Abstract
In vertebrates, hematopoietic stem cells (HSCs) regulate the supply of blood cells throughout the lifetime and help to maintain homeostasis. Due to their long lifespan, genetic integrity is paramount for these cells, and accordingly, a number of stem cell-specific mechanisms are employed. However, HSCs tend to show more DNA damage with increasing age due to an imbalance between proliferation rates and DNA damage responses. The comet assay is the most common and reliable method to study DNA strand breaks at the single-cell level. This procedure is based on the electrophoresis of agarose-embedded lysed cells. Following the electrophoretic mobilization of DNA, it is stained with fluorescent DNA-binding dye. Broken DNA strands migrate based on fragment size and form a tail-like structure called “the comet,” whereas intact nuclear DNA remains a part of the head of the comet. Since the alkaline comet assay fails to differentiate between single and double-strand breaks (DSBs), we used a neutral comet assay to quantitate the DSBs in HSCs upon aging and other physiological stresses. The protocol presented here provides procedural details on this highly sensitive, rapid, and cost-effective assay, which can be used for rare populations of cells such as HSCs.
Graphical abstract:
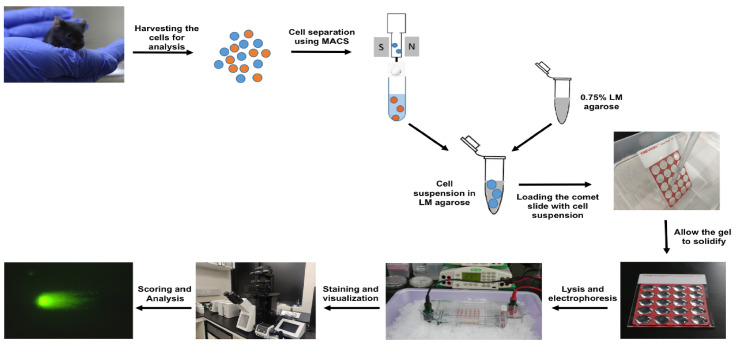
The neutral comet assay is an extremely useful tool that allows the detection and quantitation of double-strand DNA breaks at the single-cell level. The graphical abstract represents a flowchart for the neutral comet assay procedure.
Keywords: Hematopoietic stem cells, DNA damage, Double-strand breaks, Neutral comet assay
Background
During every round of cell proliferation, the genomic content is faithfully replicated, albeit with some errors. These errors lead to changes or modifications in the structure and chemical nature of genomic DNA. The replication machinery is tightly associated with DNA damage repair (DDR) pathways, thereby recovering cells from such damage (Chatterjee and Walker, 2017). Alternatively, the DNA damage response pathways can induce apoptosis (Nowsheen and Yang, 2012); however, some DNA damage is undetected and unrepaired, accumulating over a period of time. Accumulation of this DNA damage during aging poses a serious challenge to the healthy functioning of a variety of tissues, including hematopoietic systems. Recent studies have indicated that accumulation of age-related physiological changes in the stem cell population leads to alterations in immune system function ( Oh et al., 2014 ). In addition, several age-associated malignancies have been linked to DNA damage accumulation in the stem cell compartment (Beerman, 2017). Hematopoietic stem cells have cytoprotective and genoprotective mechanisms to safeguard their long-lasting functional potential; however, it has been demonstrated that DNA damage accumulates in HSCs during the aging process. Several research groups have proposed that this accumulation is due to increased proliferation rates in old HSCs. In contrast, others show that long-term dormancy may prevent rare DNA damage from accumulating due to lack of replication-associated repair ( Beerman et al., 2014 ).
