Abstract
Inteins garner significant interest from both basic and applied researchers due to their unique catalytic abilities. Herein, we describe a protocol for accurately monitoring protein splicing without purification using in-gel fluorescence immediately following Tris-Glycine SDS-PAGE. Following expression in Escherichia coli, cells are lysed by sonication, cell supernatants are separated using Tris-Glycine SDS-PAGE, and superfolder GFP (sfGFP) fluorescence is directly visualized within gels. This method is rapid, with sfGFP immediately imaged following SDS-PAGE without staining. Further, sfGFP can be specifically detected in complex samples such as E. coli cell supernatants, proteins run at expected masses, and all steps can be performed at ambient temperature. This strategy is broadly applicable beyond the study of protein splicing and can be used for sensitive and specific visualization of superfolder sfGFP-tagged proteins in-gel.
Keywords: In-gel fluorescence, Green fluorescent protein, Intein, Protein splicing
Background
Phylogenetically widespread in the microbial world, inteins (intervening proteins) have generated substantial interest as agents of autocatalysis, drivers of evolution, antibacterial and antifungal targets, and for biotechnological application (Lennon and Belfort, 2017). In the protein splicing reaction, the intein is removed by rearranging two peptide bonds and connecting the surrounding sequences, known as N-and C-exteins, with a peptide bond. While this process can occur in the absence of any external factors, its rate can be highly variable depending on conditions. Recently, compelling evidence has demonstrated that inteins can act as environmental sensors that regulate protein function in response to conditions such as metals, heat, salt, oxidative stress, and DNA damage (Belfort, 2017). Thus, significant interest from both basic and applied communities necessitate the investigation of inteins.
The study of intein catalysis is most often achieved using protein splicing reporters. The only strict requirement of these reporters is the presence of a cysteine, serine, or threonine as the first residue immediately following the intein ( Mills et al., 2014 ). We have utilized a reporter, referred to as MIG (MBP-Intein-GFP), to study the activity of a variety of inteins ( Topilina et al., 2015a and 2015b; Kelley et al., 2016 and 2018; Lennon et al., 2018 ; Green et al., 2019 ; Woods et al., 2020 ). In the MIG reporter, the intein (surrounded by 10 native extein residues) is flanked by the Maltose Binding Protein (MBP) as the N-extein and superfolder GFP (sfGFP) as the C-extein (Figure 1). Products with sfGFP attached are detected using in-gel fluorescence immediately following SDS-PAGE. sfGFP-tagged products (those containing a C-extein) allow for the monitoring of all possible intein reactions, including splicing (resulting in fluorescent ligated exteins), N-terminal cleavage (resulting in fluorescent intein-C-extein), and C-terminal cleavage (resulting in fluorescent C-extein) (Figure 1). Depending on the intein and reaction conditions, not all products may be detected. The major advantages of our sfGFP-based reporter, compared to other protein splicing reporters, is that there is no need for purification following expression and products can be visualized without staining.
Figure 1. Maltose binding protein-Intein-sfGFP (MIG) protein splicing reporter.
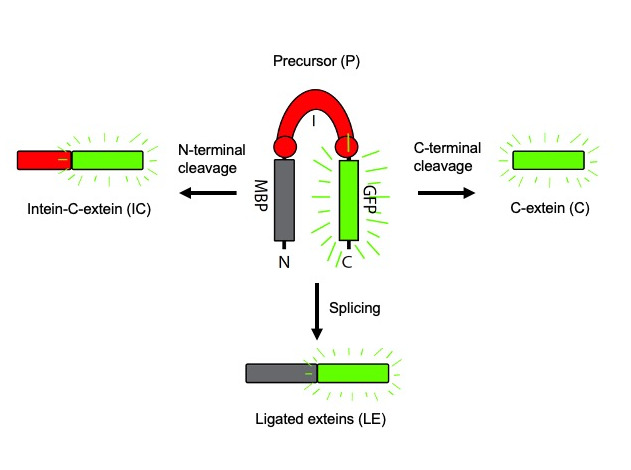
Products of splicing, N-terminal cleavage, and C-terminal cleavage that are visible (i.e., sfGFP-containing) using in-gel fluorescence are shown for each reaction.
