Abstract
Strains of Escherichia coli bearing the pks genomic island synthesize the genotoxin colibactin. Exposure of eukaryotic cells to E. coli producing colibactin induces DNA damages, ultimately leading to cell cycle arrest, senescence and death. Here we describe a simple method to demonstrate the genotoxicity of bacteria producing colibactin following a short infection of cultured mammalian cells with pks+ E. coli.
Keywords: Escherichia coli, Enterobacteria, Polyketide, Colibactin, Genotoxin, DNA damage, Cell culture, Infection
Background
Colibactin is a genotoxin discovered in extra-intestinal pathogenic, commensal and probiotic strains of Escherichia coli ( Nougayrede et al., 2006 ). Colibactin is also produced by other Enterobacteriaceae, including Klebsiella pneumonia, Enterobacter aerogenes and Citrobacter koseri ( Putze et al., 2009 ). Colibactin is a polyketide/non-ribosomal peptide hybrid compound, synthesized by a multi-enzymatic machinery, consisting of polyketide and nonribosomal peptide synthases (PKS and NRPS), tailoring and maturation enzymes, and an efflux pump (for a review: Taieb et al., 2016 ). This synthesis machinery is encoded on a 52 kb genomic locus, the ‘pks’ island. Colibactin induces DNA damages in eukaryotic cells infected with pks+ bacteria. The genotoxic effect induced by colibactin requires a direct contact of live pks+ bacteria with the eukaryotic cells. Indeed, no genotoxic effect is observed with killed bacteria, or with bacterial supernatants or lysates. Thus, to demonstrate the genotoxicity of colibactin producing E. coli, cultured mammalian cells (such as HeLa cells) are infected during 4 h with live pks+ bacteria. The dose of colibactin delivered to the cells varies with the number of infecting bacteria per cell (multiplicity of infection, or MOI). At the end of the 4 h infection, bacterial growth is monitored by optical density measurement (OD600 nm). Then, the cells are washed to remove the bacteria and further incubated three days with antibiotics, to allow the development of the cytopathic phenotype associated with the DNA damage. The cellular DNA damage response results in proliferation (cell cycle) arrest, cell death and senescence ( Secher et al., 2013 ). The microscopic observation of the cell morphology reveals the cellular response to the genotoxic insult, with reduced cell numbers and a striking giant cells phenotype (called megalocytosis) due to the cell cycle arrest and cellular senescence. The genotoxic effect can be quantified by staining the cells with methylene blue, extracting the dye and measuring the optical density at 660 nm (De Rycke et al., 1996 ).
Materials and Reagents
Tissue culture plate 96 wells flat bottom (P96) (Corning, Falcon®, catalog number: 353072 or equivalent)
Culture flask 250 ml, 75 cm2 (Corning, Falcon®, catalog number: 353136 or equivalent)
Paper towel
HeLa cells (ATCC, catalog number: CCL-2)
pks+ Escherichia coli strain (stored in LB 20% glycerol at -80 °C). Strains typically used as positive controls in the authors’ laboratory are probiotic strain Nissle 1917 or the commensal mouse strain NC101