To understand the fundamental processes involved in DNA damage accumulation and repair, quantitation of DNA damage is vital ( Lu et al., 2017 ). In 1984, Ostling and Johanson demonstrated the electrophoretic mobility of DNA fragments from the core nuclei (Ostling and Johanson, 1984). When single cells embedded in low-melting-point agarose are electrophoresed, the DNA-damaged fragments move faster, giving rise to tail-like structures resembling a comet. The intact part of the DNA is called the head of the comet and the trail produced due to impaired DNA is known as the tail of the comet. This single-cell gel electrophoresis assay, also known as the comet assay, is one of the most reliable and commonly used methods for analyzing DNA damage in cells. It has been successfully applied to understand the processes involved in genotoxicity, molecular epidemiology, and human bio-monitoring, along with solving basic biology questions regarding DNA damage and repair (Collins, 2004). This technique was further modified by Singh et al., who showed a substantial increase in reproducibility under alkaline conditions ( Lu et al., 2017 ). At alkaline pH, the sensitive sites are knocked down and the strands break due to distortion in their conformation. Therefore, the alkaline comet assay is commonly used to analyze strand breaks, DNA-protein crosslinks, and alkali-labile sites; nevertheless, the specificity for double-strand breaks is compromised ( Lu et al., 2017 ). To overcome this issue, the neutral comet assay was further developed ( Calini et al., 2002 ); it protects the DNA double strands from distortion, thus aiding in the specific detection of double-strand DNA damage. These DNA strand breaks result in the lengthening of DNA loops in genomic DNA, which form a comet-like tail in an electrophoretic mobility assay (Cortes- Gutierrez et al., 2012 ). Because double-strand DNA breaks lead to mutations and chromosomal abnormalities, it is important to detect and quantitate them. The neutral comet assay is of immense importance and is highly efficient for in vivo and ex vivo systems ( Sharma et al., 2011 ). The comets formed due to electrophoretic mobility can be visualized by fluorescence microscopy. The percentage of DNA damage can be estimated from the percentage of DNA present in the comet tail since they are directly proportional to one other. With advancements in technology such as fluorescent staining and automated comet scoring software, the comet assay has become popular for detecting DNA damage (Beedanagari, 2017). In the case of rare cell populations such as HSCs, the neutral comet assay has emerged as a promising technique to study DNA damage responses due to physiological, pathological, and age-associated stresses (Hazlehurst and Dalton, 2001; Milyavsky et al., 2010 ). The present protocol describes the method to assess DNA damage in freshly sorted hematopoietic stem cells from mice bone marrow.
Materials and Reagents
Autoclaved lint-free tissue paper
60-mm tissue culture plates (Eppendorf, catalog number: 0030701119)
26 G needle (BD PrecisionGlide, BD Medical (S) Pvt Ltd, catalog number: 302806)
1-ml syringe (BD Medical (S) Pvt Ltd, catalog number: 303060)
40-µm nylon strainer (Falcon, catalog number: 352340)
Bone marrow cells from mice
FACS tubes (Falcon, catalog number: 352054)
Comet slides (CometSlidesTM 20 well, Trevigen, catalog number: 4252-02K-01)
-
MACS apparatus
MS column (MACS, Miltenyi Biotec, catalog number: 130-042-201)
MACS multistand (MACS, Miltenyi Biotec, catalog number: 002139)
MiniMACSTM separator (MACS, Miltenyi Biotec, catalog number: 130-042-102)
-
Antibodies
APC-conjugated lineage antibody cocktail [2:100 concentration] (BD PharmingenTM, BD Biosciences, catalog number: 51-9003632)
Anti-mouse c-kit PE [1:100 concentration] (Biolegend, catalog number: 105807)
Anti-mouse Sca-1 PerCP-Cy5.5 [1:100 concentration] (Biolegend, catalog number: 122524)
1% agarose (Sigma-Aldrich, catalog number: A9539)
0.75% low-melting-point agarose (LM agarose) (Sigma-Aldrich, catalog number: A9414)
70% ethanol (Changshu Hongsheng Fine Chemical Co., Ltd, catalog number: 1170)
SYBR® Gold staining solution (ThermoFisher Scientific, Invitrogen, catalog number: S33102)
10× RBC lysis buffer (Biolegend, catalog number: 420301)
Lysis buffer (see Recipes)
1× neutral electrophoresis buffer (see Recipes)
DNA precipitation solution (see Recipes)
1× phosphate-buffered saline (PBS) (see Recipes)
Equipment
Electrophoresis unit (Mini-Sub® Cell GT, Bio-Rad, catalog number: 329BR014542)
Epifluorescence microscope (Olympus IX2-ILL100, OLYMPUS CORPORATION, catalog number: 0M04213)
Brightfield microscope (Primovert, Ziess, catalog number: 3842010199)
Software
ImageJ – Open Comet (Benjamin M. Gyori and Gireedhar Venkatachalam, www.cometbio.org)
Excel 2016 (Microsoft Corporation)
GraphPad Prism (GraphPad Software, Inc., www.graphpad.com)
Procedure
-
Cell preparation
Euthanize a C57Bl6/J mouse, dissect out the hind limbs, and remove the extra tissue from the bones to separate the femur and tibia.
Flush the bone marrow cells into a 50-ml polypropylene tube using autoclaved 1× PBS and a 26 G needle with a 1-ml syringe until the bones are cleared inside.
Mix the cells to make a single cell suspension and strain through a 40-μm nylon strainer. Centrifuge at 600 × g for 5 min at 4°C.
Lyse the RBCs by adding 500 µl 1× RBC lysis buffer (diluted in 1× PBS) to the pellet and neutralize the reaction after 2 min by adding 2 ml 1× PBS. Centrifuge at 600 × g for 5 min at 4°C.
