from acetyl-CoA and bicarbonate, with the hydrolysis of ATP (4). Acc1p is the rate-limiting enzyme of the de novo fatty acid biosynthetic pathway. Expression of the ACC1 gene is under coordinate transcriptional regulation by the phospholipid precursors inositol and choline (15). A temperature-sensitive allele of ACC1, mtr7, has been isolated in a screen for mutants that affect nuclear export of polyadenylated RNA (18; for a review, see reference 39). The nuclear transport defect of this acc1ts allele has been proposed to be due to a special lipid requirement of the nuclear membrane-nuclear pore complex (31, 33).
The basis for the synthetic lethality between hpr1Δ and the cold-sensitive acc1cs allele was investigated in more detail. We find that hpr1Δ mutant cells have a very strong defect in export of nuclear poly(A)+ RNA, a phenotype previously observed in a temperature-sensitive allele of ACC1 (31), and propose that the lethal interaction between hpr1Δ and acc1cs is due to a combined effect of the two mutations on nuclear export of polyadenylated RNA.
MATERIALS AND METHODS
Yeast strains and culture conditions.
The S. cerevisiae strains used for these experiments are listed in Table 1. Strains were grown in YEPD medium (1% yeast extract, 2% Bacto Peptone, 2% glucose) or synthetic minimal medium (35) supplemented with appropriate amino acids and glucose. Medium supplemented with fatty acids contained 1% Tween 40, 0.015% palmitic acid, and 0.015% stearic acid (28). Soraphen A, a kind gift from A. Hinnen, was added to medium from a 10-mg/ml stock solution in methanol. Optical density at 600 nm was monitored every hour for growth rate determinations. The exponential growth rate in the presence of soraphen A was established after a 3-h lag period and was expressed as a percentage of the growth rate in the absence of the inhibitor.
TABLE 1.
Yeast strain genotypes and construction
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| 479-2A | MATα acc1cs leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | H. Klein |
| 485-13A | MATa acc1cs leu2-3,112 ura3-1 ade2-1 his3-11,15 can1-100 | H. Klein |
| U768-1C | MATα hpr1Δ3::HIS3 leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | R. Rothstein |
| U768-4C | MATa hpr1Δ3::HIS3 leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 | R. Rothstein |
| W303d | MATa/MATα leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 ade2-1/ade2-1 his3-11,15/his3-11,15 can1-100/can1-100 | R. Rothstein |
| YRXS12 | MATα acc1ts leu2-Δ1 ura3-52 ade2-101 lys2-801 his3-Δ200 | Schneiter et al. (31) |
| YRXS300C-1 | MATa hpr1Δ3::HIS3 leu2 TRP1 ura3 ade2 his3 lys2-801 | This work; spore from cross between U768-4C and YRXS12 |
| YRXS300C-2 | MATα hpr1Δ3::HIS3 acc1ts leu2 trp1 ura3 ade2 his3 lys2-801 | This work; spore from cross between U768-4C and YRXS12 |
| YRXS300C-3 | MATa leu2 TRP1 ura3 ade2 his3 | This work; spore from cross between U768-4C and YRXS12 |
| YRXS300C-4 | MATα acc1ts leu2 trp1 ura3 ade2 his3 | This work; spore from cross between U768-4C and YRXS12 |
Isolation and analysis of RNA.
Total yeast RNA was extracted from exponentially growing cultures as described previously (29). Ten milligrams of RNA per well was fractionated by electrophoresis through a 0.8% agarose–3.7% formaldehyde gel and prepared for Northern blot analysis as described previously (29). For quantification of ACC1 transcription, 32P-labeled DNA probes were prepared according to the method in reference 13. Blots were serially hybridized to ACC1-specific and ACT1-specific DNA probes, in that order and without an intermediate probe-stripping step. After autoradiography of the blots, each lane in the exposed film was scanned in one dimension with the UltroScan XL system (LKB), and the ACC1/ACT1 ratios of peak areas were calculated relative to the ratio in the MATa wild-type strain. For detection of CRY1 message, filters were probed with digoxigenin-labeled probe and signal was detected by enhanced chemiluminescence with CDP-Star substrate (Tropix, Bedford, Mass.). For the analysis of rRNA processing, mutant cells were transformed with pRS316 (36) to make them uracil prototrophic. Cells were incubated at nonpermissive temperatures for 1 h and then labeled with 0.05 mCi of [3H]uridine per ml for 10 min.
Immunofluorescence and in situ hybridization.
Early-logarithmic-phase cells were prepared for immunofluorescence microscopy as previously described (31). Mouse monoclonal antibody against the yeast fibrillarin homologue, Nop1p, was applied at a 1:5,000 dilution (6). Secondary fluorescein isothiocyanate-conjugated antibody (Oncogene Science, Cambridge, Mass.) was used at a 1:200 dilution. Detection of accumulated poly(A)+ RNA was performed essentially as described elsewhere (31), with the modification that digoxigenin-labeled oligo(dT)25–30 was used for hybridization. Secondary fluorescein isothiocyanate-conjugated anti-sheep antibody was used at a 1:200 dilution. Laser scanning microscopic analysis was performed with a Leica TCS 4d confocal microscope equipped with a PL APO 100×/1.40 objective, and photographs were recorded on Ektachrome 100 film (Eastman Kodak Co., Rochester, N.Y.). Corresponding pictures were recorded with identical pinhole openings and amplification settings and were printed with the same exposure time.
