Abstract
The periodontal ligament (PDL) is an essential tissue connecting teeth and bone. It is a complex tissue specifically designed to absorb the forces of mastication; analysis of its multiple cell populations is important to understand its function and the cell changes associated with periodontal disease. Cells in the periodontal ligament are not fully understood due to their physical location and small tissue size. It is challenging to isolate thin layers of cells compared with many other more substantial tissues. Here, we provide a straightforward protocol for the isolation of periodontal ligament cells from mice.
Keywords: Mouse periodontal ligament, Single cells, Cementoblasts, Stem cells
Background
Periodontal disease is a prevalent issue and a major cause of tooth loss. Although the periodontal ligament (PDL) is known to contain resident stem cell populations, it is poorly understood. Therefore, harvesting PDL cell populations and maintaining their characteristics without in vitro culture will provide insight to guide regenerative therapies.
Periodontal ligament stem cells (PDLSCs) are capable of differentiating into osteo-/cemento-lineage cells in vitro and in vivo ( Seo et al., 2004 ; Zhao et al., 2021 ). Cementoblasts are cells responsible for the attachment of PDL fibres to the tooth. Little is known about the differentiation pathways within the PDL that lead to cementoblast formation due to difficulties in isolating this cell population (Matthews et al., 2016).
Previously, human periodontal ligament cells were isolated to evaluate their regenerative potential, which involved separation of the PDL from tooth roots and subsequent in vitro culture ( Park et al., 2011 ). A protocol was established to isolate rat PDL cells ( Kaneda et al., 2006 ), demonstrating that dissociation of PDL tissue using 2 g/ml collagenase and 0.25% trypsin for 110 min allows all attached PDL cells to be harvested. Separation of PDL tissue from mouse teeth is more difficult due to their small size. We used collagenase P for 30-45 min and found that mouse PDL cells can be collected effectively. We developed a protocol to collect sufficient PDL cells from mouse molars for single-cell transcriptomics analysis ( Zhao et al., 2021 ).
Materials and Reagents
1-ml pipette and tips (STARlabs, catalog number: S1122-1830)
(Optional) 6-well plates (CellSTAR, catalog number: 567160)
Centrifuge tubes, 15-ml and 50-ml (Corning, catalog numbers: 430719, 430829)
40-μm cell strainers (Falcon, catalog number: 352340)
(Optional) Needles 20G
Blades (Swann-Morton, catalog number: 0314 #27)
Filter (Millex, catalog number: SLGP033RS)
CD1 mice aged 8 weeks
Ice
Collagenase P (Roche, catalog number: 11213865001)
DMEM (Sigma-Aldrich, catalog number: D6429)
Fetal bovine serum (FBS) (Sigma, catalog number: F7524)
Equipment
Dissection dish
Biosafety cabinet
Stereomicroscope and light source (Zeiss Stemi 2000-C and Olympus KL1500 LCD)
Tweezers and fine tweezers
Water bath with shaking function (Grant, model: OLS 200)
Heraeus dissection hood (or any hood that can ensure control of contamination)
Procedure
-
Preparation for dissection
In a biosafety cabinet, prepare a 6-well plate with 3 ml dissection media containing 10% FBS and place on ice (2.7 ml DMEM with 300 µl FBS).
Warm the water bath to 37°C.
-
Dissection
Collect mice used for PDL cell collection.
Harvest the maxilla and mandible by separating the head from the body with a blade, followed by cutting above the tongue to separate the maxilla and the mandible. Then separate left and right maxillae or mandibles.
Carefully remove all the gingiva tissue attached to the teeth (Figures 1A and 1B).
Extract a tooth using fine tweezers by holding the crown and pulling. Be careful; the tooth can jump out of the tweezers. Discard the tooth if contaminated. If the tooth is not easy to remove, insert a 20G needle between the crown of the first and second molars until it loosens (Figure 1C).
Remove any excessive tissue-bone residue. This mostly appears at the furcation area or near the apical root (Figure 1D). The PDL tissue should be attached to the tooth surface. Make sure the roots are intact.
Store the teeth in a 6-well plate containing media on ice.
-
Dissociation
Make 3 ml fresh Collagenase P (Col P) solution (3 U/ml) and filter through a 0.22-μm filter into a 15-ml Falcon tube.
Transfer the intact teeth to a 15-ml Falcon tube and add 2 ml Col P containing 10% FBS.
Place the Falcon tube in a 37°C water bath for 30-45 min with a shaking speed of 140, and homogenise 3 times every 10-15 min by pipetting the solution up and down with a 1-ml pipette. The shaking step helps to breakup collagen and release the cells. If the shaking function cannot be used, 10-min homogenisation intervals are suggested.
To harvest single cells, when the cells are released from the tissue after 30-45 min, transfer the liquid through a 40-μm cell strainer into a 50-ml Falcon tube.
Centrifuge at 1,200 × g for 5 min. A cell pellet may not be seen if the cell number is low.
Carefully discard the supernatant.
Store cells on ice for subsequent experiments.
Figure 1. Dissection procedure.
