Abstract
Extracellular vesicles (EVs) play an important role in intercellular communication by transporting proteins and RNA. While plant cells secrete EVs, they have only recently been isolated and questions regarding their biogenesis, release, uptake and function remain unanswered. Here, we present a detailed protocol for isolating EVs from the apoplastic wash of Arabidopsis thaliana leaves. The isolated EVs can be quantified using a fluorometric dye to assess total membrane content.
Keywords: Arabidopsis thaliana, Extracellular vesicles, EVs, Apoplastic wash, DiOC6, Fluorometric quantification
Background
Extracellular vesicles (EVs) are membrane-bound structures that mediate the cell-to-cell transfer of proteins, lipids and genetic material. Interest in mammalian EVs has grown over the years due to their ability to transfer RNA and modulate immune responses. Mammalian EVs are routinely isolated for study from the medium of cultured cells, as well as a growing list of biological fluids ( Colombo et al., 2014 ). Plant EVs are also thought to have a role in the immune response but are comparatively understudied ( An et al., 2007 ; Davis et al., 2016 ). This is due, in large part, to the absence of a method of isolation.
While plant EVs have been observed since 1967 using transmission electron microscopy, methods for their isolation were not developed until 2009 (Halperin and Jensen, 1967). Regente et al. (2009) isolated small (50-200 nm in diameter) vesicle-like structures from water-imbibed sunflower (Helianthus annuus) seeds. We modified the methods presented in Regente et al. (2009) to isolate vesicles from the apoplastic wash of Arabidopsis thaliana rosettes. To determine which conditions induce or impair EV secretion, we also designed a method for staining the EV pellet with 3,3’-dihexyloxacarbocyanine iodide (DIOC6(3)), a fluorescent lipophilic dye. In the absence of sophisticated forms of nanoparticle tracking, this relatively simple approach quantifies the total membrane content and can be used to indirectly measure the concentration of EVs (Rutter and Innes, 2017). For more precise measurements, and to assess the size distributions of EVs, nanoparticle tracking can be used. Our protocols enable the study of plant EV content and composition, as well as the pathways and conditions that mediate EV biogenesis and release.
Materials and Reagents
MicroporeTM surgical tape (3M, catalog number: 1530-1)
Clear plastic domes (Hummert International, catalog number: 11-3348)
30 ml needle-less syringes (BD, catalog number: 309650)
50 ml conical tubes (VWR, catalog number: 89039-656)
Pipette tips
Paper towels
250 ml plastic bottles (w/out cap) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 3120-0250)
15 ml conical tube (VWR, catalog number: 89039-668)
1.5 ml microcentrifuge tubes (VWR, catalog number: 20170-038)
10 ml syringe
5 ml needle-less syringes (BD, catalog number: 309646)
-
Acrodisc 0.45 μm syringe filter (Pall, catalog number: 4454)
Note: An Acrodisc 0.22 μm filter (Pall, catalog number: 4192) can also be used if one is concerned about bacterial contamination.