Western blot analysis.
Whole-cell protein extracts were prepared as described elsewhere (43), and protein concentration was determined according to the method of Lowry et al. (22), with bovine serum albumin as a standard. Proteins were separated by one-dimensional sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (20), with 4% stacking and 10% separating gels. Proteins were transferred to nitrocellulose sheets (Hybond C; Amersham) and stained with Ponceau S to assess transfer efficiency. Membranes were incubated overnight in blocking solution containing 5% fat-free dry milk powder in Tris-buffered saline (TBS) (50 mM Tris-HCl, 150 mM NaCl, pH 8.0). After two washes with TTBS (0.1% Tween 20 in TBS), membranes were incubated with 10 μg of avidin-peroxidase conjugate (ExtrAvidin peroxidase; Sigma, St. Louis, Mo.), rabbit anti-Acc1p (1:700 [17]), or rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:5,000) per ml, diluted in TBS. Peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma) was used as secondary antibody. Signal detection was carried out according to the instructions provided by the manufacturer with the ECL system (Amersham) and the SuperSignal CL-HRP substrate system (Pierce, Rockford, Ill.). Densitometric scanning of X-ray films and quantification of signals were carried out with NIH Image 1.54 software.
Acc1p activity determination.
The cytosolic fraction used for Acc1p enzyme activity assays was prepared as follows. Cells were harvested at 1,200 × g for 10 min, washed with 0.1 M K2HPO4-KH2PO4 buffer (pH 6.5), mixed with breaking buffer (50 mM Tris-HCl, 100 mM NaF, 1 mM EDTA, 10 mM β-mercaptoethanol, 0.25 M sucrose, 1 mM phenylmethylsulfonyl fluoride, pH 7.5) and glass beads (0.30-mm diameter) in a ratio of 1:1:1 (wt/vol/wt), and disrupted by four 1-min bursts in a Braun-Melsungen homogenizer under CO2 cooling. Glass beads were collected by centrifugation at 3,000 × g for 5 min, and the supernatant was centrifuged at 20,000 × g for 20 min and centrifuged again at 195,000 × g for 80 min. Saturated (NH4)2SO4 was then added in three portions within 20 min to 50% saturation, and samples were stirred for an additional 30 min. The precipitate was collected by centrifugation at 15,000 × g, dissolved in HEPES buffer (50 mM HEPES, 1 mM EDTA, 0.02% Na-azide, 50% glycerol, pH 7.0), and stored frozen at −20°C. Enzymatic activities of Acc1p were found to be stable over a period of 3 weeks after freezing of samples. All steps were carried out at 4°C. Activity of acetyl-CoA carboxylase was determined by a photometric assay in a coupled enzymatic reaction as described elsewhere (23). All enzyme measurements were carried out at 24°C.
RESULTS
Synthetic lethality between acc1cs and hpr1Δ.
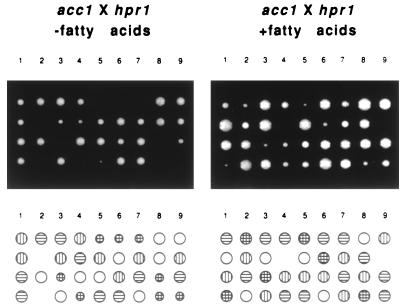
The acc1cs acc1-200cs allele was recovered in a screen for mutants that display a synthetic lethal interaction with a null allele of HPR1 (14). The synthetic lethality of the acc1cs hpr1Δ double mutant is indicated by the failure to recover spore colonies of this genotype from a cross between an acc1cs strain and an hpr1Δ strain. When the tetrads from this cross were dissected on a YEPD plate supplemented with fatty acids, the acc1cs hpr1Δ double mutant was viable but still showed reduced growth compared to the single mutant strain or the wild-type strain (Fig. 1).
FIG. 1.
acc1cs is synthetically lethal with hpr1Δ. Growth of spore segregants from an acc1cs × hpr1Δ cross. Asci were dissected on YEPD medium alone or YEPD medium supplemented with fatty acids and incubated for 3 days at 30°C. Germinated spores from each tetrad are arranged in vertical columns (1 to 9), and the corresponding genotypes are indicated by circles below as follows: wild-type spores, open circles; acc1cs spores, vertically striped circles; hpr1Δ spores, horizontally striped circles; and double mutant spores, crosshatched circles.
Soraphen A sensitivity of acc1cs and hpr1Δ mutants.
Soraphen A is a potent inhibitor of Acc1p activity in yeast (42). The acc1cs allele was found to confer hypersensitivity to soraphen A at the permissive temperature (Fig. 2A). Since the acc1cs allele is synthetically lethal with the hpr1Δ mutation, we examined the hpr1Δ mutant for soraphen A sensitivity. The hpr1Δ null mutant was found to be as soraphen A sensitive as the acc1cs mutant on solid medium (Fig. 2A) and in liquid medium (Fig. 2B).
FIG. 2.