Resuspend the cell pellet in 1 ml 1× PBS and count the cells using a hemocytometer.
Incubate the cells with the required antibodies (2 µl/5 million cells for Lin APC and 1 µl/5 million cells for c-Kit PE and Sca-1 PerCP-Cy5.5) to stain hematopoietic progenitor cells (Lin APC cocktail, c-Kit PE, Sca-1 PerCP-Cy5.5) for 30 min.
Centrifuge at 600 × g for 5 min at 4°C. Wash the cells with 1× PBS and transfer to a 5-ml FACS tube for sorting.
-
Sort the pure LSK (Lineage– Sca-1+ ckit+) population into 1× PBS and store at 4°C until further use.
Note: Alternatively, MACS sorting can be used to isolate less pure HSC progenitor populations.
-
Slide preparation
Weigh 1 g agarose and dissolve in 100 ml 1× PBS using a microwave oven.
Place the comet slides inside a plastic container in a slanted position (Figure 1A). Pipette out 1 ml agarose and evenly coat the slides and, leave to solidify for 10-15 min at room temperature (Figure 1B).
-
Freshly prepare 0.75% low-melting (LM) agarose using 1× PBS in a 100-ml beaker. Dissolve using a microwave oven.
Note: Transfer the molten 0.75% LM agarose to a storage bottle for further use. Before use, re-melt in a beaker containing hot water.
Allow the hot LM agarose to cool for at least 20 min at 37°C.
-
Add 1 × 105 cells/ml to the molten LM agarose at a ratio of 1:10 (v/v).
Note: If you take 500 µl molten LM agarose, add 50 µl cells in 1× PBS (Ca2+ and Mg2+ free) at 1 × 105 cells/ml.
-
Immediately pipette 50 µl suspension onto the comet slide. Use the tip of the pipette to spread the mixture uniformly over the surface area and allow to solidify (Figure 1C).
Note: If the sample mixture is not dispersing uniformly on the slide, heat up the LM agarose at 37°C before suspending the cells, and prepare the sample mixture again.
-
Keep the slides at 4°C for 10 min in the dark (e.g., place in a refrigerator).
Note: A clear 0.5 mm ring appears at the edge of the well (Figure 1D). To increase the adherence of the sample to the slide, the gelling time and humidity should be increased.
-
Lysis and Electrophoresis
Prepare the lysis solution (see Recipes) and place on ice for at least 20 min before use.
Pour the required amount of pre-chilled lysis solution in a tray and immerse the slides. Place the tray on ice or at 4°C for 1 h.
Take the slides from the lysis solution and drain the excess.
Wash the slides by placing them in 50 ml pre-chilled 1× neutral electrophoresis buffer (see Recipes) at 4°C for 30 min.
Pour 950 ml pre-chilled 1× neutral electrophoresis buffer into the Comet Assay® ES tank. Place the slides in the electrophoresis slide tray and cover with the slide tray overlay (Figure 2A).
Set the power supply to 21 volts and perform electrophoresis for 1 h at 4°C.
For other electrophoresis units, align the slides equidistant from the electrodes, add 1× neutral electrophoresis buffer not exceeding 0.5 cm above the slide (Figure 2B), and apply voltage for 30 min at 1 volt per cm, measuring the distance between the two opposite electrodes of the electrophoresis tank (e.g., If the distance is 21 cm then apply 21 V).
Run the electrophoresis unit either in a cold room or over an ice tray containing enough ice to partially cover the unit (Figure 2C).
-
Staining and visualization of slides
Drain the excess neutral electrophoresis buffer and immerse the slides in DNA precipitation solution for 30 min at room temperature (see Recipes).
Remove the slides and immerse them in 70% ethanol for 5 min at room temperature.
-
Allow the samples to dry for 10-15 min at room temperature.
Note: Drying the samples will allow the cells to settle in a single plane, making visualization, recording, and quantitation easier. Samples can be stored with desiccant at room temperature before scoring.
-
Add 100 µl diluted SYBR® Gold onto each sample and incubate for 30 min.
Note: Trevigen provides the CometAssay® Silver Staining Kit designed for comet staining. Silver staining helps in the visualization of comets on any transmission light microscope and permanently stains the samples for long-term storage. It is recommended that samples are dried before silver staining.
Tap the slides gently to remove excess SYBR® Gold solution. Allow the slide to dry completely at room temperature in the dark.
-
View the slides using epifluorescence microscopy and obtain unbiased images for every cell that could be visualized.