Moreover, the use of in-gel fluorescence is broadly applicable beyond the study of protein splicing. This in-gel fluorescence protocol can be used to accurately examine the size of any sfGFP-tagged protein expressed in E. coli without purification. For example, this strategy could be used to study the localization of proteins in different cellular fractions, such as the cytoplasm, periplasm, and membrane of E. coli. Similar procedures have been described for visualization of GFP-tagged proteins. The protocol described herein differs as it allows visualization without staining ( Pristov et al., 2015 ) and does not require running gels at 4°C ( Bird et al., 2015 ). Several critical parameters relating to sfGFP-detection are also addressed. As GFP structure is required to maintain its fluorescent signal, it is critical that samples are not heated prior to SDS-PAGE (Figure 2). Further, we find that the use of Tris-Glycine rather than Bis-Tris gels improves sensitivity (Figure 2).
Figure 2. sfGFP detection within SDS-PAGE.
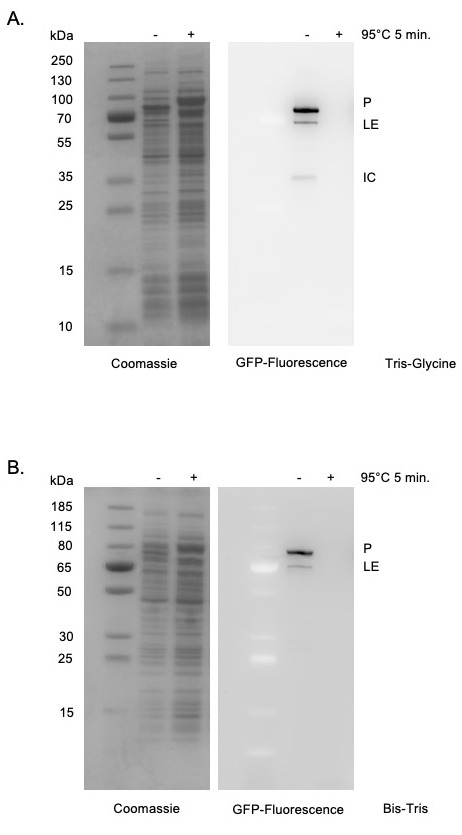
Tris-Glycine gels (Panel A) are more sensitive than Bis-Tris gels (Panel B) for detecting in-gel sfGFP signals as indicated by stronger signal, lower background, as well as detection of the minor product (IC) only in Tris-Glycine gels. sfGFP fluorescence is specifically detected in cell supernatants without purification. Heating samples at 95°C for 5 min results in a loss of fluorescent signal. Left, Coomassie stained. Right, sfGFP signal. Expected molecular weights of products are as follows: Precursor (P) 88.5 kDa; Ligated exteins (LE) 74.0 kDa; Intein-C-extein (IC) 42.6 kDa.
Materials and Reagents
10, 20, and 1,000 μl Pipette tips (USA Scientific, Inc. TipOne, catalog numbers: 1111-3800,1120-1810, and 1111-2820)
1.5 ml tubes (Eppendorf, catalog number: 022363204)
50 ml conical tubes (Corning, catalog number: 352070)
Gel loading tips (USA Scientific, catalog number: 1022-0000)
Culture tube (Fisher Scientific, catalog number: 14-961-31)
10 ml syringe (BD, catalog number: 309604)
0.22 μm syringe filter (Fisher Scientific, catalog number: 09-720-3)
Mini-PROTEAN TGX Precast Protein Gels, Tris-Glycine, 8-16% (Bio-Rad Laboratories, catalog number: 4561106)
4× Laemmli Sample Buffer (Bio-Rad Laboratories, catalog number: 1610747)