Soraphen A sensitivity of the hpr1Δ mutant. (A) Strains with indicated genotypes were streaked onto YEPD medium (top) and YEPD medium containing 0.25 mg of soraphen A per ml (bottom) and incubated for 2 days at 30°C. wt, wild type. (B) Dose-response curves to soraphen A. The exponential growth rate was established after a 3-h lag period for various concentrations of soraphen A and is expressed as a percentage of the growth rate in the absence of soraphen A. The results for two strains of each genotype were averaged and plotted against the concentration of soraphen A. Curves are the best fit calculated with respect to Hill’s equation (Hill coefficients, 3 to 4). Calculated 50% infective doses are 0.09 mg/ml for the wild type and 0.04 mg/ml for both the acc1cs and the hpr1Δ strains.
The synthetic lethal interaction between hpr1Δ and acc1 is not allele specific.
A temperature-sensitive allele of ACC1, mtr7, herein referred to as acc1ts, had previously been isolated in a screen for conditional mRNA transport mutants (31). To determine whether the synthetic lethal interaction between acc1cs and hpr1Δ is an allele-specific phenomenon, a cross between acc1ts and hpr1Δ was performed. Diploids were sporulated, and tetrads were dissected on YEPD and YEPD plates supplemented with 1 M sorbitol. Sorbitol supplementation has been found to rescue the temperature-dependent growth phenotype of both conditional acc1 alleles (data not shown). While acc1ts hpr1Δ double mutants were viable on sorbitol-supplemented medium, no acc1ts hpr1Δ double mutants were recovered from YEPD or fatty acid-supplemented plates, indicating that acc1ts is synthetically lethal with hpr1Δ (Fig. 3).
FIG. 3.
acc1ts is synthetically lethal with hpr1Δ. An acc1ts strain (YRXS12) was crossed with an hpr1Δ strain (U768-4C), diploids were sporulated, and tetrads were dissected. Four spores of a complete tetrad were replica plated on YEPD plates (top), YEPD supplemented with fatty acids (middle), and YEPD plates supplemented with 1 M sorbitol (bottom) and incubated at 30°C for 3 days. The relevant genotype of the spores is indicated on top. wt, wild type.
hpr1Δ affects Acc1p activity at the level of transcription.
The observation that hpr1Δ is synthetically lethal with two different conditional acc1 alleles and that hpr1Δ itself is hypersensitive to soraphen A suggested that Acc1p activity is affected in the hpr1Δ mutant. Acc1p enzymatic activity was determined in cytosolic fractions from the wild type and the hpr1Δ mutant strain. Cells were grown in YEPD medium at 30°C, cytosol was prepared, and Acc1p activity was determined as described previously (23). Acc1p activity in the hpr1Δ mutant was reduced approximately 15-fold compared to that in the wild type, as shown by the following data (means ± standard deviations of three determinations): enzyme activity of wild type, 38.3 × 10−5 ± 3.7 × 10−5 ΔE/min/μg; enzyme activity of the hpr1Δ mutant, 2.6 × 10−5 ± 1.7 × 10−5 ΔE/min/μg. This greatly reduced Acc1p activity in the hpr1Δ mutant background, however, did not result in an altered lipid or fatty acid composition of hpr1Δ mutant cells compared to the wild type (data not shown).
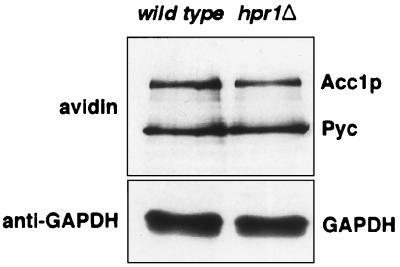
To determine at which level the lack of Hpr1p function affects the activity of Acc1p, we analyzed steady-state levels of biotinylated Acc1p. Protein was isolated from whole cells and blotted with peroxidase-labeled avidin to simultaneously detect biotinylated Acc1p and the two pyruvate carboxylase isoenzymes Pyc1p and Pyc2p (Fig. 4). To control for internal loading, GAPDH was detected by an anti-GAPDH antibody. Signal intensities of these blots were quantified, and ratios of biotinylated Acc1p to biotinylated Pyc are listed in Table 2. This analysis revealed that the percentage of biotinylated Acc1p relative to biotinylated Pyc in the hpr1Δ mutant was 50% of that of the wild type.
FIG. 4.
Western blot analysis of biotinylated Acc1p levels in wild-type and hpr1Δ cells. Equal amounts of protein (10 μg) isolated from a whole-cell lysate were separated by SDS-gel electrophoresis, blotted, and probed with peroxidase-labeled avidin to detect biotinylated Acc1p and pyruvate carboxylase (Pyc) and anti-GAPDH or anti-Acc1p antibodies. Signal intensities were quantified and are listed in Table 2.
TABLE 2.
Biotinylated levels of Acc1 protein in wild-type and hpr1Δ cellsa
| Strain | Ratio (%)
|
||
|---|---|---|---|
| Pyc/Acc | GAPDH/Acc | GAPDH/Pyc | |
| Wild type | <1.4 (100) | <2.7 (100) | <1.9 (100) |
| hpr1Δ mutant | 2.2 (50) | 3.5 (77) | 1.6 (119) |
Cells were grown in YEPD medium at 30°C; whole-cell protein was prepared; and equal amounts of protein were separated by SDS-gel electrophoresis, blotted, and probed with peroxidase-labeled avidin to detect biotinylated Acc1p and pyruvate carboxylase (Pyc1/2p) and an anti-GAPDH antibody to control for loading. Signal intensities were quantified densitometrically, and ratios are given. Percentages are relative to the ratio seen in wild-type cells.
