Abstract
We performed an assay to test the ability of different E. coli strains to survive inside amoebal cells after ingestion. In the assay we incubated bacteria together with cells of Dictyostelium discoideum for six hours. After co-incubation most of the uningested bacteria were removed by centrifugation and the remaining uningested bacteria were killed by gentamicin. Gentamicin is used because it does not penetrate into eukaryotic cells allowing the ingested bacteria to survive the antibiotic treatment, whereas bacteria outside the amoebal cells are killed.
Keywords: Bacteria, Amoebae, Protozoa, Protists, Grazing, Digestion resistance
Background
Bacteria have evolved several different strategies to avoid or resist protozoan predation ( Rønn et al., 2012 ). Mechanisms that reduce chance of ingestion such as increased cell size, aggregation, biofilm formation and increased swimming speed are well documented (Matz and Kjelleberg, 2005; Rønn et al., 2012 ) whereas mechanisms that allow bacteria to resist digestion in protozoan food vacuoles after ingestion are less studied ( Gong et al., 2016 ). Here we describe a method to investigate the ability of bacterial strains to survive after ingestion by the social amoeba Dictyostelium discoideum. We used this assay to investigate if copper resistant E. coli have higher chance of survival after ingestion than bacteria without copper resistance ( Hao et al., 2016 ). We studied copper resistance because it is known that macrophages, which are phagocytotic cells with an important role in the immune system of vertebrates, use copper to kill bacteria in the phagosome (Hodgkinson and Petris, 2012). We hypothesized that this killing mechanism originally evolved in free-living protozoa long before multicellular life arose and hence copper resistance could be a factor that protects bacteria against protozoan predation.
The assay is a modification of the Gentamicin protection assay used for evaluation of the ability of macrophages to kill bacteria (see e.g., Kaneko et al., 2016 ) and it utilizes that gentamicin is not able to penetrate into eukaryotic cells. In the original assay the number of internalized bacterial cells in the macrophages is determined at two time points. First, macrophages are incubated with bacteria for a given time period and after this co-incubation the uningested bacteria are removed and the number of internalized bacteria is estimated. Second measurement is taken after macrophages are incubated without extracellular bacteria to allow them to digest the internalized bacteria. The ratio between these two estimates is a measure of the bacterial survival rate.
Rapid digestion of some of our strains led to reduced reliability of estimates of the disappearance rate and made the original procedure unsuitable for use with Dictyostelium. We therefore modified the procedure so that we only estimated the number of surviving internalized bacteria at one time point after co-incubation of bacteria and amoebae for 6 h. This estimate reflects an equilibrium between ingestion and digestion rate of bacteria. It should be noted that this will only allow comparison of the digestion rate of different bacterial strains if it can be assumed that the bacteria are consumed with similar rates. In our case we had evidence that the different E. coli strains were taken up by the amoebae to the same extent and therefore we used the assay as an indicator of digestion rate. The assay will still be useful even if it cannot be assumed that bacteria are ingested at the same rate but of course it should be interpreted accordingly. It must also be noted that the procedure can only be used with bacteria that are sensitive to gentamicin.
Materials and Reagents
Pipettes and sterile tips
Sterile centrifuge tubes (e.g., Falcon® 50 ml conical centrifuge tubes)
Sterile cell culture flasks (e.g., 25 cm2 Nunc® EasYFlasksTM)*
24 well, cell culture plates, flat bottom (Corning, Costar®–or similar)
Sterile 1.5 ml Eppendorf tubes
50 ml Falcon tubes (Corning®–or similar)
Sterile filters (0.2 µm) for sterile filtration (Corning)
Dictyostelium discoideum AX4 (or other axenic strain; can be obtained at the Dicty Stock Center, Northwestern University, Chicago, IL, USA)
Bacterial cultures*
Gentamicin (Sigma-Aldrich, catalog number: G1264)
Dihydrostreptomycin sulfate (Sigma-Aldrich, catalog number: PHR1517)
Triton X-100 (Sigma-Aldrich, catalog number: 93443)
OxoidTM tryptone (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: LP0042)
OxoidTM yeast extract (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: LP0021)
Sodium phosphate dibasic heptahydrate (Na2HPO4.7H2O) (Sigma-Aldrich, catalog number: S9390)
Potassium phosphate monobasic (KH2PO4) (Sigma-Aldrich, catalog number: P5655)
-
Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 435570)
Note: This product has been discontinued.
Glucose (Sigma-Aldrich, catalog number: G8270)
Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C5670)
-
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 310166)
Note: This product has been discontinued.