10× Tris/Glycine/SDS electrophoresis buffer (Bio-Rad Laboratories, catalog number: 1610732)
MIG (or sfGFP) expression strain (available upon request)
Tris, 1.0 M buffer solution, pH 7.5, 0.2-μm filtered (Alfa Aesar, catalog number: J62993)
Tris, 1.0 M buffer solution, pH 8.5, 0.2-μm filtered (Alfa Aesar, catalog number: J61038)
Glycerol (Fisher Scientific, catalog number: BP229)
Zinc acetate dihydrate (Fisher Scientific, catalog number: S25634)
LB broth base (Difco, catalog number: 244610)
IPTG (Goldbio, catalog number: 12481C25)
Chloramphenicol (Sigma, catalog number: C0378)
MIG Buffer (see Recipes)
1× Tris/Glycine/SDS Running Buffer (see Recipes)
1 M Isopropyl β-D-1-thiogalactopyranoside (IPTG) (see Recipes)
25 mg/ml Chloramphenicol stock solution (i.e., 1,000×, see Recipes)
LB medium (see Recipes)
10 mM Zinc acetate solution (see Recipes)
Equipment
Cell Sonicator (Heat Systems, model: XL-2020)
FB300 Power Supply (Fisher Scientific, catalog number: S65533Q)
Mini-PROTEAN Tetra Vertical Electrophoresis Cell for Mini Precast Gels, 4-gel (Bio-Rad Laboratories, catalog number: 1658004)
Amersham Imager 600 (GE, catalog number: 29-0834-61)
P20 Pipetman (Gilson, catalog number: FA10003M)
P1000 Pipetman (Gilson, catalog number: FA10006M)
Microcentrifuge (Eppendorf, catalog number: 5420000113)
Centrifuge (Fisher Scientific, catalog number: 75005194)
Benchtop Refrigerated Shaking Incubator (Thermo Scientific, catalog number: SHKE4000)
Software
ImageJ (software free to download at https://imagej.nih.gov)
Procedure
-
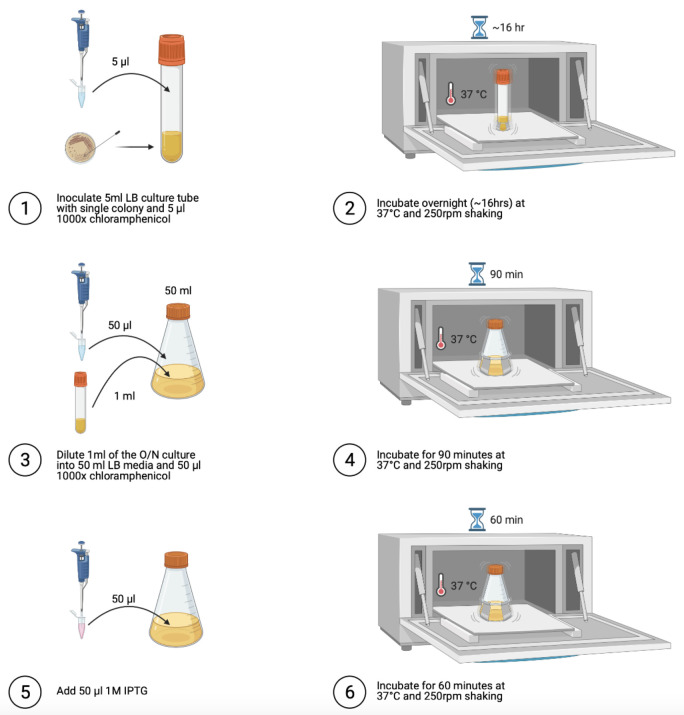
Protein expression (Figure 3)
Prepare, under sterile conditions, a saturated overnight culture of the MIG (or sfGFP) expression strain by inoculating a culture tube containing 5 ml LB media and 5 μl of 25 mg/ml (1,000×) chloramphenicol stock solution.
Incubate at 37°C and 250 RPM for approximately 16 h.
Under sterile conditions, dilute 1 ml of the overnight culture into 50 ml LB media and 50 μl 25 mg/ml (1,000×) chloramphenicol stock solution.
Incubate at 37°C and 250 RPM for 90 min.
Under sterile conditions, add 50 μl 1 M IPTG (final concentration of 1 mM IPTG).
Incubate at 37°C and 250 RPM for 60 min.
-
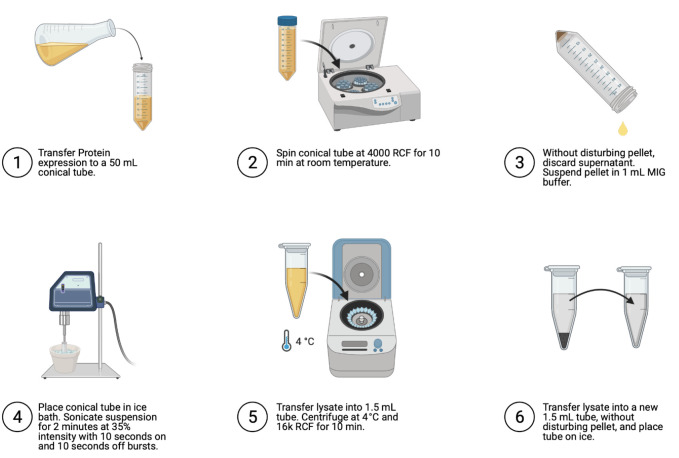
Cell lysis (Figure 4)
Transfer to 50 ml conical tube.
