Abstract
The sulfur oxygenase reductase (SOR) reaction is a dioxygen-dependent disproportionation of elemental sulfur (S0), catalyzed at optimal temperatures between 65 °C and 85 °C. Thiosulfate and sulfite are formed as oxidized products as well hydrogen sulfide as reduced product. External co-factors are not required. Usually, the SOR assay is performed in a milliliter scale in S0-containing Tris-buffer at high temperatures followed by colorimetric product quantification. In order to make the SOR assay more sensitive and better reproducible, several modifications were implemented compared to the original SOR assay (Kletzin, 1989). Here we present the modified SOR assay and the following quantification of the reaction products.
Keywords: Sulfur oxygenase reductase, Elemental sulfur, Thermozyme, Sulfur disproportionation, Thiosulfate, Sulfite, Hydrogen sulfide
Background
Sulfur oxygenase reductases (SOR) catalyze a dioxygen-dependent disproportionation of elemental sulfur with sulfite, thiosulfate and hydrogen sulfide as detectable products. The initially found SORs were derived from hyperthermophilic, sulfur-oxidizing Archaea and Bacteria. The reported temperature optima were between 65 °C and 85 °C and the pH optima between pH 5 and pH 7.4 ( Emmel et al., 1986 ; Kletzin, 1989; Sun et al., 2003 ; Pelletier et al., 2008 ). Surprisingly, SORs derived from the mesophilic bacterium Halothiobacillus neapolitanus ( Veith et al., 2012 ) and the alkalihalophilic Thioalkalivibrio paradoxus ( Rühl et al., 2017 ) also had temperature optima of around 80 °C at pH 8.4 and pH 9.0, respectively. Most SORs can be produced in E. coli by heterologous gene expression and only the first descriptions of the enzyme were derived from proteins purified from their native sources ( Emmel et al., 1986 ; Kletzin, 1989; Pelletier et al., 2008 ). Today, sor genes are found in approximately 35 different Bacteria and Archaea ( Rühl et al., 2017 ).
Usually, SORs have a stoichiometry between 4:1 and 10:1 of oxidized (thiosulfate and sulfite) and reduced products (hydrogen sulfide). The oxidation and reduction reactions could not be separated by site-directed mutagenesis, although the product stoichiometries may vary ( Veith et al., 2012 ). So far, the only exception is the Thioalkalivibrio paradoxus SOR with stoichiometries of 100-1,000:1, which makes the enzyme an oxygenase with almost no reductase activity ( Rühl et al., 2017 ). The thiosulfate:sulfite ratio increases with temperature and pH (Kletzin, 1989; Veith et al., 2012 ; Rühl et al., 2017 ). Therefore, the bulk of the thiosulfate is most likely formed rapidly by a non-enzymatic reaction between sulfur and sulfite at pH values above 6 and temperatures exceeding 70 °C.
The original SOR activity assay (Kletzin, 1989; Urich et al., 2004 ) involves shaking of an aliquot of the enzyme in a buffer containing elemental sulfur at the given reaction temperature coupled to the colorimetric determination of the amount of products at different time points. Product determination is also possible using HPLC (e.g., Rethmeier et al., 1997 ). However the number of samples that can be processed per day is higher using the colorimetric assays because of the longer time required for each HPLC run. Specific SOR activities were in the range of 10 U/mg of protein for the Acidianus ambivalens SOR and 40 U/mg for the Halothiobacillus enzyme, both determined with the original enzyme assay.
After developing several modifications, the activity assay became more sensitive, resulting in higher product formation and lower amount of the required enzyme together with a reduced incubation time of the enzyme assay (Table 1). The original assay (Kletzin, 1989) was performed in stoppered glass vials with the enzyme being added at room temperature prior to incubation. All vials were transferred simultaneously to a shaking bath with preheated heating liquid. In order to stop the reaction, the vials were transferred sequentially into an ice bath. Urich et al. (2004) modified the procedure for the use of 1.5 ml plastic reaction vials and thermomixers but kept the order of the steps. In the modified procedure, the enzyme solution is added last to the 0 min time point immediately before transfer of the entire set of vials to an ice bath. Thus, all vials remain at the assay temperature for exactly the same time minimizing background effects. Here we describe the modified SOR activity assay and the quantification of its three reaction products.