Preliminary results with a plasmid-borne ACC1-lacZ fusion as a reporter gene indicated that ACC1 expression was reduced twofold in hpr1Δ strains compared to the wild type (data not shown). To confirm that expression of the chromosomal ACC1 gene was affected in hpr1Δ strains, Northern blot analysis was carried out with total RNA. The relative levels of steady-state ACC1 mRNA were determined by normalization to ACT1 mRNA. For each genotype, the results from two strains of opposite mating type and from two separate experiments were combined. The average ACC1 mRNA level ± standard deviation (n = 4) is shown as a percentage of the wild-type value. The percentages were as follows: wild type, 100.0 ± 24.0; hpr1Δ mutant, 45.4 ± 3.8; acc1cs mutant, 90.7 ± 18.5. No significant difference was found in the levels of ACC1 message between the acc1cs mutant and the wild type. In contrast, ACC1 expression was reduced twofold in the hpr1Δ mutant compared to the wild type. These data indicate that the reduced Acc1p activity in the hpr1Δ mutant is partially due to a diminished level of steady-state ACC1 message made in the absence of Hpr1p and accounts for about half of the reduction in Acc1p activity. The fact that the biotinylated levels of Acc1p in the hpr1Δ mutant are also only 50% of the wild-type level indicates that transport and stability of the ACC1 message are not further reduced. Most likely, there is some posttranslational process that is altered in the hpr1Δ mutant that further reduced that Acc1p activity.
Cells lacking Hpr1p have recently been shown to be defective in transcriptional elongation, but transcription initiation or activation appears not to be affected (9). ACC1 is an unusually large yeast gene of approximately 7 kb. We thus reasoned that the combination of a defect in transcriptional elongation and a mutation in a large gene might cause the synthetic lethality between hpr1Δ and acc1. Conditional acc1 alleles were therefore tested for hypersensitivity towards the transcriptional elongation inhibitor 6-azauracil (5), to see if this would mimic the hpr1Δ acc1 synthetic lethality. Conditional acc1 mutants were found to be as sensitive towards 6-azauracil as were wild-type cells (data not shown). Thus, the synthetic lethal interaction between hpr1Δ and acc1 was not due to the reduced transcriptional elongation in the hpr1Δ mutant background.
hpr1Δ and acc1cs affect nuclear export of polyadenylated RNA.
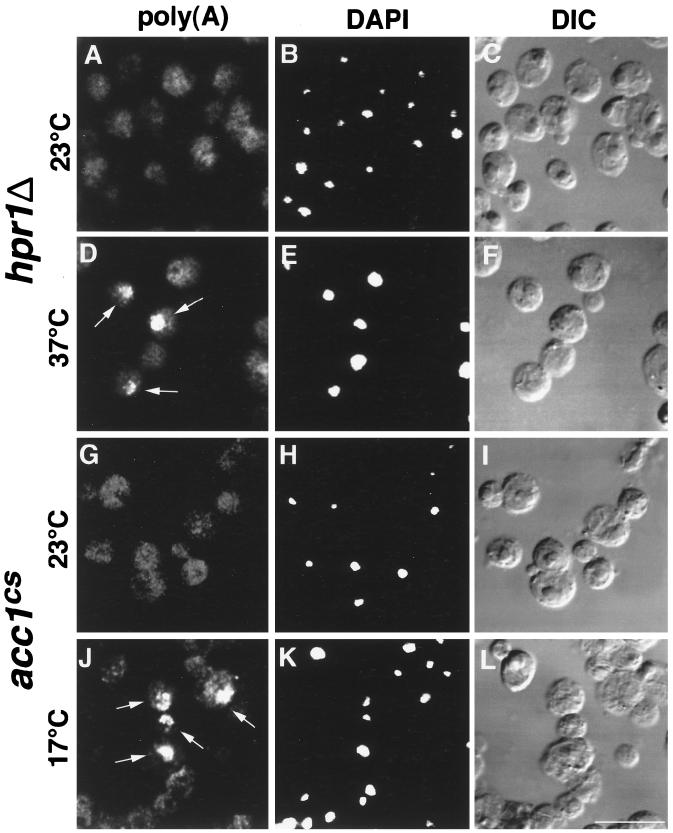
The temperature-sensitive allele of acc1, mtr7, has been isolated in a screen for mutants that affect mRNA transport (mtr [18]; for a review, see reference 39). This function of Acc1p in nuclear export of RNA is not yet fully understood, but it has been suggested that acyl chain elongation and hence the synthesis of the C26:0 very-long-chain fatty acid are required for a functional nuclear membrane-nuclear pore complex (31, 33). We thus investigated whether the synthetic lethal interaction between hpr1Δ and the conditional acc1 alleles could be due to a defect of hpr1Δ in nuclear export of polyadenylated RNA. In situ hybridization with a digoxigenin-labeled oligo(dT) probe revealed a very strong defect of hpr1Δ cells in exporting nuclear poly(A)+ RNA at the nonpermissive growth temperature of 37°C in approximately 30% of all cells. A similar albeit much weaker (comparable to that of acc1ts) defect in mRNA export was also observed in acc1cs cells (Fig. 5).
