Abstract
Puberty is the gateway to adult reproductive competence, encompassing a suite of complex, integrative, and coordinated changes in neuroendocrine functions. However, the regulatory mechanisms of transcriptional reprogramming in the arcuate nucleus (ARC) during onset of puberty are still not fully understood. To understand the role of epigenetics in regulating gene expression, mouse hypothalamic ARCs were isolated at 4 and 8 weeks, and the transcriptome, DNA hydroxymethylation, DNA methylation, and chromatin accessibility were assessed via RNA sequencing (RNA-seq), reduced representation bisulfite sequencing (RRBS-seq), reduced representation hydroxymethylation profiling (RRHP)-seq, and assay for transposase-accessible chromatin (ATAC-seq), respectively. The overall DNA hydroxymethylation and DNA methylation changes in retroelements (REs) were associated with gene expression modeling for puberty in the ARC. We focused on analyzing DNA hydroxymethylation and DNA methylation at two short interspersed nuclear elements (SINEs) located on the promoter of the 5-hydroxytryptamine receptor 6 (Htr6) gene and the enhancer of the KISS-1 metastasis suppressor (Kiss1) gene and investigated their regulatory roles in gene expression. Our data uncovered a novel epigenetic mechanism by which SINEs regulate gene expression during puberty.
Keywords: arcuate nucleus, DNA methylation, DNA hydroxymethylation, puberty, retroelement, short interspersed nuclear elements, 5-hydroxytryptamine receptor 6, KISS-1 metastasis suppressor, enhancer, Nuclear receptor subfamily 5 group A member 2
Graphical abstract

Shen et al. summarize genome-wide DNA (hydroxy)methylation of retroelements in the mouse arcuate nucleus and report novel epigenetic mechanisms of short interspersed nuclear elements for regulating gene expression across pubertal development.
Introduction
Adolescence is characterized by a myriad of physical and biological changes driven by release of pubertal hormones, which are triggered by activation of the hypothalamic-pituitary-gonadal axis.1 Hypothalamic gonadotropin-releasing hormone (GnRH), which is secreted in the preoptic area in a pulsatile manner during sleep, can stimulate the pituitary gland to release substantial amounts of follicle-stimulating hormone (FSH) and luteinizing hormone (LH), which can facilitate gonadarche and stimulate sex steroid hormone production.
The discovery of the kisspeptin system showed that the KISS-1 metastasis suppressor (Kiss1) and its receptor Kiss1r (also known as Gpr54) contribute to activation of the hypothalamic-pituitary-gonadal axis during puberty.2 The exocrine kisspeptin secreted from the arcuate nucleus (ARC), anteroventral periventricular nucleus, and the preoptic area interacts with Kiss1r on the membrane surface of GnRH neurons, which further activates canonical transient receptor potential (TRPC)-like potassium channels and continuously depolarizes GnRH neurons.3 Over the past decade, molecular insights into the Kiss1/ Kiss1r regulatory axis in GnRH activation have attracted attention for explaining the mechanism of puberty initiation.
Epigenetics is a powerful mechanism that can control gene expression in a temporal and spatial manner. Previous studies have suggested that DNA methylation, the most acknowledged epigenetic modification, shows a puberty-specific pattern in the promoters Kiss1 and Kiss1r.4 However, the genomic demethylation processes during puberty onset are largely unknown. DNA hydroxymethylation has been suggested as an intermediate of the active DNA demethylation process catalyzed by the ten-eleven translocation (TET) family of proteins.5 TET2 has been implicated in GnRH transcription activation and maintenance of reproductive function.6 In addition to epigenetic regulation of certain gene promoters, DNA methylation and demethylation contribute to maintenance of genomic integrity in somatic cells, in part through silencing or activation of transposable retroelements (REs).7 REs are found predominantly in the human genome, and approximately 13% are short interspersed nuclear elements (SINEs), 20% are long interspersed nuclear elements (LINEs), and 8% are long-terminal repeats (LTRs).8 In general, most RE promoters containing CpG islands are densely methylated in adult somatic cells, but in many types of cancer, global hypomethylation of genomic elements, including REs, is implicated in disease progression, especially metastasis,9 indicating that DNA methylation and demethylation in REs are associated with organ development and cell differentiation.
To better understand the mechanisms surrounding epigenetic regulation of REs underlying pubertal initiation, we investigated the global DNA methylation and hydroxymethylation landscape as well as the transcriptome in a female mouse ARC at the early (4 weeks of age, postnatal) and late (8 weeks of age, postnatal) pubertal stages. Our previous study described DNA methylation and hydroxymethylation of coding genes spanning puberty.10 In this study, we focused on REs associated with differentially expressed genes (DEGs) and studied the connection between DNA hydroxymethylation and DNA methylation of REs as well as their target gene expression. Additionally, the mouse hypothalamic GnRH neuron cell line GT1-7, a genetically edited cell line that has high expression of Kiss1r in response to kisspeptin stimulation and stably produces GnRH, has been used widely to study neuroendocrine regulation of the hypothalamic-pituitary-gonadal (HPG) axis.11 In this study, GT1-7 cells were employed to establish an in vitro model for deficiency of SINEs near Kiss1r using CRISPR-Cas9 and to detect transcription of Kiss1r after DNA methyltransferase (DNMT) and TET knockdown. Our data shed light on a novel mechanism of pubertal gene expression modulated by DNA hydroxymethylation of REs.
