Abstract
Glutathione is an important molecule involved in the primary and secondary metabolism of all organisms. The Glutathione redox status is an indicator of the cellular redox state. Therefore, it is important to have precise methods on hand to determine the glutathione redox status in the cell. In this protocol, we describe an improved spectrophotometric method to estimate the content of reduced (GSH) and oxidized (GSSG) forms of glutathione in the extremophilic microalga Galdieria phlegrea.
Keywords: Microalgae, Galdieria phlegrea, Glutathione, Glutathione reductase, Redox state
Background
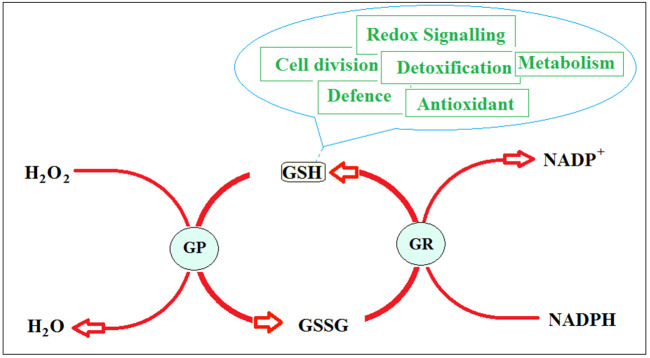
Glutathione (γ-L-glutamyl-L-cysteinyl-glycine) is an essential tripeptide existing in all known organism, ranging from bacteria to humans ( Frendo et al., 2013 ). It is involved in several different cell protective roles (Figure 1). Glutathione is considered an excellent antioxidant molecule having redox signalling function. It is also involved in response to abiotic and biotic stress, and implicated in the primary metabolism (C, N, S metabolism) of the cell ( Noctor et al., 2012 ; Hernández et al., 2015 ; Salbitani et al., 2015 ).
Figure 1. Glutathione redox cycle.
Reduced glutathione (GSH) is a tripeptide composed of cysteine, glutamic acid and glycine, which plays a key role in the control of signalling processes, detoxification and various other cell processes. Glutathione disulfide (GSSG) is the oxidized form of glutathione. It is reduced to GSH in presence of NADPH by the glutathione reductase (GR). The glutathione peroxidase (GP) converts hydrogen peroxide to water.
In plant cells, the ratio between the reduced and oxidized forms of glutathione (GSH/GSSG) plays a significant part in signalling and in the activation of numerous defence mechanisms (Foyer and Noctor, 2012; Salbitani et al., 2015 ). The GSH/GSSG redox state, which is normally tightly controlled, may transiently shift towards a slightly more oxidized value during a stress condition ( Tausz et al., 2004 ).
In the past, some researchers have developed and modified spectrophotometric methods to determine the GSH and GSSG in several organism (Anderson, 1985; Bashir et al., 2013 ). Here we describe a protocol for a simple glutathione determination, optimized on the extremophilic microalga Galdieria phlegrea. The method illustrated is based on the reaction of GSH with the thiol reagent DTNB (5,5-dithiobis(2-nitrobenzoic acid)) to form GSSG and TNB (5-thionitrobenzoic acid), which is detected spectrophotometrically at 412 nm ( Giustarini et al., 2013 ).
Materials and Reagents
Disposable plastic cuvettes (1.5 ml) (BRAND, catalog number: 759150)
5-sulfosalicylic acid hydrate (Sigma-Aldrich, catalog number: 390275)
L-glutathione reduced (GSH) (Sigma-Aldrich, catalog number: G6013)
5,5-dithiobis(2-nitrobenzoic acid) (DTNB, Ellman’s reagent) (Sigma-Aldrich, catalog number: D8130)
β-Nicotinamide adenine dinucleotide phosphate, reduced tetra(cyclohexylammonium) salt (NADPH) (Sigma-Aldrich, catalog number: N5130)
Glutathione reductase (GR) (Sigma-Aldrich, catalog number: G3664)
Sodium phosphate monobasic (NaH2PO4) (Sigma-Aldrich, catalog number: S8282)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: S7907)
Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E5134)
Reaction buffer (see Recipes)
Equipment
Bench centrifuge (Thermo Electron Corporation, model: IEC CL30)
French pressure cell press (Aminco Resources, model: FA-078)
Superspeed centrifuge (Thermo Fisher Scientific, model: SorvallTM RC-5C Plus)
Vortex mixer (Troemner, catalog number: TY-LP-945302)
Spectrophotometer (Cole-Parmer, Jenway, model: 7315)
pH-meter (Mettler-Toledo International, model: FE20)
Optical microscope (leitz laborlux K)
Bϋrker chamber (BRAND, catalog number: 719520)
Procedure
-
Preparation of microalgae extracts
-
Collect 100 ml of culture during the exponential growth phase (OD550 between 0.8 and 2.5) by low speed centrifugation (4,500 × g for 10 min) by Bench centrifuge from 100 ml of algal culture. Discard the supernatant, re-suspend the pellet in 3 ml of 5% (w/v) sulfosalicylic acid and mix by vortexing.
Note: Sulfosalicylic acid is added to the pellet for its protective effect on stability of GSH (Stempak et al., 2001) but also for the removal of proteins from samples.
-
Lyse the cells by passing twice through a French pressure cell (1,100 psi). Centrifuge at 11,000 × g for 20 min at 4 °C by Superspeed centrifuge. Use the resulting supernatant as crude extract (CE) and assay it for the glutathione content.
Note: Other methods and procedures can be used to lyse Galdieria cells; among the most common methods, there are the use of a magnetic stirrer, microwave radiation, ultrasonication and enzyme treatment (Dvoretskyet al., 2016; Farooq et al., 2016; Huang et al., 2016). The breaking of the cells can be observed with an optical microscope.