Lennox L broth base (LB medium) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 12780029)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Dulbecco’s modified Eagle medium (DMEM) with 25 mM HEPES (Thermo Fisher Scientific, GibcoTM, catalog number: 42430)
Hanks’ balanced salt solution (HBSS) (Sigma-Aldrich, catalog number: H8264)
Dulbecco’s modified Eagle medium (DMEM), high glucose, GlutaMax Supplement, pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 31966021)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106, or equivalent)
Non-essential amino acids solution (NEAA) 100x (Thermo Fisher Scientific, GibcoTM, catalog number: 11140035)
Gentamicin solution 50 mg/ml (Sigma-Aldrich, catalog number: G1397)
Paraformaldehyde (PFA) 20% (Electron Microscopy Sciences, catalog number: 15713)
Dulbecco’s phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: D8537)
10x PBS (Sigma-Aldrich, catalog number: D1408)
Methylene blue (RAL DIAGNOSTICS, catalog number: 310950)
Tris-HCl 1 M pH 8.5 (Teknova, catalog number: T1085)
Hydrochloric acid (HCl) 1 M (Merck, catalog number: 109057)
HeLa cell culture medium (see Recipes)
Fixation solution (see Recipes)
Methylene blue staining solution (see Recipes)
Methylene blue wash buffer (see Recipes)
Methylene blue extraction solution (see Recipes)
Equipment
Micropipette Research Plus 0.1-10 µl (Dutscher, Eppendorf, catalog number: 035602)
Micropipette PIPETMAN G 20-200 µl (Dutscher, Gilson, catalog number: 066811)
Multichannel micropipette 30-300 µl (Dutscher, Finnpipette, catalog number: 050667N)
Incubator for bacterial culture, with shaking (Eppendorf, New BrunswickTM, model: Innova® 42)
CO2 incubator for cell culture (Forma Scientific)
Colorimeter to measure the absorbance at 600 nm of bacterial cultures (biochrom, model: WPA CO7500)
Microplate reader for absorbance measurement at 600 and 660 nm (TECAN Infinite Pro)
Vortex (Dutscher, model: Vortex-Genie 2)
Inverted microscope (Olympus, model: CKX31)
Biological safety cabinets (Thermo Fisher Scientific, Thermo ScientificTM, model: MSC-AdvantageTM Class II)
Chemical safety hood to handle the fixative
Procedure
-
Day 1
Seed a 3 ml LB culture from a -80 °C glycerol stock or a colony on an LB agar plate, and grow overnight (16-24 h) at 37 °C with shaking (240 RPM).
Inoculate in a tissue culture 96 wells (P96) microplate, 5 x 103 HeLa cells in each well in 100 μl per well of complete cell culture medium (DMEM 10% FBS 1% NEAA 50 μg/ml gentamicin, see Recipes). Grow for 24 h in a 37 °C 5% CO2 humidified incubator. See the Data analysis section for the number of strains/conditions that can be tested per P96.
-
Day 2
-
Bacteria culture
Inoculate into 9.5 ml pre-warmed (37 °C) DMEM 25 mM HEPES in a tube with 500 μl of the bacterial overnight LB culture. Grow at 37 °C with shaking (240 RPM) to reach OD600 nm = 0.4 to 0.5 (about 2 h). Determine the number of bacteria per ml by measuring the absorbance OD600 nm of 1 ml of the culture. 1 unit of OD600 nm corresponds to 5 x 108 bacteria/ml (typical value for E. coli with a WPA colourwave CO 7500 colorimeter).
-
HeLa cell washing
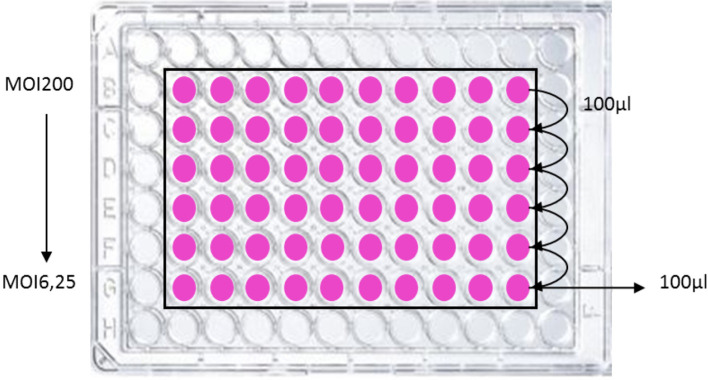
Meanwhile, wash cautiously the HeLa cells 3 times with 100 µl of warm (37 °C) HBSS then add 100 µl per well of DMEM 25 mM HEPES. For row B, add 200 µl instead of 100 µl (see Figure 1).