Results
Overview of DNA hydroxymethylation and methylation on REs in the ARC during puberty
Initially, we investigated changes in 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) enrichment at the promoters of protein-coding genes.10 However, the DNA hydroxymethylation and DNA methylation signatures in non-coding regions, such as transposons that are distributed in cis-regulatory elements, can also be implicated in transcription during embryonic development12 or disease progression,13 but their association with gene expression during puberty has not yet been revealed. First we summarized the REs with significantly different 5mC levels between ARCs of 4 and 8 weeks of age (Figure 1A). We found that 30.4% of differentially methylated regions (DMRs) (fold change [FC] > 10% or < −10%, false discovery rate [FDR] < 0.05) were distributed in repetitive elements, including 584 LINEs, 93 low-complexity sequences, 408 LTRs, 4 satellites, 200 simple repeats, and 522 SINEs, between 4- and 8-week-old ARCs. Similarly, 51.8% of differentially hydroxymethylated regions (DHMRs) (log2 FC > 1 or < −1, FDR < 10−4) containing 22,555 LINEs, 7,539 low-complexity sequences, 24,233 LTRs, 360 satellites, 17,872 simple repeats, and 46,044 SINEs were observed in 4- and 8-week-old ARCs, as shown in Figure 1B. We found that LINEs, SINEs, and LTRs comprise the majority of variable DNA-hydroxymethylated and DNA-methylated REs. Moreover, the variation in DNA methylation and hydroxymethylation had a completely opposite tendency in REs, especially SINEs (χ2 = 231.92, p < 0.001) between 4- and 8-week-old ARCs. On the other hand, 0.037% and 2.4% of the REs underwent DNA methylation and hydroxymethylation reprogramming, respectively, of a total of 4.9 million REs annotated by REPBASE (https://www.girinst.org/repbase/update/index.html) of the Genetic Information Research Institute14,15 (Figure 1C). SINEs were still the most variable RE, followed by LTRs and LINEs. These results suggest that genomic REs undergo DNA methylation and hydroxymethylation across puberty and that the variations in 5hmC and 5mC patterns in these repetitive elements might not be random events but may participate in modeling gene expression during puberty.
Figure 1.
Genome-wide DNA hydroxymethylation and DNA methylation of retroelements (REs) in the ARC during pubertal development
(A and B) Counts of different types of REs with differential DNA methylation (A) and hydroxymethylation (B) in ARCs of 4- and 8-week-old mice.
(C) The proportion of REs with differential DNA hydroxymethylation and DNA methylation.
LINE, long interspersed nuclear element; LTR, long-terminal repeat; SINE, short interspersed nuclear element.
The role of DNA methylation of REs at the coding regions in regulating gene expression
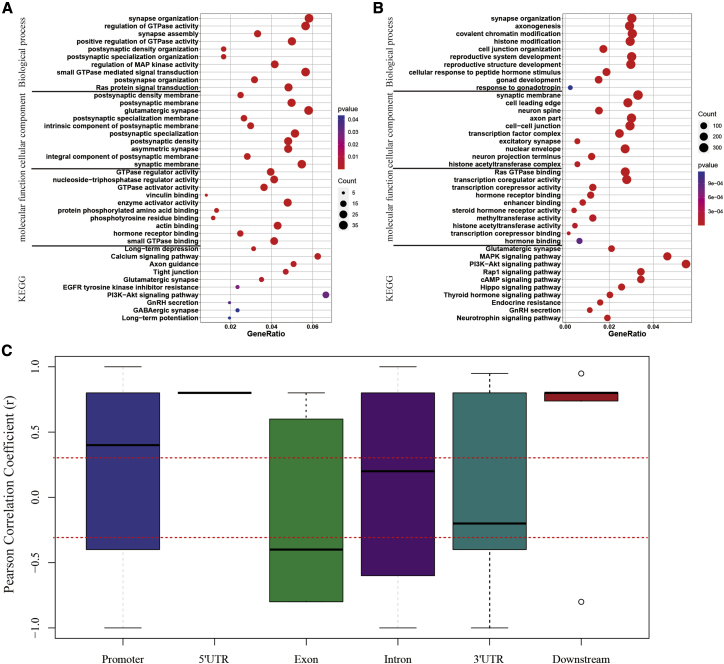
We further analyzed the positions of the REs with differential DNA hydroxymethylation (DHMREs) and DNA methylation (DMREs), and found that 46.0% of DMREs and 51.4% of DHMREs were located near or in DEGs (log2 FC > 1 or < −1, p < 0.05), which were closely associated with synapse connection, neuron development, hormone response, and epigenetic regulation (Figures 2A and 2B). Unexpectedly, DNA methylation of REs at promoters showed a strong positive correlation with gene expression (Pearson correlation coefficient [r] = 0.473, p < 0.001) (Figure 2C; Table S1), whereas the correlation between DNA methylation of REs in exons and RNA levels had no statistical significance (p > 0.05). However, 5′ UTRs and “downstream elements” (within 3 kb downstream of the 3′ UTR) were too few for the correlation coefficient to be determined statistically (Figure 2C). Thus, DNA hydroxymethylation of REs failed to correlate with gene expression (Figure S1). The positive correlation between DNA methylation of REs in gene promoters and gene expression was likely contradictory to the conventional understanding that the transcription of adjacent genes is usually inactivated by the DNA methylation of SINEs.16
Figure 2.
