Abstract
Increasing evidence has revealed a close relationship between non-coding RNAs and recurrent implantation failure (RIF). However, the role of circular RNAs (circRNAs) in RIF pathogenesis remains largely unknown. Microarray analyses were used to identify the differentially expressed circRNA-circSTK40. Functional experiments, including decidualization induction and terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay, were performed to determine the effects of circSTK40 on human endometrial stromal cells (ESCs). The interactions between circSTK40 and proteins were investigated by RNA pull-down, RNA immunoprecipitation, and co-immunoprecipitation (coIP) assays. We observed that circSTK40 expression was upregulated in the RIF midluteal-phase endometrial samples. circSTK40 overexpression in ESCs inhibited the decidualization process but concurrently enhanced cell survival during stress. Mechanistically, circSTK40 directly bound to HSP90 and CLU, thus functioning as a scaffold to block their interactions and hinder the proteasomal degradation of HSP90. The resulting high levels of HSP90 led to the activation of the AKT pathway and downregulation of FOXO1 expression. Inhibitors of AKT (MK-2206) and HSP90 (17AAG) both abolished the effects of circSTK40 overexpression in ESCs and increased the decidualization levels in a dose-dependent manner. Our findings indicate a novel epigenetic mechanism for RIF pathogenesis involving circSTK40 activity and provide a foundation for targeted treatments in patients with low endometrial receptivity.
Keywords: circular RNA, recurrent implantation failure, endometrial receptivity, heat shock protein 90
Graphical abstract
We identified a novel upregulated circRNA (circSTK40) in midluteal-phase endometrium from recurrent implantation failure (RIF) patients, which impaired endometrial receptivity in RIF via modulating the HSP90/AKT/FOXO1 axis. Our findings indicate a novel epigenetic mechanism for RIF pathogenesis and provide a foundation for targeted treatments in patients with low endometrial receptivity.
Introduction
With the development of assisted reproductive technology (ART), the obstacle of recurrent implantation failure (RIF) has been identified. This condition is generally defined as the failure to achieve clinical pregnancy after three or more transfers of high-quality embryos or multiple transfers of at least 10 embryos,1 causing large financial losses and considerable mental stress to these patients. The etiology of RIF is clinically challenging, involving embryonic, uterine, tubal, and immunological factors.2 However, the pathogenesis remains to be elucidated in most cases. Endometrial receptivity is essential for embryo implantation and can represent the ability of the endometrium to allow an embryo to successfully implant during the implantation window.2,3 The establishment of endometrial receptivity involves many biological processes, including cell proliferation, apoptosis, and differentiation; is primarily coordinated by ovarian estrogen and progesterone; and requires an elaborate interplay of different transcription factors, cytokines, and signaling pathways.4,5 Although many receptivity-associated proteins have been described,3,6 their interactions and regulatory factors in the network of endometrial development are still not fully understood, especially in the pathogenesis of unexplained RIF.
Non-coding RNAs (ncRNAs), such as microRNAs and long noncoding RNAs (lncRNAs), have been shown to be involved in endometrial receptivity and embryo development.7, 8, 9 Circular RNAs (circRNAs) are an evolutionarily conserved class of ncRNAs that are characterized by covalently closed ends and are widely expressed in a tissue-specific and cell-specific manner.10,11 circRNAs have been reported to perform biological functions via multiple mechanisms, such as acting as sponges for microRNAs,12,13 binding with cytoplasmic and nuclear proteins,14, 15, 16 or encoding peptides.17,18 Increasing evidence implicates circRNAs in the pathogenesis of various diseases, such as neurological disorders,19,20 cardiovascular diseases,12,21 and many cancers.13,22,23 Recently, the circRNA ciR8073 was reported to mediate the apoptosis of endometrial epithelial cells in dairy goats by functioning as a sponge for miR-181a, indicating a potential key role of circRNAs in endometrial receptivity.24 In humans, although variations in the expression of circRNAs have been reported in the midluteal endometrium of RIF patients,25 the functions of differentially expressed circRNAs and the potential molecular mechanisms underlying RIF have yet to be determined.
In this study, we identified an upregulated circRNA (circSTK40) in the midluteal-phase endometrium of RIF patients using microarray analyses. Using an immortalized human endometrial stromal cell line (THESCs) and primary human endometrial stromal cells (hESCs), we found that circSTK40 overexpression impaired endometrial receptivity but enhanced cell survival by directly binding to both heat shock protein 90 (HSP90) and clusterin (CLU). Functioning as a scaffold, circSTK40 blocked the interaction between these two proteins and prevented the proteasomal degradation of HSP90. The resulting high levels of HSP90 activated the AKT pathway, leading to a decrease in FOXO1 protein levels. Furthermore, inhibitors of AKT (MK-2206) and HSP90 (17AAG) both abolished the effects of circSTK40 overexpression and increased the decidualization levels in a dose-dependent manner. Our findings thus demonstrate a key contribution of circSTK40 to endometrial receptivity through the modulation of HSP90 activity, thereby providing a foundation for the development of targeted therapies to mitigate RIF during the in vitro fertilization process.
Results
circSTK40 expression is upregulated in the midluteal-phase endometrial tissues of RIF patients
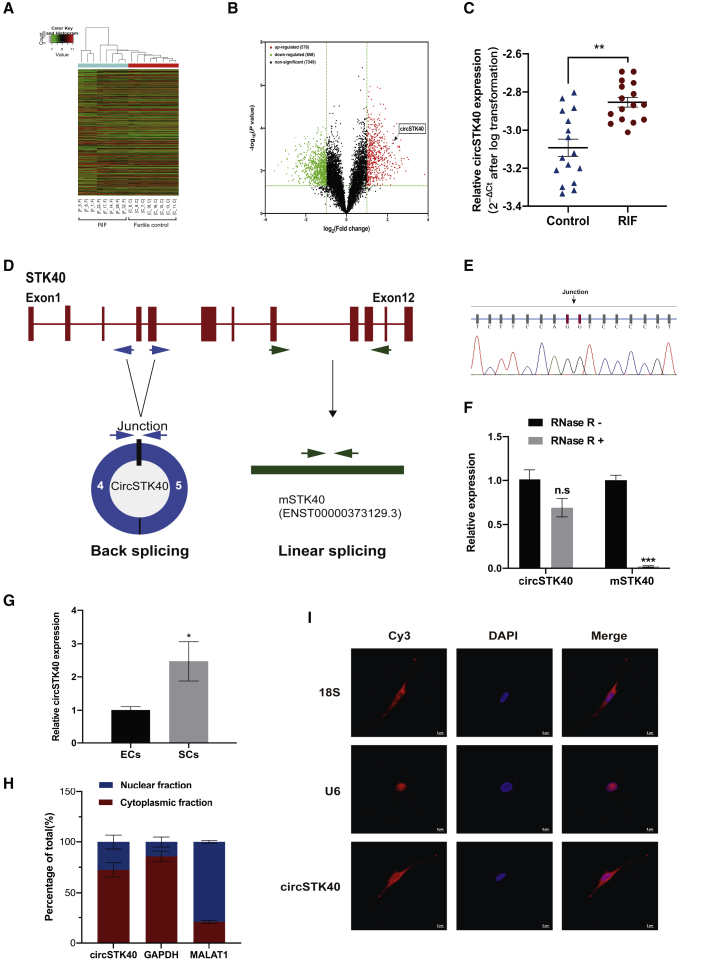
We first performed a circRNA microarray analysis using midluteal-phase endometrial samples from eight RIF patients and eight controls (Table 1) to compare the differential expression of circRNAs. Using cutoff values of p < 0.05 and fold change > 2, we found a total of 1,436 differentially expressed circRNAs between the RIF and control endometrial samples (Figures 1A and 1B). Using divergent primers and quantitative real-time PCR, several abundant, differentially expressed circRNAs were validated in the midluteal-phase endometrial samples from an independent cohort of 16 RIF and 16 control patients (Table 1; Figure S1A). In particular, circSTK40 (circBase accession: hsa_circ_0011692), a circRNA generated by back-splicing of exons 4 and 5 of the STK40 gene, was the most abundant of the validated circRNAs (Figures 1C and 1D; Figure S1A), and its sequence and back-splicing junction site were then confirmed by Sanger sequencing (Figure 1E). Subsequently, we determined that circSTK40 had greater resistance to RNase R than to the linear STK40 mRNA (mSTK40) (Figure 1F; Figure S1B). We then used cell sorting to examine the distribution of circSTK40 in the midluteal-phase endometrial samples and observed that the circSTK40 expression levels were significantly higher in the ESCs than in the endometrial epithelial cells (Figure 1G). Furthermore, the results of cytoplasmic/nuclear fractionation and fluorescence in situ hybridization (FISH) revealed that circSTK40 was localized in both the cytoplasm and nucleus of THESCs, predominantly in the cytoplasm (Figures 1H and 1I). Overall, these findings suggest that circSTK40 expression was significantly upregulated in the midluteal-phase endometrial tissues of RIF patients, especially in the ESCs.
