Abstract
The yeast Saccharomyces cerevisiae (S. cerevisiae) harboring ade1 or ade2 mutations manifest red colony color phenotype on rich yeast medium YPD. In these mutants, intermediate metabolites of adenine biosynthesis pathway are accumulated. Accumulated intermediates, in the presence of reduced glutathione, are transported to the vacuoles, whereupon the development of the red color phenotype occurs. Here, we describe a method to score for presence of oxidative stress upon expression of amyloid-like proteins that would convert the red phenotype of ade1/ade2 mutant yeast to white. This assay could be a useful tool for screening for drugs with anti-amyloid aggregation or anti-oxidative stress potency.
Keywords: Amyloid, Oxidative stress, ROS, Yeast, ade1 mutant , TDP-43, DCFDA
Background
The yeast Saccharomyces cerevisiae cells mutant for ADE1 or ADE2 genes (e.g., ade1∆, ade2∆, ade1-14, ade2-1), when grown on YPD (Yeast Peptone Dextrose) medium, accumulate red pigment in the vacuole which is an intermediate metabolite of adenine biosynthesis pathway ( Sharma et al., 2003 ). The suppressible allele ade1-14 which contains a premature stop codon, has been widely used to score for [ PSI+] prion state of the translation termination factor Sup35 protein. In a [psi-] yeast, the Sup35p remains soluble and functional, therefore translation is terminated efficiently at the premature stop codon of the ade1-14 allele leading to synthesis of truncated and non-functional Ade1 protein. Thus, the adenine biosynthesis cascade remains incomplete leading to accumulation of intermediate metabolite yielding the red phenotype of the [psi-] yeast. In contrast, in a [ PSI+] yeast, the Sup35p is aggregated and partially inactivated, thereby causing read-through of the premature non-sense codon of the ade1-14 allele that leads to functional Ade1 protein synthesis. Thus, adenine biosynthesis pathway is completed and no red intermediate metabolite is accumulated, consequently giving the [ PSI+] cells, a white phenotype ( Chernoff et al., 1993 ). In the view that the red color development from this adenine biosynthesis intermediate metabolite requires presence of reduced glutathione( Sharma et al., 2003 ), we reasoned that presence of oxidative stress which would oxidize glutathione, would also cause white color conversion alike to the white phenotype of the [ PSI+] yeast. Additionally, it is known that the aggregations of several amyloid proteins cause cellular oxidative stress, therefore we attempted to develop a red/white reporter color assay for amyloid-induced oxidative stress in the ade1/ade2 mutant yeast background similar to the widely used red/white assay for the [psi-] to [ PSI+] conversion ( Bharathi et al., 2016 ). We present here red/white color switch assay using two amyloid-like proteins, TAR DNA binding protein 43 (TDP-43) and Fused in Sarcoma (FUS), both of which are implicated in the pathogenesis of the motor neuron disease, Amyotrophic Lateral Sclerosis (ALS) ( Rossi et al., 2016 ). Such simplistic color assay for amyloid-induced oxidative stress in yeast has never been reported previous to our Bharathi et al., 2016 manuscript and has the potential to be a highly useful methodology to study for amyloid protein-induced toxicity in yeast.
Materials and Reagents
Pipette tips (Tarsons products pvt ltd., India)
Microcentrifuge tubes (Tarsons products pvt ltd., India)
Petri dishes (Genaxy scientific pvt ltd., India)
Sterile toothpick
-
Yeast Saccharomyces cerevisiae strain: 74D-694 (MATa ade1-14, his3-200, ura3-52, leu2-3, 112, trp1-289, [psi-])
Note: This assay can also be performed using an ade2-1 allele bearing mutant yeast or an ade2∆ mutant yeast as well as using an ade1∆ mutant yeast.