DNA hydroxymethylation and DNA methylation of REs and expression of proximal genes
(A and B) GO and KEGG analysis of target genes containing REs with differential DNA methylation (A) and hydroxymethylation (B).
(C) Pearson correlation analysis between DNA methylation of REs and expression of adjacent genes in pubertal ARCs.
Htr6 expression was controlled by methylation of SINEs in vitro
Previous studies have revealed that 5-hydroxytryptamine (serotonin), a metabolite of tryptophan, mediated through 5-hydroxytryptamine receptors (HTRs), acts on components of the hypothalamus-hypophysis-gonadal axis and induces a delay in sexual development in mammals, whereas HTR antagonists can exert a potent inhibitory responses to facilitate pubertal maturation during postnatal development.17,18 In the current sequencing data, Htr6 was significantly downregulated (log2FC = −2, FDR < 0.001), accompanied by an extremely hypomethylated SINE within its promoter region (chromosome 4 [chr4]: 139,075,874–139,075,961; the genomic coordinates in this study were all referred to as GRCm38) (FC = −14.5%, p = 0.005) in 8-week-old ARCs compared with that in 4-week-old ARCs.
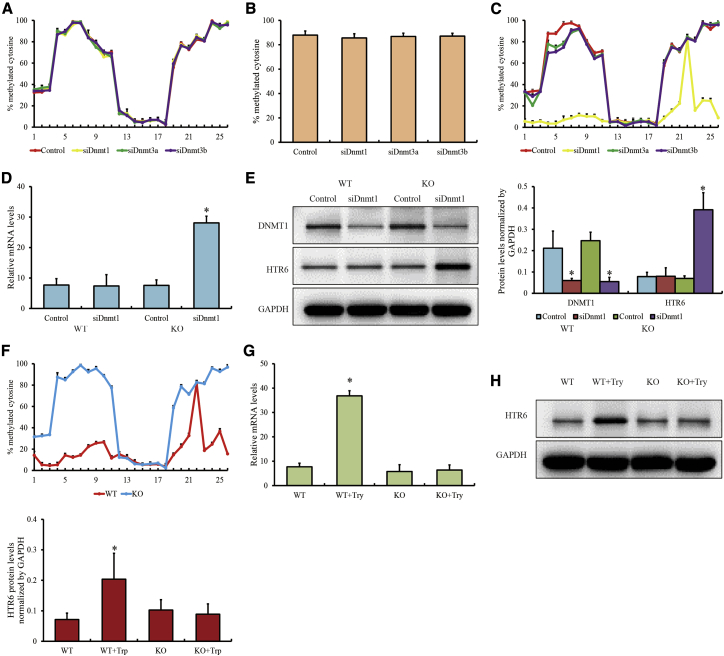
To study the epigenetic mechanism of REs during the pubertal process, transcription of Htr6, potentially regulated by DNA methylation of SINEs, was investigated in mouse hypothalamic GnRH-producing GT1-7 cells. First, GT1-7 cells were treated with Dnmt1, Dnmt3a, and Dnmt3b small interfering RNA (siRNA) (Figures S2A and S2B). We failed to find any significant changes in DNA methylation in the two CpG islands at the Htr6 promoter through a bisulfite PCR assay (Figure 3A; Figure S3). Similarly, the only CpG site of a certain SINE within the Htr6 promoter presented a slight change in methylation because of Dnmt RNA interference (Figure 3B; Figure S4), indicating that DNA methylation of this SINE might not be regulated by DNMTs. Interestingly, when GT1-7 cells with the “CG” locus deleted from this SINE (chr4: 139,075,888–139,075,889) (GT1-7SINE-CpG-KO1) were prepared using a CRISPR-Cas9 system (Figure S5A), we determined that Dnmt1 made a greater contribution to DNA methylation on promoters compared with Dnmt3a and Dnmt3b (Figure 3C; Figure S6), and a reduction in Dnmt1 treatment levels could enhance expression of Htr6 in GT1-7SINE-CpG-KO1 cells (Figures 3D and 3E). These results suggest that SINE methylation affects Dnmt1-mediated promoter methylation and Htr6 expression in GT1-7 cells. Moreover, GT1-7 cells were treated with 10 μg/mL L-tryptophan for 4 h to block GnRH secretion. We observed that, after tryptophan treatment, the promoter of Htr6 became hypomethylated in wild-type GT1-7 cells while still maintaining hypermethylation in GT1-7SINE-CpG-KO1 cells (Figure 3F; Figure S7). Htr6 expression was also upregulated in wild-type GT1-7 cells after tryptophan treatment but not in GT1-7SINE-CpG-KO1 cells (Figures 3G and 3H). DNA methylation of SINEs was likely an independent mechanism of action of Dnmt that suppressed DNA methylation of the promoter of Htr6.
Figure 3.
DNA methylation of SINEs in Htr6 promoters and Htr6 expression
(A and C) DNA methylation of two CpG islands at the Htr6 promoter affected by DNMT RNA interference in wild-type GT1-7 cells (A) and GT1-7SINE-CpG-KO1 cells (C), as observed through the bisulfite pyrosequencing assay.
(B) DNA methylation of SINEs at the Htr6 promoter affected by DNMT RNA interference, as determined through the bisulfite pyrosequencing assay.
(D and E) The levels of mRNA (D) and protein (E) in Htr6 induced by DNMT1 RNA interference in wild-type GT1-7 cells and GT1-7SINE-CpG-KO1 cells.