-
Keep the crude extract on ice before use.
Note: The CE for glutathione determination needs to be used within 2 h after the preparation; in fact, after 2 h and after freezing a drastic reduction in GSH content was observed.
-
-
Preparation of the calibration curve
Quantification of glutathione levels is based on the reduced form of glutathione (GSH). Calibration curve is performed with 0-500 μM standard GSH solution. The molar extinction coefficient (ε), calculated at 412 nm, is estimated to be 0.017 mM-1 cm-1.
-
Determination of reduced and total glutathione
-
To determine the reduced glutathione (GSH) content add to a 1.5 ml cuvette 600 µl of reaction buffer, 40 µl of 0.4% (w/v) DTNB, 10-100 µl of CE and Milli-Q water to a final volume of 1,140 µl, and mix gently. The blank solution contained all reagents except CE.
Note: The DTNB solution can be prepared, aliquoted and stored at -20 °C for six months.
Incubate the cuvettes at room temperature for 5 min and measure the absorbance at 412 nm using a spectrophotometer.
To determine the total glutathione, add to the reaction mix already present in the cuvettes, 50 µl of 0.4% NADPH and 1 µl of GR (0.5 U).
-
Incubate the cuvettes at room temperature for 30 min. Mix gently and read the absorbance by a spectrophotometer at 412 nm.
Note: This method has been improved for a cuvette assay but adaption to microplates assays is possible.
-
Data analysis
The GSSG content was calculated as the difference between the total glutathione and GSH contents.
The thiol levels were expressed as nmol ml-1 extract. The glutathione content in each sample was correlated with the cell number that was determined by Bürker chamber.
The approximate concentrations of GSH, expected in a control culture, should be around 0.8 pmol 10-5 cell-1; in addition, the concentration of total glutathione, in the same extracts, should be around 3.3 pmol 10-5 cell-1.
Recipes
-
Reaction buffer
0.1 M Na-phosphate buffer pH 7.00
1 mM EDTA
Acknowledgments
This protocol was adapted from previously published studies (Anderson, 1985; Bashir et al., 2013 ). This study was financed by Regione Campania (PON-Smart Generation and LR 5/2002).
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Anderson M. E.(1985). Determination of glutathione and glutathione disulfide in biological samples. Methods Enzymol 113: 548-555. [DOI] [PubMed] [Google Scholar]
- 2.Bashir H., Ahmad J., Bagheri R., Nauman M. and Irfan Quereshi M.(2013). Limited sulphur resource forces Arabidopsis thaliana to shift towards non-sulfur tolerance under cadmium stress . Environ Exp Bot 94:19-32. [Google Scholar]
- 3.Dvoretsky D., Dvoretsky S., Temnov M., Akulinin E., and Peshkova E.(2016). Enhanced lipid extraction from microalgae Chlorella vulgaris biomass: experiments, modelling, optimization . Chem Eng Trans 49: 175-180. [Google Scholar]
- 4.Farooq W., Mishra S. K., Moon M., Suh W. I., Shrivastav A., Kumar K., Kwon J. H., Park M. S., and Mu Y.(2016). Energy efficient process for microalgae cell disruption for oil recovery using triiodide resin. Algal Res 13: 102-108. [Google Scholar]
- 5.Foyer C. H. and Noctor G.(2012). Managing the cellular redox hub in photosynthetic organisms. Plant Cell Environ 35(2): 199-201. [DOI] [PubMed] [Google Scholar]
- 6.Frendo P., Baldacci-Cresp F., Benyamina S. M. and Puppo A.(2013). Glutathione and plant response to the biotic environment. Free Radic Biol Med 65: 724-730. [DOI] [PubMed] [Google Scholar]
- 7.Giustarini D., Dalle Donne I., Milzani A., Fanti P., Rossi R.(2013). Analysis of GSH and GSSG after derivatization with N-ethylmaleimide. Nat Protoc 8(9): 1660-1669. [DOI] [PubMed] [Google Scholar]
- 8.Hernández L. E., Sobrino-Plata J., Montero-Palmero M. B., Carrasco-Gil S., Flores-Cáceres M. L., Ortega-Villasante C. and Escobar C.(2015). Contribution of glutathione to the control of cellular redox homeostasis under toxic metal and metalloid stress. J Exp Bot 66(10): 2901-2911. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y., Qin S., Zhang D., Li L., and Mu Y.(2016). Evaluation of cell disruption of Chlorella vulgaris by pressure-assisted ozonation and ultrasonication . Energies 13: 173-184. [Google Scholar]
- 10.Noctor G., Mhamdi A., Chaouch S., Han Y., Neukermans J., Marquez-Garcia B., Queval G. and Foyer C. H.(2012). Glutathione in plants: an integrated overview. Plant Cell Environ 35(2): 454-484. [DOI] [PubMed] [Google Scholar]
- 11.Salbitani G., Vona V., Bottone C., Petriccione M. and Carfagna S.(2015). Sulfur deprivation results in oxidative perturbation in chlorella sorokiniana(211/8k). Plant Cell Physiol 56(5): 897-905. [DOI] [PubMed] [Google Scholar]
- 12.Stempak D., Dallas S., Klein J., Bendayan R., Koren G. and Baruchel S.(2001). Glutathione stability in whole blood: effect of various deproteinizing acids. Ther Drug Monit 23(5): 542-549. [DOI] [PubMed] [Google Scholar]
- 13.Tausz M., Sircelj H. and Grill D.(2004). The glutathione system as a stress marker in plant ecophysiology: is a stress-response concept valid? J Exp Bot 55(404): 1955-1962. [DOI] [PubMed] [Google Scholar]