Table 1. Incubation conditions for the original and modified SOR enzyme assays.
| Original assay | Modified assay | |
|---|---|---|
| Total incubation time (min) | 10 | 2.5 |
| Incubation time points for product determination (min) | 0; 2; 4; 6; 8; 10 | 0; 0.5; 1; 1.5; 2; 2.5 |
| Pre-heating time of assay mixture on thermomixer before enzyme addition (min) | 0 | 5 |
| Enzyme addition to assay buffer at … | Room temperature | Assay temperature |
Materials and Reagents
Safe-lock reaction vials, 1.5 ml (SARSTEDT, catalog number: 72.690.001)
Micro cuvettes, polystyrene (SARSTEDT, catalog number: 67.742)
37% [wt/vol] formaldehyde (Merck, catalog number: 104003)
Double deionized water (ddH2O; 18.2 MΩ at 25 °C)
Tris(hydroxymethyl)aminomethane (Tris base) (Carl Roth, catalog number: 5429.2)
32% [wt/vol] hydrochloric acid (HCl) (Carl Roth, catalog number: P074.4)
Tween 20 (Carl Roth, catalog number: 9127.2)
-
Sulfur flower (AppliChem, catalog number: A1687)
Note: This product has been discontinued; alternative product: Merck, catalog number: 107983.
Methylene blue (Merck, catalog number: 115943)
Sodium thiosulfate pentahydrate (Merck, catalog number: 106513)
Fuchsine (Merck, catalog number: 105226)
Sulfuric acid (Carl Roth, catalog number: 9316.2)
Sodium sulfite (Merck, catalog number: 106657)
Zinc acetate dihydrate (Merck, catalog number: 108802)
Acetic acid (Carl Roth, catalog number: 3738.5)
Dimethyl-4-phenylenediamine dihydrochloride (Merck, catalog number: 103067)
Iron-(III)-chloride hexahydrate (Carl Roth, catalog number: 7119.1)
-
20% [wt/vol] ammonium sulfide solution (Sigma-Aldrich, catalog number: A1925)
Note: This product has been discontinued; alternative product: Merck, catalog number: 105442.
SOR assay buffer (see Recipes)
Methylene blue solution (see Recipes)
Sodium thiosulfate solution (1 mM) (see Recipes)
Fuchsine solution (see Recipes)
Sodium sulfite solution (1 mM) (see Recipes)
Zinc acetate solution (see Recipes)
Dimethyl-4-phenylenediamine dihydrochloride solution (see Recipes)
Iron-(III)-chloride solution (see Recipes)
Ammonium sulfide solution (1 mM) (see Recipes)
Equipment
Polypropylene Griffin beaker 250 ml (Carl Roth, catalog number: 2875.1)
-
Pipettes L20, L200, L1000 (Abimed LABMATE Optima)
Note: This product has been discontinued; alternative product: Gilson, catalog number: F167350.
Magnetic stirrer (e.g., IKAMAG RET; IKA)
Thermomixer basic (shaking heat block; CellMedia, Elsteraue, Germany)
-
pH meter (Xylem, WTW, model: inoLab pH 720) with SenTix 41 pH electrode (Xylem, WTW, catalog number: 103635)
Note: The product “Xylem, WTW, model: inoLab pH 720” has been discontinued.
Centrifuge Heraeus Pico 17 Microcentrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraeusTM PicoTM 17, catalog number: 75002410)
Spectrophotometer (e.g., Beckmann Coulter, model: DU-640)
Ultrasound device (Emerson Electric, Branson, model: Sonifier 250, catalog number: 100-132-868) equipped with a macrotip
Software
Microsoft Excel 365 (Microsoft)
Procedure
-
Preparation of the SOR assay buffer
-
Prepare a 70 mM Tris solution in ddH2O in a plastic beaker and adjust the pH value with 32% [wt/vol] hydrochloric acid, usually to 7.2.
Note: The actual pH depends on the pH optimum of the respective SOR.