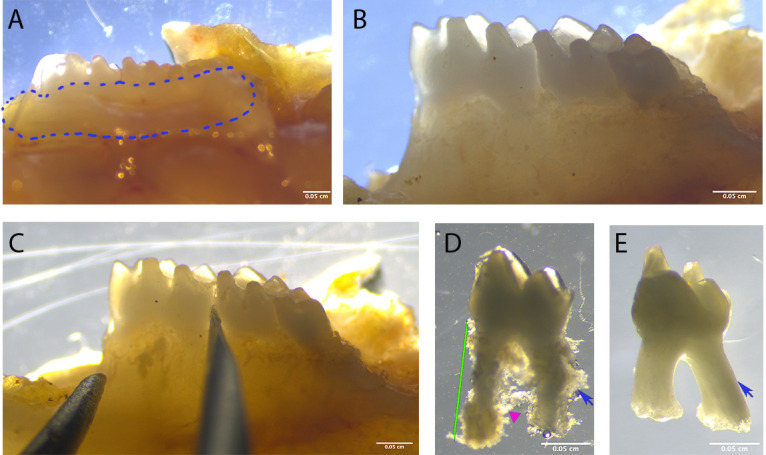
A. Dissect the maxilla and mandible and remove the soft tissue around the tooth (circled area). B. Make sure there is no soft tissue left around the teeth. C. If the tooth is hard to extract, place a needle between the teeth and move around to loosen the tooth, then attempt to extract again. D. The extracted tooth, before enzyme digestion, should show PDL tissue on the root surface (blue arrow); make sure there is no bone tissue in the furcation area (magenta triangle). Keep only the intact teeth (green lines). E. Extracted teeth show a smooth root surface with no PDL tissue attached.
Data analysis
Anticipated results
PDL tissue should be dissociated as shown in Figure 1E. Before enzyme digestion, the PDL tissue is attached to the tooth surface (Figure 2). After enzyme digestion, the PDL tissue is detached, including the cementoblasts on the root surface. Notably, although pulp cells are connected to PDL cells via the apical foramen, they are not released during the digestion process. This only applies to teeth with fully formed roots; if developing teeth are used, pulp cells will be released and collected together with PDL cells (Figure S1).
Figure 2. Harvested cells from adult periodontal ligament.
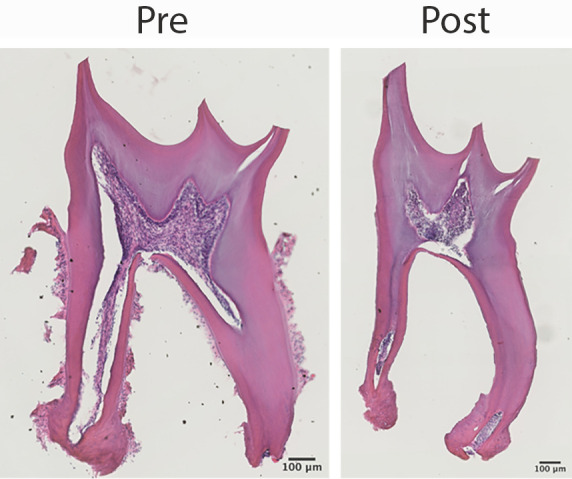
H&E staining shows that extracted teeth have PDL tissue attached to the root surface before enzyme digestion (left). However, after digestion, cells on the tooth surface are released without contamination from pulp cells (right).
Cells isolated from periodontal tissue dissected using this protocol have been subjected to single-cell RNA sequencing and the data used to investigate cell population heterogeneity, cell changes in periodontitis, and cell-cell signalling interactions ( Jing et al., 2021 ).
Acknowledgments
We thank Dr. Lucia Zaugg, Dr. Arshiya Banu, and Dr. Jan Krivanek for their help. Jing Zhao holds training support from the China Scholarship Council Studentship with Kings College London. Funding from the NIHR GSTFT/KCL Biomedical Research Centre is acknowledged. The protocol was first described in our original research paper, “Stem cell contributions to cementoblast differentiation in healthy periodontal ligament and periodontitis” published in Stem Cells ( Zhao et al., 2021 ).
Competing interests
There are no conflicts of interest or competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Supplementary Data.
References
- 1.Kaneda T., Miyauchi M., Takekoshi T., Kitagawa S., Kitagawa M., Shiba H., Kurihara H. and Takata T.(2006). Characteristics of periodontal ligament subpopulations obtained by sequential enzymatic digestion of rat molar periodontal ligament. Bone 38(3): 420-426. [DOI] [PubMed] [Google Scholar]
- 2.Matthews B. G., Roguljic H., Franceschetti T., Roeder E., Matic I., Vidovic I., Joshi P., Kum K.Y. and Kalajzic I.(2016). Gene-expression analysis of cementoblasts and osteoblasts. J Periodontal Res 51(3): 304-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park J. C. Kim J. M., Jung I. H., Kim J. C., Choi S. H., Cho K.S. and Kim C. S.(2011) Isolation and characterization of human periodontal ligament(PDL) stem cells(PDLSCs) from the inflamed PDL tissue: in vitro and in vivo evaluations . J Clin Periodontol 38(8): 721-731. [DOI] [PubMed] [Google Scholar]
- 4.Seo B.M., Miura M., Gronthos S., Bartold P. M., Batouli S., Brahim J., Young M., Robey P.G., Wang C. Y. and Shi S.(2004). Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet 364(9429): 149-155. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J., Faure L., Adameyko I. and Sharpe P. T.(2021). Stem cell contributions to cementoblast differentiation in healthy periodontal ligament and periodontitis. Stem Cells 39(1): 92-102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