Table 1.
Clinical characteristics of patients with RIF and controls
| Variables | Microarray analysis |
Separate verification |
||||
|---|---|---|---|---|---|---|
| Controls (n = 8) | RIFs (n = 8) | p∗ | Controls (n = 16) | RIFs (n = 16) | p∗ | |
| Age (years) | 28.88 ± 4.70 | 33.25 ± 3.96 | 0.064 | 30.81 ± 5.023 | 33.22 ± 2.922 | 0.093 |
| Infertility duration (years) | 3.69 ± 2.19 | 6.13 ± 3.5 | 0.117 | 4.97 ± 3.11 | 4.56 ± 3.13 | 0.703 |
| BMI (kg/m2) | 24.64 ± 3.80 | 24.19 ± 3.41 | 0.811 | 24.23 ± 3.49 | 23.43 ± 2.86 | 0.467 |
| Basal FSH (IU/L) | 6.44 ± 0.97 | 7.74 ± 3.99 | 0.387 | 6.59 ± 1.15 | 7.43 ± 2.81 | 0.271 |
| Right AFC | 8.13 ± 2.90 | 8.13 ± 3.91 | 1 | 8.38 ± 2.42 | 8.00 ± 3.31 | 0.711 |
| Left AFC | 6.88 ± 3.00 | 8.25 ± 3.99 | 0.449 | 7.75 ± 2.70 | 8.28 ± 3.58 | 0.634 |
| Gravida, median (25th, 75th) | 0 (0, 1) | 0 (0, 1) | 0.25 | 1 (0, 2) | 0 (0, 1) | 0.525 |
| Parity, median (25th, 75th) | 0 | 0 | 0.25 | 0 (0, 1) | 0 | 0.263 |
| Implantation failure times, median (25th, 75th) | 0 | 4 (4, 5.75) | 0 | 4 (3.75, 5) | – | |
Statistical methods: two-tailed Student’s t test or Kolmogorov-Smirnov test. BMI, body mass index; FSH, follicle-stimulating hormone; AFC, antral follicle count. ∗p < 0.05 was considered to be statistically significant.
Figure 1.
circSTK40 is upregulated in the midluteal-phase endometrial tissues of patients with RIF
(A and B) Microarray cluster and volcano plot of differentially expressed circular RNAs (circRNAs) in midluteal-phase endometrial tissues between controls (n = 8) and RIF patients (n = 8). (C) The expression levels of circSTK40 validated in the midluteal-phase endometrial tissues of controls (n = 16) and RIF patients (n = 16). Ct values were normalized to β-ACTIN. (D) Schema illustrating the production of circSTK40. circSTK40 was formed by back-splicing of exons 4 and 5 of the STK40 gene. (E) The back-splicing junction site of circSTK40 was validated by Sanger sequencing (arrow). (F) The relative levels of circSTK40 and mSTK40 after RNase R treatment (n = 3). (G) The expression levels of circSTK40 in primary human endometrial epithelial cells (ECs) and stromal cells (SCs) (n = 3). (H) Separation of cytoplasmic and nuclear RNA from THESCs and measurement of circSTK40 expression levels by quantitative real-time PCR (n = 4). (I) Subcellular localization of circSTK40 was detected by RNA fluorescence in situ hybridization assay in THESCs. U6 and 18S RNA served as the nuclear and cytoplasmic localization controls, respectively. Scale bars represent 5 μm. Data are presented as the mean ± SEM. Student’s t test. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; n.s., non-significant.
Overexpression of circSTK40 inhibits the decidualization process of ESCs while enhancing cell survival
To explore the potential role of circSTK40 in RIF pathogenesis, we constructed an adenovirus vector for circSTK40 overexpression. The efficiency of circSTK40 overexpression was confirmed by quantitative real-time PCR (Figure S1C).
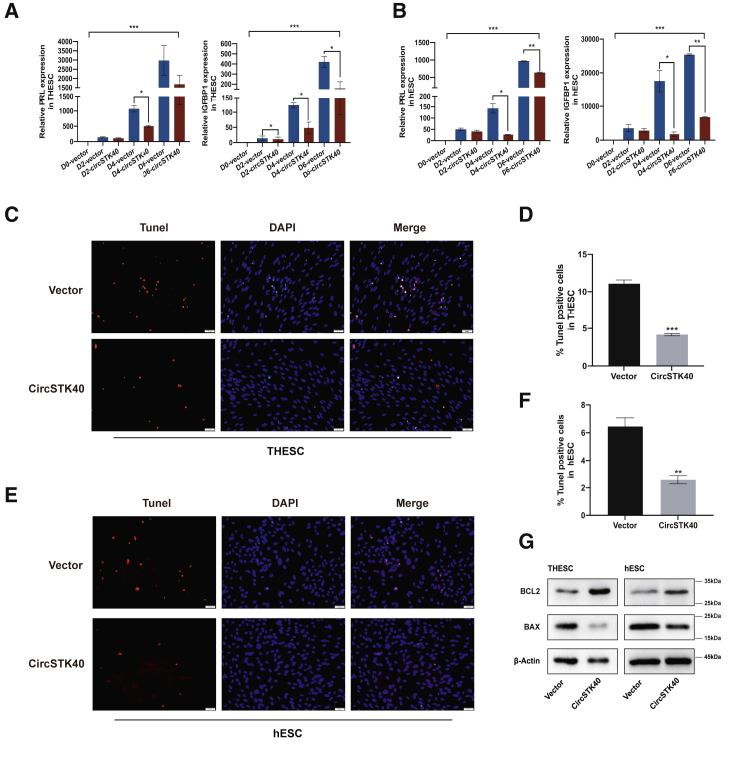
We next investigated the effects of circSTK40 overexpression on the decidualization of ESCs, which is essential for endometrial receptivity. After circSTK40 overexpression in both THESCs and primary hESCs and subsequent in vitro induction of decidualization for 2–6 days, the mRNA and protein levels of the decidualization markers prolactin (PRL) and insulin-like growth factor-binding protein 1 (IGFBP1) showed that the decidualization level had elevated gradually and significantly with the increase in treatment time and that circSTK40 overexpression significantly reduced decidualization at day 4 in both THESCs (Figure 2A; Figure S1D) and primary hESCs (Figure 2B). We therefore decided to use day 4-induced THESCs (i-THESCs) and day 4-induced primary hESCs (i-hESCs) for further experiments.
Figure 2.
circSTK40 overexpression inhibits decidualization of human endometrial stromal cells while enhancing cell survival
(A and B) The mRNA levels of PRL (left) and IGFBP1 (right) on days 0, 2, 4, and 6 (indicated as D0, D2, D4, and D6, respectively, in the figure) of induced decidualization after infection with the control vector or circSTK40-overexpression adenovirus in THESCs/primary hESCs. The relative expression levels of these genes at different times were compared with their expression levels in cells with the control vector at D0 (n = 3). (C–F) Representative photomicrographs of TUNEL staining in day 4-induced, circSTK40-overexpressing THESCs/primary hESCs. Quantification of TUNEL-positive cells presented as the percentage of TUNEL-positive cells among DAPI-stained nuclei (n = 4–5). Scale bars represent 50 μm. (G) The protein levels of BCL2 and BAX in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs. Data are presented as the mean ± SEM. Student’s t test was used for determining the differences between two independent samples; an ANOVA was used for determining the overall differences within multiple samples. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; n.s., non-significant.
We also observed that circSTK40-overexpressing ESCs exhibited higher viability with fewer dead cells during decidualization compared with controls. We then assessed the role of circSTK40 in ESC apoptosis, a process that is also associated with endometrial receptivity. Our results showed that circSTK40 overexpression significantly reduced apoptosis in i-THESCs and i-hESCs exposed to 200 μM H2O2 compared with the controls, as indicated by a decreased number of terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL)-positive cells (Figures 2C–2F) and an increase in the BCL2/BAX ratio detected using western blots (Figure 2G). Interestingly, we also found that the expression of both circSTK40 and mSTK40 was significantly upregulated after exposing i-THESCs and i-hESCs to 400 μM H2O2 and with a larger increment of circSTK40 than mSTK40 (Figure S1E). Collectively, these results strongly suggested that circSTK40 hindered the decidualization process while concurrently enhancing cell survival.