(F) DNA methylation of two CpG islands at the Htr6 promoter affected by L-tryptophan in wild-type GT1-7 cells and GT1-7SINE-CpG-KO1 cells, as determined through the bisulfite pyrosequencing assay.
(G and H) The levels of mRNA (G) and protein (H) in Htr6 induced by L-tryptophan in wild-type GT1-7 cells and GT1-7SINE-CpG-KO1 cells.
si, siRNA; WT, wild-type GT1-7 cells; KO, GT1-7SINE-CpG-KO1 cells; Try, L-tryptophan; 1–25, 25 CpG sites distributed within two CpG islands in the Htr6 promoter as detected in this study. The qPCR data are presented as the mean ± SEM of three separate experiments. ∗p < 0.05 in Student’s t test.
Dnmt1 recruitment to the promoter of Htr6 excluded by methylated SINE via NR5A2
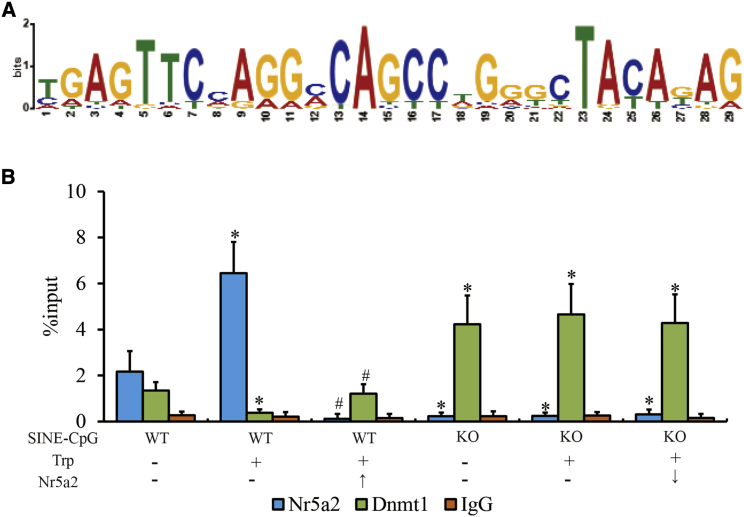
The consensus DNA motifs of 83 REs whose DNA methylation was positively correlated with transcription of adjacent genes were analyzed. Given the SINE sequence (Figure 4A), nuclear receptor subfamily 5 group A member 2 (NR5A2) was considered the top binding protein by a search of the JASPAR database (http://jaspar.genereg.net/).19 Previous studies have reported that members of the NR5A family of proteins are early gonadal markers that regulate adrenal and gonadal development and function in vivo.20,21 Enrichment of NR5A2 and DNMT1 at the promoter of Htr6 was investigated via a chromatin immunoprecipitation (ChIP)-qPCR assay (Figure 4B). We determined that NR5A2 was substantially enriched in the promoter of Htr6 but completely lost in GT1-7SINE-CpG-KO1 cells compared with wild-type GT1-7 cells, and the change in DNMT1 occupancy was opposite that of NR5A2. Moreover, tryptophan could enhance enrichment of NR5A2 and reduce the affinity of DNMT1 for the promoter of Htr6 in wild-type GT1-7 cells but not in GT1-7SINE-CpG-KO1 cells. Additionally, knockdown of NR5A2 (Figures S8A and S8B) could compromise the effect of tryptophan and rescue DNMT1 affinity in wild-type GT1-7 cells, whereas ectopic overexpression of NR5A2 (Figures S8A and S8B) failed to affect enrichment of DNMT1 on the Htr6 promoter in GT1-7SINE-CpG-KO1 cells treated with tryptophan. These results indicated that DNA methylated SINEs could be recognized by NR5A2 and played an inhibitory role in DNMT1 recruitment of the Htr6 promoter.
Figure 4.
Effect of NR5A2 on SINE methylation at the Htr6 promoter
(A) DNA motif analysis of SINEs whose DNA methylation was positively correlated with transcription of adjacent genes using the JASPAR tool.
(B) The occupancy of NR5A2 and DNMT1 on the Htr6 promoter in GT1-7 cells by ChIP-qPCR assay.
↑, ectopic overexpression; ↓, RNA interference. The qPCR data are presented as the mean ± SEM of three separate experiments. ∗p < 0.05 compared with L-tryptophan treatment. #p <0.05, representing significant differences between NR5A2 overexpression or blocking treatment.
