Abstract
Prohibitins comprise a protein family in eukaryotic cells with potential roles in senescence and tumor suppression. Phb1p and Phb2p, members of the prohibitin family in Saccharomyces cerevisiae, have been implicated in the regulation of the replicative life span of the cells and in the maintenance of mitochondrial morphology. The functional activities of these proteins, however, have not been elucidated. We demonstrate here that prohibitins regulate the turnover of membrane proteins by the m-AAA protease, a conserved ATP-dependent protease in the inner membrane of mitochondria. The m-AAA protease is composed of the homologous subunits Yta10p (Afg3p) and Yta12p (Rca1p). Deletion of PHB1 or PHB2 impairs growth of Δyta10 or Δyta12 cells but does not affect cell growth in the presence of the m-AAA protease. A prohibitin complex with a native molecular mass of approximately 2 MDa containing Phb1p and Phb2p forms a supercomplex with the m-AAA protease. Proteolysis of nonassembled inner membrane proteins by the m-AAA protease is accelerated in mitochondria lacking Phb1p or Phb2p, indicating a negative regulatory effect of prohibitins on m-AAA protease activity. These results functionally link members of two conserved protein families in eukaryotes to the degradation of membrane proteins in mitochondria.
The mechanisms which underlie the turnover of membrane proteins in the cell remain poorly understood. Various plasma membrane proteins are internalized in a ubiquitin-dependent manner and degraded in lysosomes and vacuoles (14). Proteolysis of membrane proteins of the endoplasmic reticulum requires their retrograde transport to the cytosol and proceeds via the cytoplasmic ubiquitin-proteasome pathway (5, 30). Another proteolytic system has emerged from studies on the turnover of mitochondrial inner membrane proteins. Proteolysis is mediated by membrane-embedded ATP-dependent AAA proteases, which comprise a conserved protein family with homologues in various organisms, including bacteria, plants, yeasts and humans (18, 28, 31).
Two AAA proteases have been identified in the mitochondrial inner membrane of Saccharomyces cerevisiae (11, 26, 34, 35). Both proteases are composed of homologous subunits and form high-molecular-mass complexes of approximately 1 MDa (3, 20). Their catalytic sites, however, are exposed to opposite membrane surfaces: the m-AAA protease containing Yta10p (Afg3p) and Yta12p (Rca1p) acts on the matrix side, while the i-AAA protease containing Yme1p is active in the intermembrane space (3, 20). In S. cerevisiae, AAA proteases fulfill crucial roles in mitochondrial biogenesis. Deletion of YME1 impairs the respiratory competence of the cells at high temperature and results in changes in the morphology of mitochondria (6, 34). Cells lacking Yta10p or Yta12p, on the other hand, lose respiratory competence and exhibit deficiencies in the assembly of the F1F0-ATP synthase and respiratory chain complexes in the inner membrane (11, 27, 32, 35). The m-AAA protease controls the expression of the intron-containing mitochondrial genes COB and COX1, affecting the splicing of the respective pre-mRNAs (2). In addition, evidence is accumulating for posttranslational functions of the m-AAA protease during the biogenesis of the respiratory chain (2, 27). Mutations in a human homologue of Yta10p and Yta12p, paraplegin, have recently been reported to cause a hereditary form of spastic paraplegia, pointing to important functions of AAA proteases also in higher eukaryotes (7).
Prohibitin was originally identified in mammalian cells for its decreased expression in tumor cells and for its ability to negatively regulate cell proliferation (21, 25). Subsequent studies established that prohibitin belongs to a ubiquitous protein family in eukaryotes which is highly conserved from yeast to human (9). In eukaryotic cells there are two homologues which were initially found in association with the plasma membrane in B lymphocytes (33) but later localized to the inner membrane of mitochondria in various cell types (4, 8, 15). The S. cerevisiae prohibitin homologues Phb1p and Phb2p have been implicated in the regulation of the replicative life span of the cells (4, 8). Moreover, the observation of a genetic interaction of PHB1 or PHB2 with mitochondrial inheritance components points to a role for the maintenance of mitochondrial morphology (4). The function of prohibitins on the molecular level, however, is unknown.
Here we describe a role of the prohibitin family members Phb1p and Phb2p of S. cerevisiae for the degradation of membrane proteins by the m-AAA protease. Phb1p and Phb2p are anchored to the mitochondrial inner membrane via an N-terminal transmembrane segment and expose their C termini to the intermembrane space. They form a high-molecular-mass complex of approximately 2 MDa which physically interacts with the m-AAA protease but not with the homologous i-AAA protease. Proteolysis of nonassembled Cox3p and Atp6p by the m-AAA protease proceeds at an increased rate in mitochondria lacking Phb1p or Phb2p, suggesting an inhibitory effect of prohibitins on m-AAA protease activity.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast strains used in this study are derivatives of W303. Cells were grown at 30°C on YP medium (1% yeast extract, 2% peptone) containing 2% glucose or, for isolation of mitochondria, 2% galactose and 0.5% lactate.
Δyta10 (YHA101), Δyta12 (YHA201), and Δyme1 strains were described previously (2, 20). PHB1 and PHB2 were disrupted in wild-type, Δyta10, Δyta12, and Δyme1 cells by PCR-targeted homologous recombination (36) using the heterologous marker HIS3MX6 (Δphb1 [YGS410], Δphb1 Δyta10 [YGS404], and Δphb1 Δyme1 [YGS406]) or KanMX4 (Δphb1 Δyta12 [YGS416], Δphb2 [YGS501], Δphb2 Δyta10 [YGS504], Δphb2 Δyme1 [YGS505], and Δphb2 Δyta12 [YGS506]). The complete open reading frames of PHB1 or PHB2 were replaced by the disruption cassettes. COX4 was disrupted in W303 by homologous recombination using the auxotrophic marker TRP1. PHB1 and PHB2 were deleted in Δcox4 and Δatp10 cells (1) by PCR-targeted homologous recombination using the heterologous markers HIS3MX6 and KanMX4, respectively (36). Homologous recombination was verified in each case by PCR.
Gel filtration analysis.