-
HeLa cells infection
Inoculate the cells with the bacteria, starting in row B, to achieve the maximum required number of bacteria per cell (multiplicity of infection, MOI). For dose/effect examination, prepare serial two-fold dilutions from row B to G, by transferring between each row 100 µl (see Figure 1). Typically, the highest MOI is 200 in first row B, and the MOI in the last row G is 6.25. Include on the plate a positive (bacteria synthetizing colibactin) and a negative control (bacteria not synthetizing colibactin). The plate is incubated at 37 °C with 5% CO2 for 4 h. Measure the absorbance at 600 nm with a microplate reader to monitor bacterial growth.
-
Post-infection
Wash the cells at least three times with 100 µl of pre-warmed (37 °C) HBSS. Check under an inverted microscope whether most of the bacteria are removed. Perform additional washes if required. Then, 100 µl of complete medium (DMEM GlutaMax, 10% FBS, 1% NEAA) supplemented with 200 µg/ml of gentamicin is added to each well. Incubate the P96 at 37 °C with 5% CO2 for 3 days.
-
-
Day 3-4
Check the cells under an inverted microscope every day for bacterial overgrowth, replace with fresh medium supplemented with gentamicin if required.
-
Day 5
-
Cell fixation
The plate is rinsed two times with PBS. Under a chemical safety hood proceed to fixation with 100 µl of fixation solution (PBS 4% formaldehyde, see Recipes) for 20 min. Throw out the fixative in a toxic waste container, and rinse 2 times with PBS then once with methylene blue wash buffer (see Recipes).
-
Methylene blue coloration
Stain with 100 µl/well of methylene blue staining solution (see Recipes) for 1 h. Rinse several times with methylene blue wash buffer (see Recipes), tapping the plate between each wash on a paper towel to remove all the liquid. Repeat until no more blue stain is visible on the paper towel. Air dry overnight at room temperature or a few hours at 37 °C.
-
Microscopic observation
Observe the cells with an inverted microscope. Control cells should be confluent with a normal cellular morphology. In contrast, cells infected with high MOI of E. coli producing colibactin (MOI around 100) should exhibit a cytopathic phenotype characterized by low numbers of cells, megalocytosis (giant cells) and signs of cell death (apoptotic bodies, cell debris) (see Figure 2). At lower MOI of E. coli producing colibactin (around MOI 25), the phenotype is less marked, and islets of normal cells (that were able to repair the moderate DNA damage and resumed proliferation) should be visible. At MOI < 12 the cells are similar to control cells.
-
Genotoxicity quantification
The methylene blue is extracted in 100 µl per wells of methylene blue extraction solution, 15 min under agitation. Transfer 75 µl to a new plate and measure the absorbance at 660 nm with a microplate reader. Prepare a graph with OD660 nm in function of the MOI and the strain, with control strains as references. The more the strain is genotoxic, the less coloration will be measured, due to cell growth inhibition, senescence (megalocytosis) and cell death.
-
Figure 1. Example of plate infection scheme.
For dose/effect examination, bacteria are inoculated to HeLa cells at various multiplicity of infection (MOI, i.e., the number of bacteria per cell at the onset of the infection). Typically, bacteria are inoculated in row B with 200 µl per well (preferentially in duplicate or triplicate) then serially diluted (2-fold as shown) to achieve an infectious dose-effect. The final interaction medium volume is 100 µl. Peripheral wells should not be used to avoid edge effects that alter HeLa cell growth and response.
Figure 2. Megalocytosis phenotype observed three days after a 4 h exposure of HeLa cells to a pks+ E. coli strain at a multiplicity of infection (MOI) of 12 to 100 bacteria per cell.