Effect of 5hmC of SINEs on enhancers and gene expression in the ARC during pubertal maturation
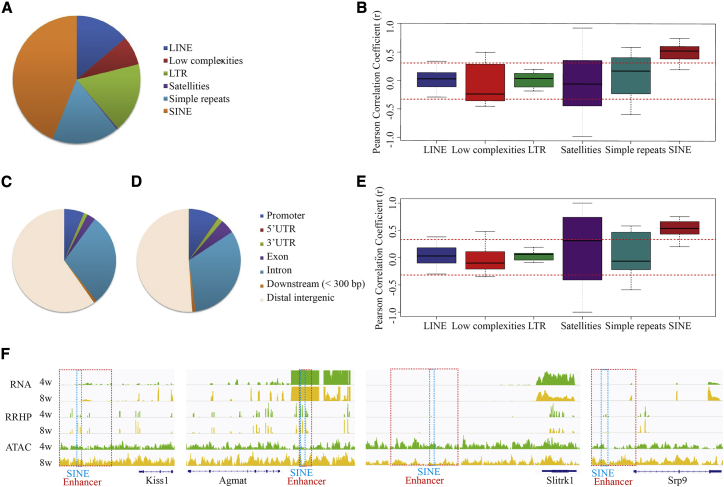
In addition to the transcribed genes, enhancers are also considered to play a distal regulatory role in gene expression. Neuron-specific enhancers were annotated by “Enhanceratlas.”22 For DMREs, only five SINEs and one simple repeat linked with the promoters of UCK2, selenoprotein (SELENOW), PRDM10, and MAP1B were likely to be involved with the annotated enhancers. PRDM10 has been reported previously as an essential transcription factor for maintaining global translation during cell differentiation and ontogenesis.23,24 MAP1B is crucial for synaptic maturation, including dendritic spine and axon development.25,26 However, for DHMREs, 10,685 enhancers containing 38,405 REs (5,302 LINEs, 6,802 LTRs, 2,812 low complexity sequences, 114 satellites, 6,531 simple repeats, and 16,844 SINEs) showed significant differential hydroxymethylation (Figure 5A). In particular, the change in 5hmC in SINEs rather than other REs at the enhancers was positively correlated with the nearest gene expression between 8-week and 4-week ARCs (r = 0.615, p = 0.016) (Figure 5B).
Figure 5.
DNA hydroxymethylation and DNA methylation of REs within enhancers and expression of their target genes
(A) Differentially DNA hydroxymethylated REs within enhancers compared between 4- and 8-week-old pubertal ARCs.
(B) Pearson correlation analysis between DNA hydroxymethylation of REs within enhancers and expression of target genes (<10 kB) in the pubertal ARC.
(C and D) Annotated genomic regions of chromatin accessibility in 4-week-old (C) and 8-week-old (D) ARCs.
(E) Pearson correlation analysis between DNA hydroxymethylation of REs and chromatin accessibility in enhancer regions in pubertal ARCs.
(F) Genome browser views on DNA hydroxymethylation and chromatin accessibility on Kiss1, Agmat, Slitrk1, and Srp9 as well as the enhancers near these genes.
Correlation between chromatin accessibility and hydroxymethylated SINEs in enhancers
To further investigate the role of hydroxymethylated REs at the enhancers in epigenetic regulation of gene transcription, assay for transposase-accessible chromatin (ATAC-seq) was employed to study the change in chromatin accessibility during puberty. The insert fragment size distribution of the ATAC-seq libraries indicates good quality control of these samples (Figure S9). The coverage of most genomic peaks in the 4- and 8-week ARCs were annotated in the distal intergenic regions (Figures 5C and 5D; Figure S10). Combined with reduced representation hydroxymethylation profiling (RRHP) data, we focused on REs in enhancers and determined that the significant changes in chromatin accessibility (log2 FC > 0.585 or < −0.585, p < 0.05) of SINEs were positively correlated with changes in 5hmC (r = 0.638, p = 0.009) (Figure 5E). In other words, an increase in DNA hydroxymethylated REs was usually accompanied by open chromatin in enhancer regions, such as Kiss1, Agmat, Slitrk1, and Srp9 (Figure 5F; Table S2).
Kiss1 transcription activated by hydroxymethylated SINEs in upstream enhancers in GT1-7 cells
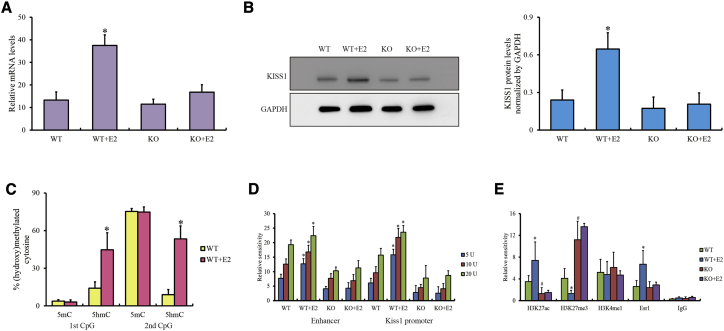
To verify the effects of the DNA hydroxymethylation of SINEs on the transcription activity of enhancers, two known distal enhancers (chr1: 133,308,985–133,319,710; chr1:133,339,139–133,339,571) around Kiss1 modulated by estrogen27,28 were considered in our study. Coincidentally, the upstream enhancer of Kiss1 also contains two SINEs (chr1: 133,304,850–133,305,026 and 133,305,119–133,305,238), the first of which had significantly higher hydroxymethylation at 8-week-old than at 4-week-old ARCs (log2FC = 1.86, FDR < 0.0001). We also created GT1-7SINE-CpG-KO2 cells with two CpG deletions at the first SINE via CRISPR-Cas9 (Figure S5B). The expression of Kiss1 was slightly downregulated and was not upregulated after estradiol induction in GT1-7SINE-CpG-KO2 cells compared with wild-type cells (Figures 6A and 6B). The levels of DNA methylation and hydroxymethylation at these two CpGs, as detected with a bisulfite assay, showed that these CpGs were always stably methylated, whereas they were substantially hyper-hydroxymethylated in wild-type GT1-7 cells treated with estrogen (Figure 6C; Figure S11), indicating that only 5hmC might be attributed to the enhancer activity in this system. The results of the DNase I-qPCR assay indicated that the chromatin accessibility of the entire enhancer region and Kiss1 promoter was compromised in GT1-7SINE-CpG-KO2 cells (Figure 6D). Likewise, ChIP-qPCR assay results also revealed decreased levels of acetylation of histone H3 at lysine 27 (H3K27ac) and estrogen receptor α (ESR1) as well as increased levels of tri-methylation of histone H3 at lysine 27 (H3K27me3) in the enhancer region, indicating that loss of hydroxymethylation in SINEs excluded ESR1 recruitment and resulted in primed enhancer and silenced Kiss1 transcription activity in GT1-7SINE-CpG-KO2 cells (Figure 6E). Our data show that hydroxymethylated SINEs might facilitate transcriptional activation of enhancers in the case of Kiss1.