Agar
Sterile HL5 medium (see Recipes)
SorC buffer (see Recipes)
Sterile Luria Broth (LB) (see Recipes)
Agar plates with Luria agar (see Recipes)
*Note: This protocol describes the procedure we used to compare survival rate of different E. coli strains, but other bacteria may be used. It is a requirement that the bacteria are sensitive to gentamicin.
Equipment
Pipette
Laminar flow hood (Heraeus, model: Air HLB 2472)
125 ml Erlenmeyer flasks
Rotary shaker for incubation at 22 °C
37 °C constant incubator (Thermo Fisher Scientific)*
Rotary shaker for incubation at 37 °C*
Benchtop centrifuge for Eppendorf tubes (cooled at 4 °C) (Eppendorf, model: 5424 R)
Centrifuge for 50 ml Falcon tubes (HERMLE LABORTECHNIK, model: Z 326 K)
Spectrophotometer and respective cuvettes (600 nm)
Autoclave (STIK, model: MJ-Series)
Vortex mixer (Scientific Industries, model: Vortex-Genie 2, catalog number: SI-0236)
pH meter and calibration buffers
Inverted microscope (Olympus, model: CKX31)**
Microscope (Nikon Instruments, model: ECILPSE 80i)
Neubauer hemocytometer blood count with double counting chamber (Gizmo Supply, Germany)
*Note: Incubators set at 37 °C are necessary for work with E. coli but if other bacteria are used, growth temperature conditions should of course be modified accordingly.
**Note: An inverted microscope is very useful for inspection of culture flasks and cell culture plates during the experiment.
Procedure
Notes:
All work is carried out under sterile conditions using a laminar flow hood.
The experiments are carried out with liquid axenic cultures of Dictyostelium discoideum. Strains of D. discoideum are usually provided on plates (e.g., from the Dicty stock center). Before start of the experiments, the amoebae should be brought into liquid culture. Detailed and useful information about culturing Dictyostelium can be found at the homepage of the Dicty stock center, http://dictybase.org/ (Fey et al., 2007).
Preculturing of Dictyostelium in cell culture flasks has the advantage that it allows direct inspection of the cultures, but it is also possible to grow the cultures in shaking liquid cultures e.g., in 125 ml Erlenmeyer flasks. This can make it easier to obtain high cell densities in the cultures.
Growth of Dictyostelium can be improved if it is supplied with washed heat-killed bacteria. We have previously used overnight cultures of E. coli which were washed by centrifugation, resuspended in SorC and heat-killed at 65 °C for 10 min.
In this protocol we used different strains of E. coli. The procedure could be used for other bacteria; in this case bacteria-specific growth conditions, optical densities (OD) for estimation of bacterial cell density etc. should be modified accordingly.
-
Day 1. Start D. discoideum culture
Add 500 µl of a 2-3 day old D. discoideum culture to 10 ml fresh sterile HL5 in sterile culture flasks.
Incubate flasks at 22 °C (without shaking).
-
Day 2(-3). Start bacterial cultures
Start bacterial cultures by inoculating 25 ml sterile LB medium with 1% inoculum of E. coli culture (overnight growth) in 125 ml Erlenmeyer flasks.
Incubate overnight on a rotary shaker at 37 °C, 200 rpm.
-
Day 3(-4). Survival assay
-
Harvest bacteria
Harvest 1 ml of bacteria by centrifugation at 12,000 × g for 2 min.
Remove supernatant by carefully pipetting the liquid without disturbing the pellet.
Resuspend bacterial pellet in 1 ml of 1:4 HL5:SorC solution.
Centrifuge at 12,000 × g for 2 min.
Remove the supernatant and resuspend bacterial pellet in 1 ml of 1:4 HL5:SorC solution.
Adjust bacterial density to OD600 of 0.1 (around 108 CFU ml-1).
-
Harvest D. discoideum cells
Add amoebal culture to 50 ml Falcon tubes and centrifuge at 500 × g for 4 min.
Remove supernatant by carefully pipetting the liquid without disturbing the pellet.
Resuspend amoebae in 25 ml of 1:4 HL5:SorC solution.
Centrifuge at 500 × g for 4 min.
Resuspend amoebae in 1:4 HL5:SorC solution.
Estimate amoebal density in the suspension by counting in a hemocytometer.
Adjust amoebal density to 1 x 106 cells ml-1.
-
Start survival assay
-
Add 900 µl amoebal suspension (from step C2g) to a number of wells in 24-well culture plates.
Note: We use at least 6 replicates for each bacterial strain.
Add 100 µl bacterial suspension adjusted to approximately 4.5 x 107 cells ml-1 in order to obtain a multiplicity of infection (MOI) of 5.
To obtain other levels of MOI, the bacterial cell densities can be adjusted accordingly.