Add 2% [wt/vol] elemental sulfur (sulfur flower) and 0.1% [vol/vol] Tween 20 for a better dispersal of the sulfur. Stir mixture on a magnetic stirrer for approximately 1 min.
-
Transfer the beaker to an ice bath and disperse the elemental sulfur by sonication for 5 min with a macrotip at level 10 and 100% duty cycle.
Note: The ultrasonic treatment of the almost insoluble sulfur flower in the presence of detergent will result in a finely dispersed yellow sulfur suspension.
-
-
SOR activity assay
Stir the sonicated enzyme buffer on a magnetic stirrer and transfer 6 x 1 ml to 1.5 ml reaction tubes while continuing to stir.
-
Preheat the reaction tubes with the enzyme buffer to the appropriate temperature for 5 min in a thermomixer with continuous shaking at 800 rpm.
Note: The incubation temperature usually depends on the optimum of the respective SOR and is mostly in the range between 65 °C and 85 °C.
-
Add 2 µg of enzyme to the first reaction tube and repeat this step every 30 sec for each consecutive tube (Figure 1).
Notes:
The first addition of your enzyme to the reaction buffer corresponds to the 150 sec time point, the last addition corresponds to the 0 sec value of the enzyme assay.
The amount of enzyme can be raised up to 100 µg/ml or lowered to 0.5 µg/ml depending on the specific activity of the individual SOR preparation, the assay temperature or pH in order to provide a suitable amount of product for the colorimetric assays (see below).
Immediately after the enzyme addition to the last reaction tube, transfer all reaction tubes to an ice/water bath to quench the reaction. Keep the tubes for approximately 2-3 min on ice.
Transfer the reaction tubes to a microcentrifuge and sediment the elemental sulfur (13,000 × g, 1 min).
Use the supernatant for the three colorimetric assays (Figures 1 and 2) to quantify the reaction products.
As a negative control, perform the same incubation with enzyme reaction buffer by adding water instead of enzyme solution to the reaction vials and perform the colorimetric assays accordingly.
-
Thiosulfate determination
The quantification of thiosulfate is based on the discoloration of methylene blue (Pachmayr, 1960).
Label one micro cuvette for each of the time points to be analyzed.
Transfer 250 µl of the enzyme reaction mixture into an empty cuvette.
Add 750 µl methylene blue solution (see Recipes).
Incubate at room temperature for 30 min.
Determine the absorption spectrophotometrically at 670 nm against ddH2O as reference.
-
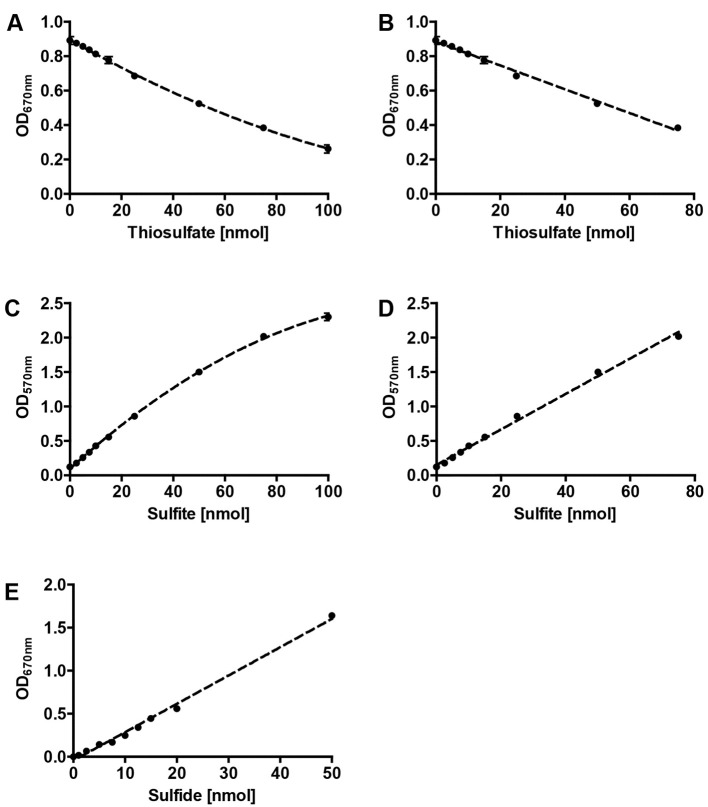
Prepare a standard curve based on a 1 mM freshly prepared sodium thiosulfate solution (see Recipes, see Figures 2A and 2B). Perform the colorimetric assay as described above and replace the reaction mixture with different volumes of the sodium thiosulfate solution (0, 1, 5, 10, 20, 50, 75 and 100 µl; these values correspond to the same amount of thiosulfate in nmol). Make up the volume to 250 µl with ddH2O.