Figure 6.
DNA methylation of SINEs in the enhancer for Kiss1 expression
(A and B) The levels of mRNA (A) and protein (B) in Kiss1 in WT GT1-7 cells and GT1-7SINE-CpG-KO2 cells induced by estradiol.
(C) DNA (hydroxyl)methylation levels for SINEs within the enhancers in WT GT1-7 cells induced by estradiol.
(D) Chromatin accessibility of the enhancer and Kiss1 promoter regions in estradiol-induced WT GT1-7 cells and GT1-7SINE-CpG-KO2 cells by DNase I-qPCR.
(E) Histone modifications on the enhancer regions in estradiol-induced WT GT1-7 cells and GT1-7SINE-CpG-KO2 cells assessed via ChIP-qPCR.
E2, estradiol. The qPCR data are presented as the mean ± SEM of three experiments. ∗,#p < 0.05 compared with the wild-type group.
Discussion
REs are important components in mammalian genomes that profoundly affect chromatin structure and gene expression activity. Although a vast majority of REs are termed “inactive molecular fossils,”29 a small part can be rapidly amplified via RNA intermediates throughout the genome. In the current study, we determined the changes in DNA methylation and hydroxymethylation of REs, especially SINEs, during the pubertal process (Figure 1). As an intermediate of the DNA demethylation process, DNA hydroxymethylation usually contributes to activation of gene expression, in contrast to DNA methylation. Although we analyzed the functional enrichment of DHMRs and DMRs containing REs across puberty and found that most associated Gene Ontology and signaling pathways overlapped (Figures 2A and 2B), a controversial observation after further analysis revealed that DNA methylation of REs located at the promoters of adjacent genes is beneficial for gene expression (Figure 2C), whereas DNA hydroxymethylation of intragenic REs does not seem to affect gene expression levels (Figure S1). Thus, the presence of a large number of REs with DNA hypomethylation in 8-week-old compared with 4-week-old ARCs indicates that only a small proportion of REs may indeed affect gene expression in neuronal cells during sexual maturation. We still have no idea about the function and biological significance of other REs, although they show remarkably different (hydroxyl)methylation patterns.
In this study, we show two cases of Htr6 and Kiss1 expression regulated by (hydroxyl)methylation of SINEs. From the results, we conclude that a complicated regulatory machinery of DNA (hydroxyl)methylation on SINEs coordinates with transcription factors and affects chromatin structure. SINEs are a heterogeneous group of genetic elements derived from transfer RNA (tRNA),30 characterized by an internal RNA polymerase III (RNA Pol III) promoter within their 5′ terminus as well as a poly(A) repeat at their 3′ terminus. When SINEs integrate into new sites, the regulatory sequences from SINEs may simultaneously affect the transcription activity of adjacent genes in addition to controlling RNA Pol III for the SINE per se. Epigenetic modifications such as DNA methylation on SINEs are likely to be linked with gene expression but are still obscure and controversial.31,32
Our study reveals a novel mechanism for DNA methylation of SINEs for gene expression. Initially, we found that DNA methylation of the SINE at the Htr6 promoter was positively correlated with Htr6 expression, which is unexpected and contrary to common sense. Further molecular experiments revealed that SINE methylation may protect the Htr6 promoter from DNMT1 (Figure 3C), maintaining control of Htr6 expression beyond DNA methylation (Figures 3D and 3E). Until the SINE loses its methylation, Htr6 expression can be normally controlled by DNMT1 (Figure 3F). Moreover, SINE methylation per se was not affected by DNMT knockdown (Figures 3A and 3B). These results suggest that dramatic DNA methylation is expected to affect the activities of SINEs, and a certain level of DNA methylation at SINEs may particularly stabilize and maintain DNA methylation in surrounding genomic areas. As observed in previous studies, because persistent hypermethylation of SINEs may inhibit their mobilization in mice and humans,33, 34, 35 SINEs and other types of REs need to keep themselves from being methylated by mammalian DNMTs. Instead, SINE methylation was thought to be necessary for tryptophan-induced Htr6 expression coordinated by NR5A2 (Figures 3G, 3H, and 4B), which cannot be mechanistically addressed by current data. In this light, epigenetic regulation of SINEs (or other types of REs) and their effects on expression of nearby genes are still rather complicated, with the exception of DNA methylation.