FIG. 5.
hpr1Δ and acc1cs cells are defective in nuclear export of polyadenylated RNA. Nuclear accumulation of polyadenylated RNA in hpr1Δ and acc1cs cells at the nonpermissive temperature is shown. Cells were grown to early logarithmic phase at 23°C and shifted to 37 and 17°C, respectively, for 4 h, and fixed and processed for in situ hybridization with a digoxigenin-labeled oligo(dT)25–30 probe. Strong accumulation of nuclear poly(A)+ RNA was visible in hpr1Δ and acc1cs cells shifted to nonpermissive temperatures (arrows in panels D and J) but not in cells grown at permissive temperatures (A and G). DAPI (4′,6-diamidino-2-phenylindole) staining and differential interference contrast (DIC) pictures of the same visual field are shown to the right. Bar, 10 μm.
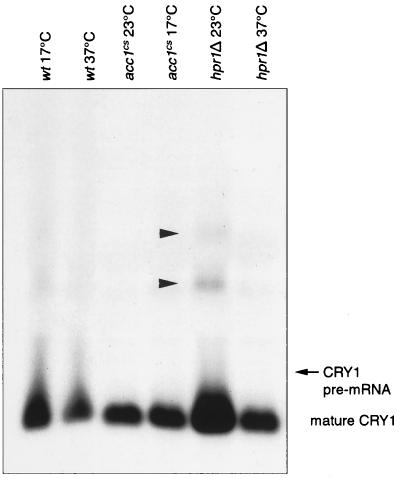
To characterize the transport defect in more detail, we analyzed pre-mRNA splicing and mRNA processing (transcription initiation and 3′-end formation) in hpr1Δ and acc1cs mutant cells. Equal amounts of total RNA isolated from cells incubated at permissive and nonpermissive temperatures were subjected to Northern blot analysis with CRY1, which codes for a ribosomal protein (21), as a probe (Fig. 6). As has previously been described for a number of temperature-sensitive mutants that block nuclear export of polyadenylated RNA (e.g., mtr1, mtr3, mtr4, and mtr17 [18]), the synthesis of oversized CRY1 transcripts was observed for hpr1Δ cells. Oversized transcripts were not detected in any of the other lanes shown in Fig. 6 even on overexposure of the blot (data not shown). Detection of aberrant transcripts confirms the previously proposed function of Hpr1p in some steps of transcription. Similar to acc1ts (mtr7 [18]), acc1cs did not affect CRY1 processing.
FIG. 6.
Northern analysis of CRY1 mRNA processing in wild-type, hpr1Δ, and acc1cs cells. Ten micrograms of total RNA isolated from cells grown at the permissive temperature (23°C) or shifted for 4 h to a nonpermissive temperature (37 or 17°C) was analyzed by Northern hybridization with CRY1 as a probe. The position of mature CRY1 mRNA (540 bases) is shown. The position of CRY1 pre-mRNA (847 bases) is indicated by the arrow. Oversized transcripts detected in hpr1Δ cells are indicated by the arrowheads pointing to the right. wt, wild type.
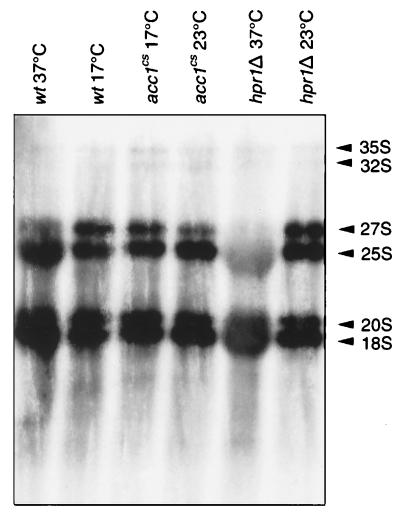
To determine whether the observed block in mRNA export also affects processing of rRNA, mutant strains were incubated at either permissive or nonpermissive temperatures and then pulse-labeled with [3H]uridine for 10 min at the same temperature. The efficiency of labeling in hpr1Δ was greatly reduced at 37°C (2.6%) compared to 23°C, suggesting that in the hpr1Δ mutant either the synthesis of RNA, [3H]uridine uptake, or [3H]UTP synthesis is decreased at nonpermissive temperatures. To normalize this effect, the labeled RNA samples analyzed on agarose gels each were loaded with equal amounts of radioactivity. As shown in Fig. 7, rRNA processing appeared to be disturbed in the hpr1Δ, but not the acc1cs, mutant. The labeling of all rRNA species was greatly reduced in the hpr1Δ mutant.
FIG. 7.
rRNA processing in wild-type, hpr1Δ, and acc1cs strains. Strains were preincubated for 1 h at permissive or nonpermissive temperatures and pulse-labeled for 10 min with [3H]uridine. RNA was isolated, and equal amounts of incorporated radioactivity were loaded on a denaturing agarose gel. The positions of 35, 32, 27, 25, 20, and 18S rRNA are shown by arrowheads. wt, wild type.