Mitochondria were isolated from wild-type, Δphb1, and Δyta10 cells as described previously (13, 37) and resuspended (0.4 mg) at a concentration of 1 mg/ml in buffer A (phosphate-buffered saline [pH 7.4], 10% glycerol, 1 mM ATP, 4 mM magnesium acetate, 0.5 mM phenylmethylsulfonyl fluoride) containing 1% digitonin or 0.2% Triton X-100 as indicated. After incubation for 30 min at 4°C under vigorous mixing, mitochondrial extracts were centrifuged for 20 min at 109,000 × g. The supernatant was analyzed by fast protein liquid chromatography using a Superose 6 column equilibrated with buffer A containing 1% digitonin or 0.2% Triton X-100 (flow rate, 0.3 ml/min). Fractions of 0.5 or 0.25 ml were collected as indicated, precipitated with trichloroacetic acid (12.5%), and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Yta10p, Yta12p, Phb1p, and Phb2p were detected in the eluate by immunostaining using a chemiluminescence detection system. Protein amounts were determined by laser densitometry and are given as percentage of the specified protein in the eluate.
Coimmunoprecipitation experiments.
Mitochondria were isolated from the strains indicated and lysed as described for the gel filtration experiments, using buffer A containing 0.2% Triton X-100. Virtually identical results were obtained in the presence of 1% digitonin. After lysis, an aliquot was withdrawn from the supernatant for control. Supernatants were incubated under gentle shaking for 60 min at 4°C with preimmune or antiserum directed against Phb1p, Phb2p, Yta10p, Yta12p, or Yme1p as indicated. Each serum had been coupled to protein A-Sepharose. The beads were then washed three times with buffer A. The immunocomplexes were dissociated in SDS sample buffer by vigorous shaking for 10 min at room temperature and incubated for 3 min at 95°C. Precipitated proteins and control preparations were analyzed by SDS-PAGE and immunostaining.
Determination of the topology of Phb1p and Phb2p in the inner membrane.
Mitochondria were resuspended in buffer B (50 mM HEPES-KOH [pH 7.2], 0.75% bovine serum albumin, 0.5 M sorbitol, 80 mM KCl, 2.5 mM magnesium acetate, 1 mM potassium phosphate, 0.5 mM MnCl2) at a concentration of 100 μg/ml or, for osmotic disruption of the outer membrane (swelling), in buffer C (20 mM HEPES-KOH [pH 7.4], 0.3% bovine serum albumin, 50 mM sorbitol, 8 mM KCl, 1 mM magnesium acetate, 0.8 mM potassium phosphate, 0.2 mM MnCl2) at a concentration of 40 μg/ml. Samples were supplemented with proteinase K (100 μg/ml) or trypsin (100 μg/ml) when indicated and incubated for 30 min at 4°C. The proteases were inhibited by incubating the samples for 3 min at 4°C in the presence of phenylmethylsulfonyl fluoride (0.5 mM) or a 20-fold molar excess of soybean trypsin inhibitor, respectively. Mitochondrial fractions were analyzed by SDS-PAGE and immunostaining using antisera directed against Phb1p, Phb2p, d-lactate dehydrogenase, and Mge1p.
Labeling of mitochondrial translation products in vivo.
Mitochondrially encoded proteins were labeled in intact cells with [35S]methionine in the presence of cycloheximide for 5 min at 30°C as previously described (10, 19, 22). To examine the stability of newly synthesized polypeptides, unlabeled methionine was added to a final concentration of 30 mM and cells were further incubated at 30°C. Aliquots were withdrawn at the time points indicated and analyzed by SDS-PAGE and fluorography. The amount of newly synthesized proteins present in the cells was quantified with a phosphorimaging system. The kinetics of degradation were analyzed in logarithmic plots by assuming a simple first-order reaction.
Generation of Phb1p- and Phb2p-specific antisera.
To generate polyclonal antisera directed against Phb1p and Phb2p, 0.8-kb DNA fragments of PHB1 and PHB2, corresponding to amino acids 27 to 287 of Phb1p and to amino acids 61 to 315 of Phb2p, respectively, were amplified by PCR. The fragments were cloned into the BamHI/PstI sites of the bacterial expression vector pQE9 (Qiagen). After expression in Escherichia coli XL1-Blue, the proteins were purified according to standard procedures and used for generation of antibodies in rabbits.
RESULTS
A high-molecular-mass complex in the inner membrane containing m-AAA protease and prohibitins.
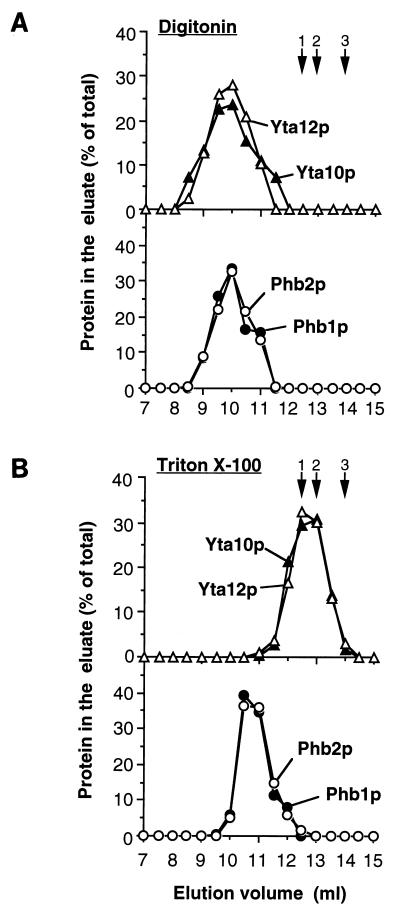
Upon determination of the native molecular mass of the m-AAA protease by gel filtration, strikingly different results were obtained in the presence of different detergents: when mitochondrial membranes were extracted with digitonin, the m-AAA protease subunits Yta10p and Yta12p eluted from the column in a single peak which corresponded to a molecular mass greater than 2 MDa (Fig. 1A). In contrast, after solubilization of mitochondria with the nonionic detergent Triton X-100, both m-AAA protease subunits coeluted from the sizing column in fractions corresponding to a native molecular mass of approximately 1 MDa as reported previously (Fig. 1B) (3). These results suggest that Yta10p and Yta12p are part of a supercomplex with an unexpected large molecular mass which remains stable when mitochondria are treated with the mild detergent digitonin.