There is a lack of studies on DNA hydroxymethylation in SINEs, and here we report that SINE hydroxymethylation is beneficial for transcriptional activation and chromatin opening (Figure 5B, 5E, and 5F), which is similar to the results from other studies on embryonic development and pluripotent stem cells.36,37 Our results show that DNA hydroxymethylation of SINEs at intergenic regions is independent of DNA methylation (Figure 6C). Knockout of CpG sites to block DNA hydroxymethylation caused Kiss1 silencing via enhancer activity and histone modification (Figures 6D and 6E). The alteration of chromosome spatial architecture and DNA looping structure affecting the distal interaction between enhancers and target genes likely explains this outcome. The coordination between DNA hydroxymethylation of SINEs and the chromatin accessibility organized by epigenetic regulators, such as CCCTC binding factor (CTCF), cohesion complexes, and non-coding RNAs, and multiple transcription factors needs to be explored in future research.
Our results showed a novel regulatory mechanism of REs, especially for SINEs with DNA methylation and hydroxymethylation modifications, emphasizing the importance of epigenetic alterations in regulating gene transcription for pubertal onset and development. We also provide a safer and more efficient strategy for in situ genetic editing at CpG sites on REs compared with epigenetic drug utilization for puberty-associated diseases and treatment of other diseases.
Materials and methods
Experimental animals
C57BL/6 female mice purchased from Shanghai SLAC Laboratory Animal Co. (Shanghai, China) were housed in clean cages and maintained at 22°C ± 2°C with a constant 12 h light/dark cycle. The animals were allowed free access to food and water. Four- and eight-week-old mice (n = 10 per group) were used in this study. Preliminary experiments employing dye injection were performed to target the location of the ARC using its initial orientation (0.4 mm lateral, 1.60 mm posterior to the bregma, 7.40 mm below the surface of the dura), as described previously.38,39 Mice were sacrificed via cervical dislocation, and their whole brains were immediately isolated. The hypothalamic ARC tissues in each group were harvested and collected for subsequent experiments according to the previously mentioned dye staining procedure. All procedures were performed in accordance with the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University.
RNA-seq library construction and data analysis
ARC tissues were stored in 1 mL TRIZOL (Thermo Fisher Scientific, Waltham, MA, USA). They were then ground in liquid nitrogen. 100 μL chloroform was added to the ground tissue, and the mixture was centrifuged at the highest speed at 4°C for 10 min. The supernatant was transferred into a new tube, one volume of isopropanol was added, and the solution was centrifuged again at the highest speed at 4°C for 10 min. The precipitate was washed with cold 75% ethanol and dissolved in diethylpyrocarbonate (DEPC) water. The concentration and quality of RNA were measured using a Nanodrop 2000 (Thermo Fisher Scientific) and an Agilent Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Four micrograms of RNA from each group was used for library preparation using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) following the manufacturer’s instructions and sequenced on an Illumina Hiseq platform.
In the raw data, adaptors were trimmed, and low-quality reads were filtered out using Trimmomatic,40 and the quality of the clean reads was checked using FastQC.41 Next, the clean reads were aligned to the latest mouse genome assembly, mm10, using Hisat2.42 The transcripts were assembled, and the expression levels were estimated through fragments per kilobase per million (FPKM) values using the StringTie algorithm with default parameters43. Differential mRNA and long non-coding RNA (lncRNA) expression among the groups was evaluated using the R package Ballgown.44 The statistical significance of the differences was computed using the Benjamini and Hochberg (BH) p value adjustment method (log2 FC > 1 or < −1, p < 0.05). Gene annotation was performed using the Ensembl Genome Browser database (http://www.ensembl.org//useast.ensembl.org/index.html?redirectsrc=//www.ensembl.org%2Findex.html). The R package ClusterProfiler was used to annotate DEGs using Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.45
Reduced representation bisulfite sequencing (RRBS), RRHP library construction, and data analysis
The genomic DNA of the ARCs in the two groups was extracted using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany). High-quality DNA (200 ng) was then digested by the restriction endonuclease MspI (NEB) and subjected to 3′ end blunting, single-nucleotide (A) addition, and adaptor ligation. For RRHP, the 5hmC positions at the adaptor junctions were modified by T4 phage β-glucosyltransferase (NEB), and non-glucosyl-5hmCs were removed through another round of MspI digestion. The 250- 500-bp fragments were then selected and treated through bisulfite conversion using an Epitect Bisulfite Kit (QIAGEN) according to the manufacturer’s instructions.
A bisulfite pyrosequencing assay was performed using PyroMark Q24 Tests (QIAGEN), following the protocol provided in the PyroMark Q24 CpG Line-1 handbook.
For high-throughput sequencing, the converted DNA was eluted, and PCR amplification was performed to enrich for fragments with adapters on both ends. The constructed libraries were quantified using an Agilent Bioanalyzer 2100 (Agilent Technologies, Carlsbad, CA, USA) and subjected to high-throughput sequencing using an Illumina Hiseq 2500 platform with paired-end 50-bp sequencing (PE50).
Trim Galore v.0.5.0, Cutadapt v.1.18 (non-default parameters:–max-n 0–minimum-length 35), and Trimmomatic v.0.38 (non-default parameters: SLIDINGWINDOW:4:15 LEADING:10 TRAILING:10 MINLEN:35) were used to filter adapters, short reads (length < 35 bp), and low-quality reads. FastQC (with default parameters) was used to ensure high read quality. The trimmed reads of RRBS data were aligned to the reference genome (assembly GRCm38) using Bismark v.0.7.0, and the DNA methylation profiles were analyzed using the methylKit package.46 DMRs were selected when their FDR value was less than 0.05 and their methylation percentage change between the control and test groups was more than 10%. For RRHP, clean reads were mapped to the mouse genome (assembly GRCm38) using Bowtie2 v.2.3.4.1 software (with default parameters). Aligned reads with a CCGG tag at the 5′ end were counted. DMR analysis was performed using methylKit. DHMRs were determined using the diffReps software. DHMRs were characterized as having a log2 FC greater than 1 or less than −1 and FDR less than< 10−4.