Pryme PCR 8 strip 0.2 ml tubes (MIDSCI, catalog number: AVSST)
Kim-wipes (KCWW, Kimberly-Clark, catalog number: 34120)
COSTAR EIA/RIA Plate, 96 well, half area, no lid, flat bottom, non-treated, black polystyrene plate (Corning, Costar®, catalog number: 3694)
5.8 ml transfer pipets (Thermo Fisher Scientific, catalog number: 222-1S)
Petri dishes, 100 x 15 mm (VWR, catalog number: 25384-302)
Arabidopsis seeds
Bleach (Austin’s, catalog number: 90000360)
PRO-MIX PGX Biofungicide potting mix (Premier Tech Horticulture, catalog number: 10382RG)
OptiprepTM density gradient medium (Sigma-Aldrich, catalog number: D1556)
Murashige Skoog basal salt mixture (Sigma-Aldrich, catalog number: M5524)
Sodium hydroxide (NaOH) pellets (Avantor Performance Materials, MACRON, catalog number: 7708-10)
Potassium hydroxide (KOH) pellets (Fisher Scientific, catalog number: P250-500)
Agar (Sigma-Aldrich, catalog number: 05040)
MES hydrate (Sigma-Aldrich, catalog number: M8250)
Calcium chloride (CaCl2) (Sigma-Aldrich, catalog number: C1016)
Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653)
Trizma® base (Sigma-Aldrich, catalog number: T1503)
Hydrochloric acid (HCl) (EMD Millipore, catalog number: HX0603-3)
3,3’-Dihexyloxacarbocyanine iodide (DiOC6(3)) (Thermo Fisher Scientific, InvitrogenTM, catalog number: D273)
Plant protease inhibitor cocktail (Sigma-Aldrich, catalog number: P9599)
2,2’-Dipyridyl disulfide (DPDS) (Sigma-Aldrich, catalog number: 149225)
0.5x Murashige and Skoog agar (see Recipes)
Vesicle isolation buffer (VIB) (see Recipes)
20 mM Tris-HCl, pH 7.5 (see Recipes)
DiOC6 staining solution (see Recipes)
Vesicle resuspension buffer (see Recipes)
Equipment
Metal forceps
Scissors
Stainless steel tweezers, type 3 (Techni-Tool, catalog number: 758TW474)
Scale
1 L plastic beakers
Titanium French press coffee maker, 24 fl oz (Snow Peak, catalog number: CS-111)
Vacuum chamber, 0.20 Cu. Ft. (SP Scienceware - Bel-Art Products - H-B Instrument, catalog number: F42031-0000)
Vacuum pump, 3.5 CFM (Ideal Vacuum Products, catalog number: P101532)
JA-14 fixed-angle rotor (Beckman Coulter, model: JA-14, catalog number: 339247)
Pipettes
MJ research PTC-200 thermal cycler (MJ Research, catalog number: 8252-30-0001)
-
Fluorometric microplate reader
Note: We use an AppliskanTM fluorometric plate reader (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 5230000) to measure DiOC6 fluorescence, but any fluorometer capable of detecting multiple wavelengths could work.
SW 41 Ti rotor, swinging bucket, titanium, 6 x 13.2 ml, 41,000 rpm, 288,000 × g (Beckman Coulter, model: SW 41 Ti, catalog number: 331362)
Thickwall polycarbonate centrifuge tubes (3.5 ml, 13 x 51 mm) (Beckman Coulter, catalog number: 349622)
Thinwall, Ultra-ClearTM 13.2 ml, 14 x 89 mm ultracentrifuge tubes (Beckman Coulter, catalog number: 344059)
TLA100.3 fixed-angle rotor (Beckman Coulter, model: TLA-100.3, catalog number: 349481)
Avanti J-26S XP centrifuge (Beckman Coulter, model: Avanti J-26S XPI, catalog number: B22989)
Fluorescent light microscope
Growth chamber
Ice bucket (schuett-biotech, Spongex, catalog number: 3.680 052)
Optima TLX ultracentrifuge (Beckman Coulter, model: OptimaTM TLX, catalog number: 361545)
Nanoparticle trackers (Particle Metrix, model: ZetaView®, or Malvern Instruments, model: Nanosight NS300)
Procedure
-
Preparing the plants
Sterilize Arabidopsis seeds with 50% Bleach for no more than 5 min and wash three times with sterile dH2O. Each wash step should last 1-2 min.
Plate the seeds on 0.5x Murashige and Skoog (MS) medium containing 0.8% agar (see Recipes).
Wrap medical tape around the circumference of each MS plate and store the plates in the dark at 4 °C for 2 days.
After 2 days, move the plates to short day conditions (9-h days, 22 °C, 150 μEm-2). Store the plates vertically and allow the seeds to germinate and grow for 7 days.
Transfer the seedlings to PRO-MIX PGX. Cover the seedlings with a clear plastic dome for one week. On day 7, crack the dome. The next day, remove the dome completely. Water twice a week or as needed. Allow the plants to grow for 6 weeks total before harvest.
Note: Growing seedlings on MS plates first and transferring them to soil later ensures an exact number of healthy plants for each experiment. A typical experiment requires ~36 plants for each genotype/treatment. We have found that six-week-old plants yield the optimal amount of intercellular wash fluid with the smallest risk of cellular contamination.