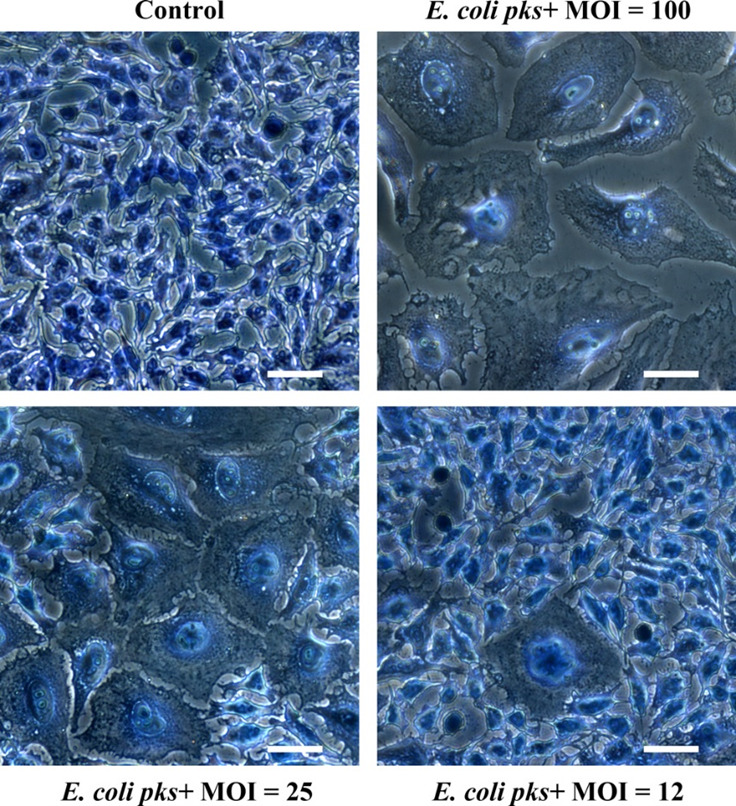
The cells were stained with methylene blue and photographed with a 20x objective. Scale bars = 25 µm.
Data analysis
The cells should be first examined visually under the microscope (Figure 2), then by quantifying the methylene blue (Figure 3). Quantify the stain to measure the protein content in the cell layer, and thus represent the global cell viability inversely correlated to the genotoxicity. We suggest testing each strain at least in technical duplicate wells for each MOI tested. Thus, five independent conditions (strains) with a dose-effect can be tested per plate. If a statistical analysis is required, perform at least three independent experiments to obtain a biological triplicate, and compare the groups by 1-way ANOVA after log-transformation of the data.
Figure 3. Quantification of methylene blue extracted from HeLa cells 3 days after a 4 h exposition to various MOI of pks+ E. coli , the isogenic clbH mutant (that does not produce the genotoxin), or the clbH mutant complemented with pClbH plasmid.
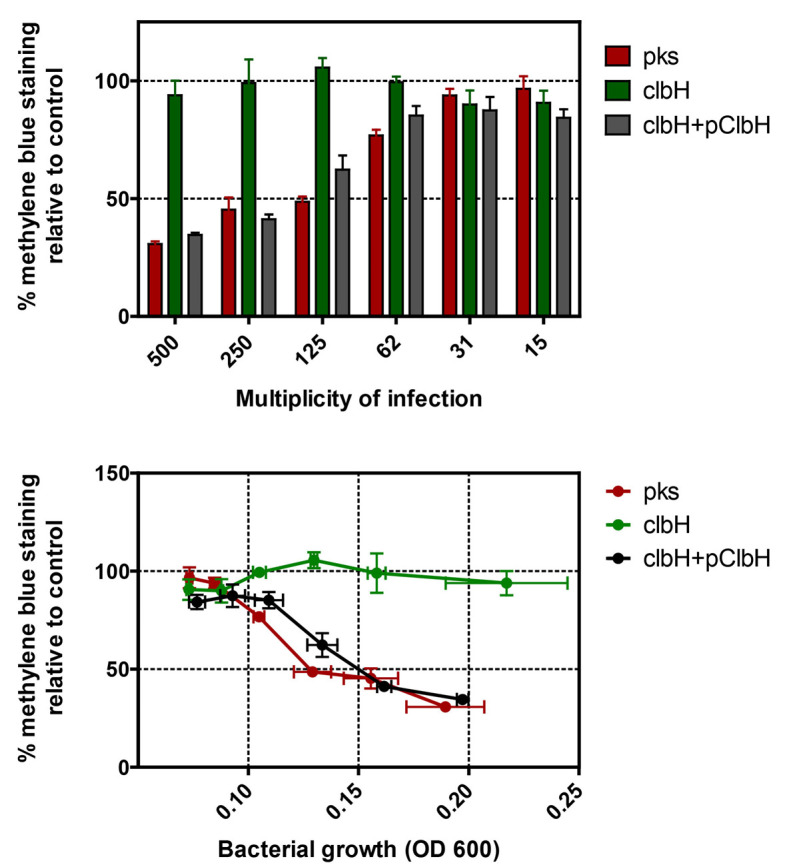
The percentage of methylene blue staining relative to control was calculated as the mean optical density in triplicate wells (from one experiment) divided by that in control (untreated) wells. The data can be plotted in function of the multiplicity of infection (MOI: number of infecting bacteria per cultured cell at the onset of infection), or in function of the bacterial growth at the end of the infection (as measured by optical density at 600 nm in the interaction wells). The error bars shown here represent the standard deviation of the mean in the triplicate wells.
Notes
All the HeLa cells washes should be performed with caution to avoid damaging or removing the cells from the monolayer; aspirate and add the medium slowly (about 1-2 sec).
Recipes
-
HeLa cell culture medium (complete medium)
500 ml DMEM high glucose with pyruvate and GlutaMax
55 ml fetal bovine serum5.5 ml NEAA
50 µg/ml gentamicin
-
