Abstract
A novel method to assess the dissolution of the core pluripotency transcription-factor circuit of mouse Embryonic Stem Cells (mESCs) has been developed ( Ying et al., 2003 ; Betschinger et al., 2013 ). In order to efficiently identify genes essential for the break-down of the pluripotency network in mutant mESCs with proliferation defects, we adapted this ‘exit from pluripotency assay’ ( Bodak et al., 2017 ; Cirera-Salinas et al., 2017 ). The protocol described here has been successfully applied to several mESC lines and is easily transposable from one laboratory to another.
Keywords: Mouse embryonic stem cells, Pluripotency, Commitment, 2i media, RNA interference, Alkaline phosphatase, in vitro assay
Background
For decades, scientists have tried to identify the mechanisms underlying the differentiation potential of mESCs with general (e.g., Embryoïd Body) or directed (e.g., Neuronal Precursor Cells) differentiation protocols. Recently, the 2i culture media was discovered allowing the captivation of the naïve stem cells state in vitro ( Ying et al., 2008 ). Betschinger and colleagues took advantage of the medium and developed a new ‘exit from pluripotency’ assay allowing the identification of novel factors involved in the commitment of mESCs ( Betschinger et al., 2013 ). Briefly, mESCs maintained in 2i media conditions, are placed for two days in permissive media. Subsequently, the 2i media is reintroduced allowing only the survival of naïve mESCs. In this assay, wild type (WT) mESCs commit to differentiation during the two days of permissive media and die after reintroduction of 2i medium. Indeed, indicating that only two days of permissive media are sufficient to break-down the pluripotency network and commit to differentiation. Unfortunately, mutant mESCs for RNA interference pathways, i.e., Dicer and Dgcr8 genes, proliferate much slower than their WT counterparts making the assessment of the exit from pluripotency in two days with the original protocol less suitable. We decided to extend the presence of the cells in the permissive media to four days and then to reintroduce the 2i media for three more days. Only cells that do not commit during the four days in permissive media conditions will be able to survive and proliferate during the final three days in 2i media. Finally, cell survival and stemness are measured with Alkaline Phosphatase (AP) staining, as in the initial protocol.
Materials and Reagents
10 cm plate (TPP Techno Plastic Products, catalog number: 93100)
6-well plate (TPP Techno Plastic Products, catalog number: 92006)
10 ml pipettes (Bioswisstec, catalog number: 515210)
5 ml pipettes (Bioswisstec, catalog number: 515205)
15 ml Falcon tube (Greiner Bio One International, catalog number: 188271)
Glass Pasteur pipette (HUBERLAB, catalog number: 1.1127.01)
Mouse embryonic stem cells (E14TG2a mESC line obtained from ATCC: ES-E14TG2a) (ATCC, catalog number: CRL-1821)
Phosphate-buffered saline (PBS) 1x (Thermo Fisher Scientific, GibcoTM, catalog number: 10010015)
Trypsin-EDTA 0.05% (Thermo Fisher Scientific, GibcoTM, catalog number: 25300054)
Alkaline phosphatase Kit (Sigma-Aldrich, catalog number: 86R-1KT)
-
ddH2O (Sigma-Aldrich, catalog number: 99053)
Note: This product has been discontinued.