-
Preparing tools for apoplastic wash collection
-
Using metal forceps heated in a flame, melt 8 additional holes at the end of a 30 ml syringe (Figure 1 and Video 1).
Note: These additional channels allow for the optimal collection of apoplastic wash and provide an alternate route should the primary hole become plugged.
-
Use the heated forceps to perforate the cap of a 50 ml conical tube multiple times, creating a circle ~2.5 cm in diameter. Punch out the circle to create a hole in the middle of the cap. Soften the cap in a flame and slide it onto the 30 ml syringe until it is ~1.5-2 cm from the top. The syringe can now be screwed securely onto a 50 ml conical tube (Figure 1 and Video 1).
Note: This prevents the end of the syringe from becoming submerged in the apoplastic wash.
-
-
Collecting the apoplastic wash
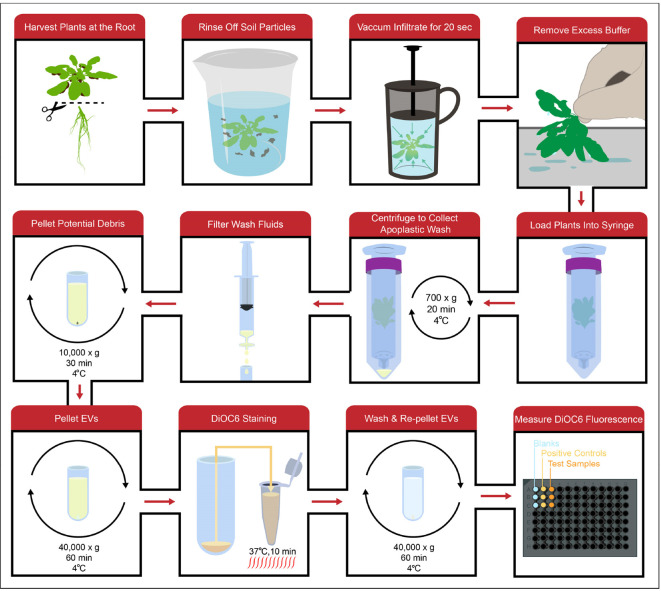
Note: An overview of the workflow from apoplastic wash collection to DiOC6 quantification can be seen in Figure 2.
-
Using scissors cut each Arabidopsis plant off at the root.
Note: Leaving a little of the root will help later on with manipulating the rosettes. See Video 1.
-
Collect the harvested rosettes in a 1 L plastic beaker and record the mass.
Note: Include three biological replicates for each treatment/genotype. The replicates should all be the same volume and should not be less than 0.5 ml. It takes ~3 g of rosettes to collect ~0.5 ml of apoplastic wash.
Rinse the rosettes 3 times with water to remove soil particles.
Carefully place the rosettes in the French press with 300-500 ml of vesicle isolation buffer (VIB) (see Recipes). Place the lid on the French press and gently lower the plunger until the plants are fully submerged.
Place the French press, plants and buffer in a vacuum chamber. Apply the vacuum for 20 sec.
-
Remove the French press from the vacuum chamber, take the lid off and pour the VIB into a separate 1 L plastic beaker. Remove the rosettes from the French press.
Note: The VIB can be reused multiple times on the same day, but, once used for vacuum infiltration, the VIB should not be stored for use on another day.
Remove excess VIB from the plants by gently shaking them by the roots and then brushing the leaves across a paper towel.
-
Place the rosettes root down on top of the opening of a 30 ml syringe (Figure 3A). Gently prod and coax the rosettes into the syringe without crushing or tearing the leaves (Figure 3B). Once the rosettes are in far enough, tapping the syringe on a hard surface will force them fully into the syringe (Figure 3C).
Note: The rosettes can be inserted in stacks of 2 or 3. Typically, 4-6 six-week-old rosettes will fit inside a 30 ml syringe. The rosettes should not be so tightly packed as to damage the tissues or obstruct the flow of apoplastic wash out of the syringe. See Video 1.
Screw the syringe into a 50 ml conical tube (Figure 3D) and place the tube and syringe in a 250 ml plastic bottle (Figure 3E). The cap secured around the top of the syringe should prevent the tube from falling into the bottle.