Note: Note that there is a decrease in absorbance with an increase of the amount of thiosulfate (or concentration).
-
Sulfite determination
The quantification of sulfite is based on the reaction of fuchsine with sulfite. The addition of formaldehyde leads to the formation of a stable purple compound (Pachmayr, 1960).
Label one 1.5 ml reaction tube for each of the time points.
Transfer 50 µl fuchsine solution (see Recipes) into each reaction tube and add 200 µl ddH2O.
Add 250 µl of the enzyme reaction mixture and incubate for 5 min at room temperature.
Pipet exactly 5 µl of 37% [wt/vol] formaldehyde into the inner lid of the reaction tube.
Carefully close the reaction tube and briefly spin down the formaldehyde.
Transfer the whole mixture to a cuvette using an L1000 pipette and incubate for 60 min at room temperature.
Determine the absorption spectrophotometrically at 570 nm against ddH2O as reference.
Prepare a standard curve based on a freshly prepared 1 mM sodium sulfite solution (see Recipes, see Figures 2C and 2D). Perform the colorimetric assay as described above and replace the reaction mixture with different volumes of the sodium sulfite solution as described above for the thiosulfate calibration curve (0, 1, 5, 10, 20, 50, 75 and 100 µl).
-
Hydrogen sulfide determination
The quantification of sulfide is based on the formation of methylene blue (King and Morris, 1967).
Label one micro cuvette for each of the time points.
Transfer 250 µl zinc acetate solution (see Recipes) into each of the cuvettes for fixation of the volatile hydrogen sulfide.
Add 350 µl reaction mixture, 125 µl dimethyl-4-phenylenediamine dihydrochloride solution (see Recipes) and 50 µl iron-(III)-chloride solution (see Recipes) in the order provided.
-
Incubate at room temperature for at least 12 h.
Note: The incubation time can be extended up to 72 h if desired, e.g., over the weekend.
Determine the absorption spectrophotometrically at 670 nm against ddH2O as reference.
Prepare a standard curve based on a freshly prepared 1 mM ammonium sulfide solution (see Recipes, see Figure 2E). Perform the colorimetric assay as described above and replace the reaction mixture with different volumes of the ammonium sulfide solution as described above for the thiosulfate calibration curve (0, 1, 2.5, 5, 7.5, 10, 15 and 20 µl).
Figure 1. Schematic overview for the SOR activity assay and the colorimetric reactions.
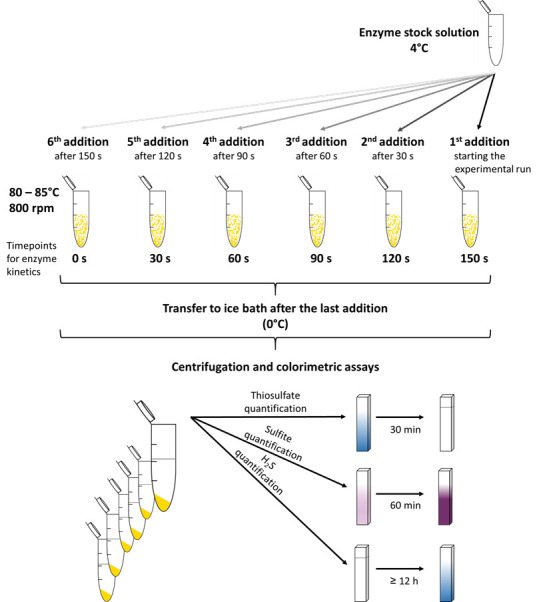
Figure 2. Representative calibrations curves for colorimetric assays.