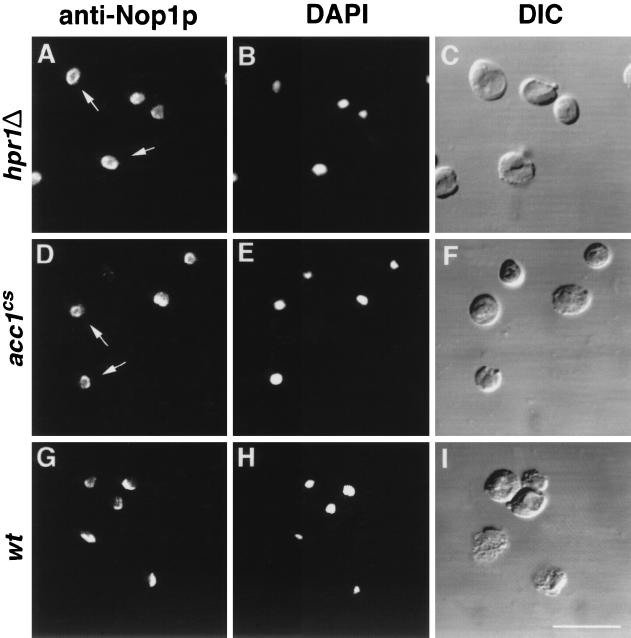
hpr1Δ and acc1cs cells display an aberrant ring-shaped nucleolus.
The yeast nucleolus is a crescent-shaped structure that makes extensive contact with the nuclear envelope. A defect in nuclear export of polyadenylated RNA is frequently accompanied by an altered structure of the nucleolus, i.e., fragmentation and/or enlargement (18, 32). We thus analyzed the structure of the nucleolus in hpr1Δ and acc1cs mutant cells by immunofluorescence microscopy with an antibody against an abundant nucleolar protein, Nop1p (6). As shown in Fig. 8, at permissive temperatures the nucleolus in hpr1Δ and acc1cs cells displayed a ring-shaped structure rather than the typical crescent-shaped structure seen in wild-type cells. This ring-like structure collapses into a normal-looking, crescent-shaped nucleolus upon shifting of hpr1Δ and acc1cs cells to nonpermissive temperatures (data not shown).
FIG. 8.
Ring-shaped structure of the nucleolus in hpr1Δ and acc1cs cells. Shown is immunofluorescence localization of the nucleolar antigen Nop1p in hpr1Δ, acc1cs, and wild-type cells. Strains were cultivated in YEPD to early logarithmic growth phase and fixed and processed for immunofluorescence detection of Nop1p as described in Materials and Methods (A, D, and G). Arrows in panels A and D point to ring-shaped nucleoli. DAPI (4′,6-diamidino-2-phenylindole) staining (B, E, and H) and differential interference contrast (DIC) pictures (C, F, and I) of the same visual field are shown to the right. Bar, 10 μm. wt, wild type.
DISCUSSION
We have described a synthetic lethal interaction between a loss-of-function allele of HPR1 and two conditional alleles of ACC1, encoding yeast acetyl-CoA carboxylase. Acc1p is an essential cytoplasmic enzyme that catalyzes the rate-limiting step of de novo synthesis of fatty acids. In the hpr1Δ mutant background, Acc1p activity was reduced approximately 15-fold. This reduced activity did not result in gross alterations of the lipid or fatty acid composition of the hpr1Δ mutant but rendered the mutant hypersensitive to soraphen A, an inhibitor of Acc1p activity. Analysis of steady-state levels of biotinylated Acc1p and ACC1 mRNA revealed that levels of biotinylated Acc1p and ACC1 transcripts were reduced twofold in the absence of Hpr1p, indicating that the lack of Hpr1p affected cellular Acc1p activity already at the level of transcription.
Several lines of evidence suggested that the synthetic lethal interaction of hpr1Δ with conditional acc1 alleles was not simply due to the combination of a twofold reduction of ACC1 transcription with an enzymatically challenged mutant Acc1p allele. First, like other genes encoding lipid biosynthetic enzymes, ACC1 transcription is under the regulatory control of the transcriptional activators Ino2p and Ino4p. In the absence of Ino2p or Ino4p, ACC1 transcription is reduced two- to threefold (15). acc1cs ino2 or acc1cs ino4 double mutants, grown in the presence of 11 μM inositol to rescue the inositol auxotrophy, however, did not show any reduced growth compared to the acc1cs mutant alone (data not shown). Moreover, the hpr1Δ mutation did not result in an inositol auxotrophic phenotype or a reduction in INO1 mRNA levels (10), indicating that HPR1 does not function like Ino2p and Ino4p in coordinate regulation by inositol (26). Second, ACC1 transcription was reduced twofold in a top1Δ mutant background (data not shown). Unlike hpr1Δ, however, top1Δ was not synthetically lethal with acc1cs or acc1ts. As is the case for the ino2 and ino4 mutants, a twofold reduction of acc1cs levels in a top1Δ mutant is thus not sufficient to cause synthetic lethality. These data argue that the interaction between hpr1Δ and conditional acc1 alleles is characteristic for the way that hpr1Δ affects ACC1 expression and that the synthetic lethality is not solely due to a twofold reduction of ACC1 transcription in the hpr1Δ mutant background.