-
Place the syringe, 50 ml conical tube and bottle into the slot of a centrifuge rotor and centrifuge for 20 min at 700 × g, 4 °C.
Note: This step is designed for a JA-14 fixed-angle rotor.
Remove each syringe and conical tube from the centrifuge while carefully maintaining the tube’s angle. Unscrew the syringe and remove the apoplastic wash from the bottom of the tube. Make sure to avoid any pelleted soil particles. Transfer the apoplastic wash to a sterile 15 ml conical tube and keep the sample on ice.
-
Using a 10 ml syringe, filter the apoplastic wash through a 0.45 μm filter into a new, sterile 15 ml conical tube.
Note: The filtration step is a precaution meant to remove large pieces of debris. This step can result in the loss of apoplastic wash and can be skipped if the volume of apoplastic wash is small (~500 μl). In these cases, the 10,000 x g centrifugation should be enough to remove any debris.
-
-
EV isolation and DiOC6 staining
Transfer each volume of apoplastic wash to ultracentrifuge tubes. Bring the volume in each tube up to the tube’s maximum volume using chilled VIB.
Centrifuge for 30 min at 10,000 × g, 4 °C.
-
Carefully, transfer the supernatants to new ultracentrifuge tubes using a pipet.
Note: Maintain the position of the tube after centrifugation and be careful to angle the pipet away from any debris that may have pelleted.
Centrifuge for 60 min at 40,000 × g, 4 °C.
-
Decant the supernatant and remove any excess buffer from the opening of the tube with a pipet. Resuspend the pellet in 100 μl of 100 μM DiOC6 in 20 mM Tris-HCl pH 7.5 (see Recipes). Add 1% plant protease inhibitor cocktail (PIC) and 0.2 mM DPDS to the DiOC6 staining solution (see Recipes) if the EVs will be used for an immunoblot after DiOC6 quantification.
Note: When using small volumes of apoplastic wash, the EV pellet is invisible. A thin, translucent pellet can appear when using large volumes of apoplastic wash (≥ 12 ml). To effectively resuspend the pellet, use a pipet to repeatedly wash the side of the tube containing the pellet with half the added DiOC6 solution.
Transfer the resuspended vesicles to a PCR tube and heat at 37 °C for 10 min in a thermal cycler.
Transfer the sample to a new, clean ultracentrifuge tube and bring the volume up to the tube’s maximum volume using chilled 20 mM Tris-HCl pH 7.5.
Centrifuge for 60 min at 40,000 × g, 4 °C.
-
Decant the supernatant and use a pipet followed by a Kim-wipe to remove excess buffer for the opening and upper regions of the tube.
Note: It is important to wipe out remaining buffer immediately after decanting, while the centrifuge tube is still upside down. Carefully removing all excess buffer ensures that each pellet is resuspended in the same total volume.
-
Resuspend the pellet in 52 μl of vesicle resuspension buffer (20 mM Tris-HCl pH 7.5, see Recipes).
Note: Add 1% plant protease inhibitor cocktail (PIC) and 0.2 mM DPDS to the vesicle resuspension buffer if the EVs will be used for an immunoblot after DiOC6 quantification.
-
Collecting fluorometric data
-
Add 50 μl of each sample to wells in a black polystyrene, 96-well, half-area, flat-bottom EIA/RIA plate.
Note: For a typical experiment, there are three kinds of samples: a negative control of buffer (20 mM Tris-HCl + 1% plant PIC and 0.2 mM DPDS) to account for background signal, a positive control to determine vesicle secretion under mock conditions or in a genetically wild-type background, and the test sample to determine the effect of a treatment or mutation on vesicle secretion. As previously stated, include three biological replicates for each sample.
-
Insert the plate into the fluorometric microplate reader. Excite the samples at 485 nm and record fluorescent emissions at 535 nm. Take three readings of each plate.
Note: After recording fluorescence, the samples remain useful for a number of different assays, including staining for transmission electron microscopy, fluorescent light microscopy and immunoblots.
