Abstract
Streptococcus pneumoniae (pneumococcus) is an important human pathogen that causes pneumonia, meningitis, sepsis, and otitis media. This bacterium normally resides in the nasopharynx as a commensal, but sometimes disseminates to sterile sites of humans and causes local or systemic inflammation. This biphasic behavior of S. pneumoniae is correlated with a reversible switch between the opaque and transparent colony forms on agar plates, a phenomenon referred to as phase variation. The opaque variants appear to be more virulent in animal models of bacteremia but are deficient in nasopharyngeal colonization animal models. In contrast, the transparent variants display higher levels of nasopharyngeal colonization but relatively lower virulence in animal models. We have recently demonstrated that pneumococcal phase variation between these two colony types is caused by a reversible switch of genome DNA methylation (or epigenetic) patterns, which is driven by DNA inversions in the DNA methyltransferase genes. Observation of colony morphology is a simple and useful method to differentiate colonies with different characteristics, such as size, color, and opacity. This protocol describes how to study pneumococcal phase variation in colony morphology with a dissection microscope.
Keywords: Streptococcus pneumoniae, Colony morphology, Phase variation, Epigenetic switch, DNA inversion, Dissection microscope
Background
Streptococcus pneumoniae is a leading cause of bacterial pneumonia, meningitis, and sepsis in children worldwide ( Walker et al., 2013 ). The success of this pathogen in its adaptation to various ecological niches of human host depends on its remarkable phenotypic plasticity ( Croucher et al., 2013 ; Johnston et al., 2014a ), which has been reflected by the inter-strain antigenic variation in the capsular polysaccharides and surface proteins ( Croucher et al., 2013 and 2011), acquisition of new virulence factors ( Park et al., 2012 ), extensive drug resistance ( Croucher et al., 2014 ) within the species. Natural genetic transformation is a well-known mechanism contributing to this phenotypic plasticity ( Johnston et al., 2014b ). Moreover, S. pneumoniae is also capable of spontaneous phase variation between opaque and transparent colony phenotypes, a widespread phenomenon in microbial pathogens (van der Woude, 2011). The opaque variants, which have more capsule and less teichoic acid, are more virulent in the lung and bloodstream; the transparent counterparts, which express less capsule and have more teichoic acid, are more adaptive to the nasopharynx (Kim and Weiser, 1998; Weiser et al., 1994 ; Manso et al., 2014 ; Li et al., 2016 ). Our recent studies have revealed that pneumococcal phase variation between two colony phenotypes is determined by DNA inversion between the methyltransferase hsdS genes in the colony opacity determinant (cod) locus ( Feng et al., 2014 ; Li et al., 2016 ).
The protocol we present here describes the complete experimental procedure from preparing a bacterial stock to obtaining an image of colony morphology as described in our recent study ( Li et al., 2016 ). Animal blood is commonly added to agar medium to promote growth of S. pneumoniae, but the color of the blood makes it difficult to differentiate opaque and transparent colonies by microscopic approach. Instead, catalase is supplemented to agar plates to culture S. pneumoniae by neutralizing the inhibition effect of hydrogen peroxide (produced by the pneumococci themselves) on pneumococcal growth when it comes to observe and document colony morphologies by dissection microscope (Kim and Weiser, 1998; Weiser et al., 1994 ; Manso et al., 2014 ; Li et al., 2016 ). Since colony morphology phenotypes can be indicative of bacterial physiological and pathogenic properties, this protocol may offer a valuable method to study the impact of genetic and epigenetic elements or environmental conditions on bacterial biology and disease pathogenesis.
Materials and Reagents
Pipette tips
Petri dishes (100 mm) (Thermo Fisher Scientific, Thermo Scientific TM, catalog number: 263991)
50 ml tubes (Thermo Fisher Scientific, Thermo Scientific TM, catalog number: 339652)
0.2 μm filter (Pall, catalog number: 4612)
1.5 ml tubes (Eppendorf, catalog number: 022363204)
Spreading rods (Sigma-Aldrich, catalog number: Z376779)
S. pneumoniae strain ST556 ( Li et al., 2012 )
Todd Hewitt broth (Sigma-Aldrich, catalog number: T1438)
Yeast extract (Sigma-Aldrich, catalog number: Y1625)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Tryptic soy agar (TSA) (BD, DifcoTM, catalog number: 236950)
Catalase from bovine liver (Sigma-Aldrich, catalog number: C9322)
Phosphate buffered saline (PBS) (Mediatech, catalog number: 21-040-CV)
Todd Hewitt broth with yeast extract (THY) medium (see Recipes)
TSA medium (see Recipes)
Catalase working solution (see Recipes)
Equipment
Pipettes
Dissection microscope (ZEISS, model: Stemi 2000-C)
37 °C water bath
Spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: GENESYSTM 30)
37 °C 5% CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HeracellTM 150i)
Autoclave (SANYO, model: MLS-3780)
Analytical balance (Mettler-Toledo International, model: NewClassic ML802)
Vortex mixer
Centrifuge (Eppendorf, model: 5417 R)
Class II biological safety cabinet (Thermo Fisher Scientific, Thermo ScientificTM, model: MSC-AdvantageTM Class II)
Digital single lens reflex camera (Canon, model: EOS 550D)
Small mirror with a diameter of ~5 cm
Procedure
-
Setup of the microscope (Video 1)
Position the dissection microscope ~5 cm off the bench by placing two boxes under both sides of the dissection microscope base.