Gelatin (Sigma-Aldrich, catalog number: G1890)
Dulbecco’s modified Eagle medium (DMEM) high glucose (Sigma-Aldrich, catalog number: D6429)
Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106)
Leukemia inhibitory factor (LIF) 10 millions unit/ml (Merck, catalog number: ESG1107)
2-Mercaptoethanol (βM) 50 mM (Thermo Fisher Scientific, GibcoTM, catalog number: 31350010)
Mixture of penicillin and streptomycin (PS) (Sigma-Aldrich, catalog number: P0781)
N2B27 (Takara Bio, catalog number: Y40002)
MAPK/ERK inhibitor 0.4 μM (PD03) (Stemolecule PD0325901) (STEMCELL Technologies, catalog number: 72184)
GSK3β inhibitor 3 μM (Chiron) (Stemolecule CHIR99021) (STEMCELL Technologies, catalog number: 72054)
Acetone (Merck, catalog number: 1.00014.1000)
Formaldehyde 37% (Sigma-Aldrich, catalog number: 47608-1L-F)
0.2% gelatin solution (see Recipes)
MES medium (see Recipes)
MEF medium (see Recipes)
2i + LIF medium (see Recipes)
Fixative solution (see Recipes)
Substrate solution (see Recipes)
Equipment
HeracallTM 150i incubator (37 °C and 8% CO2) (Thermo Fischer Scientific, Thermo ScientificTM, model: HeracallTM 150i)
Centrifuge (Eppendorf, model: 5810)
Tissue culture hood (FASTER, model: Safe FAST Premium 209, catalog number: F00024900000)
Millipore ScepterTM 2.0 cell counter (EMD Millipore, model: ScepterTM 2.0, catalog number: PHCC00000) (or any cell counter)
Inverted microscope (Nikon Instruments, model: Eclipse TS100)
Software
ImageJ software 1.48v
Adobe Photoshop CS6
GraphPad Prism 6.0a software
Procedure
-
Maintenance of mESCs
mESCs are routinely cultured on 10 cm dish pre-coated with 0.2% gelatin (see Recipes) in 10 ml of MES medium (see Recipes), in an 8% CO2 incubator at 37 °C (Figure1A).
-
Medium is changed every other day.
Note: Special attention should be taken to avoid confluency (> 70%) of mESCs (Figure 1B) in maintenance cultures as it might result in detrimental differentiation of the cells.
-
Exit from pluripotency assay
Coat a 6-well plate with 0.2% gelatin (500 μl per well) and verify that the gelatin is well distributed. Place the plate in the incubator.
Remove the media from a 50% confluent mESCs 10 cm dish. Wash once with 4 ml of 1x PBS. Add 2 ml of trypsin-EDTA to the cells and verify that the trypsin is well distributed. Place the cells back in the incubator for 5 min. Verify under the microscope that the mESC colonies are detached from the dish.
-
Add 4 ml of MEF media (see Recipes) to the dish to inactivate the trypsin. Use a 5 ml pipette gently up and down to achieve single cells and move them in a 15 ml Falcon tube.
Note: Try to achieve a single cell suspension and be careful to avoid too many bubbles.
Count the cell density using a cell counter or equivalent method.
-
Centrifuge 2 x 104 cells in a new tube at 190 × g for 5 min.
Note: All centrifugations are carried at room temperature.
Carefully resuspend the cell pellet using 2 ml of 2i + LIF media (see Recipes).
-
Plate the 2 x 104 mESCs in 2 ml of 2i + LIF medium in the 6-well pre-coated plate with 0.2% gelatin. Medium is changed every other day.
Notes:
Do not forget to remove the gelatin before plating the cells.
The day on which the culture is started is designated as Day 0.
After 3 days, aspirate the 2i + LIF media without perturbing the mESC colonies (Day 3) and add N2B27 media (without LIF nor inhibitors) for 4 days (permissive media). Medium is changed every other day.
At Day 7, remove the N2B27 (permissive media) media without perturbing the mESC colonies and reintroduce 2i + LIF media for 3 additional days. Medium is changed every other day.
On Day 10 alkaline phosphatase staining can be performed (Figure 2).
-
Alkaline phosphatase staining
Aspirate the 2i medium with a Pasteur pipette.
-
Wash once the cells with 1 ml of 1x PBS per well.
Note: Be careful not to wash off the colonies from the plate.
-
Fix the cells carefully for 45 sec with 0.5 ml fixative solution (see Recipes).
Note: Prolonged fixation can destroy alkaline phosphatase activity.
-
Wash the cells twice with 0.5 ml ddH2O.