circSTK40 interferes with HSP90-CLU interaction to reduce HSP90 degradation
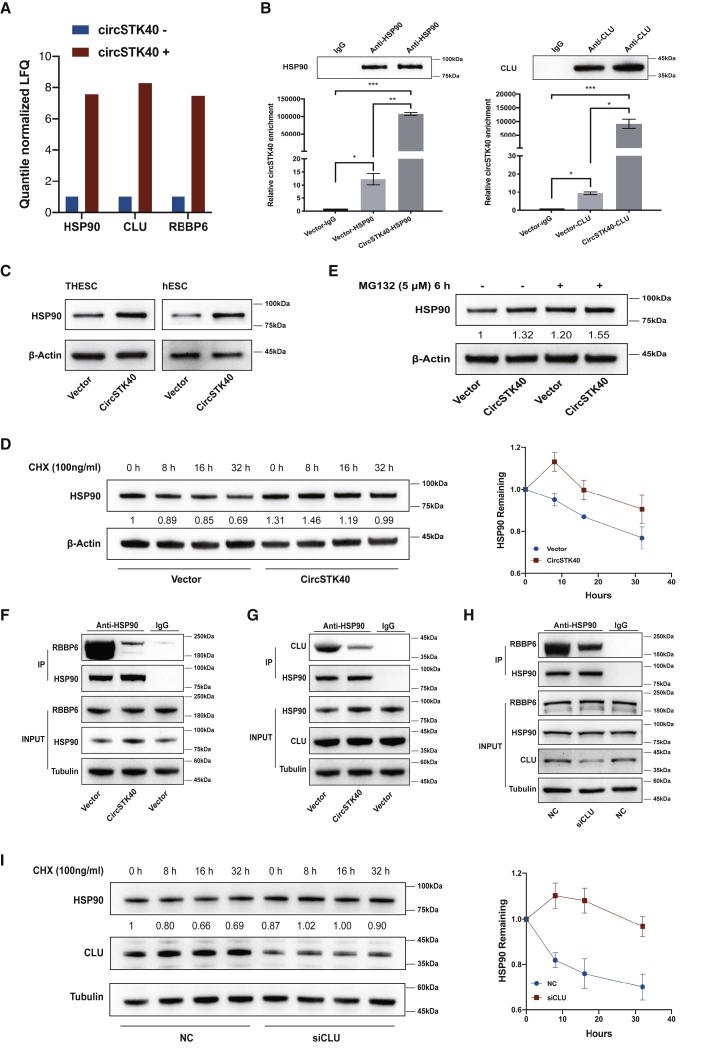
To elucidate the molecular mechanisms by which circSTK40 functions in cell decidualization and apoptosis, we performed an RNA pull-down assay and subsequent liquid chromatography-mass spectrometry to search for and identify potential circSTK40-binding proteins. Compared with the circSTK40-negative probes with the antisense transcript, we found that stress-associated proteins HSP90, CLU, and E3 ubiquitin ligase RBBP6 were all exclusively enriched among the pull-down products using the circSTK40-positive probes (Figure 3A). Subsequently, an RNA immunoprecipitation (RIP) assay was performed to confirm the endogenous direct associations between HSP90, CLU, and circSTK40. The findings revealed that circSTK40 was enriched among the immunoprecipitates of both anti-HSP90 and anti-CLU antibodies in i-THESCs carrying an empty vector and that the enrichment was significantly higher in lysates of cells carrying a circSTK40-overexpression adenovirus vector (Figure 3B).
Figure 3.
circSTK40 blocks the interaction between HSP90 and CLU, thus reducing the proteasomal degradation of HSP90
(A) Fold changes of HSP90 and CLU detected through a liquid chromatography-based mass spectrometry analysis after circSTK40 pull-down assays. The antisense transcript of circSTK40 was used as the negative probe. (B) Enrichment of circSTK40 in the RNA immunoprecipitates of anti-HSP90 (left) or anti-CLU (right) antibodies in day 4-induced THESCs infected with a control vector or a circSTK40-overexpression adenovirus (n = 3). (C) The protein levels of HSP90 in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs. (D) The protein levels of HSP90 at 0, 8, 16, and 32 h after treatment with CHX (a protein synthesis inhibitor, 100 ng/mL) in day 4-induced, circSTK40-overexpressing THESCs (left). The results of relative quantification are expressed as the mean ± SEM (n = 3) and the degradation curves (right). (E) The protein levels of HSP90 in day 4-induced, circSTK40-overexpressing THESCs after treatment with or without MG132 (a proteasome inhibitor, 5 μM) for 6 h. (F) The association between HSP90 and RBBP6 detected using coIP with anti-HSP90 antibody in day 4-induced, circSTK40-overexpressing THESCs. (G) The association between HSP90 and CLU detected using coIP with anti-HSP90 antibody in day 4-induced, circSTK40-overexpressing THESCs. (H) The association between HSP90 and RBBP6 detected using coIP with anti-HSP90 antibody in day 4-induced THESCs after CLU knockdown with siRNA. (I) The protein levels of HSP90 at 0, 8, 16, and 32 h after treatment with CHX (100 ng/mL) in day 4-induced THESCs after CLU knockdown with siRNA (left). The results of relative quantification are expressed as the mean ± SEM (n = 3) and the degradation curves (right). Data are presented as the mean ± SEM. Student’s t test was used for determining the differences between two independent samples; an ANOVA was used for determining the overall differences within multiple samples. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; n.s., non-significant.
We next examined the effect of circSTK40 overexpression on the expression of HSP90 and CLU. A western blot analysis showed that circSTK40 overexpression resulted in a significant upregulation of HSP90 protein levels (Figure 3C), without altering CLU protein levels (Figure S2B), in i-THESCs and i-hESCs. As no significant alterations in the HSP90 mRNA levels were found after circSTK40 overexpression (Figure S2A), we then checked for changes in HSP90 degradation. After cycloheximide (CHX)-mediated inhibition of protein synthesis, we found that circSTK40 overexpression retarded the degradation of HSP90 and prolonged its half-life (Figure 3D). Moreover, treatment with MG132 to inhibit proteasomal degradation led to the stabilization of HSP90 protein levels (Figure 3E), thus suggesting that circSTK40 interferes with the proteasomal degradation of HSP90.
CLU is a stress-activated, cytoprotective heat shock chaperone involved in many physiological processes and interacts with HSP90.26 In addition, studies have shown that CLU can interact with E3 ubiquitin ligases to promote proteasome-mediated degradation of target proteins.27 The presence of an E3 recognition motif and network loops and the enrichment of GO term pairs (http://ubibrowser.ncpsb.org.cn/ubibrowser/) together suggested that RBBP6 likely functions as an E3 ubiquitin ligase in the degradation of HSP90, but not CLU. Thus, we hypothesized that CLU interacts with RBBP6 and promotes the proteasomal degradation of HSP90. To investigate this possibility, we first confirmed the interactions between HSP90, CLU, and RBBP6 in ESCs using co-immunoprecipitation (coIP) assays (Figures 3F and 3G; Figures S2D–S2F). Further, we found that circSTK40 overexpression led to a reduction in the protein levels of both RBBP6 and CLU among the immunoprecipitates with anti-HSP90 antibody (Figures 3F and 3G; Figures S2E and S2F), thus indicating that circSTK40 potentially interfered in HSP90-RBBP6 and HSP90-CLU interactions. We next investigated whether CLU and RBBP6 play a regulatory role in the proteasomal degradation of HSP90. In agreement with our results showing the effects of circSTK40 overexpression, we found that knockdown of CLU via small interfering RNA (siRNA) interference reduced the interaction between HSP90 and RBBP6 (Figure 3H; Figure S2G) and that knockdown of CLU or RBBP6 via their respective siRNA interference (Figure S2C) attenuated the degradation of HSP90 after CHX-mediated protein synthesis inhibition (Figure 3I; Figure S2H). Taken together, these results indicate that circSTK40 directly binds to HSP90, thereby reducing its proteasomal degradation by blocking its interactions with CLU and RBBP6.