Polynomial (A) and linear (B) fits of thiosulfate calibration , polynomial (C) and linear (D) fits of sulfite calibration and linear fit of sulfide calibration (E); error bars from 3 replicates.
Data analysis
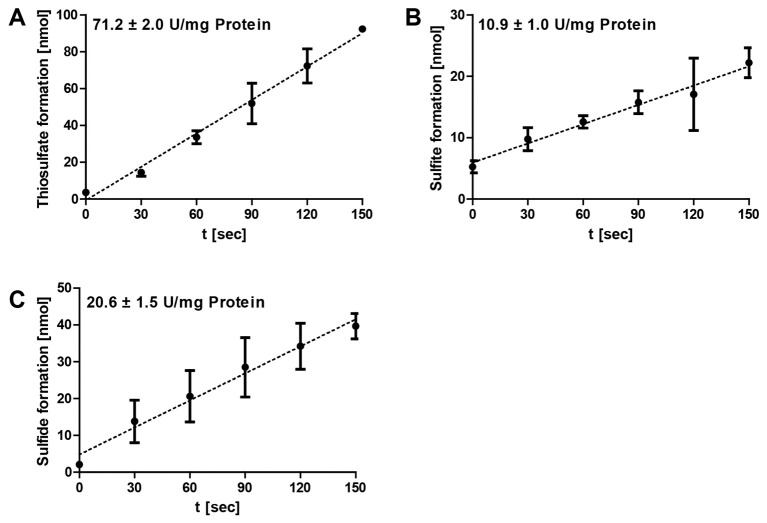
The data analysis was performed using Microsoft Excel. Specific enzymatic activities are determined from the range of the linear increase/decrease of the optical densities of the colorimetric assays (Figure 3). The corresponding amounts of the respective products are calculated and plotted against the time. The gradient corresponds to the product formation per minute and the amount of enzyme added to each reaction mixture (= enzyme activity). The specific SOR activities are defined as 1 µmol of sulfite plus thiosulfate (oxygenase activity) or 1 µmol of hydrogen sulfide (reductase activity) formed per minute and mg of protein (= 16.7 µkat/g).
Figure 3. Representative data of Acidianus ambivalens SOR enzyme reaction.
The formation of thiosulfate (A), sulfite (B) and sulfide (C) at different time points were followed at 85 °C and pH 7.2 and with 2 µg of purified enzyme in 1 ml SOR assay buffer; the error bars are from triplicate measurements. Note the differences in the scaling of the y-axes.
Notes
The volume of reaction mixture during colorimetric assays can be scaled down to 50 µl, if the coloration or discoloration is too strong for the linear range of the calibration curve.
The amount of enzyme can be increased up to 100 µg/ml during the enzymatic assay to enhance product formation and sensitivity of the colorimetric assays, e.g., with low temperature measurements and/or high/low pH values.
Sulfur flower can contain minor amounts of thiosulfate and/or form thiosulfate non-enzymatically during the pre-heating step of the assay at alkaline pH. The background levels in the colorimetric assay were 8-16 nmol/ml at pH 7.2 and 28-40 nmol/ml at pH 9.
Ammonium sulfide and sodium sulfite solutions should be always prepared with freshly boiled water (microwave) in order to remove dissolved oxygen, which chemically oxidizes the sulfur compounds.
Recipes
Notes:
Use ddH2O for all solutions unless stated otherwise.
Sodium thiosulfate solution and especially the sodium sulfite and ammonium sulfide solutions are unsuitable for long-term storage and should be prepared freshly.
All other solutions could be stored at room temperature for up to one month and even longer, if the calibration curves are done. Although the solutions are not especially light-sensitive, direct sunlight should be avoided.