-
Plasmids
pRS416-GAL1p-FUS-YFP (URA3) (Addgene, catalog number: 29593)
pRS416-GAL1p-TDP43-YFP (URA3) (Addgene, catalog number: 27447)
pRS416 (URA3)
pAG416 GAL1p-ccdB-EGFP (URA3) (Addgene, catalog number: 14195)
0.1 M phosphate buffer (pH 7.4)
2’,7’-Dichlorofluoroscein diacetate [DCFDA] (Sigma-Aldrich, catalog number: D6883)
Lithium acetate (Sigma-Aldrich, catalog number: 517992)
Peptone (HiMedia Laboratories, catalog number: RM001)
D-glucose [Dextrose] (AMRESCO, catalog number: 0188)
Yeast extract (HiMedia Laboratories, catalog number: RM027)
Bacteriological agar (HiMedia Laboratories, catalog number: RM026)
D-raffinose (Sigma-Aldrich, catalog number: 83400)
D-galactose (Sigma-Aldrich, catalog number: 48260)
Yeast nitrogen base (HiMedia Laboratories, catalog number: G090)
Sodium dodecyl sulphate [SDS] (Sigma-Aldrich, catalog number: L3771)
Chloroform (Sigma-Aldrich, catalog number: 372978)
Ammonium sulfate (Sigma-Aldrich, catalog number: A2939)
Amino acids: Arginine, Histidine, Isoleucine, Valine, Lysine, Methionine, Adenine, Phenylalanine, and Tyrosine (HiMedia Laboratories, India); Leucine and Tryptophan (Sigma-Aldrich, USA)
YPD medium (see Recipes)
SRaf-Ura + 0.1% gal + ¼ YP plate (see Recipes)
SRaf-Ura broth (see Recipes)
SD-Ura + ¼ YP plate (see Recipes)
SD-Ura + YP plate (see Recipes)
DCF extraction buffer (see Recipes)
Equipment
Pipettes (Corning, USA)
250 ml conical flasks (Borosil Glass Works ltd., India)
Microcentrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: MicroCL 21R)
Fluorescence microscope (Leica Microsystems, model: Leica DM2500)
Laminar flow biosafety cabinet (Esco Micro, model: ACB-4A1)
Temperature controlled incubator (JS Research, model: JSGI-100T)
Temperature controlled orbital shaker (Eppendorf, New BrunswickTM, model: Excella® E24)
UV-Vis spectrophotometer (Hitachi High-Technologies, model: U-3900)
Vortex mixer (Remi, model: CM-101)
Autoclave sterilizer (JS Research, model: JSAC-100)
Multimode microplate reader (Molecular Devices, model: Spectramax M5e)
Camera
Software
-
GraphPad QuickCalcs software (from GraphPad Software Inc., USA)
Procedure
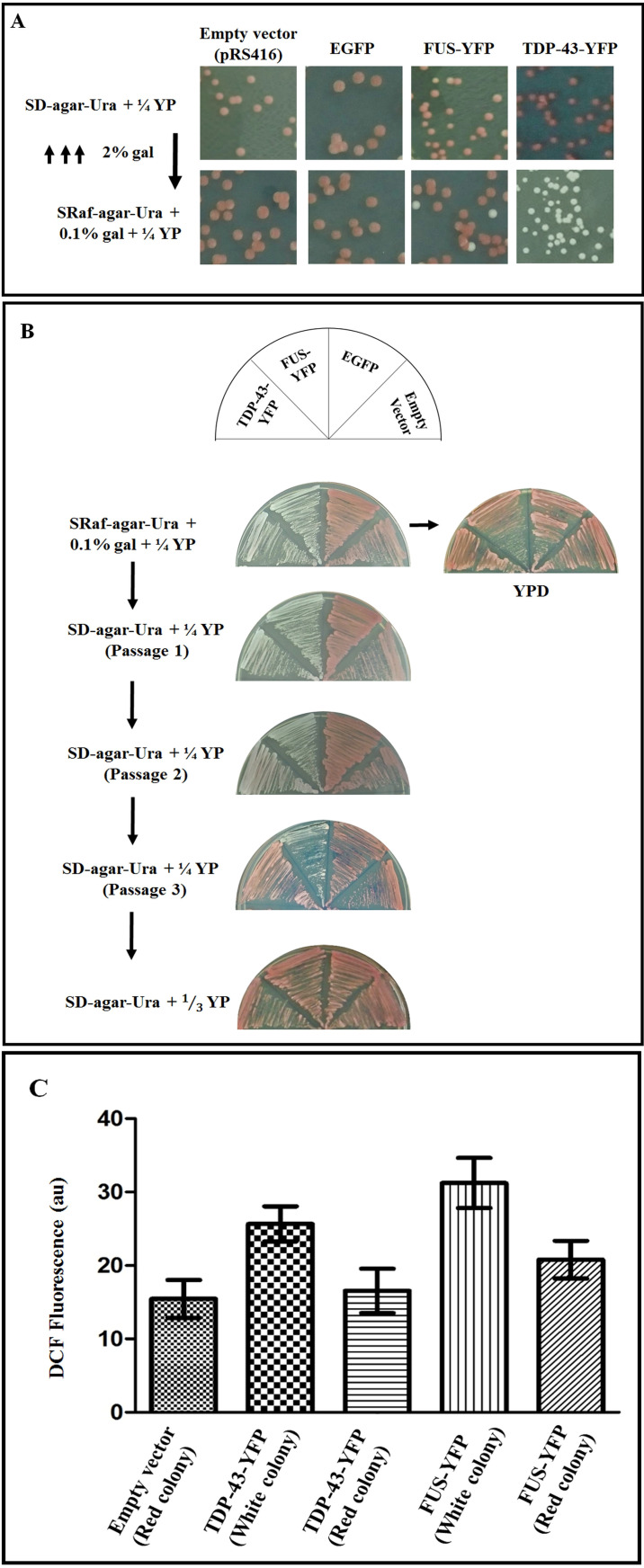
Yeast cells bearing ade1-14 allele (or those with ade1∆, or ade2∆ or ade2-1 allele) are pre-transformed with URA3 marked, galactose-inducible GAL1 promoter driven plasmid encoding amyloid forming proteins (for example: TDP-43-YFP or FUS-YFP). These cells are spread on plasmid selective SD-Ura + ¼ YP plates (see Recipes) and incubated at 30 °C for 4-5 days to obtain independent single colonies. Please note that the presence of ¼ YP in the medium allows for the red colony color development even on the drop-out medium, which otherwise is normally scored on YPD medium (see Recipes).