circSTK40 enhances HSP90-AKT interaction and activates the AKT/FOXO1 pathway
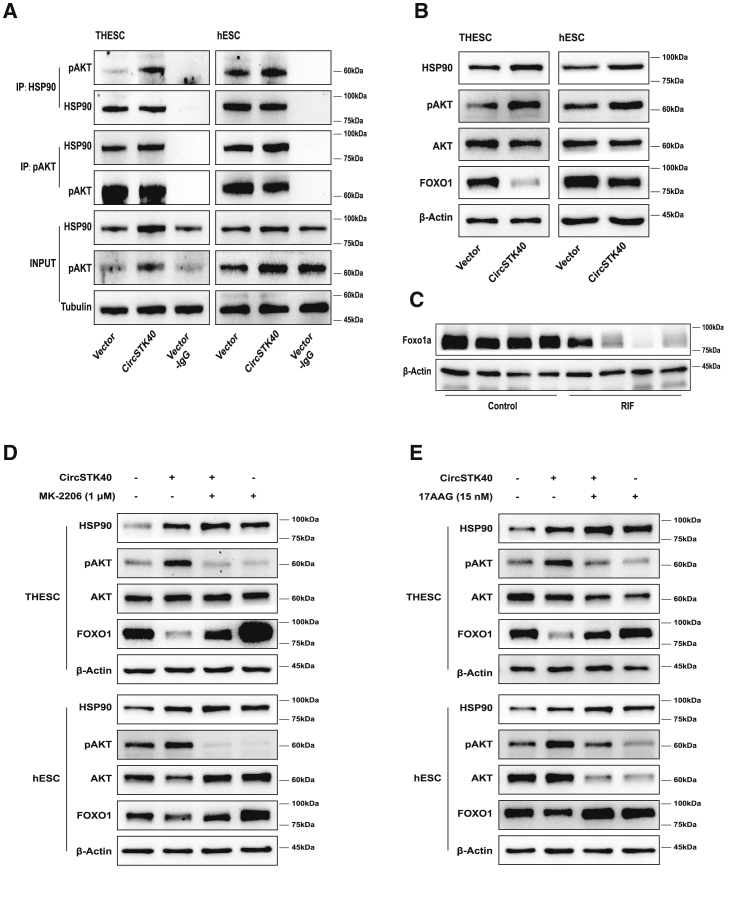
Given that HSP90 is a ubiquitous molecular chaperone that can regulate the conformation, activation/deactivation, and degradation of client proteins,28,29 we next investigated how circSTK40 overexpression affects the association of HSP90 and its well-established client protein, AKT, a protein kinase associated with proliferation, decidualization, and apoptosis in ESCs. coIP assays using antibodies against HSP90 or phosphorylated AKT (pAKT) showed that circSTK40 overexpression enhanced the interaction between HSP90 and pAKT (Figure 4A). We then observed that the AKT pathway was activated, indicated by increasing levels of pAKT concurrent with decreasing levels of FOXO1 in circSTK40-overexpressing i-THESCs and i-hESCs (Figure 4B). Notably, lower FOXO1 levels were also identified by a western blot analysis in RIF endometrial samples in comparison with control endometrial samples (Figure 4C).
Figure 4.
circSTK40 enhances HSP90–AKT interaction and activates the AKT/FOXO1 pathway
(A) The association between phosphorylated AKT (pAKT) and HSP90 detected using coIP with anti-HSP90 and anti-pAKT antibodies in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs. (B) The protein levels of HSP90, pAKT, AKT, and FOXO1 in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs. (C) The protein levels of FOXO1 in the midluteal-phase endometrial tissues of controls (n = 4) and patients with recurrent implantation failure (n = 4). (D) The protein levels of HSP90, pAKT, AKT, and FOXO1 in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs after rescue with or without MK-2206 (1 μM). (E) The protein levels of HSP90, pAKT, AKT, and FOXO1 in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs after rescue with or without 17AAG (15 nM).
To further confirm that the decline in FOXO1 levels was induced by an increase in pAKT and HSP90 levels, the specific inhibitors MK-2206 for AKT and 17AAG for HSP90 were used to rescue the expression of FOXO1. The reduction of FOXO1 levels in circSTK40-overexpressing i-THESCs and i-hESCs was significantly mitigated after AKT phosphorylation was inhibited by MK-2206 and 17AAG (Figures 4D and 4E). Thus, these results suggest that circSTK40 enhances HSP90-AKT interaction and subsequently decreases FOXO1 expression through AKT pathway activation.
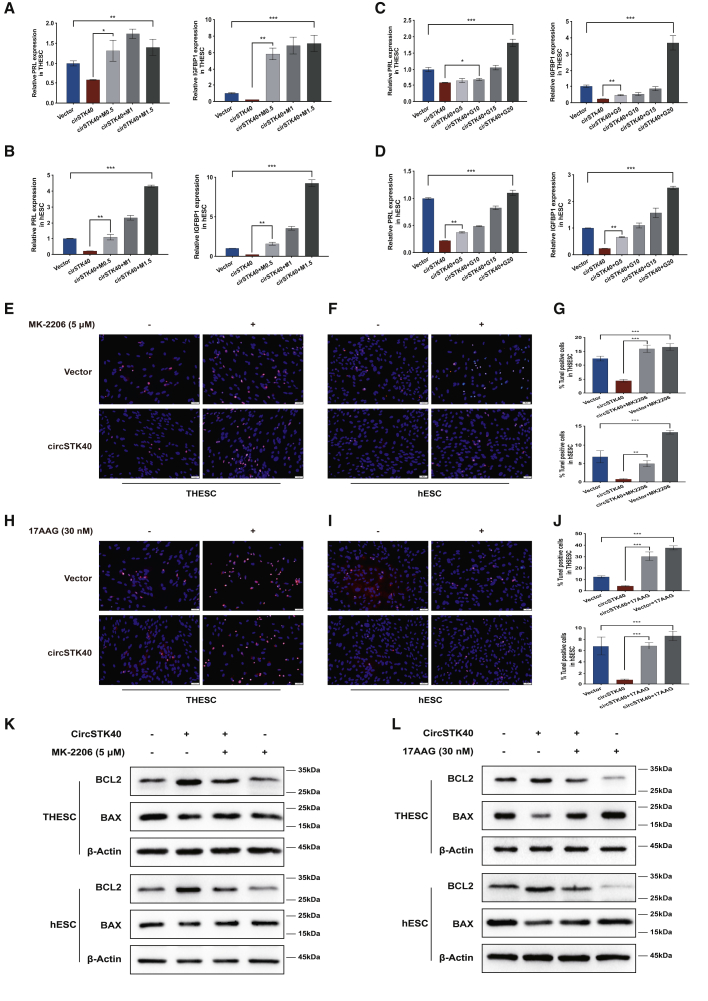
circSTK40 overexpression-mediated decreases in cell apoptosis and decidualization can be abolished by MK-2206 and 17AAG
To further confirm that circSTK40 inhibits cell decidualization and apoptosis via the HSP90/AKT/FOXO1 axis, we treated i-THESCs and i-hESCs with both the AKT inhibitor MK-2206 and the HSP90 inhibitor 17AAG. We applied a range of doses of MK-2206 (0.5, 1, and 1.5 μM) and 17AAG (5, 10, 15, and 20 nM) to circSTK40-overexpressing ESCs and found that the reduced decidualization level was not only significantly restored but also gradually elevated in a dose-dependent manner (Figures 5A–5D). In addition, these inhibitors also restored the wild-type level of cell apoptosis that was reduced by circSTK40 overexpression, as shown by the increased number of TUNEL-positive cells (Figures 5E–5J) and by the inversion of the BCL2/BAX ratio in comparison with that in circSTK40-overexpressing cells, as detected by western blots (Figures 5K and 5L). Together, these results indicate that the effect of circSTK40 overexpression on decidualization and cell apoptosis can be completely abolished by treatment with the inhibitors of AKT and HSP90.
Figure 5.
Inhibition of decidualization and apoptosis resulting from circSTK40 overexpression can be abolished by MK-2206 and 17AAG
(A and B) The mRNA levels of PRL (left) and IGFBP1 (right) in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs after treatment with different doses of MK-2206 (0.5, 1, and 1.5 μM, indicated as M0.5, M1, and M1.5, respectively) (n = 3). (C and D) The mRNA levels of PRL (left) and IGFBP1 (right) in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs after treatment with different doses of 17AAG (5, 10, 15, and 20 nM, indicated as G5, G10, G15, and G20, respectively) (n = 3). (E–G) Representative photomicrographs of TUNEL staining in day 4-induced, circSTK40-overexpressing THESCs/primary hESCs after treatment with or without MK-2206 (5 μM) (n = 3–4). Quantification of TUNEL-positive cells presented as the percentage of TUNEL-positive cells among DAPI-stained nuclei (n = 4–5). Scale bars represent 50 μm. (H–J) Representative photomicrographs of TUNEL staining in day 4-induced, circSTK40-overexpressing THESCs/primary hESCs after treatment with or without 17AAG (30 nM) (n = 3–4). Quantification of TUNEL-positive cells presented as the percentage of TUNEL-positive cells among DAPI-stained nuclei (n = 4–5). Scale bars represent 50 μm. (K) The protein levels of BCL2 and BAX in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs after treatment with or without MK-2206 (5 μM). (L) The protein levels of BCL2 and BAX in day 4-induced, circSTK40-overexpressing THESCs and primary hESCs after treatment with or without 17AAG (30 nM). Data are presented as the mean ± SEM. Student’s t test was used for determining the differences between two independent samples; an ANOVA was used for determining the overall differences within multiple samples. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; n.s., non-significant.
Discussion
Decidual transformation and the establishment of endometrial receptivity represent a complex and coordinated process, in which ncRNAs play essential roles.7 To the best of our knowledge, this is the first study to comprehensively investigate the functions and underlying mechanisms of a differentially expressed circRNA involved in the impaired endometrial receptivity that is responsible for RIF in humans. Using microarray analyses, we identified an upregulated circRNA (circSTK40) in the midluteal-phase endometrium of RIF patients. Functional studies revealed that circSTK40 overexpression inhibited the decidualization process and cell apoptosis in ESCs. Mechanistically, circSTK40 reduced the degradation of HSP90 by hindering its interactions with CLU and RBBP6, which then resulted in the activation of the AKT pathway and the downregulation of FOXO1 expression.