Spin to pellet at 4,000 × g for 10 min at room temperature (~22°C).
Discard supernatant without disturbing pellet.
Resuspend pellet in 1 ml MIG buffer.
Sonicate on ice (35% intensity) for 10 s on, then 10 s off for 2 min.
Transfer lysed cells into 1.5 ml tube.
Spin at 4°C and 16,000 × g for 10 min.
Without disturbing pellet, transfer supernatant into 1.5 ml tube and place on ice.
-
Protein splicing
-
Pipette 45 μl of supernatant into a 1.5 ml tube, add 15 μl 4× Laemmli dye and mix, and place at -20°C.
Note: This is time point 0.
Pipette 495 μl of supernatant and 5 μl ddH2O into a 1.5 ml tube and incubate under foil at room temperature (~21°C).
In another 1.5 ml tube, pipette 99 μl supernatant and 1 μl 10 mM zinc acetate dihydrate and incubate under foil at room temperature (~21°C).
-
At 1, 2, 4, 8, and 24 h time points, take 45 μl from the tube without zinc acetate dihydrate and pipette into a new 1.5 ml tube, add 15 μl 4× Laemmli dye and mix, and place at -20°C.
Note: The rate of protein splicing is highly variable depending on the intein and incubation conditions. Adjust time points accordingly. These conditions are for the Mycobacterium smegmatis DnaBi1 intein.
At the 24-hour time point, take 45 μl from the tube with supernatant and zinc acetate dihydrate and pipette into new 1.5 ml tubes, add 15 μl 4× Laemmli dye and mix, and place at -20°C.
-
-
Tris-Glycine SDS-PAGE
Thaw samples at room temperature.
-
Do not heat samples prior to electrophoresis.
Note: Heating will denature sfGFP and result in loss of fluorescent signal (Figure 2).
Add 1× running buffer to electrophoresis tank.
Insert Tris-Glycine gel into the electrophoresis gel holder and add gel holder to tank.
Add 1× running buffer inside of electrophoresis gel holder to cover gel wells.
Load 4 μl ladder and 6 μl of each sample.
Connect tank to power supply and run gel at 200 V for 40 min.
Remove gel from plastic shell and place in ddH2O.
-
Image Gel
Measure in-gel fluorescence using a Amersham 600 Imager or an equivalent imager capable of detecting sfGFP fluorescence (Figure 5).
Lay gel flat on imaging tray.
Place tray in imager.
Set imager to Epi-RGB: Blue light (460 nm, Filter Cy2).
Set exposure to auto with high dynamic range.
Save picture of gel as tiff or jpeg. Relative fluorescence can be calculated by densitometry using ImageJ.
Figure 3. Steps for protein expression.
Image generated using Biorender.
Figure 4. Steps for cell lysis.
Image generated using Biorender.
Figure 5. Monitoring protein splicing of Mycobacterium smegmatis DnaBi1 intein using in-gel fluorescence with Tris-Glycine SDS-PAGE.
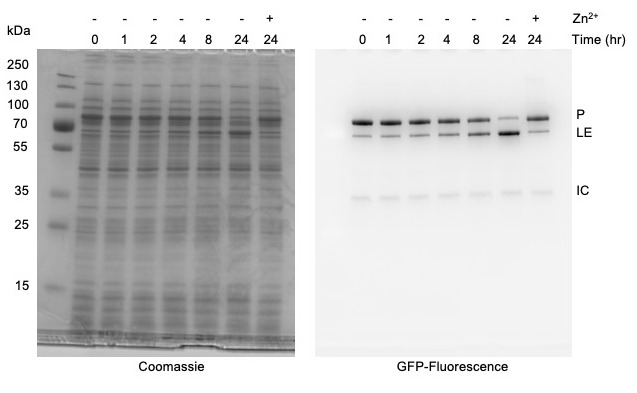
Presence of 100 μM zinc acetate inhibits splicing. Left, Coomasssie stained. Right, sfGFP signal. sfGFP-tagged products are indicated as Precursor (P), Ligated exteins (LE), and Intein-C-extein (IC).