The phenotypes that have been studied for hpr1Δ strains are all related to nuclear events: hyperrecombination of direct repeats in chromosomal DNA (3), synthetic lethality with a mutant allele of DNA topoisomerase genes (3), or a mutation in one of the histone genes (10). The temperature-sensitive growth of hpr1Δ is suppressed by mutations in SOH genes that play a role in transcription (10). Some of the soh mutants suppress the soraphen A sensitivity of hpr1Δ and the synthetic phenotype of the acc1cs hpr1Δ double mutant (11). The presence of a nuclear localization sequence in Hpr1p suggests that this protein is most likely nuclear, and recent studies using a FLAG-tagged Hpr1p have confirmed this localization (11). Whether Hpr1p is confined to the chromatin-rich nucleoplasm or to the nucleolus is currently being investigated.
We now report that hpr1Δ cells are conditionally defective in nuclear export of polyadenylated RNA, synthesize oversized CRY1 transcripts, are impaired in rRNA processing, and display an aberrant structure of the nucleolus. These are phenotypic changes that previously have been analyzed to characterize a set of conditional mutants that block mRNA transport (mtr [18]). A defect in nuclear export of polyadenylated RNA has previously also been observed in a temperature-sensitive acc1ts (mtr7) mutant (31), and we now report that a second, cold-sensitive acc1cs mutant also has an Mtr phenotype. We thus propose that the basis for the synthetic phenotype between hpr1Δ and conditional acc1 alleles lies in their common defect in mRNA export and that the combination of mutations that affect nuclear mRNA export is lethal. This would explain the absence of a synthetic phenotype in the acc1cs top1Δ and acc1cs ino2/4 double mutants. The synthetic phenotype would indicate either that each mutant slightly decreases mRNA export and that the additive effect of the two mutations is lethal or that each mutant blocks transport through a different pathway (for a review, see reference 39). Whether Hpr1p acts directly or indirectly to affect transport is not known, but it is likely that Hpr1p affects expression of a gene(s) that is involved in transport and that Acc1p affects nuclear membrane synthesis and hence nuclear pore complex function. M. Chang et al. have suggested that the RNA polymerase II complex in which Hpr1p is found is involved in the expression of cell wall genes (7). The cell wall defects of mutants encoding components of the RNA polymerase II complex may be analogous to the nuclear transport phenotype that we see in hpr1Δ mutants, which may be caused by a reduced expression of nuclear transport factors.
Interestingly, hpr1Δ and acc1cs mutant cells display an aberrant circle-shaped nucleolus. The nucleolus is the site of ribosomal DNA (rDNA) transcription by RNA polymerase I, processing of rDNA transcripts, and assembly of ribosomes (for reviews, see references 24, 30, and 34). In wild-type yeast cells, the nucleolus is a crescent-shaped region of the nucleus that stands in close contact with the nuclear envelope (16). A rounded nucleolar structure that often lacked extensive contact with the nuclear envelope has recently been observed for strains that express polymerase II-transcribed 35S rRNA (25). Furthermore, many of the previously characterized mRNA transport mutants display a fragmented or enlarged nucleolus (18), an observation that led us to propose an involvement of the nucleolus or nucleolar proteins in the mRNA export pathway (32). More recently, a correlation between structural alterations of the nucleolus and aging of yeast cells has also been observed (19). In this case, nucleolar changes appear to be due to the accumulation of extrachromosomal rDNA circles in old cells (37, 38). The significance of the observed morphological alterations of the nucleolus in hpr1Δ and acc1cs mutant cells is not clear at present. Hpr1p affects nuclear events that may be connected to rDNA transcription and/or recombination, but the hpr1Δ mutant has not been found to have any altered rate of rDNA recombination (3). Acc1p, on the other hand, affects the lipid composition of all cellular membranes, including the nuclear envelope that contacts the nucleolus. How this contact between nucleolar structures and the nuclear envelope is maintained is not known. The two proteins thus clearly have distinct functions, and there is no obvious overlap between them. Nevertheless, both mutants affect nuclear export of polyadenylated RNA and display an altered nucleolar morphology. We propose that the two mutations affect two different, but overlapping, functions required for efficient nucleocytoplasmic transport: the lipid composition of the nuclear envelope in the case of the acc1cs mutation and transcription-packaging of nascent transcripts into a transport-competent state in the case of the hpr1Δ mutation. Reducing the efficiency of both processes at the same time is lethal.
ACKNOWLEDGMENTS
We thank S. A. Henry and R. Rothstein for strains, A. Hinnen for the gift of soraphen A, J. P. Aris for the anti-Nop1p antibody, and A. Leber for critically reading the manuscript.
This work was supported by the Swiss National Science Foundation (823A-046702 to R.S.), the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich (project 11731 to S.D.K.), and the National Institutes of Health (grant GM30439 to H.L.K.).