Human endometrial decidualization is triggered by elevated progesterone and cAMP in the secretory phase of the menstrual cycle,30 which induce the nuclear accumulation of the transcription factor FOXO1 in ESCs and subsequently lead to cell differentiation, observed as a significant increase in the expression levels of the decidualization markers PRL and IGFBP1.7,31 Activation of the AKT pathway initiates the kinase cascade to phosphorylate FOXO1, which promotes FOXO1 export from the nucleus and blocks nuclear translocation, finally leading to ubiquitin-proteasomal degradation of FOXO1 and decrease of the decidualization level.32,33 We found that the suppression of decidualization marker expression by circSTK40 was most prominent on day 4, and it became limiting on day 6, possibly because of the presence of other compensation or feedback mechanisms to compete with the effects of ectopically expressed circSTK40 when the decidualization level increased. It is worth noting that apoptosis is the terminal step in cell decidualization, of which BCL2 family proteins are regulators.34, 35, 36 It has been documented that both the activation of the AKT pathway and the reduction of FOXO1 expression in the nucleus can inhibit cell apoptosis through the inactivation of pro-apoptotic molecules such as BAD and BAX37 or by decreasing the expression of the apoptotic regulator BIM (BCL2-interacting mediator of cell death).32 In this study, FOXO1 expression was also shown to be significantly downregulated in the midluteal-phase endometrium of RIF patients. Collectively, these results demonstrate that the dysregulation of circSTK40 partially accounts for the deficient endometrial receptivity in RIF through interference with the decidualization process and inhibition of cell apoptosis via the activation of the AKT pathway and the reduction of FOXO1 expression.
Previous proteomic analyses have identified many differentially expressed proteins associated with apoptosis and stress response in the mid-secretory-phase endometrial fluid compared with the early-secretory-phase fluid38,39 or the non-implantative cycle fluid,40 suggesting that it is common for the endometrium to encounter different stresses during the implantation window and that the regulation of cell apoptosis plays an important role. Notably, our results revealed that when ESCs were subjected to H2O2 insult, circSTK40 transcription significantly increased while cell apoptosis decreased, suggesting that the upregulation of circSTK40 expression to modulate the proteasomal degradation of HSP90 could be a protective mechanism to promote cell survival under stress while concurrently impairing endometrial receptivity. Further investigation is needed to identify the mechanisms underlying the stress-associated upregulation of circSTK40 expression.
Recent studies have revealed that lncRNAs can function as RNA scaffolds to recruit and modify proteins.41, 42, 43, 44 Direct protein interaction to regulate function is also a common mechanism by which circRNAs act as regulators of protein activity.14, 15, 16,45 Strikingly, the pull-down/mass spectrometry and RIP assays performed in this study demonstrated that circSTK40 independently binds to two proteins, HSP90 and CLU, thereby blocking the interaction between them. Although ncRNA-protein interactions have been established as a means of enhancing enzyme activity, our study provides evidence of circRNAs functioning as negative regulators of protein activity. It has been reported that lncRNAs can facilitate the combination of HSP90 with its client proteins (P53, AKT, and IKK) and thus competitively inhibit their ubiquitination.46,47 However, only the AKT/FOXO1 pathway regulated by HSP90 was investigated in this study; therefore, whether other HSP90 client proteins,28 which may also be involved in endometrial receptivity, such as steroid receptors,4 P53,48 and proteins of the ERK and JNK pathways,49 are affected by the dysregulation of circSTK40 requires further clarification. In addition, given that circSTK40 is localized in both the cytoplasm and nucleus of ESCs, further investigation is needed to determine whether this circRNA also has nuclear functions.
Previous work has uncovered the interactions between HSP90 and CLU and their synergistic effects in modulating the functions of client proteins.26 Furthermore, CLU inhibition has been shown to decrease the transcription of heat shock factor 1 (HSF1), thus leading to the downregulation of the expression of heat shock proteins.50 Our study provides new insight into the association between HSP90 and CLU, revealing that CLU interacts with the E3 ligase and promotes the proteasomal degradation of HSP90, thus indicating a dual role of CLU in HSP90 expression and increasing the complexity of the interactions between stress-associated proteins.
As reported in numerous preclinical and clinical studies, HSP90 inhibitors, such as 17AAG, 17DMAG, and BIIB021, have been developed for treating various cancers with higher safety and efficacy.51, 52, 53 In our work, HSP90 inhibition by 17AAG in a certain range of doses resulted in not only a complete rescue of the effects of circSTK40 but also a significant, dose-dependent increase in decidualization levels, suggesting a potential therapeutic strategy for RIF patients. Given that the improvement in endometrial receptivity remains an intractable problem for clinicians, this finding may provide a foundation for future development of treatments for RIF. However, HSP90 inhibitors are mostly applied in cancer treatment combined with other antitumor drugs (such as cytotoxins, radiation, and antiangiogenics agents) and inevitably cause some dose-limiting side effects, including gastrointestinal reaction, ocular toxicity, and hepatotoxicity.51,52 Therefore, the application of these drugs in improving the endometrial environment still needs further investigation from animal testing to human trials, especially from the aspects of drug dosage, administration route, and safety concerns regarding maternal and fetal health. Advances in novel drug development and nanotechnology are expected to broaden the choice of HSP90 inhibitor-based treatments.
Conclusion and future directions
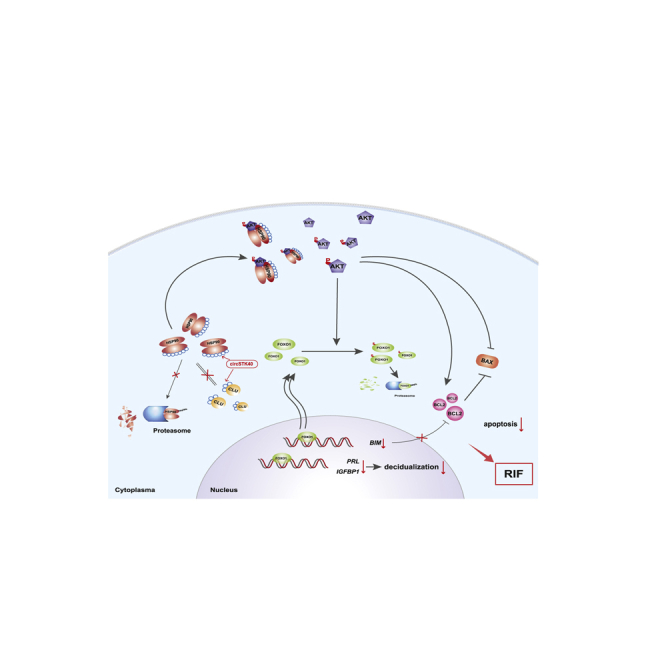
In summary, our study demonstrated that a previously unknown circRNA, circSTK40, participates in RIF pathogenesis through impairing endometrial receptivity. We determined that circSTK40 functions as a scaffold for proteins and reduces the degradation of HSP90 by hindering its interactions with CLU and RBBP6 and that the resulting high levels of HSP90 lead to the activation of the AKT pathway and the downregulation of FOXO1 expression. Inhibitors of AKT (MK-2206) and HSP90 (17AAG) both abolished the effects of circSTK40 overexpression in ESCs and increased the decidualization levels in a dose-dependent manner (Figure 6). This work describes a novel epigenetic mechanism for RIF pathogenesis. To construct a complete regulatory network of different ncRNAs and proteins involved in the modulation of endometrial receptivity, other mechanisms of circSTK40 and the effects of other differentially expressed circRNAs warrant further investigation. Our findings also provide a potentially attractive option for targeted therapies based on HSP90 inhibitors in patients with low endometrial receptivity; nevertheless, further animal testing and human trials are still warranted.
Figure 6.
Schematic summary of the role of circSTK40 in inhibiting the decidualization process and cell apoptosis through the modulation of the HSP90/AKT/FOXO1 axis
Materials and methods
Patients and sample collection
This study was approved by the Ethics Committee of the Reproductive Medicine Center of Shandong University, and informed consent was obtained from all participants.
Midluteal-phase endometrial samples were obtained from 24 RIF patients and 24 age-matched controls by Pipelle biopsy at the implantation window of a monitored natural cycle 7 days after ovulation.4,54 All participants (24 controls and 24 RIF patients) were younger than 40 years and underwent in vitro fertilization (IVF)/intracytoplasmic sperm injection (ICSI) treatment from January 2015 to January 2017 at the Reproductive Medical Center of Shandong University. Patients were diagnosed with unexplained RIF when clinical pregnancy was still absent after three or more transfers of high-quality embryos or transfer of at least 10 embryos over several cycles1 and no clear causes were found. The fertile control group included patients who underwent their first IVF/ICSI treatment for purely male factor and achieved live birth after the first or second transfer cycle.55 The exclusion criteria were as follows: (1) diagnosis of uterine cavity abnormalities, such as congenital uterine anomalies, fibroids, endometrial polyps, and adenomyosis; (2) diagnosis of hydrosalpinx, chromosome anomalies, or endocrine disorders; (3) receipt of hormone therapy or intrauterine operations during the last three menstrual cycles.