FIG. 1.
Cofractionation of Phb1p and Phb2p with the m-AAA protease upon sizing chromatography. Wild-type mitochondria were solubilized in buffer A containing 1% digitonin (A) or 0.2% Triton X-100 (B) as described in Materials and Methods. Extracts were fractionated by Superose 6 chromatography. Eluate fractions (0.5 ml) were analyzed by SDS-PAGE and immunostaining with antisera directed against Yta10p (▴), Yta12p (▵), Phb1p (●), and Phb2p (○). Hsp60 (12.5 ml; arrow 1), thyroglobulin (13 ml; arrow 2), apoferritin (14 ml; arrow 3), alcohol dehydrogenase (15.5 ml; not shown) and bovine serum albumin (16.5 ml; not shown) were used for calibration.
In an attempt to identify other proteins which might interact with Yta10p and Yta12p, we examined the native molecular mass of the prohibitin homologues Phb1p and Phb2p in the inner membrane. Eluate fractions of the gel filtration columns were analyzed by immunostaining using Phb1p- and Phb2p-specific polyclonal antisera. Phb1p and Phb2p coeluted with Yta10p and Yta12p from the column if mitochondrial extracts were fractionated by gel filtration in the presence of digitonin (Fig. 1A). After solubilization of mitochondria with Triton X-100, Phb1p and Phb2p also coeluted in a single peak which was only slightly shifted toward the lower-molecular-mass region (Fig. 1B). They eluted in fractions corresponding to a native molecular mass of approximately 2 MDa under these conditions. Nonassembled Phb1p and Phb2p, which have molecular masses of 31 and 34 kDa, respectively, were not detectable in the eluate. In contrast, peak fractions containing Yta10p and Yta12p were significantly shifted toward a lower molecular mass after solubilization of mitochondria with Triton X-100 (Fig. 1B).
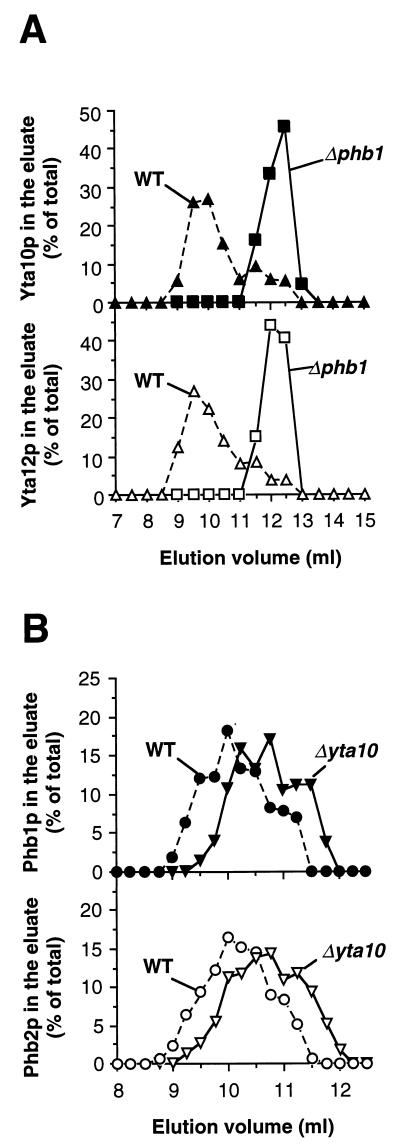
The cofractionation of Phb1p and Phb2p with each other and with the m-AAA protease subunits Yta10p and Yta12p in the presence of digitonin points to an interaction of these proteins. To further investigate this possibility, phb1- and phb2-null strains were constructed by disruption of the PHB1 and PHB2 genes, respectively. Notably, Phb1p was not detectable in Δphb2 cells by immunostaining, nor was Phb2p detectable in Δphb1 cells (data not shown) (4). The cellular levels of Yta10p and Yta12p were not altered by mutations in PHB1 or PHB2 (data not shown). Mitochondria were isolated from Δphb1 cells and solubilized in digitonin, and extracts were analyzed by gel filtration. In contrast to wild-type mitochondria, Yta10p and Yta12p were exclusively detected in eluate fractions corresponding to a molecular mass of approximately 1 MDa (Fig. 2A). The assembly of Yta10p and Yta12p was apparently not affected in Δphb1 mitochondria, but the formation of the larger complex depended on the presence of Phb1p. Likewise, when the native molecular mass of prohibitins was determined in digitonin extracts of Δyta10 mitochondria lacking m-AAA protease, Phb1p and Phb2p still coeluted from the column (Fig. 2B). The peak fractions, however, were slightly shifted compared to wild-type mitochondria, corresponding to a somewhat lower molecular mass of approximately 2 MDa (Fig. 2B).
FIG. 2.
Gel filtration analysis of mitochondrial extracts lacking prohibitins or m-AAA protease. (A) Wild-type (WT) and Δphb1 mitochondria or (B) wild-type (WT) and Δyta10 mitochondria were lysed in buffer A containing 1% digitonin and fractionated by sizing chromatography as described in Materials and Methods. In panel B, fractions of 0.25 ml were collected to increase the resolution. Eluate fractions were analyzed by SDS-PAGE and immunostaining with antisera directed against (A) Yta10p (▴ and ■) and Yta12p (▵ and □) or (B) Phb1p (● and ▾) and Phb2p (○ and ▿). Proteins used for calibration are described in the legend to Fig. 1.
Taken together, these results provide initial evidence for an interaction of the prohibitin homologues Phb1p and Phb2p with the m-AAA protease subunits Yta10p and Yta12p. Moreover, we conclude from these experiments that Phb1p and Phb2p have similar native molecular masses of approximately 2 MDa in the inner membrane regardless of the presence of the m-AAA protease.
Interaction of a Phb1p-Phb2p complex with the m-AAA protease.