REFERENCES
- 1.Aguilera A, Klein H L. Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics. 1989;122:503–517. doi: 10.1093/genetics/122.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera A, Klein H L. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera A, Klein H L. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Feel W, Chirala S S, Wakil S J. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1992;89:4534–4538. doi: 10.1073/pnas.89.10.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archambault J, Lacroute F, Ruet A, Friesen J D. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aris J P, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang M, French-Cornay D, Fan H, Klein H, Denis C L, Jaehning J A. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol. 1999;19:1056–1067. doi: 10.1128/mcb.19.2.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang M, Jaehning J A. A multiplicity of mediators: alternative forms of transcription complexes communicate with transcriptional regulators. Nucleic Acids Res. 1997;25:4861–4865. doi: 10.1093/nar/25.24.4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan H, Klein H L. Characterization of mutations that suppress the temperature sensitive growth of the hpr1Δ mutant of Saccharomyces cerevisiae. Genetics. 1994;137:945–956. doi: 10.1093/genetics/137.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, H.-Y., and H. L. Klein. Unpublished observation.
- 12.Fan H Y, Cheng K K, Klein H L. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 delta of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 14.Guerra C E, Klein H L. Mapping of the ACC1/FAS3 gene to the right arm of chromosome XIV of Saccharomyces cerevisiae. Yeast. 1995;11:697–700. doi: 10.1002/yea.320110711. [DOI] [PubMed] [Google Scholar]
- 15.Hasslacher M, Ivessa A S, Paltauf F, Kohlwein S D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 16.Hurt E C, Mutvei A, Carmo-Fonseca M. The nuclear envelope of the yeast Saccharomyces cerevisiae. Int Rev Cytol. 1992;136:145–184. doi: 10.1016/s0074-7696(08)62052-5. [DOI] [PubMed] [Google Scholar]
- 17.Ivessa A S, Schneiter R, Kohlwein S D. Yeast acetyl-CoA carboxylase is associated with the cytoplasmic surface of the endoplasmic reticulum. Eur J Cell Biol. 1997;74:399–406. [PubMed] [Google Scholar]
- 18.Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff A M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy B K, Gotta M, Sinclair D A, Mills K, McNabb D S, Murthy M, Pak S M, Laroche T, Gasser S M, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Larkin J C, Thompson J R, Woolford J L., Jr Structure and expression of the Saccharomyces cerevisiae CRY1 gene: a highly conserved ribosomal protein gene. Mol Cell Biol. 1987;7:1764–1775. doi: 10.1128/mcb.7.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 23.Matsuhashi M. Acetyl-CoA carboxylase from yeast. Methods Enzymol. 1969;14:3–8. [Google Scholar]
- 24.Melese T, Xue Z. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7:319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 25.Oakes M, Aris J P, Brockenbrough J S, Wai H, Vu L, Nomura M. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol. 1998;143:23–34. doi: 10.1083/jcb.143.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paltauf F, Kohlwein S D, Henry S A. Regulation and compartmentalization of lipid synthesis in yeast. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 415–500. [Google Scholar]
- 27.Piruat J I, Aguilera A. Mutations in the yeast SRB2 general transcription factor suppress hpr1-induced recombination and show defects in DNA repair. Genetics. 1996;143:1533–1542. doi: 10.1093/genetics/143.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roggenkamp R, Numa S, Schweizer E. Fatty acid-requiring mutant of Saccharomyces cerevisiae defective in acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1980;77:1814–1817. doi: 10.1073/pnas.77.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M D, Winston F, Hieter F. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 30.Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 31.Schneiter R, Hitomi M, Ivessa A S, Fasch E V, Kohlwein S D, Tartakoff A M. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneiter R, Kadowaki T, Tartakoff A M. mRNA transport in yeast: time to reinvestigate the functions of the nucleolus. Mol Biol Cell. 1995;6:357–370. doi: 10.1091/mbc.6.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneiter R, Kohlwein S D. Organelle structure, function, and inheritance in yeast: a role for fatty acid synthesis? Cell. 1997;88:431–434. doi: 10.1016/s0092-8674(00)81882-6. [DOI] [PubMed] [Google Scholar]
- 34.Shaw P J, Jordan E G. The nucleolus. Annu Rev Cell Dev Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- 35.Sherman F, Fink G R, Hicks J B N. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 36.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinclair D A, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 38.Sinclair D A, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem Sci. 1998;23:131–134. doi: 10.1016/s0968-0004(98)01188-8. [DOI] [PubMed] [Google Scholar]
- 39.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 40.Trash C, Bankier A T, Barrell B G, Sternglanz R. Cloning, characterization, and sequence of the yeast DNA topoisomerase I gene. Proc Natl Acad Sci USA. 1985;82:4374–4378. doi: 10.1073/pnas.82.13.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uemura H, Pandit S, Jigami Y, Sternglanz R. Mutations in GCR3, a gene involved in the expression of glycolytic genes in Saccharomyces cerevisiae, suppress the temperature-sensitive growth of hpr1 mutants. Genetics. 1996;142:1095–1103. doi: 10.1093/genetics/142.4.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vahlensieck H F, Pridzun L, Reichenbach H, Hinnen A. Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr Genet. 1994;25:95–100. doi: 10.1007/BF00309532. [DOI] [PubMed] [Google Scholar]
- 43.Yaffe M P, Schatz G. Two nuclear mutations that block mitochondrial protein import. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Y, Peterson C L, Christman M F. HPR1 encodes a global positive regulator of transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1698–1708. doi: 10.1128/mcb.15.3.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]