CircRNA microarray
The circRNA expression profiles were detected on the endometrial samples of eight RIF patients and eight age-matched control women. Briefly, total RNA was digested with RNase R (Epicenter, Madison, WI, USA) to remove linear RNAs and enrich circRNAs. Subsequently, after amplification and transcription into fluorescent cRNAs using a random priming method (Arraystar Super RNA Labeling Kit; Arraystar, Rockville, MD, USA), RNA array hybridization was performed using the Arraystar Human circRNA Arrays V2 (8 × 15K, Arraystar). Microarray scanning and data analyses were conducted by KangChen Bio-tech (China). Differential analysis between the groups was performed using t tests. The cutoffs were fold changes > 2 and p values < 0.05.
RNA preparation and quantitative real-time PCR
Total RNA from endometrial samples and cells was extracted using TRIzol Reagent (TaKaRa, China) according to the manufacturer’s protocol. For RNase R treatment, 1 μg of total RNA was incubated with RNase R (Geneseed, China) at 37°C for 10 min. Reverse transcription (RT) was performed using the PrimeScript RT reagent kit (TaKaRa, China) with random hexamers for circRNA or oligo (dT) for mRNA. Thereafter, RNA expression was determined by quantitative real-time PCR using TB Green Premix Ex Taq (TaKaRa, China). Using TA cloning, the PCR products of circRNAs were inserted into pMD18-T vector (TaKaRa, China) and subjected to Sanger sequencing to confirm the back-splicing junctions. The primer sequences are listed in Table S1.
Cell culture and treatment
THESCs and hESCs were used for functional and mechanistic research. THESCs (ATCC CRL-4003) were kindly provided by Professor H.B. Wang (Xiamen University, China), and hESCs were isolated from the biopsies of proliferative-phase endometrial tissues from fertile volunteers of reproductive age who had regular cycles and no hormonal medication or history of gynecological malignancy.
ESCs were cultured in phenol-red-free DMEM/F-12 maintenance medium (Gibco, Waltham, MA, USA) containing 10% charcoal-stripped fetal bovine serum (CS-FBS).56,57 The ESCs were then treated with 0.5 mM db-cAMP (cAMP; Sigma, St. Louis, MO, USA) and 1 μM medroxyprogesterone acetate (MPA; Sigma, St. Louis, MO, USA) for 2–6 days to induce in vitro decidualization in DMEM/F-12 differentiation medium containing 2% CS-FBS,57,58 and the medium was changed every 48 h. Cells and supernatants were collected at different times (days 0, 2, 4, and 6) to measure the mRNA and protein levels of the decidualization markers (PRL and IGFBP1). Different doses of 17AAG (Selleck, Houston, TX, USA), the specific inhibitor of HSP90, and MK-2206 (Selleck, Houston, TX, USA), the selective inhibitor of AKT1/2/3, were added to the above-mentioned differentiation medium to investigate the effects of HSP90 and the AKT pathway on cell functions. THESCs were treated with 100 ng/mL CHX (a protein synthesis inhibitor; Meilunbio, China) for 0, 8, 16, and 32 h to evaluate the half-life of HSP90 and with 5 μM MG132 (a proteasome inhibitor; APExBIO, Houston, TX. USA) for 6 h to explore the degradation pattern of the proteins.
Adenoviral infection
circSTK40 overexpression was achieved using an adenovirus vector, which was synthesized and packaged by HanBio Biotechnology (China). At 70% confluency, the ESCs were infected with an adenovirus expressing circSTK40 and a control virus harboring an empty vector at an MOI of 150:1 when initiating decidualization induction.
Sorting of hESCs and endometrial epithelial cells
Midluteal-phase endometrial tissues from fertile control women were collected, minced, and rinsed repeatedly with phosphate-buffered saline (PBS). The tissues were mixed with an appropriate amount of collagenase I (Gibco, Waltham, MA, USA; 0.5 mg/mL) and incubated at 37°C for 30 min. The digested suspension was first filtered through a 150-μm mesh to remove the fibers and undigested tissues and subsequently through a 40-μm mesh to separate ESCs and endometrial epithelial cells. These two types of cells were collected and subjected to RNA extraction and quantitative real-time PCR to determine circSTK40 expression.
Cytoplasmic/nucleus fraction
Separation of cytoplasmic and nuclear RNA was performed using a PARIS kit (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. GAPDH and MALAT1 were used as the positive controls for cytoplasmic and nuclear RNA, respectively.
FISH
THESCs were hybridized overnight with a mixture of Cy3-labeled circSTK40, U6, or 18S probes (RiboBio, China). All operations were completed using a Fluorescence in situ Hybridization Kit (RiboBio, China) according to the manufacturer’s instructions. Specimens were visualized using a laser confocal microscope (Andor, UK).
TUNEL
After inducing decidualization for 3 days, ESCs were subjected to H2O2 insult (200 μM) and serum starvation for 18 h. Cell apoptosis was then detected with TUNEL staining (KeyGEN, China) according to the manufacturer’s instructions.
Western blots
Proteins from the endometrial samples were extracted using a Minute Total Protein Extraction Kit (Invent, Plymouth, MN, USA) according to the manufacturer’s instructions. Cells were lysed with sodium dodecyl sulfate (SDS) lysis buffer (Beyotime, China) containing protease and phosphatase inhibitor cocktail (Cell Signaling Technology, Danvers, MA, USA). Protein concentrations were quantified using a BCA Protein Assay Kit (Beyotime, China).
Proteins were separated by SDS-polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane (Millipore, Burlington, MA, USA). After blocking with 5% skim milk for 1 h, the membranes were incubated with primary antibodies, including anti-HSP90 (4877, Cell Signaling Technology, Danvers, MA, USA), anti-AKT (4691, Cell Signaling Technology, Danvers, MA, USA), anti-phospho-AKT (4060, Cell Signaling Technology, Danvers, MA, USA ), anti-FOXO1 (2880, Cell Signaling Technology, Danvers, MA, USA), anti-BCL2 (12789-1-AP, Proteintech, China), anti-BAX (ab32503, Abcam, UK), anti-RBBP6 (abs139373, Absin, China), anti-α-Tubulin (66031-1-Ig, Proteintech, China), and anti-β-Actin (A5441, Sigma, St. Louis, MO, USA) antibodies, at 4°C overnight and then incubated with secondary antibodies conjugated to horseradish peroxidase (Proteintech, China) at room temperature for 1 h. The immunostained bands were detected with an ECL chemiluminescence kit (Millipore, Burlington, MA, USA) and a ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA). The details of the antibodies are listed in Table S2.
Enzyme-linked immunosorbent assay (ELISA)
Cell supernatants were collected and centrifuged at 4°C for 10 min to remove cell fragments. Next, 100 μL of supernatants or standards were added into each well of an ELISA plate. All steps were performed according to the manufacturer’s instructions in the ELISA kits for PRL and IGFBP1 (Cusabio, China). The optical density was determined using a microplate reader at 450 nm/540 nm to generate standard curves and measure the protein levels.
RNA pull-down assay and mass spectrometry analysis
Biotin-labeled positive and negative probes of circSTK40 (Table S3) were synthesized and purified by Cloudseq (China). Streptavidin magnetic beads were incubated with biotinylated probes at room temperature for 1 h. THESC lysates were then added to the two mixtures and incubated at room temperature for 2 h. After thoroughly washing the mixtures five times, the retrieved proteins were digested with trypsin and subjected to protein label-free quantitation (LFQ) by liquid chromatography-based mass spectrometry. The mass spectrometry data were analyzed using the MaxQuant computational platform (MaxQuant v.1.6.0.16, Germany) and quantified as LFQ and quantile-normalized LFQ59,60 at Cloudseq (China).
RIP
RIP was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Burlington, MA, USA) according to the manufacturer’s instructions. THESCs (1 × 107) were collected with an RIP lysis buffer containing RNase inhibitor and protease inhibitor cocktail. Magnetic beads were incubated with 5 μg of anti-HSP90 (ab13492, Abcam, UK), anti-CLU (sc-5289, Santa Cruz, Dallas, TX, USA), or IgG antibodies (CS200621, Millipore, Burlington, MA, USA) at room temperature for 30 min. Subsequently, cell lysates were incubated with the bead-antibody complexes at 4°C overnight. After thoroughly washing the mixtures six times, the co-precipitated RNAs were extracted, reverse-transcribed, and quantified with quantitative real-time PCR. Simultaneously, immunoprecipitated proteins were confirmed using an immunoblotting analysis. The details of the antibodies are listed in Table S2.