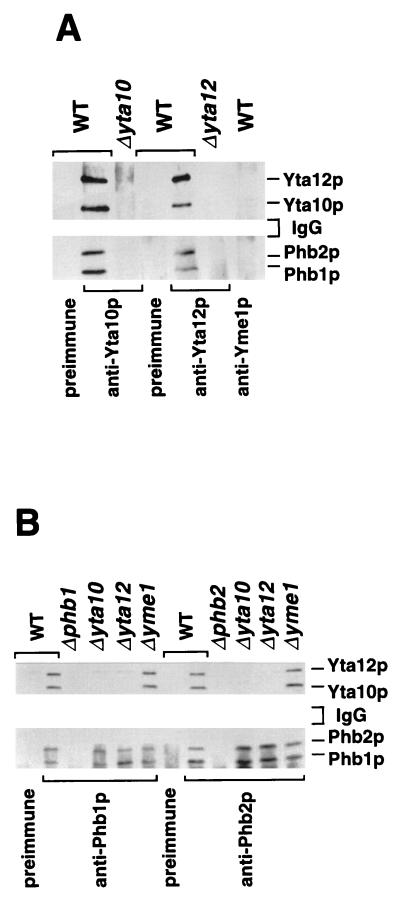
To demonstrate a direct physical interaction of Phb1p and Phb2p with each other and with Yta10p and Yta12p in an unambiguous fashion, coimmunoprecipitation experiments were performed. Mitochondria were solubilized and treated with a Yta10p- or Yta12p-specific antibody or preimmune serum (Fig. 3A). Yta10p and Yta12p were specifically coprecipitated under these conditions (Fig. 3A), confirming their previously reported association as a complex of the inner membrane (3). Moreover, both Phb1p and Phb2p were coprecipitated with antibodies directed against Yta10p and Yta12p but not with preimmune serum (Fig. 3A). Precipitation of prohibitins with the Yta10p- and Yta12p-specific antiserum was not observed in Δyta10 or Δyta12 mitochondria, respectively (Fig. 3A). Likewise, when the Phb1p- or Phb2p-specific antiserum was used for immunoprecipitation, Yta10p and Yta12p were present in the immunoprecipitates from wild-type mitochondria but not from Δphb1 or Δphb2 mitochondria (Fig. 3B). Notably, Phb2p was coprecipitated with an antibody directed against Phb1p, as was Phb1p with an antibody against Phb2p under these conditions (Fig. 3B). These experiments establish complex formation of Phb1p and Phb2p with subunits of the m-AAA protease in mitochondria and suggest that the two prohibitin homologues interact directly with each other. We also used a Yme1p-specific antiserum in these experiments to test for a possible association of prohibitins with the i-AAA protease subunit Yme1p, which is homologous to Yta10p and Yta12p (Fig. 3A). However, neither Phb1p or Phb2p nor Yta10p or Yta12p was detected in the precipitate by Western blot analysis, indicating that prohibitins specifically interact with the m-AAA protease.
FIG. 3.
Coimmunoprecipitation of the m-AAA protease with the Phb1p-Phb2p complex. Mitochondria isolated from the indicated strains were solubilized and subjected to immunoprecipitation as described in Materials and Methods, using antisera directed against Yta10p, Yta12p, and Yme1p (A) or Phb1p and Phb2p (B) and the respective preimmune sera. The immunoprecipitates were analyzed by SDS-PAGE and immunostaining. WT, wild type.
Does the Phb1p-Phb2p complex interact only with the assembled m-AAA protease, or does it also bind to nonassembled Yta10p and Yta12p subunits? Nonassembled Yta10p or Yta12p subunits accumulated at wild-type levels in mitochondria lacking Yta12p or Yta10p, respectively (data not shown). Therefore, coimmunoprecipitation experiments with Phb1p- and Phb2p-specific antisera were performed with detergent extracts derived from Δyta12 or Δyta10 mitochondria (Fig. 3B). In contrast to wild-type mitochondria, Yta12p was not precipitated either with the Phb1p- or Phb2p-specific antiserum in Δyta10 mitochondria (Fig. 3B), nor was Yta10p precipitated by either antibody in Δyta12 mitochondria (Fig. 3B). On the other hand, deletion of YME1 did not affect the interaction of the prohibitins with Yta10p and Yta12p (Fig. 3B). We conclude from these experiments that Phb1p and Phb2p interact only with the assembled m-AAA protease. It should also be noted that neither the coprecipitation of Phb2p with the Phb1p-specific antibody nor the coprecipitation of Phb1p with the Phb2p-specific antibody was affected in the absence of Yta10p and Yta12p (Fig. 3B). This observation is in agreement with the observed cofractionation of both prohibitins as a high-molecular-weight complex upon sizing chromatography under these conditions (Fig. 2B).
Topology of Phb1p and Phb2p in the inner membrane.
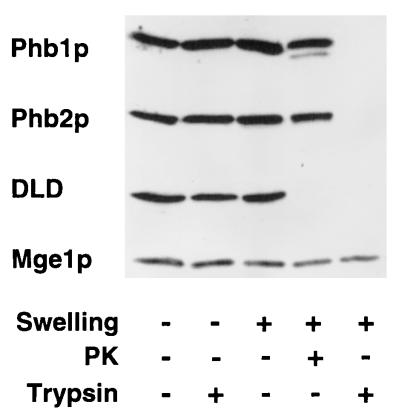
Phb1p and Phb2p both contain single hydrophobic stretches at their N termini which may serve as membrane-spanning segments. Submitochondrial fractionation identified Phb1p and Phb2p as integral components of the mitochondrial inner membrane (data not shown) (4). After osmotic disruption of the outer membrane, Phb1p and Phb2p were accessible to trypsin when added at high concentrations (Fig. 4). The protease sensitivity is shared with d-lactate dehydrogenase, an inner membrane protein with a large domain exposed to the intermembrane space (Fig. 4) (29). The matrix protein Mge1p, on the other hand, was not accessible to the protease under these conditions (Fig. 4). These results indicate that an N-terminal domain anchors Phb1p and Phb2p to the inner membrane, while the C terminus of each protein protrudes into the intermembrane space. Phb1p and Phb2p were not degraded in mitoplasts by proteinase K, suggesting that the intermembrane space segments of both proteins form a tightly folded domain (Fig. 4). The topology of the prohibitins is opposite that of Yta10p and Yta12p, both of which have a large C-terminal domain, containing the catalytic sites, exposed to the matrix space (3, 26). Thus, two large complexes which expose large domains to opposite membrane surfaces exist in the inner membrane and interact with each other: the Phb1p-Phb2p complex with an apparent molecular mass of approximately 2 MDa, and the m-AAA protease with an apparent molecular mass of approximately 1 MDa.