Recipes
-
MIG Buffer
2.5 ml Tris, 1.0 M buffer solution, pH 7.5, 0.2 μm filtered
2.5 ml Tris, 1.0 M buffer solution, pH 8.5, 0.2 μm filtered
10 ml Glycerol
85 ml ultrapurified H2O
-
1× Tris/Glycine/SDS running buffer (25 mM Tris, pH 8.3, 0.1% SDS, 192 mM Glycine)
100 ml 10× Tris/Glycine/SDS electrophoresis buffer
900 ml ultrapurified H2O
-
10 mM zinc acetate
109 mg zinc acetate dihydrate
50 ml ultrapurified H2O
-
LB Medium
25 g LB broth base
1,000 ml ddH2O
Autoclave to sterilize
-
1 M Isopropyl β-D-1-thiogalactopyranoside (IPTG)
2.38 g IPTG
10 ml ultrapurified H2O
Place in 10-ml syringe and pass through 0.22-μm filter to sterilize
Aliquot in 1.5-ml tubes and store at -20°C
-
25 mg/ml chloramphenicol stock solution
250 mg chloramphenicol
10 ml 100% ethanol
Place in 10-ml syringe and pass through 0.22-μm filter to sterilize
Aliquot in 1.5-ml tubes and store at -20°C
Acknowledgments
This work was supported by National Institutes of Health grants P20GM103436 to C.W.L. and GM44844 to Marlene Belfort. This protocol is based on our previous publication ( Woods et al., 2020 ).
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Belfort M. J.(2017). Mobile self-splicing introns and inteins as environmental sensors. Curr Opin Microbiol 38: 51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird L. E., Rada H., Verma A., Gasper R., Birch J., Jennions M., Lwe J., Moraes I. and Owens R. J.(2015). Green fluorescent protein-based expression screening of membrane proteins in Escherichia coli. J Vis Exp(95): e52357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green C. M., Li Z., Smith A. D., Novikova O., Bacot-Davis V. R., Gao F., Hu S., Banavali N. K., Thiele D. J. and Li H. J.(2019). Spliceosomal Prp8 intein at the crossroads of protein and RNA splicing. PLoS Biol 17(10): e3000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley D. S., Lennon C. W., Li Z., Miller M. R., Banavali N. K., Li H. and Belfort M.(2018). Mycobacterial DnaB helicase intein as oxidative stress sensor. Nat Commun 9(1): 4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelley D. S., Lennon C. W., SEA-PHAGES, Belfort M. and Novikova O.(2016). Mycobacteriophages as Incubators for Intein Dissemination and Evolution. mBio 7(5): e01537-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lennon C. W. and Belfort M.(2017). Inteins. Curr Biol 27(6): R204-R206. [DOI] [PubMed] [Google Scholar]
- 7.Lennon C. W., Stanger M., Banavali N. K. and Belfort M.(2018). Conditional Protein Splicing Switch in Hyperthermophiles through an Intein-Extein Partnership. mBio 9(1): e02304-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills K. V., Johnson M. A. and Perler F. B.(2014). Protein splicing: how inteins escape from precursor proteins. J Biol Chem 289(21): 14498-14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pristov J. B., Opacic M., Dimitrijevic M., Babic N. and Spasojevic I.(2015). A method for in-gel fluorescent visualization of proteins after native and sodium dodecyl sulfate polyacrylamide gel electrophoresis. Anal Biochem 480: 6-10. [DOI] [PubMed] [Google Scholar]
- 10.Topilina N. I., Green C. M., Jayachandran P., Kelley D. S., Stanger M. J., Piazza C. L., Nayak S. and Belfort M.(2015). SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses. Proc Natl Acad Sci U S A 112(33): 10348-10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topilina N. I., Novikova O., Stanger M., Banavali N. K. and Belfort M.(2015). Post-translational environmental switch of RadA activity by extein-intein interactions in protein splicing. Nucleic Acids Res 43(13): 6631-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woods D., Vangaveti S., Egbanum I., Sweeney A. M., Li Z., Bacot-Davis V., LeSassier D. S., Stanger M., Hardison G. E., Li H., Belfort M. and Lennon C. W.(2020). Conditional DnaB Protein Splicing Is Reversibly Inhibited by Zinc in Mycobacteria. mBio 11: e01403-20. [DOI] [PMC free article] [PubMed] [Google Scholar]