Note: The efficiency of the method can be tested using cells challenged with genotoxic stress as a positive control. Here, hematopoietic stem and progenitor cells from young adults (8 weeks old) (Figure 3A) were compared with cells from aged mice (2 years old) (Figure 3B). Comet tail formation in cells from aged mice is clearly visible. Comparison between the two cell types for the proportion of genomic DNA in the head versus the tail can be used as a reliable quantitation of the extent of DNA damage.
-
Scoring and analysis of the cells
Scoring of the comet is performed in the ImageJ plugin – Open Comet. Launch the plugin in ImageJ (Figure 4A).
Input your image files to the Input files option by clicking on the Browse button (Figure 4B).
Choose an Output directory where the output files will be stored, along with the name of the file that it should be saved under (Figure 4C and 4D).
Choose the default analysis option and Click RUN.
The analyzed output data will appear in the Output directory.
The Output file consists of an Output Image and an Output Spreadsheet. Review all the Output images for false analysis and outliers. Note the Number given specifically for each image (Figure 5A).
Open the Spreadsheet and manually delete the outlier numbers.
Obtain the values for Olive moment and Tail moment from the Spreadsheet and graphically represent the data using GraphPad PRISM (Figure 5B).
Figure 1. Preparation of a comet slide.
A. A clean Trevigen® CometSlideTM HT is placed inside a plastic container. B. The comet slide is pre-coated with molten 1% agarose. This ensures that cells adhere to the slide efficiently. After pre-coating with agarose, allow the gel to solidify. C. The 0.75% LM agarose containing the cells is added gradually to the slide. Allow the gel to solidify at 4°C. D. The comet slide after solidification of the gel.
Figure 2. Electrophoresis under neutral conditions.
A. After lysis, the comet slide is placed in the electrophoretic unit and neutral electrophoresis buffer is added to the buffer tank. B. The neutral electrophoresis buffer should not exceed 0.5 cm above the slide. C. Perform electrophoresis at 21 volts for 1 h at 4°C.
Figure 3. Staining and visualization of cells.

A. HSC from young mice; the nucleus in the cell is intact, indicating that there is no DNA damage. B. HSC from old mice; the nucleus in the cell is not intact and is forming a head and a comet tail, indicating that there is DNA damage.
Figure 4. Comet scoring using ImageJ.

A. Open the ImageJ software and click on Plugins. Select OpenComet_src_v1.3.1 and click on OpenComet. B. A dialogue box will open, where you need to browse the Input files and Output directory. C. Select the images for scoring the input file. D. Choose the Output directory.
Figure 5. Results of comet scoring.

A. Identification of the comet head and tail. B. Excel spreadsheet containing the computed comet parameters.
Recipes
-
Lysis buffer (Ingredients/1,000 ml)
3.5 M NaCl – 146.1 g
100 mM EDTA – 37.2 g
100 mM Trizma Base – 1.2 g
Dissolve the ingredients in 700 ml dH2O.
Add 8 g NaOH pellets and adjust the pH to 10.0 using HCl or NaOH (concentrated).
50 ml is required for each assay.
Add 1% Triton X-100 – 0.5 ml.
10% dimethyl sulphoxide (DMSO) – 5 ml
-
1× neutral electrophoresis buffer
To prepare 10× neutral electrophoresis buffer:
Tris base (mol. wt. = 121.14) – 60.57 g
Sodium acetate (mol. wt. = 136.08) – 204.12 g
Dissolve in 450 ml dH2O and adjust the pH to 9.0 using glacial acetic acid.
Adjust the volume to 500 ml and filter sterilize. Store at room temperature.
Dilute the 10× stock to 1× in dH2O to prepare 1 L working strength buffer and pre-chill at 4°C.
-
DNA precipitation solution
Prepare a 10 ml stock solution of 7.5 M ammonium acetate: NH4Ac (mol. wt. = 77.08) = 5.78 g
dH2O (after NH4Ac is dissolved) – 10 ml
For 50 ml DNA precipitation solution:
7.5 M NH4Ac (mol. wt. = 77.08) – 6.7 ml
95% EtOH (reagent grade) – 43.3 ml
-
SYBR® Gold staining solution
From SYBR® Gold concentrate (10,000× in DMSO)
SYBR® Gold – 1 µl
Electrophoresis Buffer – 10 ml
Store at 4°C in the dark
-
1× phosphate-buffered saline (PBS)
To prepare 10× PBS stock solution:
NaCl – 80 g
KCl – 2 g
Na2HPO4 – 14.4 g
KH2PO4 – 2.4 g
Take 800 ml Milli-Q water, dissolve the salts and adjust the pH to 7.4, and make up the volume to 1,000 ml
Dilute the 10× stock to 1× in dH2O
Acknowledgments
This work was supported by the Wellcome Trust/DBT India Alliance Fellowship (IA/I/15/2/502061) awarded to SK and intramural funds from the Indian Institute of Science Education and Research Thiruvananthapuram (IISER TVM). The institutional animal facility is supported by funds from the Department of Science and Technology, Government of India (under FIST scheme; SR/FST/LS-II/2018/217). IMR is supported by a Senior Research Fellowship from the University Grants Commission (UGC), India. This protocol was taken from the article by Biswas et al. (2020) “The Periostin/Integrin-av Axis Regulates the Size of Hematopoietic Stem Cell Pool in the Fetal Liver” and was modified from the protocol described in the Trevigen comet assay kit ( Biswas et al., 2020 ).