FIG. 4.
Topology of Phb1p and Phb2p in the inner membrane. Mitochondria were subfractionated by osmotic disruption of the outer membrane, and accessibility to externally added trypsin or proteinase K (PK) was examined as described in Materials and Methods. Mitochondrial fractions were analyzed by SDS-PAGE and immunostaining using antisera directed against Phb1p, Phb2p, d-lactate dehydrogenase (DLD; as a marker for the intermembrane space), and Mge1p (as a marker for the mitochondrial matrix).
Accelerated degradation of nonassembled inner membrane proteins by the m-AAA protease in the absence of prohibitins.
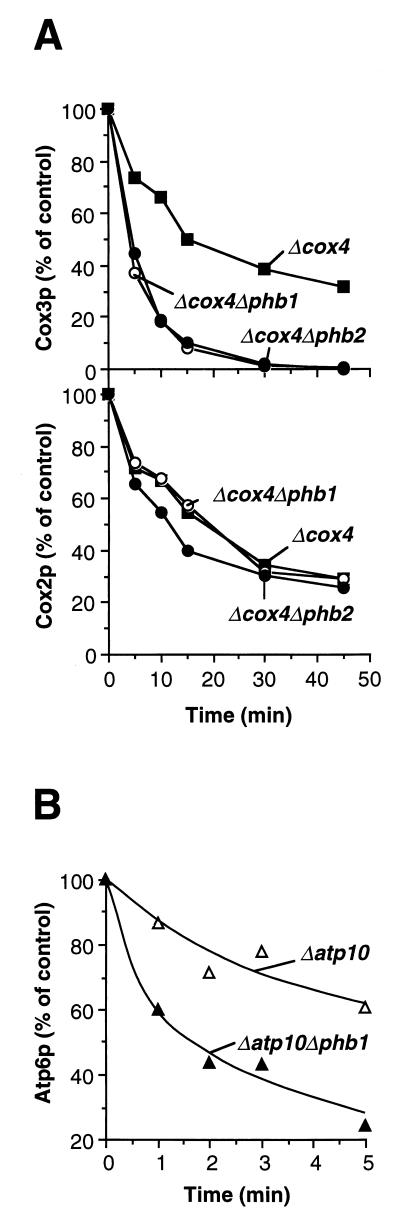
The effect of prohibitins on the proteolytic activity of the m-AAA protease was assessed by taking advantage of the observation that deletion of the nuclear gene COX4, encoding subunit 4 of the cytochrome c oxidase, results in the proteolysis of the nonassembled, mitochondrially encoded subunits Cox2p and Cox3p in the inner membrane (23). Cox2p is degraded by the i-AAA protease containing Yme1p (24), whereas Cox3p is a substrate of the m-AAA protease (3, 12). After deletion of PHB1 or PHB2 in Δcox4 cells, mitochondrial translation products were labeled with [35S]methionine. The cells were then further incubated to examine the stability of nonassembled Cox2p and Cox3p. Cox3p was degraded with a half-life of ∼16 min in Δcox4 cells (Fig. 5A). In Δcox4 cells lacking Phb1p or Phb2p, proteolysis of Cox3p was accelerated approximately threefold; it occurred with a half-life of ∼5 min (Fig. 5A). These results suggest that prohibitins regulate the proteolytic activity of the m-AAA protease in a negative manner. The effect of prohibitins is apparently restricted to the m-AAA protease. Degradation of nonassembled Cox2p by the i-AAA protease was not affected in Δcox4 cells harboring a deletion of PHB1 or PHB2 (Fig. 5A).
FIG. 5.
Accelerated degradation of nonassembled inner membrane proteins by the m-AAA protease in the absence of prohibitins. Mitochondrial translation products were labeled in the strains indicated, and cells were further incubated at 30°C to assess the stability of newly synthesized polypeptides as described in Materials and Methods. (A) Proteolysis of Cox3p and Cox2p in Δcox4 cells. Rate constants for proteolysis of Cox3p: k = 0.044 min−1 (in Δcox4 cells), k = 0.13 min−1 (in Δcox4 Δphb1 cells), and k = 0.15 min−1 (in Δcox4 Δphb2 cells). Rate constants for proteolysis of Cox2p: k = 0.036 min−1 (in Δcox4 cells), k = 0.037 min−1 (in Δcox4 Δphb1 cells), and k = 0.033 min−1 (in Δcox4 Δphb2 cells). (B) Proteolysis of Atp6p by the m-AAA protease in Δatp10 cells. Rate constants for proteolysis of Atp6p: k = 0.12 min−1 (in Δatp10 cells) and k = 0.29 min−1 (in Δatp10 Δphb1 cells).
To substantiate these conclusions, we analyzed the role of prohibitins in the degradation of the mitochondrially encoded subunit 6 of the F1F0-ATPase (Atp6p) which is mediated by the m-AAA protease (3, 12). In the absence of the nucleus-encoded protein Atp10p (1), assembly of the F0 moiety of the ATPase complex is defective, resulting in the proteolytic breakdown of the nonassembled F0-subunit Atp6p. Pulse-chase experiments after labeling of mitochondrial translation products in Δatp10 cells revealed a half-life of ∼6 min for this process (Fig. 5B). Deletion of PHB1 caused a significantly increased rate of degradation. Proteolysis occurred with a half-life of ∼2 min in Δatp10 Δphb1 cells (Fig. 5B). These results indicate that prohibitins exert a general effect on the turnover of membrane proteins by the m-AAA protease.
Impaired growth of Δphb1 and Δphb2 cells lacking m-AAA protease.