Fixation solution, 1x PBS, 4% PFA 5 ml 20% PFA stock solution
5 ml 10x PBS
40 ml distilled H2O
Aliquot and store at -20 °C
-
Methylene blue staining solution
5 g methylene blue
500 ml 0.01 M Tris pH 8.5 (Methylene blue wash buffer)
Mix during 3-4 h
Store at room temperature
-
Methylene blue wash buffer
5 ml 1 M Tris pH 8.5
495 ml distilled H2O
-
Methylene blue extraction solution
50 ml 1 M HCl
450 ml distilled H2O
Acknowledgments
This protocol is based on previously published cultured cell infection method with pks+ E. coli (De Rycke et al., 1996 ; Nougayrede et al., 2006 ). This work was supported by the French Institut National du Cancer (grant INCAPLBIO13-123).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.De Rycke J., Mazars P., Nougayrede J. P., Tasca C., Boury M., Herault F., Valette A. and Oswald E.(1996). Mitotic block and delayed lethality in HeLa epithelial cells exposed to Escherichia coli BM2-1 producing cytotoxic necrotizing factor type 1 . Infect Immun 64(5): 1694-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nougayrede J. P., Homburg S., Taieb F., Boury M., Brzuszkiewicz E., Gottschalk G., Buchrieser C., Hacker J., Dobrindt U. and Oswald E.(2006). Escherichia coli induces DNA double-strand breaks in eukaryotic cells . Science 313(5788): 848-851. [DOI] [PubMed] [Google Scholar]
- 3.Putze J., Hennequin C., Nougayrede J. P., Zhang W., Homburg S., Karch H., Bringer M. A., Fayolle C., Carniel E., Rabsch W., Oelschlaeger T. A., Oswald E., Forestier C., Hacker J. and Dobrindt U.(2009). Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae . Infect Immun 77(11): 4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Secher T., Samba-Louaka A., Oswald E. and Nougayrede J. P.(2013). Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells . PLoS One 8(10): e77157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taieb F., Petit C., Nougayrede J. P. and Oswald E.(2016). The enterobacterial genotoxins: cytolethal distending toxin and colibactin. EcoSal Plus 7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]