Competing interests
The authors have no competing financial or non-financial interests relevant to this study.
Ethics
The animals used to isolate the HSCs for DNA damage analysis (C57BL/6J-CD45.2) were bred and maintained in the animal facility at IISER Thiruvananthapuram. The animals were maintained as per the guidelines provided by the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Ministry of Environment and Forests (MoEF), Government of India. During the experiments, mice were maintained in isolator cages and allowed access to autoclaved acidified water and irradiated food ad libitum. All animal experiments were approved by the Institutional Animal Ethics Committee (IAEC) of the institute.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Beedanagari S.(2017). 4.11-Genetic Toxicology. In: Chackalamannil, S., Rotella, D. and Ward, S. E.(Eds.). Comprehensive Medicinal Chemistry III. pp 195-203. Elsevier, Oxford. [Google Scholar]
- 2.Beerman I.(2017). Accumulation of DNA damage in the aged hematopoietic stem cell compartment. Semin Hematol 54(1): 12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beerman I., Seita J., Inlay M. A., Weissman I. L. and Rossi D. J.(2014). Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell 15(1): 37-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas A., Roy I. M., Babu P. C., Manesia J., Schouteden S., Vijayakurup V., Anto R. J., Huelsken J., Lacy-Hulbert A., Verfaillie C. M. and Khurana S.(2020). The Periostin/Integrin-alphav Axis Regulates the Size of Hematopoietic Stem Cell Pool in the Fetal Liver. Stem Cell Reports 15(2): 340-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calini V., Urani C. and Camatini M.(2002). Comet assay evaluation of DNA single- and double-strand breaks induction and repair in C3H10T1/2 cells. Cell Biol Toxicol 18(6): 369-379. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee N. and Walker G. C.(2017). Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen 58(5): 235-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins A. R.(2004). The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol 26(3): 249-261. [DOI] [PubMed] [Google Scholar]
- 8.Cortes-Gutierrez E. I., Hernandez-Garza F., Garcia-Perez J. O., Davila-Rodriguez M. I., Aguado-Barrera M. E. and Cerda-Flores R. M.(2012). Evaluation of DNA single and double strand breaks in women with cervical neoplasia based on alkaline and neutral comet assay techniques. J Biomed Biotechnol 2012: 385245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazlehurst L. A. and Dalton W. S.(2001). Mechanisms associated with cell adhesion mediated drug resistance(CAM-DR) in hematopoietic malignancies. Cancer Metastasis Rev 20(1-2): 43-50. [DOI] [PubMed] [Google Scholar]
- 10.Lu Y., Liu Y. and Yang C.(2017). Evaluating In Vitro DNA Damage Using Comet Assay . J Vis Exp (128). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milyavsky M., Gan O. I., Trottier M., Komosa M., Tabach O., Notta F., Lechman E., Hermans K. G., Eppert K., Konovalova Z., Ornatsky O., Domany E., Meyn M. S. and Dick J. E.(2010). A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell 7(2): 186-197. [DOI] [PubMed] [Google Scholar]
- 12.Nowsheen S. and Yang E. S.(2012). The intersection between DNA damage response and cell death pathways. Exp Oncol 34(3): 243-254. [PMC free article] [PubMed] [Google Scholar]
- 13.Oh J., Lee Y. D. and Wagers A. J.(2014). Stem cell aging: mechanisms, regulators and therapeutic opportunities. Nat Med 20(8): 870-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostling O. and Johanson K. J.(1984). Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem Biophys Res Commun 123(1): 291-298. [DOI] [PubMed] [Google Scholar]
- 15.Sharma A., Shukla A. K., Mishra M. and Chowdhuri D. K.(2011). Validation and application of Drosophila melanogaster as an in vivo model for the detection of double strand breaks by neutral Comet assay . Mutat Res 721(2): 142-146. [DOI] [PubMed] [Google Scholar]