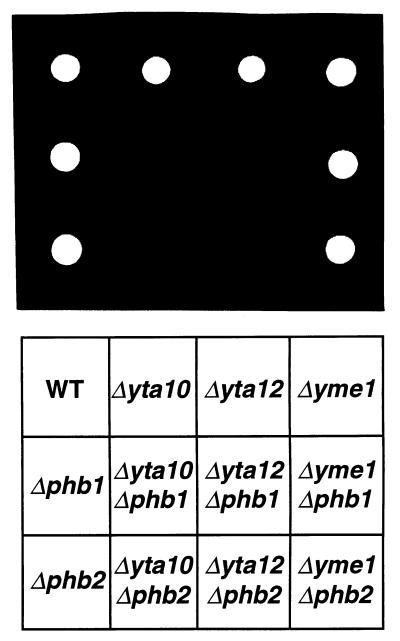
The mitochondrial m-AAA protease is required for respiratory growth of S. cerevisiae but is dispensable for cell growth on fermentable carbon sources. In contrast, deletion of PHB1 or PHB2 individually or deletion of both genes together does not cause a detectable growth phenotype in wild-type cells (Fig. 6) (4, 8). To provide genetic evidence for a functional interaction of prohibitins with the m-AAA protease, we examined the effects of deletions in PHB1 or PHB2 in cells lacking subunits of the m-AAA protease. Strikingly, the growth of Δyta10 or Δyta12 cells on fermentable carbon sources was strongly impaired in the absence of Phb1p or Phb2p (Fig. 6). Thus, prohibitins interact also genetically with subunits of the m-AAA protease. On the other hand, deletion of PHB1 or PHB2 did not affect the growth of Δyme1 cells lacking i-AAA protease on fermentable or nonfermentable carbon sources (Fig. 6 and data not shown). This observation is in agreement with our biochemical evidence and suggests a specific functional interaction of prohibitins with the m-AAA protease.
FIG. 6.
Impaired cell growth of Δphb1 and Δphb2 strains lacking m-AAA protease. PHB1 and PHB2 were disrupted in wild-type (WT), Δyta10, Δyta12, and Δyme1 cells by PCR-targeted homologous recombination (36). Disruptants were grown on YPD medium (rich medium containing 2% glucose) and harvested in exponential phase. Equal amounts of cells were spotted onto YPD agar plates and incubated at 30°C for 16 h. Slow growth of Δyta10 and Δyta12 cells lacking PHB1 or PHB2 was detected upon prolonged incubation of the plates (data not shown).
DISCUSSION
Prohibitins have been implicated in diverse cellular processes ranging from cell proliferation, senescence, and the maintenance of mitochondrial morphology. Their activity on the molecular level, however, remained obscure. Our results reveal an unexpected role of prohibitins in the degradation of inner membrane proteins in mitochondria. A large complex containing the prohibitin homologues Phb1p and Phb2p of S. cerevisiae associates with the m-AAA protease. The prohibitin complex appears to modulate the activity of the protease, thus exerting a regulatory function during proteolysis.
A physical interaction of Phb1p and Phb2p with each other and with the m-AAA protease was established by coimmunoprecipitation. Gel filtration analysis demonstrates that the m-AAA protease subunits Yta10p and Yta12p are part of a supercomplex which contains the prohibitins and has a molecular mass greater than 2 MDa. This observation raises the possibility that Phb1p and Phb2p represent novel subunits of the m-AAA protease. However, while proteolysis by the m-AAA protease is required for the respiratory competence of the cells (2, 3, 11, 35), deletion of Phb1p or Phb2p does not cause any detectable growth phenotype (4, 8), demonstrating the activity of Yta10p and Yta12p in the absence of prohibitins. Consistently, the assembly of Yta10p and Yta12p is not affected in Δphb1 cells, nor is the formation of the Phb1p-Phb2p complex affected in cells lacking m-AAA protease. Thus, in the inner membrane there are two large protein complexes, the prohibitin complex and the m-AAA protease, which are formed independently but assemble with each other.
In mitochondria lacking Phb1p or Phb2p, proteolysis of nonassembled inner membrane proteins by the m-AAA protease is enhanced. Although different effects on other substrate polypeptides cannot at this point be excluded, our findings indicate a negative regulatory role of the prohibitins in this process. The m-AAA protease is most likely the direct target of prohibitin action. This inference is suggested by the physical association of prohibitins with the m-AAA protease as well as by the apparent substrate specificity of prohibitin action. The turnover of Cox3p and Atp6p, both substrates of the m-AAA protease (3, 12), was accelerated in the absence of prohibitins, whereas the rate of Cox2p degradation by the i-AAA protease was not affected.
How could the prohibitin complex affect the proteolysis of membrane proteins by the m-AAA protease? The large C-terminal domains of both Phb1p and Phb2p protrude into the intermembrane space, while Yta10p and Yta12p expose their catalytic sites to the matrix. This topology in the inner membrane suggests that the prohibitin complex exerts its effect via the membrane-embedded N-terminal part of the m-AAA protease which includes segments exposed to the intermembrane space. Prohibitins may stabilize the m-AAA protease in a conformation with lowered proteolytic activity. Alternatively, prohibitins may modulate the accessibility or conformation of membrane-embedded proteolytic substrates and thereby regulate the association of substrate polypeptides with the protease. By this means, the prohibitins could prevent the premature degradation of nonassembled membrane proteins by the m-AAA protease and thereby ensure their proper assembly. Mitochondrial preproteins, which are present in a nonnative conformation during membrane translocation, might be protected against proteolytic attack in a similar manner. In mammalian cells, little prohibitin was found in association with cristae by immunoelectron microscopy, whereas it was enriched in the periphery of mitochondria (15). Taking into account the biochemically established localization of prohibitins to the inner membrane (4), this observation might point to an enrichment of prohibitins near the outer membrane, i.e., in the inner boundary membrane. A local inhibition of the m-AAA protease by prohibitins in proximity to the import sites is therefore conceivable.
The role of prohibitins in the degradation of mitochondrial inner membrane proteins by the m-AAA protease is reminiscent of previous findings in prokaryotes. The activity of the E. coli AAA protease FtsH has been demonstrated to be negatively regulated by a complex of two homologous membrane proteins, HflK and HflC, which were found to interact directly with substrate polypeptides (16, 17). Interestingly, both HflK and HflC are distantly related to prohibitin family members of various organisms (Fig. 7). Like Phb1p and Phb2p, HflK and HflC are anchored to the plasma membrane of E. coli via an N-terminal membrane-spanning segment (17). They expose a large domain to the periplasmic side of the plasma membrane of E. coli, i.e., opposite that of FtsH (17). The overall sequence identity of HflK and HflC to eukaryotic prohibitins, however, is significantly lower than within the prohibitin family. Nevertheless, the existence of distantly related proteins in bacteria and eukaryotes suggests that regulatory mechanisms for AAA proteases are conserved and derived from an early common ancestor.