Note: Be careful not to wash out the colonies from the plate.
Add 0.5 ml substrate solution (see Recipes) for 15 min at room temperature in the dark (either drawer or aluminum foil).
Wash with 0.5 ml ddH2O.
Let the plate air dry overnight in a drawer.
-
Clonal alkaline phosphatase positive number quantification
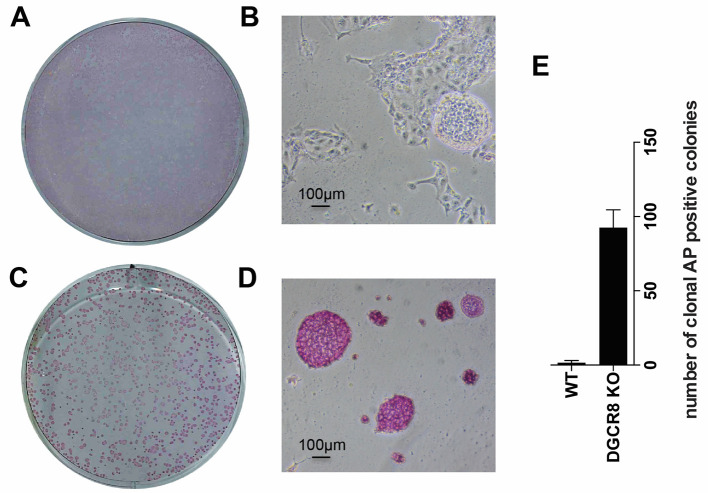
Entire 6-well plates used for alkaline phosphatase staining are scanned with a printer/scanner machine to capture the total plate area in a single image (Figures 2A and 2C). More detailed images are also acquired with the microscope to better observe the morphology and alkaline phosphatase staining intensity in the different cell lines (Figures 2B and 2D).
Scanned PDF images are converted into TIFF images with Adobe Photoshop CS6.
-
Images are processed using ImageJ software 1.48v as follows:
Number of alkaline phosphatase positive colonies is calculated on threshold intensity of inverted regions that are user–selected (full well–identical areas between conditions)
Using the Analyze Particles tool (default parameters, ImageJ) the total number of alkaline phosphatase colonies is counted.
Total number of alkaline phosphatase positive colonies is depicted in a graph as a clonal assay (Figure 2E).
Figure 1. Typical morphologies of mESCs.
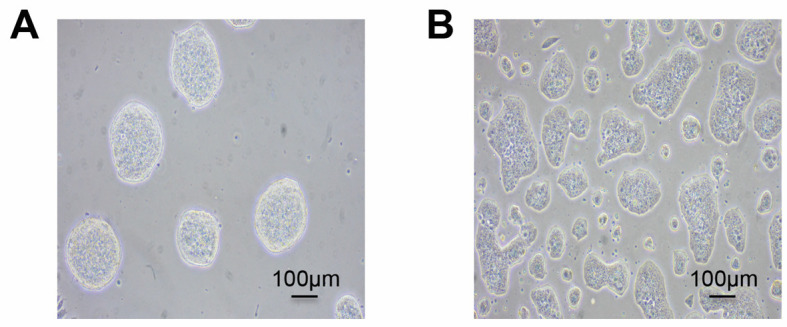
Feeder-free undifferentiated mESCs are shown at low 20-30% (A) and high 60-70% (B) density. Healthy, emergent mESCs grow as tightly formed, refractile colonies throughout passages (A). Over confluency (> 70%) of mESCs (B) creates flat colony morphology and potential differentiation.
Figure 2. Alkaline phosphatase (AP) staining after exit from pluripotency assay.
A. Whole 6-well after AP staining of wildtype mESCs; B. Wildtype mESCs are colorless and/or differentiated; C. Whole 6-well after AP staining of Dgcr8_KO mESCs; D. Dgcr8_KO mESCs cannot exit pluripotency and appear purple and undifferentiated; E. Number of positive clonal AP staining of cells after exit from pluripotency assay.