-
-
EV purification
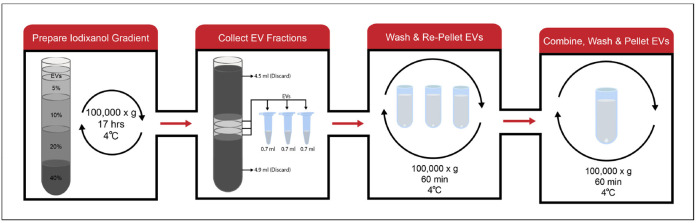
Note: Differential centrifugation yields a crude EV preparation that may contain other components of the apoplast. In-depth studies of EV composition require a further purification. The following protocol is designed to separate EVs out from other contaminants using a discontinuous iodixanol density gradient. An overview of the purification process can be seen in Figure 4.
Bring the volume of the isolated, crude EV preparation to 0.5 ml using sterile, cold VIB. Leave the sample on ice.
Prepare iodixanol solutions of 40% (3 ml, v/v), 20% (3 ml, v/v), 10% (3 ml, v/v), and 5% (2 ml, v/v) using aqueous 60% iodixanol (OptiPrep) and sterile, cold VIB.
Using a transfer pipet, add the iodixanol solutions to an ultracentrifuge tube. Carefully layer the solutions on top of one another, starting with the 60% solution and ending with the 5% solution. Lastly, add the 0.5 ml of crude EVs to the top of the gradient.
Centrifuge in a swinging-bucket rotor for 17 h at 100,000 × g, 4 °C.
Using a pipet, remove and discard the first 4.5 ml from the top of the gradient.
Collect the next three volumes of 0.7 ml as individual samples.
Transfer each sample to a fixed-angle ultracentrifuge tube. Bring each sample up to 3.5 ml using sterile, cold 20 mM Tris-HCl, pH 7.5.
Centrifuge for 60 min at 100,000 × g, 4 °C.
Decant the supernatant and resuspend and combine the three pellets in 3 ml of sterile, cold 20 mM Tris-HCl, pH 7.5.
Centrifuge for 60 min at 100,000 × g.
Decant the supernatant and remove excess buffer from the mouth of the tube using a Kim-wipe.
-
Resuspend the pellet in 50 μl of sterile, cold 20 mM Tris-HCl.
Notes: Purifying plant EVs requires a larger starting volume of apoplastic wash (~12-50 ml). A pale, milky pellet is visible when using larger volumes of apoplastic wash. However, a band is not visible in the discontinuous iodixanol gradient because the EVs are spread over a wider area.
Figure 1. Tool for apoplastic wash collection.

Apoplastic wash is collected by packaging whole Arabidopsis thaliana rosettes in a 30 ml syringe. Eight additional holes are melted into the end of the syringe with forceps to create channels for the wash to flow through. A 2.5 cm hole is punched out of a 50 ml conical tube cap. The cap is then slid onto the syringe 2 cm from the top. This allows the syringe to be screwed securely into a 50 ml conical tube. See Video 1.
Video 1. EV isolation demonstration.

This video demonstrates how to prepare the syringe and conical tube apparatus shown in Figure 3, and the procedures for extracting apoplastic wash fluid from leaves and subsequent purification of EVs from this fluid.
Figure 2. General workflow.
Apoplastic wash is collected by vacuum infiltrating full Arabidopsis rosettes with an isotonic buffer, washing out the infiltrated buffer using a low-speed centrifugation spin and filtering the wash to remove any large debris. The wash fluid is then processed using differential centrifugation to isolate EVs, which can be stained with the lipophilic dye DiOC6. After the EVs are washed and pelleted again to remove excess dye, DiOC6 fluorescence in the EV pellet can be measured to indirectly quantify the amount of EVs.
Figure 3. Packaging rosettes for apoplastic wash extraction.
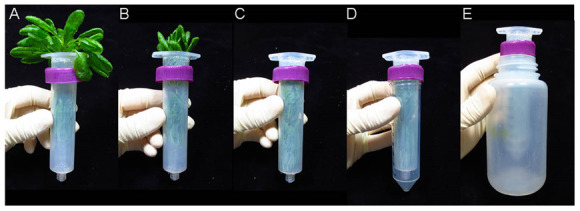
A. The rosettes are placed root-down on the top of the syringe. B. The rosettes are gently prodded into the syringe. C. The syringe is tapped on a hard surface to force the rosettes into it. D. The syringe is screwed into a 50 ml conical tube. E. The rosettes, syringe and 50 ml conical tube is placed into a 250 ml plastic bottle, which is then placed into a JA-14 rotor for centrifugation. See Video 1.