Take down the illuminator from the microscope.
Fix the illuminator to the surface of the bench between the two boxes using a piece of tape, and make the direction of the light source vertically towards the observer.
Remove the lens of the camera and install the camera on the top of the microscope.
Remove the object stage that is embedded in the base of the microscope.
-
Preparation of pneumococcal colonies for observing colony morphology
Thaw a frozen stock of pneumococcal strain ST556 in a water bath at 37 °C.
Inoculate 5 ml THY medium with 50 μl of the thawed frozen stock.
Incubate the culture in a 5% CO2 incubator at 37 °C for about 5 h.
-
Check the optical density (OD) of the culture at a wavelength of 620 nm using a spectrophotometer until the culture reaches mid-log phase (OD620 = 0.4-0.6).
Note: The phase-variable colonies can be formed by either bacterial cells from fresh cultures or frozen stocks. If you want to use the freshly grown culture as described in 2d to make colonies, proceed to step 2g.
Mix the bacterial culture with an equal volume of 30% glycerol in THY, and store the stocks at -80 °C for future use.
Thaw a frozen stock of S. pneumoniae strain ST556 in a water bath at 37 °C.
Dilute the culture or thawed stock with PBS at 1:10,000 dilution to approximately 104 colony forming unit (CFU)/ml.
Thaw a frozen stock of catalase working solution at room temperature.
Mix 50 μl of bacterial diluted stock with 100 μl of the catalase working solution, and spread the mixture on a TSA plate.
-
Incubate the plate in the 5% CO2 incubator at 37 °C for 16 h.
Note: In this protocol, we only detected the phase variation in colony opacity of pneumococcal strain ST556. Based on our previous observations (Li et al., 2016), 6 out of 8 strains showed opaque and transparent colony phenotypes at the 16-h time point, such as ST556 (serotype 19F), TH2901 (serotype 6B), and TH2835 (serotype 14). However, TIGR4 (serotype 4) and D39 (serotype 2) displayed different colony phenotypes at the 24-h time point. The incubation time of the plates should be adjusted according to the strain you use.
-
Observation of pneumococcal colonies (Video 2)
Open the lid of the plate (Procedure 2), and put the plate with its open side down on the stage of the dissection microscope.
Turn on the illuminator, and use a small mirror under the stage of the microscope to reflect the light onto the plate.
Adjust the intensity of the light and angle of reflected light by the mirror until you can see the colonies.
Adjust the magnification of the microscope based on your needs.
Adjust the focus of the microscope to make your field clear.
Turn on the camera, and turn off the flash and automatic brightness adjustment system of the camera.
Fix the angle of the mirror by holding it steadily.
Photograph the colonies.
Video 1. Setup of the dissection microscope.
Video 2. Observation of pneumococcal colonies.
Data analysis
The representative image (Figure 1) illustrates that pneumococcal strain ST556 is capable of spontaneous phase variation in colony opacity, resulting in the formation of opaque and transparent two types of colony morphologies on the TSA plate supplemented with catalase.
Figure 1. A photograph of the phase variation between opaque and transparent colonies in a clonal population of S. pneumoniae strain ST556.
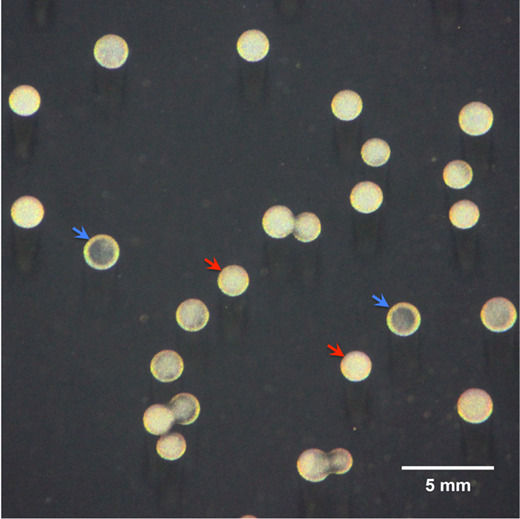
Colonies were produced on a TSA plate supplemented with catalase after 16-h incubation and observed under a dissection microscope as described above. The representative opaque and transparent colonies are indicated by red and blue arrowheads, respectively. The scale bar was shown at the bottom right of this image ( Li et al., 2016 ).