CoIP
ESCs were collected with Pierce IP Lysis Buffer (Pierce, Waltham, MA, USA) supplemented with protease and phosphatase inhibitor cocktail (Cell Signaling Technology, Danvers, MA, USA) according to the manufacturer’s instructions. A total of 0.5 mg of proteins were incubated with 1–4 μg of anti-HSP90 (ab13492, Abcam, UK), anti-phospho-AKT (4060, Cell Signaling Technology, Danvers, MA, USA), anti-CLU (sc-5289, Santa Cruz, Dallas TX, USA), or IgG (CS200621, Millipore, Burlington, MA, USA) antibodies at 4°C overnight and then with PureProteome Protein A/G Magnetic Beads (Millipore, Burlington, MA, USA) at room temperature for 30 min. The immunoprecipitates were washed three times with 0.1% Tween-20 in PBS and then subjected to immunoblotting with the indicated antibodies.
Statistics
All experiments were performed independently three or more times under similar conditions. Graphs were generated using GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA), and figures were assembled using Adobe Illustrator 23 (Adobe, San Jose, CA, USA). All statistical analyses were performed using SPSS version 20.0 (IBM, Armonk, NY, USA). Data for normally distributed variables are expressed as the mean ± SD/SEM, and statistical differences were analyzed by the two-tailed Student’s t test and an analysis of variance (ANOVA). In contrast, data for non-normally distributed variables are expressed as medians and quartiles, and statistical differences were analyzed by a non-parametric test. Significant differences are represented as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001, and n.s. indicates non-significance.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The microarray data that support the findings of this study are available at Gene Expression Omnibus (GEO), with the accession number GEO: GSE147442 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147442).
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFC1002804 and 2016YFC1000202) and the Youth Program of National Natural Science Foundation of China (81901556).
Author contributions
T.N. and Q.Z. have made substantial contributions to acquisition, analysis, interpretation of data, and drafting the manuscript; Y.L. has made substantial contributions to revising the manuscript critically for important intellectual content; C.H. and T.Z. have made substantial contributions to acquisition and interpretation of part of the data; J.Y. and Z.C. have made substantial contributions to conception and design of the work.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.06.021.
Supplemental information
References
- 1.Thornhill A.R., deDie-Smulders C.E., Geraedts J.P., Harper J.C., Harton G.L., Lavery S.A., Moutou C., Robinson M.D., Schmutzler A.G., Scriven P.N., et al. ESHRE PGD Consortium ESHRE PGD Consortium ‘Best practice guidelines for clinical preimplantation genetic diagnosis (PGD) and preimplantation genetic screening (PGS)’. Hum. Reprod. 2005;20:35–48. doi: 10.1093/humrep/deh579. [DOI] [PubMed] [Google Scholar]
- 2.Coughlan C., Ledger W., Wang Q., Liu F., Demirol A., Gurgan T., Cutting R., Ong K., Sallam H., Li T.C. Recurrent implantation failure: definition and management. Reprod. Biomed. Online. 2014;28:14–38. doi: 10.1016/j.rbmo.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Lessey B.A., Young S.L. What exactly is endometrial receptivity? Fertil. Steril. 2019;111:611–617. doi: 10.1016/j.fertnstert.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Cha J., Sun X., Dey S.K. Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 2012;18:1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X., Jin X., Liu L., Man C.W., Huang J., Wang C.C., Zhang S., Li T.C. Differential expression of vascular endothelial growth factor angiogenic factors in different endometrial compartments in women who have an elevated progesterone level before oocyte retrieval, during in vitro fertilization-embryo transfer treatment. Fertil. Steril. 2015;104:1030–1036. doi: 10.1016/j.fertnstert.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Cakmak H., Taylor H.S. Implantation failure: molecular mechanisms and clinical treatment. Hum. Reprod. Update. 2011;17:242–253. doi: 10.1093/humupd/dmq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu H., Huang X., Mor G., Liao A. Epigenetic modifications working in the decidualization and endometrial receptivity. Cell. Mol. Life Sci. 2020;77 doi: 10.1007/s00018-019-03395-9. 20191–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouckenheimer J., Assou S., Riquier S., Hou C., Philippe N., Sansac C., Lavabre-Bertrand T., Commes T., Lemaître J.-M., Boureux A., De Vos J. Long non-coding RNAs in human early embryonic development and their potential in ART. Hum. Reprod. Update. 2016;23:19–40. doi: 10.1093/humupd/dmw035. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q., Ni T., Dang Y., Ding L., Jiang J., Li J., Xia M., Yu N., Ma J., Yan J., Chen Z.J. MiR-148a-3p may contribute to flawed decidualization in recurrent implantation failure by modulating HOXC8. J. Assist. Reprod. Genet. 2020;37:2535–2544. doi: 10.1007/s10815-020-01900-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patop I.L., Wüst S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 12.Hall I.F., Climent M., Quintavalle M., Farina F.M., Schorn T., Zani S., Carullo P., Kunderfranco P., Civilini E., Condorelli G., Elia L. Circ_Lrp6, a Circular RNA Enriched in Vascular Smooth Muscle Cells, Acts as a Sponge Regulating miRNA-145 Function. Circ. Res. 2019;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 13.Han D., Li J., Wang H., Su X., Hou J., Gu Y., Qian C., Lin Y., Liu X., Huang M., et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 14.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 16.Wang L., Long H., Zheng Q., Bo X., Xiao X., Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol. Cancer. 2019;18:119. doi: 10.1186/s12943-019-1046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dube U., Del-Aguila J.L., Li Z., Budde J.P., Jiang S., Hsu S., Ibanez L., Fernandez M.V., Farias F., Norton J., et al. Dominantly Inherited Alzheimer Network (DIAN) An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat. Neurosci. 2019;22:1903–1912. doi: 10.1038/s41593-019-0501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Huang R., Cheng M., Wang L., Chao J., Li J., Zheng P., Xie P., Zhang Z., Yao H. Gut microbiota from NLRP3-deficient mice ameliorates depressive-like behaviors by regulating astrocyte dysfunction via circHIPK2. Microbiome. 2019;7:116. doi: 10.1186/s40168-019-0733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K., Gan T.Y., Li N., Liu C.Y., Zhou L.Y., Gao J.N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., Li P.F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarnerio J., Zhang Y., Cheloni G., Panella R., Mae Katon J., Simpson M., Matsumoto A., Papa A., Loretelli C., Petri A., et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019;29:628–640. doi: 10.1038/s41422-019-0192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Liu X., Che S., Cui J., Ma X., An X., Cao B., Song Y. Endometrial Epithelial Cell Apoptosis Is Inhibited by a ciR8073-miR181a-Neurotensis Pathway during Embryo Implantation. Mol. Ther. Nucleic Acids. 2019;14:262–273. doi: 10.1016/j.omtn.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L., Li L., Ma X., Yue F., Wang Y., Wang L., Jin P., Zhang X. Altered Circular RNA Expression in Patients with Repeated Implantation Failure. Cell. Physiol. Biochem. 2017;44:303–313. doi: 10.1159/000484887. [DOI] [PubMed] [Google Scholar]
- 26.Xiong J., Wang S., Chen T., Shu X., Mo X., Chang G., Chen J.-J., Li C., Luo H., Lee J.-D. Verteporfin blocks Clusterin which is required for survival of gastric cancer stem cell by modulating HSP90 function. Int. J. Biol. Sci. 2019;15:312–324. doi: 10.7150/ijbs.29135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zoubeidi A., Ettinger S., Beraldi E., Hadaschik B., Zardan A., Klomp L.W.J., Nelson C.C., Rennie P.S., Gleave M.E. Clusterin facilitates COMMD1 and I-kappaB degradation to enhance NF-kappaB activity in prostate cancer cells. Mol. Cancer Res. 2010;8:119–130. doi: 10.1158/1541-7786.MCR-09-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoter A., El-Sabban M.E., Naim H.Y. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci. 2018;19:2560. doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biebl M.M., Buchner J. Structure, Function, and Regulation of the Hsp90 Machinery. Cold Spring Harb. Perspect. Biol. 2019;11:a034017. doi: 10.1101/cshperspect.a034017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gellersen B., Brosens J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014;35:851–905. doi: 10.1210/er.2014-1045. [DOI] [PubMed] [Google Scholar]