Subsequently, a red color phenotype manifesting single yeast colony from the SD-Ura + ¼ YP plates (Figure 1A) is picked and grown overnight in 5 ml of SRaf-Ura broth in a shaking incubator maintained at 200 rpm and 30 °C to overcome glucose repression.
Next morning, suitable aliquots of yeast cells from the overnight culture are inoculated to 5 ml of fresh SRaf-Ura broth to obtain starting OD of A600 nm = 0.2 and induced with 2% galactose final concentration to overexpress the GAL1 promoter driven genes and then allowed to grow at 30 °C for 8 h for the expressions of the FUS-YFP or TDP-43-YFP proteins (or until the amyloid aggregation is observed, which can vary for different amyloid proteins). Amyloid-like aggregation is scored by examining the yeast cells in fluorescence microscope for formation of punctate fluorescent foci of the FUS-YFP or TDP-43-YFP proteins.
Next, the cells are harvested by centrifugation at 3,000 × g at 25 °C in a table-top microcentrifuge and re-suspended in sterile water and then appropriately diluted, after recording A600 nm in a spectrophotometer, to spread ~200 cells per plate evenly on SRaf-Ura + 0.1% gal + ¼ YP plates (see Recipes) (1 OD600 nm = 1 x 107 cells/ml). Plates are then incubated at 30 °C for 4-5 days to grow single yeast colonies.
After four days, plates are examined for colony color and the appearance of white colonies due to oxidative-stress induced by the prior over-expression of the amyloid proteins (TDP-43-YFP and FUS-YFP) using 2% galactose (step 3) and the continued moderate expression of the amyloid proteins on the SRaf-Ura + 0.1% gal + ¼ YP plate (step 4) (Figure 1A). Please note that the percentage of white colonies obtained may vary for different amyloid proteins. When TDP-43-YFP was expressed, 100% white colonies were observed whereas for FUS-YFP, only 17% of the colonies were white and the remaining were red color. In controls (carrying only empty vector plasmid or any non-amyloid protein expressing plasmid like EGFP), only red color colonies were observed on SRaf-Ura + 0.1% gal + ¼ YP plate as amyloid-induced oxidative stress is not present.
-
Next, using sterile toothpick, small amount of cells from the white colonies on SRaf-Ura + 0.1% gal + ¼ YP plates are patched to SRaf-Ura + 0.1% gal + ¼ YP, SD-Ura + ¼ YP and YPD plates. Yeast cells continue to look white on the SRaf-Ura + 0.1% gal + ¼ YP and SD-Ura + ¼ YP plates but turn red on YPD plates (Figure 1B).
Note: Normalizations of cells are not required while passaging.