-
Prepare a set of replicates with 900 µl amoebal suspension and 100 µl sterile 1:4 HL5:SorC.
Note: This treatment serves as a negative control to ensure that the initial amoebal cultures are free from bacteria.
-
For each of the bacterial strains, prepare a set of replicates with 100 µl bacterial suspension and 900 µl 1:4 HL5:SorC solution (without amoebae).
Note: This treatment serves as a control for the efficiency of gentamicin to kill extracellular bacteria.
Incubate at 22 °C for six hours (without shaking).
-
-
Add gentamicin
Transfer content of each well in the 24-well cell culture plates to a 1.5 ml Eppendorf tube.
Centrifuge at 500 × g for 4 min.
Remove supernatant by carefully pipetting the liquid without disturbing the pellet.
Add 1 ml SorC buffer.
Centrifuge at 500 × g for 4 min.
Remove supernatant by carefully pipetting the liquid without disturbing the pellet.
Repeat steps C4d-C4f.
Add 1 ml SorC containing 400 µg ml-1 gentamicin.
Incubate for 1 h at room temperature.
-
Lyse amoebal cells and count internalized bacteria
Centrifuge at 500 × g for 4 min.
Remove supernatant by carefully pipetting the liquid without disturbing the pellet.
Add 1 ml SorC buffer.
Centrifuge at 500 × g for 4 min.
Remove supernatant by carefully pipetting the liquid without disturbing the pellet.
Repeat steps C5c-C5e.
-
Add 1 ml of a solution with 0.4% Triton X-100 diluted in SorC buffer.
Note: This will lyse the amoebal cells.
-
Mix and prepare 10-fold dilution series of the lysate. The dilutions are performed in SorC buffer using a vortex mixer.
Note: Preparation of dilution series and plating should be performed immediately after addition of Triton-X since longer exposure to the solvent may kill bacteria. The most important thing is that the different replicates are exposed for the solvent for approximately the same length of time.
-
Plate 100 µl of relevant dilutions on LB agar plates.
Note: We recommend using at least two replicate agar plates per dilution level in order to reduce variability at this step. In our experiment we had relatively low bacterial density so for our lowest dilution we simply plated 100 µl of sample directly from the experimental wells.
Incubate for 16 h at 37 °C.
Count colonies on agar plates.
-
Data analysis
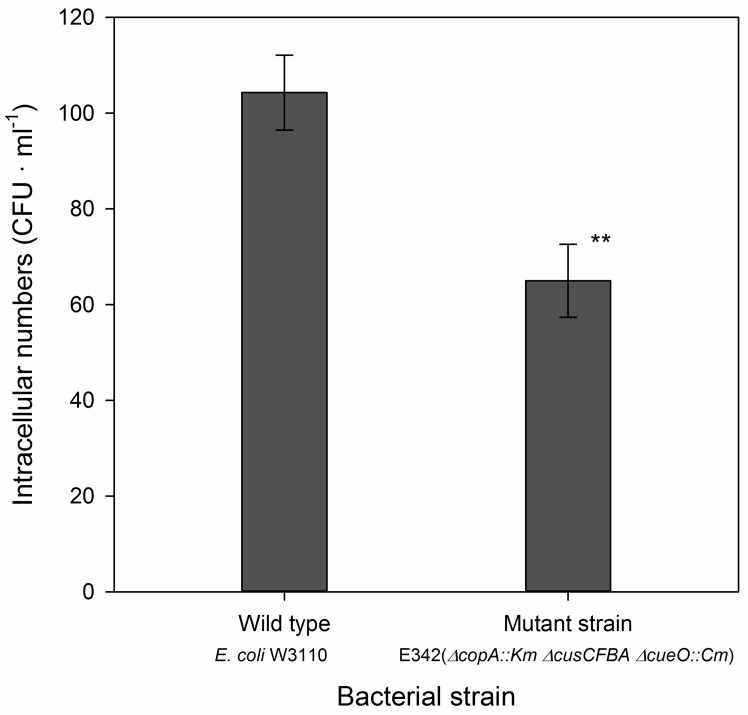
Use agar plates with ~20-200 colonies to calculate the number of colony-forming units (CFU) per ml for each of the replicate wells. The number of CFU’s in different treatments is analyzed with one-way analysis of variance, followed by a test to compare means. We used Duncan’s test ( Hao et al., 2016 ) but other tests would be equally suitable. In the example above we only compared two strains and here we used a t-test (Figure 1).
Figure 1. Number of intracellular bacteria in Dictyostelium cells co-incubated with a wild type strain of E. coli (W3110) and with the mutant, E345, with triple gene deletion.