Figure 4. Workflow for purification of EVs.
Crude EVs are loaded on top of a discontinuous iodixanol gradient and centrifuged for 17 h at 100,000 × g, 4 °C. Following centrifugation, three EV-containing fractions are isolated. These fractions are washed and re-pelleted to remove the iodixanol. All three samples of EVs are then combined into a single tube, washed and pelleted to create one purified EV sample.
Data analysis
Table 1 and Figure 5 show a typical data analysis for DiOC6 fluorescence, which includes the following steps:
Figure 5. Example graph of DiOC6 fluorescence.
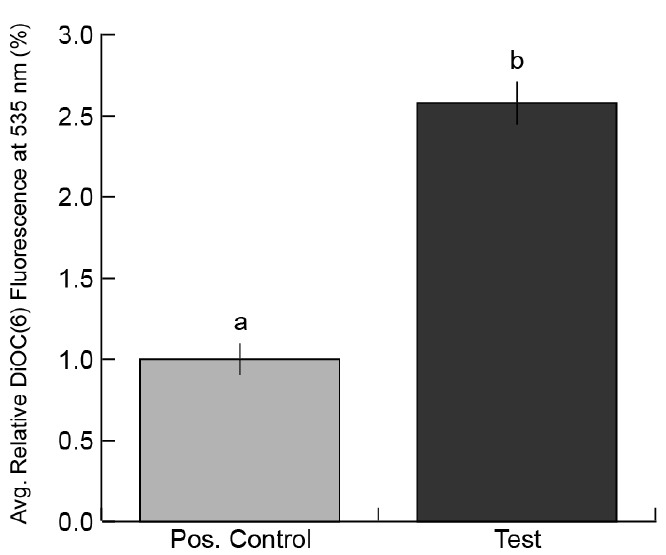
The above graph was generated from a replicate of an experiment presented in Rutter and Innes (2017). The experiment compared EV secretion between mock-treated Arabidopsis thaliana plants (Positive Control) and plants infected with a virulent strain of Pseudomonas syringae (Test).
Average the three readings for each sample. Calculate the mean value for the blank (avg. blank) by determining the mean of the three sample means (i.e., the average of all nine blank readings).
Subtract the mean value for the blank from the mean values for each positive control sample and each test sample.
After subtracting the blank value, determine the mean value of the three positive control samples, and then divide each individual value by this mean (each should be close to 1.0). These numbers will be used to calculate the standard deviation of the positive control samples.
Divide each of the test sample values (after subtracting the blank) by the mean of the positive control (this normalizes the test samples relative to the positive control).
Determine the mean of these values to determine the average ratio of DIOC6 fluorescence in the test samples compared to the positive control.
-
Calculate standard deviation and determine statistical significance using a two-tailed unpaired Student’s t-test.
Table 1. Example of raw data from DiOC6 fluorescence quantification.
The following data was generated from a replicate of an experiment presented in Rutter and Innes (2017). The experiment compared EV secretion between mock-treated Arabidopsis thaliana plants (Positive Control) and plants infected with a virulent strain of Pseudomonas syringae (Test).Sample replicate reading 1 reading 2 reading 3 avg. avg. blank –avg. blank (avg. pos.–avg. blank)/(avg. pos.–avg. blank)avg. X/(avg. pos. –avg. blank) Blank 1 457.5 439.5 433 443.3333333 444.5444444 2 456.9 451.2 442.6 450.2333333 3 457.1 431.6 431.5 440.0666667 Positive Control 1 2096.7 1653.1 2523.2 2091 1646.455556 1515.655556 1.086299291 1 2 2085 1516 2323.3 1974.766667 1530.222222 1.009610803 3 1556.7 1570.7 2317.1 1814.833333 1370.288889 0.904089906 Test 1 4073.9 5160.6 3786.1 4340.2 3895.655556 2.570277621 2.584294291 2 4385.9 5328.7 3916.9 4543.833333 4099.288889 2.704630926 3 4030.8 4762.8 3807.3 4200.3 3755.755556 2.477974327
P value = 4.74 x 10-5
Note: Although DiOC6 staining provides an estimate of EV yield, it does not provide information on EV concentration and sizes. To determine these values, one must have access to a nanoparticle tracker, which, as the name implies, are instruments that can track individual particles in a solution. By monitoring the Brownian motion of individual EVs, these instruments can calculate EV sizes, and can count the number of EVs in a defined volume. There are currently two manufacturers of nanoparticle trackers, Particle Metrix ( https://www.particle-metrix.de/en/products/zetaview-nanoparticle-tracking.html) and Malvern Instruments (http://www.malvern.com/en/products/technology/nanoparticle-tracking-analysis/).