Subsequently, the cells from the white patches are sequentially passaged from the SD-Ura + ¼ YP plates to the same SD-Ura + ¼ YP medium plates as patches to observe the yeast color switch from white to red. This indicates that in the absence of protein expression and aggregate formation, the oxidative stress in the yeast cells gradually ceases and causes the white to red color switch. Please note that the amount of added YP required for obtaining red phenotype may vary for different amyloid proteins. For example, the white colonies of TDP-43-YFP turn red after the three passages on SD-Ura + ¼ YP plate, whereas white colonies of FUS-YFP require additional passage on SD-Ura + YP plate (see Recipes) for conversion to the red phenotype (Figure 1B).
To ascertain that the white color conversion is indeed due to oxidative stress, presence of reactive oxygen species (ROS) is subsequently analyzed. For this quantification of the ROS levels in the white and red color colonies, the yeast cells from the plates (or even from same liquid broth) are first washed twice with 0.1 M phosphate buffer (pH 7.4, temperature 25 °C) using centrifugation at 3,000 × g at 25 °C in a table-top microcentrifuge and then re-suspended in 450 µl of the phosphate buffer. To this cell suspension, 50 µl of 2’,7’-dichlorofluoroscein diacetate (DCFDA) dye is then added from a 100 µM stock (stored at -20 °C) to obtain 10 µM final DCFDA followed by incubation in the dark for 30 min at 25 °C. Then, the cells are centrifuged at 3,000 × g for 5 min at 25 °C.
Resulting pellet is re-suspended in 500 µl of 2 M lithium acetate (temperature 25 °C) and agitated gently on vortex mixer for 2 min. Then cells are pelleted by centrifugation at 3,000 × g at 25 °C for 5 min. Next, the DCF molecules, produced due to cleavage of DCFDA by cellular esterases followed by oxidation by cellular ROS, are extracted from the cells by re-suspending the pellet in 500 µl of DCF extraction buffer (see Recipes) and agitating vigorously for 2 min on vortex mixer. Finally, samples are centrifuged at 3,000 × g for 5 min at 25 °C and 300 µl of supernatant is used for DCF fluorescence measurement.
The DCF fluorescence is recorded using Spectramax M5e multimode microplate reader by recording emission at 524 nm after excitation at 485 nm (Eruslanov and Kusmartsev, 2010; James et al., 2015 ). The presence of elevated ROS levels is correlated by comparing the DCF fluorescence of appropriate control samples assayed similarly and simultaneously (Figure 1C).
Figure 1. Assay of amyloid protein-induced oxidative stress in S. cerevisiae yeast model.
A. Red phenotype bearing 74-D694 [psi-] yeast S. cerevisiae cells were pre-transformed either with empty vector plasmid pRS416 or those encoding EGFP, TDP-43-YFP or FUS-YFP proteins driven by GAL1 promoter. Overexpression of these proteins was induced with 2% galactose and cells were then plated on SRaf-Ura + 0.1% gal + ¼ YP plates to obtain single colonies. All the obtained colonies of empty vector and EGFP expressing cells were red, whereas ~17% FUS-YFP expressing colonies and 100% TDP-43-YFP expressing colonies displayed white phenotype. Images were taken by using a smartphone camera (13 MP). B. White phenotype displaying colonies obtained from FUS-YFP and TDP-43-YFP expressions on the SRaf-Ura + 0.1% gal + ¼ YP plates were sequentially passaged as patches on SD-Ura + ¼ YP plates. After the third passage, the TDP-43-YFP plasmid-bearing patches were found to turn red. However, the white patches of FUS-YFP plasmid-bearing cells remained white even after three passages on SD-Ura + ¼ YP plates but swiftly turned red on SD-Ura + YP plates. The white patches from both TDP-43-YFP and FUS-YFP converted to red phenotype even after the first passage on the rich complex medium containing YPD plates. Patches from empty vector or EGFP expressing plasmid transformants remained red on SRaf-Ura + 0.1% gal + ¼ YP, SD-Ura + ¼ YP, SD-Ura + YP as well as YPD plates. Images were taken by using a smartphone camera (13 MP). C. Relative ROS levels between the red and white phenotype manifesting yeast transformants were examined using DCFDA assay (Eruslanov and Kusmartsev, 2010; James et al., 2015 ). The yeast cells from TDP-43-YFP white patches (from first passage on SD-Ura + ¼ YP) yielded statistically significant [P = 0.0148, n = 3] increase in DCF fluorescence compared to the TDP-43-YFP red patches (from third passage on SD-Ura + ¼ YP plate) suggesting higher ROS in the white patches. Likewise, when white and red colonies from FUS-YFP expression from Figure 1A were passaged once as patches on SD-Ura + ¼YP plate (where they still retained their respective original phenotypes) and then assayed for ROS levels, the white patches yielded statistically significant [P = 0.0133, n = 3] higher DCF fluorescence compared to the red patches. As expected, the DCF fluorescence levels from white patches of TDP-43-YFP or FUS-YFP expressing cells were also significantly higher when compared with the cells transformed with the empty vector [P = 0.0072, n = 3; P = 0.0031, n = 3]. In all cases, unpaired t-test was used for the statistical comparisons.