Recipes
-
THY medium
Dissolve the powder of Todd Hewitt broth (TH) and yeast extract (Y) in ddH2O (30 g TH and 5 g Y/L)
Autoclave the medium at 121 °C for 15 min
Store the medium at 4 °C
-
TSA agar plates
Dissolve the TSA powder in ddH2O (40 g/L)
Autoclave the medium at 121 °C for 15 min
Pour 15-20 ml TSA medium into each dish
After solidification, use plates directly or store at 4 °C
-
Catalase working solution (40,000-100,000 U/ml)
Weigh 0.2 g catalase using an analytical balance
Dissolve the catalase from 3a in 10 ml PBS in a 50 ml tube by vortex for 1 min
Centrifuge the solution at 12,000 × g for 10 min at 4 °C
Sterilize the supernatant from 3c by filtration using a 0.2 μm filter
Aliquot the solution into 1.5 ml tubes
-
Store the tubes at -80 °C
Note: Avoid using the freeze-thawed catalase solution.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (No. 31530082; No. 31530082) and the Grand Challenges Exploration of the Bill and Melinda Gates Foundation (No. OPP1021992). We thank Dr. Jeffrey N. Weiser (Department of Microbiology, New York University School of Medicine, New York, USA) for sharing his experience about observation of pneumococcal colony morphology.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Croucher N. J., Chewapreecha C., Hanage W. P., Harris S. R., McGee L., van der Linden M., Song J. H., Ko K. S., de Lencastre H., Turner C., Yang F., Sa-Leao R., Beall B., Klugman K. P., Parkhill J., Turner P. and Bentley S. D.(2014). Evidence for soft selective sweeps in the evolution of pneumococcal multidrug resistance and vaccine escape. Genome Biol Evol 6(7): 1589-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Croucher N. J., Finkelstein J. A., Pelton S. I., Mitchell P. K., Lee G. M., Parkhill J., Bentley S. D., Hanage W. P. and Lipsitch M.(2013). Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45(6): 656-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Croucher N. J., Harris S. R., Fraser C., Quail M. A., Burton J., van der Linden M., McGee L., von Gottberg A., Song J. H., Ko K. S., Pichon B., Baker S., Parry C. M., Lambertsen L. M., Shahinas D., Pillai D. R., Mitchell T. J., Dougan G., Tomasz A., Klugman K. P., Parkhill J., Hanage W. P. and Bentley S. D.(2011). Rapid pneumococcal evolution in response to clinical interventions. Science 331(6016): 430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Z., Li J., Zhang J. R. and Zhang X.(2014). qDNAmod: a statistical model-based tool to reveal intercellular heterogeneity of DNA modification from SMRT sequencing data. Nucleic Acids Res 42(22): 13488-13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnston C., Campo N., Berge M. J., Polard P. and Claverys J. P.(2014). Streptococcus pneumoniae, le transformiste. Trends Microbiol 22(3): 113-119. [DOI] [PubMed] [Google Scholar]
- 6.Johnston C., Martin B., Fichant G., Polard P. and Claverys J. P.(2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol 12(3): 181-196. [DOI] [PubMed] [Google Scholar]
- 7.Kim J. O. and Weiser J. N.(1998). Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis 177(2): 368-377. [DOI] [PubMed] [Google Scholar]
- 8.Li G., Hu F. Z., Yang X., Cui Y., Yang J., Qu F., Gao G. F. and Zhang J. R.(2012). Complete genome sequence of Streptococcus pneumoniae strain ST556, a multidrug-resistant isolate from an otitis media patient . J Bacteriol 194(12): 3294-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J., Li J. W., Feng Z., Wang J., An H., Liu Y., Wang Y., Wang K., Zhang X., Miao Z., Liang W., Sebra R., Wang G., Wang W. C. and Zhang J. R.(2016). Epigenetic switch driven by DNA inversions dictates phase variation in Streptococcus pneumoniae . PLoS Pathog 12(7): e1005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manso A. S., Chai M. H., Atack J. M., Furi L., M. De Ste Croix, Haigh R., Trappetti C., Ogunniyi A. D., Shewell L. K., Boitano M., Clark T. A., Korlach J., Blades M., Mirkes E., Gorban A. N., Paton J. C., Jennings M. P. and Oggioni M. R.(2014). A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat Commun 5: 5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park I. H., Kim K. H., Andrade A. L., Briles D. E., McDaniel L. S. and Nahm M. H.(2012). Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. MBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Woude M. W.(2011). Phase variation: how to create and coordinate population diversity. Curr Opin Microbiol 14(2): 205-211. [DOI] [PubMed] [Google Scholar]
- 13.Walker C. L., Rudan I., Liu L., Nair H., Theodoratou E., Bhutta Z. A., O'Brien K. L., Campbell H. and Black R. E.(2013). Global burden of childhood pneumonia and diarrhoea. Lancet 381(9875): 1405-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiser J. N., Austrian R., Sreenivasan P. K. and Masure H. R.(1994). Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun 62(6): 2582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]