Notes
Because EV secretion is enhanced during stress, it is important to use healthy plants. It is also important to limit cytoplasmic contamination by avoiding damaged plants and handling the rosettes gently.
We recommend following up total membrane quantification with an immunoblot for known plant EV markers, such as PEN1 or PATL1 (Rutter and Innes, 2017). Parallel increases or decreases in EV proteins bolster the DiOC6 data and help rule out the presence of other contaminating membranes.
When isolating large volumes of apoplastic wash fluid, 1% plant PIC and 0.2 mM DPDS should be added immediately to wash samples to prevent degradation by apoplastic or cellular proteases.
Recipes
-
0.5x Murashige and Skoog agar (0.5 L)
Dissolve 1.1 g Murashige Skoog basal salt mixture in 500 ml of deionized water
Adjust pH to 5.7-5.9 with 1 N KOH
Add 4 g (0.8%) agar (w/v)
Autoclave at 121 °C for 20 min
Store the medium at room temperature
Poured plated can be stored at 4 °C
-
Vesicle isolation buffer (VIB)
20 mM MES hydrate
2 mM CaCl2
0.1 M NaCl
Adjust pH to 6 with 10 N NaOH
Autoclave at 121 °C for 20 min
Note: To avoid possible contamination, use two sources of VIB. VIB for infiltrating plants is made in advance and stored at room temperature. VIB used for washing or resuspending vesicles is filtered through a 0.2 μm filter and stored at 4 °C.
-
20 mM Tris-HCl, pH 7.5
1 M Tris stock, pH to 7.5 with concentrated HCl
Dilute to 20 mM with distilled water
Autoclave at 121 °C for 20 min
-
DiOC6 staining solution
10 μM DiOC6 diluted in sterile 20 mM Tris-HCl pH 7.5
1% PIC (optional)
0.2 mM DPDS (optional)
-
Vesicle resuspension buffer
Sterile 20 mM Tris-HCl pH 7.5
1% PIC (optional)
1.2 mM DPDS (optional)
Acknowledgments
This work was supported by a grant from the United States National Science Foundation (IOS-1645745) to R.W.I. A condensed version of this protocol was presented in Rutter and Innes (2017).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.An Q., van Bel A. J. and Huckelhoven R.(2007). Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal Behav 2(1): 4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colombo M., Raposo G. and Thery C.(2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30: 255-289. [DOI] [PubMed] [Google Scholar]
- 3.Davis D. J., Kang B. H., Heringer A. S., Wilkop T. E. and Drakakaki G.(2016). Unconventional protein secretion in plants. Methods Mol Biol 1459: 47-63. [DOI] [PubMed] [Google Scholar]
- 4.Halperin W. and Jensen W. A.(1967). Ultrastructural changes during growth and embryogenesis in carrot cell cultures. J Ultrastruct Res 18(3): 428-443. [DOI] [PubMed] [Google Scholar]
- 5.Regente M., Corti-Monzon G., Maldonado A. M., Pinedo M., Jorrin J. and de la Canal L.(2009). Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett 583(20): 3363-3366. [DOI] [PubMed] [Google Scholar]
- 6.Rutter B. D. and Innes R. W.(2017). Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol 173(1): 728-741. [DOI] [PMC free article] [PubMed] [Google Scholar]