Data analysis
DCF fluorescence was used for relative ROS level measurements in the red versus white phenotype manifesting yeast cells. To compare the statistical significance of the observed DCF fluorescence (and hence the relative ROS levels) between the red and white yeast cells after expression of FUS-YFP or TDP-43-YFP, data from three independent colonies of red and white phenotype each, were used and the P values were obtained by unpaired t-test using GraphPad QuickCalcs software (from GraphPad Software Inc., USA).
Notes
DCFDA assay should always be performed in the dark and amber colored vials should be used to store the stock solution in the view that DCFDA is light-sensitive and can readily auto-oxidize when directly exposed to light.
Recipes
-
YPD plate (100 ml)
2 g peptone
2 g dextrose
1 g yeast extract
2 g agar
-
SRaf-Ura + 0.1% gal + ¼ YP plate (100 ml)
0.7 g yeast nitrogen base with ammonium sulfate and amino acids but lacking uracil
1 g raffinose
0.1 g galactose
0.5 g peptone
0.25 g yeast extract
2 g agar
-
SRaf-Ura broth (100 ml)
0.7 g yeast nitrogen base with ammonium sulfate and amino acids but lacking uracil
1 g raffinose
-
SD-Ura + ¼ YP plate (100 ml)
0.7 g yeast nitrogen base with ammonium sulfate and amino acids but lacking uracil
2 g dextrose
0.5 g peptone
0.25 g yeast extract
2 g agar
-
SD-Ura + YP plate (100 ml)
0.7 g yeast nitrogen base with ammonium sulfate and amino acids but lacking uracil
2 g dextrose
0.66 g peptone
0.33 g yeast extract
2 g agar
-
DCF extraction buffer
5 µl 10% SDS
5 ml sterile distilled water
100 µl chloroform
Acknowledgments
This protocol is adapted from our original manuscript published previously as Bharathi et al. (2016). We thank Prof. Susan Liebman, University of Nevada Reno, USA, for yeast strains and plasmids. We thank IIT-Hyderabad, funded by MHRD, Government of India, for consumable support and research infrastructure. Junior Research Fellowship to Vidhya Bharathi from DBT, Government of India, and Senior Research Fellowship to Amandeep Girdhar from MHRD, Government of India, are also being duly acknowledged.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Bharathi V., Girdhar A., Prasad A., Verma M., Taneja V. and Patel B. K.(2016). Use of ade1 and ade2 mutations for development of a versatile red/white colour assay of amyloid-induced oxidative stress in Saccharomyces cerevisiae . Yeast 33: 607-620. [DOI] [PubMed] [Google Scholar]
- 2.Chernoff Y. O., Derkach I. L. and Inge-Vechtomov S. G.(1993). Multicopy SUP35 gene induces de-novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae . Curr Genet 24(3): 268-270. [DOI] [PubMed] [Google Scholar]
- 3.Eruslanov E. and Kusmartsev S.(2010). Advanced Protocols in Oxidative Stress II. In: Armstrong, D.(Ed.). Humana Press 57-72. [Google Scholar]
- 4.James, J., Fiji, N., Roy, D., Andrew, MG. D., Shihabudeen, M. S., Chattopadhyay, D. and Thirumurugan, K.(2015). A rapid method to assess reactive oxygen species in yeast using H2DCF-DA . Analytical Methods 7: 8572-8575. [Google Scholar]
- 5.Rossi S., Cozzolino. M. and Carri, M. T. (2016). Old versus new mechanisms in the pathogenesis of ALS. Brain Pathol 26: 276-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma K. G., Kaur R. and Bachhawat A. K.(2003). The glutathione-mediated detoxification pathway in yeast: an analysis using the red pigment that accumulates in certain adenine biosynthetic mutants of yeasts reveals the involvement of novel genes. Arch Microbiol 180(2): 108-117. [DOI] [PubMed] [Google Scholar]