FIG. 7.
Sequence similarity of E. coli HflC with prohibitin family members. Amino acid sequences of S. cerevisiae Phb1p (ScPhb1p; P40961) and prohibitin from Arabidopsis thaliana (AtPhb; U69155) and Homo sapiens (hsPhb; P35232) were aligned with E. coli HflC (EcHflC; P25661) by using the Clustal W program, version 1.7. Identical amino acid residues are shaded in black. Sequence identity between prohibitins of different organisms is over 50%. Predicted transmembrane segments at the N termini are underlined. E. coli HflC show 20% identical and 33% homologous residues with S. cerevisiae Phb1p and 13% identical and 29% homologous residues with S. cerevisiae Phb2p. We observed a similar degree of similarity between E. coli HflK and S. cerevisiae Phb1p and Phb2p (data not shown).
The functional interaction of prohibitins with the m-AAA protease raises the intriguing question of whether cellular activities previously attributed to prohibitins reflect their regulatory roles in membrane protein degradation. The observed requirement of prohibitins for efficient cell growth in the absence of the m-AAA protease provides genetic evidence for a functional relationship of both complexes but cannot be explained by a physical interaction. Therefore, additional functions of prohibitins in mitochondria, likely linked to the degradation of inner membrane proteins and the activity of the m-AAA protease, must exist. Prohibitins have recently been implicated in the regulation of mitochondrial morphology, as they were found to genetically interact with the mitochondrial inheritance components Mdm10p, Mdm12p, and Mmm1p in the outer membrane (4). In view of our results, it is conceivable that this genetic interaction is caused by alterations in the turnover of inner membrane proteins in the absence of prohibitins. In contrast to cells lacking the i-AAA protease subunit Yme1p (6), however, evidence for defects in mitochondrial morphology in the absence of the m-AAA protease, or after its overexpression, is lacking. Alternatively, the genetic interaction of prohibitins with mitochondrial inheritance components may reflect nonproteolytic functions of prohibitins. In view of the size of the prohibitin complex in the inner membrane, one can envision a scaffolding function of prohibitins which may allow for the assembly of a variety of mitochondrial proteins. Further insights into this question will require the identification of additional subunits of the prohibitin complex.
In view of the sequence conservation of prohibitins and AAA proteases from yeast to human (sequence identity of >50%), similar functions in all eukaryotic cells are likely. Another intriguing question raised by our findings is, therefore, how the effects of prohibitins on the m-AAA protease are linked to their roles in proliferation of mammalian cells and for cellular senescence. Alterations in mitochondrial physiology may change the energy level in the cell or its redox balance and thereby affect cell proliferation and aging. Prohibitins may affect mitochondrial activity by modulating the turnover of a short-lived regulatory protein by the m-AAA protease. It is therefore of interest to identify such putative substrates of the m-AAA protease and further characterize the specificity of prohibitin action during proteolysis.
ACKNOWLEDGMENTS
We thank B. Guiard for the COX4 disruption construct, S. Ackermann for the Δatp10 strain, A. Tzagoloff for critically reading the manuscript, and P. Coates for yeast strains and stimulating discussions during early phases of the project. The technical assistance of Petra Robisch and Alexandra Stiegler is gratefully acknowledged.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (La918/1-2; SFB 184/B21) and the Medizinische Wochenschrift to T.L.
REFERENCES
- 1.Ackermann S H, Tzagoloff A. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem. 1990;265:9952–9959. [PubMed] [Google Scholar]
- 2.Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA-protease. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- 4.Berger K H, Yaffe M P. Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4043–4052. doi: 10.1128/mcb.18.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky J L, McCracken A A. ER-associated and proteasome-mediated protein degradation: how two topologically restricted events came together. Trends Cell Biol. 1997;7:151–156. doi: 10.1016/S0962-8924(97)01020-9. [DOI] [PubMed] [Google Scholar]
- 6.Campbell C L, Tanaka N, White K H, Thorsness P E. Mitochondrial morphological and functional defects in yeast caused by yme1 are suppressed by mutation of a 26S protease subunit homologue. Mol Biol Cell. 1994;5:899–905. doi: 10.1091/mbc.5.8.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casari G, De-Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, DeMichele G, Filla A, Cocozza S, Marconi R, Durr A, Fontaine B, Ballabio A. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 8.Coates P J, Jamieson D J, Smart K, Prescott A R, Hall P A. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr Biol. 1997;7:R607–R610. doi: 10.1016/s0960-9822(06)00261-2. [DOI] [PubMed] [Google Scholar]
- 9.Dell’Orco R T, McClung J K, Jupe E R, Liu X-T. Prohibitin and the senescent phenotype. Exp Gerontol. 1996;31:245–252. doi: 10.1016/0531-5565(95)02009-8. [DOI] [PubMed] [Google Scholar]
- 10.Douglas M, Finkelstein D, Butow R A. Analysis of products of mitochondrial protein synthesis in yeast: genetic and biochemical aspects. Methods Enzymol. 1979;56:58–66. doi: 10.1016/0076-6879(79)56009-1. [DOI] [PubMed] [Google Scholar]
- 11.Guélin E, Rep M, Grivell L A. Sequence of the AFG3 gene encoding a new member of the FtsH/Yme1/Tma subfamily of the AAA-protein family. Yeast. 1994;10:1389–1394. doi: 10.1002/yea.320101016. [DOI] [PubMed] [Google Scholar]