Data analysis
Number of clonal AP positive colonies is expressed as ± SEM. Statistical differences can be measured by Student’s t-test. A value of P < 0.05 is considered significant. Data analysis can be performed using GraphPad Prism 6.0a software. AP staining must be performed in triplicate. The original research papers containing the aforementioned protocol are Cirera-Salinas et al., 2017 and Bodak et al., 2017 .
Notes
The aforementioned protocol has been designed for RNAi mutant mESCs with strong proliferative defects. If other mouse ESC lines are used, we recommend to first adapting the cell lines to the 2i media. The adaption time varies from 3 days for good adherent cell lines, to 5-7 days if the cells have difficulties to attach to the plate. If the mESC line did not attach well, treat the plate with 0.2% gelatin for 24 h in the incubator before to plate them. Morphology of the cells is a good indicative for the adaptation as the cells become very round when adapted to the 2i media. Usually mESCs need 2 days in permissive media to exit from pluripotency. Finally, independently of the cell line used, 3 days in 2i media are necessary before staining with alkaline phosphatase.
Recipes
-
0.2% gelatin
Dilute 1 g of gelatin from porcine skin with 500 ml of ddH2O, mix well and then autoclave solution
Reagent Volume Autoclaved water 500 ml Gelatin from porcine skin 1 g Vtotal = 500 ml -
MES medium
Reagent Volume DMEM 42.5 ml FBS (decomplemented) 7.5 ml (15%) LIF 5 µl βMPS100 µl500 µlVtotal = 50 ml -
MEF medium
Reagent Volume DMEM 45 ml FBS (decomplemented) 5 ml (10%) βMPS100 µl500 µlVtotal = 50 ml -
2i + LIF medium
Reagent Volume N2B27 45 ml LIF 5 µl PSPD03Chiron500 µl5 µl (1 µM Fc)15 µl (3 µM Fc)Vtotal = 45.5 ml -
Fixative solution (15 ml)
12.25 ml acetone
3.92 ml citrate (provided 25 mM solution from the Sigma-Aldrich kit)
1.5 ml 37% formaldehyde
-
Substrate solution (3 ml)
2.81 ml ddH2O
62.5 µl nitrite (provided by the Sigma-Aldrich kit)
62.5 µl FRV (provided by the Sigma-Aldrich kit)
62.5 µl of Naphthol (provided by the Sigma-Aldrich kit)
Note: The order of the substrates nitrite (1st), FRV (2nd) and Naphthol (3rd) is important. Liquid has to be yellow before adding it to the cells.
Acknowledgments
This protocol was originally published as part of Cirera-Salinas et al. (2017). The authors wish to thank M. Bodak for fruitful discussions. D. C-S is supported by a Post-doctoral fellowship from the Peter and Traudl Foundation.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Betschinger J., Nichols J., Dietmann S., Corrin P. D., Paddison P. J. and Smith A.(2013). Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153(2): 335-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodak M., Cirera-Salinas D., Yu J., Ngondo R. P. and Ciaudo C.(2017). Dicer, a new regulator of pluripotency exit and LINE-1 elements in mouse embryonic stem cells. FEBS Open Bio 7(2): 204-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cirera-Salinas D., Yu J., Bodak M., Ngondo R. P., Herbert K. M. and Ciaudo C.(2017). Non-canonical function of DGCR8 controls mESCs exit from pluripotency. J Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ying Q. L., Stavridis M., Griffiths D., Li M. and Smith A.(2003). Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol 21(2): 183-186. [DOI] [PubMed] [Google Scholar]
- 5.Ying Q. L., Wray W., Nichols J., Battle-Morera L., Doble B., Woodgett J., Cohen P. and Smith A.(2008). The ground state of embryonic stem cell self-renewal. Nature 453(7194): 519-23. [DOI] [PMC free article] [PubMed] [Google Scholar]