- 31.Gellersen B., Brosens J. Cyclic AMP and progesterone receptor cross-talk in human endometrium: a decidualizing affair. J. Endocrinol. 2003;178:357–372. doi: 10.1677/joe.0.1780357. [DOI] [PubMed] [Google Scholar]
- 32.Ma J., Matkar S., He X., Hua X. FOXO family in regulating cancer and metabolism. Semin. Cancer Biol. 2018;50:32–41. doi: 10.1016/j.semcancer.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 33.Lee S.Y., Lee Y.Y., Choi J.S., Yoon M.-S., Han J.-S. Phosphatidic acid induces decidualization by stimulating Akt-PP2A binding in human endometrial stromal cells. FEBS J. 2016;283:4163–4175. doi: 10.1111/febs.13914. [DOI] [PubMed] [Google Scholar]
- 34.Katz S., Abrahamsohn P.A. Involution of the antimesometrial decidua in the mouse. An ultrastructural study. Anat. Embryol. (Berl.) 1987;176:251–258. doi: 10.1007/BF00310059. [DOI] [PubMed] [Google Scholar]
- 35.Akcali K.C., Khan S.A., Moulton B.C. Effect of decidualization on the expression of bax and bcl-2 in the rat uterine endometrium. Endocrinology. 1996;137:3123–3131. doi: 10.1210/endo.137.7.8770938. [DOI] [PubMed] [Google Scholar]
- 36.Akcali K.C., Gibori G., Khan S.A. The involvement of apoptotic regulators during in vitro decidualization. Eur. J. Endocrinol. 2003;149:69–75. doi: 10.1530/eje.0.1490069. [DOI] [PubMed] [Google Scholar]
- 37.Matheny R.W., Jr., Adamo M.L. Current perspectives on Akt Akt-ivation and Akt-ions. Exp. Biol. Med. (Maywood) 2009;234:1264–1270. doi: 10.3181/0904-MR-138. [DOI] [PubMed] [Google Scholar]
- 38.Scotchie J.G., Fritz M.A., Mocanu M., Lessey B.A., Young S.L. Proteomic analysis of the luteal endometrial secretome. Reprod. Sci. 2009;16:883–893. doi: 10.1177/1933719109337165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasvandik S., Saarma M., Kaart T., Rooda I., Velthut-Meikas A., Ehrenberg A., Gemzell K., Lalitkumar P.G., Salumets A., Peters M. Uterine Fluid Proteins for Minimally Invasive Assessment of Endometrial Receptivity. J. Clin. Endocrinol. Metab. 2020;105:dgz019. doi: 10.1210/clinem/dgz019. [DOI] [PubMed] [Google Scholar]
- 40.Azkargorta M., Escobes I., Iloro I., Osinalde N., Corral B., Ibañez-Perez J., Exposito A., Prieto B., Elortza F., Matorras R. Differential proteomic analysis of endometrial fluid suggests increased inflammation and impaired glucose metabolism in non-implantative IVF cycles and pinpoints PYGB as a putative implantation marker. Hum. Reprod. 2018;33:1898–1906. doi: 10.1093/humrep/dey274. [DOI] [PubMed] [Google Scholar]
- 41.Hu W.L., Jin L., Xu A., Wang Y.F., Thorne R.F., Zhang X.D., Wu M. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol. 2018;20:492–502. doi: 10.1038/s41556-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 42.Sun M., Nie F., Wang Y., Zhang Z., Hou J., He D., Xie M., Xu L., De W., Wang Z., Wang J. LncRNA HOXA11-AS Promotes Proliferation and Invasion of Gastric Cancer by Scaffolding the Chromatin Modification Factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–6310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 43.Sun T.-T., He J., Liang Q., Ren L.-L., Yan T.-T., Yu T.-C., Tang J.-Y., Bao Y.-J., Hu Y., Lin Y., et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 44.Wang X., Zhang X., Dang Y., Li D., Lu G., Chan W.-Y., Leung P.C.K., Zhao S., Qin Y., Chen Z.-J. Long noncoding RNA HCP5 participates in premature ovarian insufficiency by transcriptionally regulating MSH5 and DNA damage repair via YB1. Nucleic Acids Res. 2020;48:4480–4491. doi: 10.1093/nar/gkaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garikipati V.N.S., Verma S.K., Cheng Z., Liang D., Truongcao M.M., Cimini M., Yue Y., Huang G., Wang C., Benedict C., et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat. Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou C.C., Yang F., Yuan S.X., Ma J.Z., Liu F., Yuan J.H., Bi F.R., Lin K.Y., Yin J.H., Cao G.W., et al. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016;63:850–863. doi: 10.1002/hep.28393. [DOI] [PubMed] [Google Scholar]
- 47.Guo H., Zhao L., Shi B., Bao J., Zheng D., Zhou B., Shi J. GALNT5 uaRNA promotes gastric cancer progression through its interaction with HSP90. Oncogene. 2018;37:4505–4517. doi: 10.1038/s41388-018-0266-4. [DOI] [PubMed] [Google Scholar]
- 48.Pohnke Y., Schneider-Merck T., Fahnenstich J., Kempf R., Christian M., Milde-Langosch K., Brosens J.J., Gellersen B. Wild-type p53 protein is up-regulated upon cyclic adenosine monophosphate-induced differentiation of human endometrial stromal cells. J. Clin. Endocrinol. Metab. 2004;89:5233–5244. doi: 10.1210/jc.2004-0012. [DOI] [PubMed] [Google Scholar]
- 49.Zhou W.-J., Hou X.-X., Wang X.-Q., Li D.-J. Fibroblast Growth Factor 7 Regulates Proliferation and Decidualization of Human Endometrial Stromal Cells via ERK and JNK Pathway in an Autocrine Manner. Reprod. Sci. 2017;24:1607–1619. doi: 10.1177/1933719117697122. [DOI] [PubMed] [Google Scholar]
- 50.Lamoureux F., Thomas C., Yin M.-J., Kuruma H., Beraldi E., Fazli L., Zoubeidi A., Gleave M.E. Clusterin inhibition using OGX-011 synergistically enhances Hsp90 inhibitor activity by suppressing the heat shock response in castrate-resistant prostate cancer. Cancer Res. 2011;71:5838–5849. doi: 10.1158/0008-5472.CAN-11-0994. [DOI] [PubMed] [Google Scholar]
- 51.Talaei S., Mellatyar H., Asadi A., Akbarzadeh A., Sheervalilou R., Zarghami N. Spotlight on 17-AAG as an Hsp90 inhibitor for molecular targeted cancer treatment. Chem. Biol. Drug Des. 2019;93:760–786. doi: 10.1111/cbdd.13486. [DOI] [PubMed] [Google Scholar]
- 52.Mellatyar H., Talaei S., Pilehvar-Soltanahmadi Y., Barzegar A., Akbarzadeh A., Shahabi A., Barekati-Mowahed M., Zarghami N. Targeted cancer therapy through 17-DMAG as an Hsp90 inhibitor: Overview and current state of the art. Biomed. Pharmacother. 2018;102:608–617. doi: 10.1016/j.biopha.2018.03.102. [DOI] [PubMed] [Google Scholar]
- 53.He W., Hu H. BIIB021, an Hsp90 inhibitor: A promising therapeutic strategy for blood malignancies (Review) Oncol. Rep. 2018;40:3–15. doi: 10.3892/or.2018.6422. [DOI] [PubMed] [Google Scholar]
- 54.Franasiak J.M., Holoch K.J., Yuan L., Schammel D.P., Young S.L., Lessey B.A. Prospective assessment of midsecretory endometrial leukemia inhibitor factor expression versus ανβ3 testing in women with unexplained infertility. Fertil. Steril. 2014;101:1724–1731. doi: 10.1016/j.fertnstert.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lédée N., Munaut C., Aubert J., Sérazin V., Rahmati M., Chaouat G., Sandra O., Foidart J.M. Specific and extensive endometrial deregulation is present before conception in IVF/ICSI repeated implantation failures (IF) or recurrent miscarriages. J. Pathol. 2011;225:554–564. doi: 10.1002/path.2948. [DOI] [PubMed] [Google Scholar]
- 56.Salker M.S., Christian M., Steel J.H., Nautiyal J., Lavery S., Trew G., Webster Z., Al-Sabbagh M., Puchchakayala G., Föller M., et al. Deregulation of the serum- and glucocorticoid-inducible kinase SGK1 in the endometrium causes reproductive failure. Nat. Med. 2011;17:1509–1513. doi: 10.1038/nm.2498. [DOI] [PubMed] [Google Scholar]
- 57.Wei M., Gao Y., Lu B., Jiao Y., Liu X., Cui B., Hu S., Sun L., Mao S., Dong J., et al. FKBP51 regulates decidualization through Ser473 dephosphorylation of AKT. Reproduction. 2018;155:283–295. doi: 10.1530/REP-17-0625. [DOI] [PubMed] [Google Scholar]
- 58.Matsuoka A., Kizuka F., Lee L., Tamura I., Taniguchi K., Asada H., Taketani T., Tamura H., Sugino N. Progesterone increases manganese superoxide dismutase expression via a cAMP-dependent signaling mediated by noncanonical Wnt5a pathway in human endometrial stromal cells. J. Clin. Endocrinol. Metab. 2010;95:E291–E299. doi: 10.1210/jc.2010-0619. [DOI] [PubMed] [Google Scholar]
- 59.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 60.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The microarray data that support the findings of this study are available at Gene Expression Omnibus (GEO), with the accession number GEO: GSE147442 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147442).