- 12.Guélin E, Rep M, Grivell L A. Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in the degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 1996;381:42–46. doi: 10.1016/0014-5793(96)00074-9. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann J M, Fölsch H, Neupert W, Stuart R A. Isolation of yeast mitochondria and study of mitochondrial protein translation. In: Celis D E, editor. Cell biology: a laboratory handbook. San Diego, Calif: Academic Press; 1994. pp. 538–544. [Google Scholar]
- 14.Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 1997;11:1215–1225. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 15.Ikonen E, Fiedler K, Parton R G, Simons K. Prohibitin, an antiproliferative protein, is localized to mitochondria. FEBS Lett. 1995;358:273–277. doi: 10.1016/0014-5793(94)01444-6. [DOI] [PubMed] [Google Scholar]
- 16.Kihara A, Akiyama Y, Ito K. A protease complex in the Escherichia coli plasma membrane: HflKC (HflA) forms a complex with FtsH (HflB), regulating its proteolytic activity against SecY. EMBO J. 1996;15:6122–6131. [PMC free article] [PubMed] [Google Scholar]
- 17.Kihara A, Akiyama Y, Ito K. Host regulation of lysogenic decision in bacteriophage λ: transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA) Proc Natl Acad Sci USA. 1997;94:5544–5549. doi: 10.1073/pnas.94.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langer T, Leonhard K, Arlt H, Perryman R, Neupert W. Degradation of membrane proteins by AAA proteases in mitochondria. In: Hopsu-Havu V K, Järvinen M, Kirschke H, editors. Proteolysis in cell functions. Amsterdam, The Netherlands: IOS Press; 1997. pp. 323–331. [Google Scholar]
- 19.Langer T, Pajic A, Wagner I, Neupert W. Proteolytic breakdown of membrane-associated polypeptides in mitochondria of Saccharomyces cerevisiae. Methods Enzymol. 1995;260:495–503. doi: 10.1016/0076-6879(95)60161-9. [DOI] [PubMed] [Google Scholar]
- 20.Leonhard K, Herrmann J M, Stuart R A, Mannhaupt G, Neupert W, Langer T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996;15:4218–4229. [PMC free article] [PubMed] [Google Scholar]
- 21.McClung J K, Danner D B, Stewart D A, Smith J R, Schneider E L, Lumpkin C K, Dell’Orco R T, Nuell M J. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem Biophys Res Commun. 1989;164:1316–1322. doi: 10.1016/0006-291x(89)91813-5. [DOI] [PubMed] [Google Scholar]
- 22.McKee E E, Poyton P. Mitochondrial gene expression in Saccharomyces cerevisiae. Optimal conditions for protein synthesis in isolated mitochondria. J Biol Chem. 1984;259:9320–9331. [PubMed] [Google Scholar]
- 23.Nakai T, Mera Y, Yasuhara T, Ohashi A. Divalent metal ion-dependent mitochondrial degradation of unassembled subunits 2 and 3 of cytochrome c oxidase. J Biochem (Tokyo) 1994;116:752–758. doi: 10.1093/oxfordjournals.jbchem.a124592. [DOI] [PubMed] [Google Scholar]
- 24.Nakai T, Yasuhara T, Fujiki Y, Ohashi A. Multiple genes, including a member of the AAA family, are essential for the degradation of unassembled subunit 2 of cytochrome c oxidase in yeast mitochondria. Mol Cell Biol. 1995;15:4441–4452. doi: 10.1128/mcb.15.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuell M J, Stewart D A, Walker L, Friedman V, Wood C M, Owens G A, Smith J R, Schneider E L, Dell’Arco R, Lumpkin C K, Danner D B, McClung J K. Prohibitin, an evolutionary conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol Cell Biol. 1991;11:1372–1381. doi: 10.1128/mcb.11.3.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pajic A, Tauer R, Feldmann H, Neupert W, Langer T. Yta10p is required for the ATP-dependent degradation of polypeptides in the inner membrane of mitochondria. FEBS Lett. 1994;353:201–206. doi: 10.1016/0014-5793(94)01046-3. [DOI] [PubMed] [Google Scholar]
- 27.Paul M F, Tzagoloff A. Mutations in RCA1 and AFG3 inhibit F1-ATPase assembly in Saccharomyces cerevisiae. FEBS Lett. 1995;373:66–70. doi: 10.1016/0014-5793(95)00979-j. [DOI] [PubMed] [Google Scholar]
- 28.Rep M, Grivell L A. The role of protein degradation in mitochondrial function and biogenesis. Curr Genet. 1996;30:367–380. doi: 10.1007/s002940050145. [DOI] [PubMed] [Google Scholar]
- 29.Rojo E E, Guiard B, Neupert W, Stuart R A. Sorting of d-lactate dehydrogenase to the inner membrane of mitochondria. J Biol Chem. 1998;273:8040–8047. doi: 10.1074/jbc.273.14.8040. [DOI] [PubMed] [Google Scholar]
- 30.Sommer T, Wolf D H. Endoplasmic reticulum degradation: reverse protein flow of no return. FASEB J. 1997;11:1227–1233. doi: 10.1096/fasebj.11.14.9409541. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki C K, Rep M, Van Dijl J M, Suda K, Grivell L A, Schatz G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem Sci. 1997;22:118–123. doi: 10.1016/s0968-0004(97)01020-7. [DOI] [PubMed] [Google Scholar]
- 32.Tauer R, Mannhaupt G, Schnall R, Pajic A, Langer T, Feldmann H. Yta10p, a member of a novel ATPase family in yeast, is essential for mitochondrial function. FEBS Lett. 1994;353:197–200. doi: 10.1016/0014-5793(94)01045-5. [DOI] [PubMed] [Google Scholar]
- 33.Terashima M, Kim K-M, Adachi T, Nielsen P J, Reth M, Köhler G, Lamers M C. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994;13:3782–3792. doi: 10.1002/j.1460-2075.1994.tb06689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorsness P E, White K H, Fox T D. Inactivation of YME1, a member of the ftsH-SEC18-PAS1-CDC48 family of putative ATPase-encoding genes, causes increased escape of DNA from mitochondria in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:5418–5426. doi: 10.1128/mcb.13.9.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tzagoloff A, Yue J, Jang J, Paul M F. A new member of a family of ATPase is essential for assembly of mitochondrial respiratory chain and ATP synthetase complexes in Saccharomyces cerevisiae. J Biol Chem. 1994;269:26144–26151. [PubMed] [Google Scholar]
- 36.Wach A, Brachat A, Poehlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 37.Zinser E, Daum G. Isolation and biochemical characterization of organelles from the yeast Saccharomyces cerevisiae. Yeast. 1995;11:493–536. doi: 10.1002/yea.320110602. [DOI] [PubMed] [Google Scholar]