-
SOR assay buffer (250 ml)
70 mM Tris (2.1 g) adjusting the pH value with 32% HCl
0.1% [vol/vol] Tween 20 (250 µl)
2% [wt/vol] elemental sulfur (sulfur flower; 5 g)
Methylene blue solution (1 L)12 mg methylene blue dissolved in 5 N HCl
Sodium thiosulfate solution (1 mM)Dissolve 0.248 g sodium thiosulfate pentahydrate in 10 ml ddH2O for a 100 mM solutionPrepare a 1:100 dilution to give the final 1 mM thiosulfate solution (0.1 ml 100 mM sodium thiosulfate solution with 9.9 ml ddH2O)
Fuchsine solution (100 ml)40 mg fuchsine dissolved in 87.5 ml ddH2O and 12.5 ml concentrated sulfuric acid
Sodium sulfite solution (1 mM)Dissolve 0.126 g sodium sulfite in 10 ml freshly boiled (microwave) ddH2O for a 100 mM solutionPrepare a 1:100 dilution with freshly boiled ddH2O to give the final 1 mM sulfite solution (0.1 ml 100 mM sodium sulfite solution with 9.9 ml ddH2O)
Zinc acetate solution (250 ml)6.5 g zinc acetate dissolved in 249.75 ml ddH2O and 250 µl 100% [wt/vol] acetic acid
Dimethyl-4-phenylenediamine dihydrochloride solution (100 ml)0.1 g dimethyl-4-phenylenediamine dihydrochloride dissolved in 5 M HCl
Iron-(III)-chloride solution (50 ml)0.16 g iron-(III)-chloride dissolved in 0.6 M HCl
Ammonium sulfide solution (1 mM)Add 3.4 µl 20% [wt/vol] ammonium sulfide solution to 10 ml freshly boiled ddH2O
Acknowledgments
Patrick Rühl was supported by a fellowship of the Carlo und Karin Giersch-Stiftung an der TU Darmstadt, Darmstadt, Germany and by a grant of the Deutsche Forschungsgemeinschaft to Arnulf Kletzin (Az Kl885-7/1). This protocol was adapted from Kletzin (1989). The original enzyme assay for sulfite formation from sulfur was developed by Emmel et al. (1986).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Emmel T., Sand W., König W. A. and Bock E.(1986). Evidence for the existence of a sulfur oxygenase in Sulfolobus brierleyi . J Gen Microbiol 132: 3415-3420. [Google Scholar]
- 2.King T. E. and Morris R.(1967). Determination of acid-labile sulfide and sulfhydryl groups. Meth Enzymol 10: 634-641. [Google Scholar]
- 3.Kletzin A.(1989). Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens . J Bacteriol 171(3): 1638-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pachmayr F.(1960). Vorkommen und Bestimmung von Schwefelverbindungen in Mineralwasser. Dissertation, Justus-Maximilian Universität, München.
- 5.Pelletier N., Leroy G., Guiral M., Giudici-Orticoni M. T. and Aubert C.(2008). First characterisation of the active oligomer form of sulfur oxygenase reductase from the bacterium Aquifex aeolicus . Extremophiles 12(2): 205-215. [DOI] [PubMed] [Google Scholar]
- 6.Rethmeier J., Rabenstein A., Langer M., and Fischer U.(1997). Detection of traces of oxidized and reduced sulfur compounds in small samples by combination of different high-performance liquid chromatography methods. J Chromatogr A 760(2): 295-302. [Google Scholar]
- 7.Rühl P., Pöll U., Braun J., Klingl A. and Kletzin A.(2017). A sulfur oxygenase from the haloalkaliphilic bacterium Thioalkalivibrio paradoxus with atypically low reductase activity . J Bacteriol 199(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun C. W., Chen Z. W., He Z. G., Zhou P. J. and Liu S. J.(2003). Purification and properties of the sulfur oxygenase/reductase from the acidothermophilic archaeon, Acidianus strain S5 . Extremophiles 7(2): 131-134. [DOI] [PubMed] [Google Scholar]
- 9.Urich T., Bandeiras T. M., Leal S. S., Rachel R., Albrecht T., Zimmermann P., Scholz C., Teixeira M., Gomes C. M. and Kletzin A.(2004). The sulphur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron centre . Biochem J 1): 137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veith A., Botelho H. M., Kindinger F., Gomes C. M. and Kletzin A.(2012). The sulfur oxygenase reductase from the mesophilic bacterium Halothiobacillus neapolitanus is a highly active thermozyme . J Bacteriol 194(3): 677-685. [DOI] [PMC free article] [PubMed] [Google Scholar]