The assay was performed at MOI 1. Data were analyzed with a t-test and the two strains were significantly different at the 1% level (P = 0.0045, n = 6 for each strain). Further details and examples can be found in Hao et al., 2016 .
Note: The two control treatments (from steps C3d and C3e) should be free from bacteria. Presence of bacteria in the negative control without bacteria (step C3d) indicate that the amoebal cultures are contaminated with bacteria and experiments will have to be repeated with newly established axenic amoebal cultures without contaminant bacteria. Presence of bacteria in control treatments without amoebae (step C3e) indicates that the bacteria were not efficiently killed by gentamicin and data cannot be used to evaluate bacterial survival.
Recipes
-
Sterile modified HL5
14.0 g tryptone (Oxoid)
7.0 g yeast extract (Oxoid)
0.35 g Na2HPO4.7H2O
1.2 g KH2PO4
Dissolve components in 937.5 ml distilled water, adjust pH with HCl to pH 6.4-6.7, and autoclave for 20 min to sterilize
Prepare a sterile solution of glucose (224 g L-1) by filtering through 0.2 µm sterile membrane filters. Add 62.5 ml of this solution to 937.5 ml of the autoclaved solution above
(0.05 g L-1 dihydrostreptomycin-sulfate*)
*Note: Addition of dihydrostreptomycin-sulfate can be useful when cultures are initiated or have been contaminated with bacteria. However, for the precultures for the actual experiments we avoided the use of antibiotics, since antibiotics could interact with our assay of bacterial survival.
-
SorC buffer
2.0 g L-1 KH2PO4
0.29 g L-1 Na2HPO4
Dissolve in distilled water, check that pH is 6.0 ± 0.1, if not adjust with HCl
Add 1 ml 50 mM CaCl2 to 1 L SorC solution, and autoclave
-
Sterile Luria Broth (LB)
10.0 g L-1 tryptone
5.0 g L-1 yeast extract
10.0 g L-1 NaCl
Dissolve components in distilled water, adjust pH with HCl to pH 6.8-7.2, and autoclave for 20 min
-
Agar plates with Luria agar
Add 15.0 g L-1 agar to Luria Broth
Autoclave and pour plates
Note: Make sure to calculate the number of agar plates you need for the experiment. Remember that for each bacterial strain you need to plate samples both from the replicates with bacteria added (steps C3a-C3b) and for the control plates without amoebae (step C3e). Furthermore, in addition to the treatments with bacterial strains, you should also prepare plates for the control treatment with amoebae but without added bacteria (step C3d).
Acknowledgments
Research in the laboratory of CR was funded by startup funds from Fujian Agriculture and Forestry University and the 100 Talents program from Fujian province. Authors would like to thank the International Postdoctoral Exchange Fellowship Program (No. 20150079), and Twasol Research Excellence Program, ‘TRE Program’, King Saud University, Saudi Arabia for support. DLH thanks the Fulbright Kommission for a Fulbright Scholar grant at the University of Copenhagen. This protocol was adapted from the publication by Hao et al., 2016 .
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Fey P., Kowal A. S., Gaudet P., Pilcher K. E. and Chisholm R. L.(2007). Protocols for growth and development of Dictyostelium discoideum . Nat Protoc 2(6): 1307-1316. [DOI] [PubMed] [Google Scholar]
- 2.Gong J., Qing Y., Zou S., Fu R., Su L., Zhang X. and Zhang Q.(2016). Protist-bacteria associations: Gammaproteobacteria and Alphaproteobacteria are prevalent as digestion-resistant bacteria in ciliated protozoa . Front Microbiol 7: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao X., Luthje F., Ronn R., German N. A., Li X., Huang F., Kisaka J., Huffman D., Alwathnani H. A., Zhu Y. G. and Rensing C.(2016). A role for copper in protozoan grazing- two billion years selecting for bacterial copper resistance. Mol Microbiol 102(4): 628-641. [DOI] [PubMed] [Google Scholar]
- 4.Hodgkinson V. and Petris M. J.(2012). Copper homeostasis at the host-pathogen interface. J Biol Chem 287(17): 13549-13555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneko M., Emoto Y. and Emoto M.(2016). A simple, reproducible, inexpensive, yet old-fashioned method for determining phagocytic and bactericidal activities of macrophages. Yonsei Med J 57(2): 283-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matz C. and Kjelleberg S.(2005). Off the hook--how bacteria survive protozoan grazing. Trends Microbiol 13(7): 302-307. [DOI] [PubMed] [Google Scholar]
- 7.Rønn R., Vestergård M. and Ekelund F.(2012). Interactions between bacteria, protozoa and nematodes in soil. Acta Protozool 51: 223-235. [Google Scholar]