Assay for transposase-accessible chromatin (ATAC) library construction and data analysis
Freshly harvested ARC tissues (1 × 105) were washed with cold PBS, ground in liquid nitrogen, filtered using a 40-μm cell strainer (Corning, New York City, NY, USA), and centrifuged at 12,000 × g at 4°C for 10 min. The supernatant was transferred to a new Eppendorf tube and incubated with 50 μL cold hypotonic buffer (10 mM NaCl, 3 mM MgCl2, 10 mM Tris-HCl [pH 7.5]) containing 0.1% NP40, 0.1% Tween 20, and 0.01% digitonin on ice for 3 min, after which 1 mL cold hypotonic buffer containing 0.1% Tween 20 was added, the resulting mixture was centrifuged at 500 × g at 4°C for 5 min, and the supernatant was discarded. The precipitate was incubated with 50 μL transposition mix (Vazyme, Nanjing, China) at 37°C and then centrifuged at 1,000 rpm. Transposed DNA of the mixture was isolated using a QIAGEN MinElute PCR Purification Kit (QIAGEN, Hilden, Germany) and amplified under appropriate PCR conditions. The library was subjected to high-throughput sequencing using an Illumina Hiseq 2500 platform with paired-end 150-bp sequencing (PE150).
Data quality control and pre-treatment procedures were similar to those done in RRHP. Peak calling was performed using MACS2.47 Credible peaks were screened using irreproducible discovery rate (IDR)48 and annotated using ChIPseeker.49 Motifs was analyzed using Multiple Em for Motif Elicitation (MEME, https://meme-suite.org/meme/tools/memeg),50 and the differential peaks (log2 FC > 0.585 or < −0.585, p < 0.05) were analyzed using MAnorm.51
Data deposits
The raw sequencing data were deposited to ArrayExpress with accession numbers E-MTAB-9420, E-MTAB-9421, and E-MTAB-10263.
Cell culture
GT1-7 cells purchased from the Cell Bank of Shanghai Institutes of Biological Sciences (Shanghai, China) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific). The siRNA sequences for DNMTs and NR5A2 and the pEGFP-NR5A2 plasmid were purchased from Sangon Biotech (Shanghai, China). One million cells were harvested and transfected with 25 pmol siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific) for 18 h. The spent medium was replaced with regular DMEM, and the cells were cultured for another 48 h. The oligonucleotides of guide RNA (gRNA) were designed (https://zlab.bio/guide-design-resources) and inserted into the px330 plasmid (73131) obtained from Addgene (Watertown, MA, USA). GT1-7 cells were transfected with px330 linked with gRNA and donor DNA using Lipofectamine 3000 (Thermo Fisher Scientific) and screened with 10 μg/mL puromycin. The target genomic region was verified via PCR. The cells were treated with 10 μg/mL L-tryptophan (Sigma-Aldrich, St. Louis, MO, USA) for 4 h,52 and GT1-7 cells were treated with 1 nM 17β estradiol (Sigma-Aldrich) for another 48 h.53
Western blot (WB) assay
Cells (5 × 106) were harvested, 500 μL radio immunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) containing a protease inhibitor cocktail (Beyotime Biotechnology, Nantong, Jiangsu, China) was the added to the cells, and the total protein concentration was quantified using the bicinchoninic acid (BCA) method. Aliquots of proteins (40 μg) were added to the lanes of a 10% SDS polyacrylamide gel, and the proteins were separated via electrophoresis and transferred onto nitrocellulose membranes. Subsequently, the membranes were blocked with 5% nonfat dry milk in 0.01 M PBS buffer (pH 7.4) and 0.05% Tween 20 for 1 h at room temperature (RT). The blocked membranes were then incubated with primary antibodies against Dnmt1 (1:2,000, NB100-56519, Novus Biologicals, Centennial, CO, USA), Htr6 (1:1,500, NBP1-46557, Novus), Kiss1 (1:2,000, NBP1-45672, Novus), and Gapdh (1:5,000, 5174, Cell Signaling Technology, Beverly, MA, USA) overnight at 4°C, followed by incubation with the appropriate secondary antibodies (horseradish peroxidase-conjugated rabbit anti-mouse diluted at 1:10,000 and donkey anti-rabbit diluted 1:5,000, Cell Signaling Technology) for 30 min at RT. The protein bands were observed through enhanced chemiluminescence using the Amersham Imager 600 system (GE Healthcare Life Sciences, Pittsburgh, PA, USA), and the density of the immunoblots was measured with Quantity One 4.62 software (Bio-Rad, Hercules, CA, USA).
ChIP assay
Briefly, 5 × 106 cells were fixed with 1% formaldehyde, quenched with 0.125 M glycine at RT, and then lysed in 500 μL of lysis buffer (10 mM Tris-HCl [pH] 8.0, 10 mM NaCl, and 0.2% IGEPAL CA-630 [Thermo Fisher Scientific]) on ice for 30 min. Genomic DNA was sonicated to 200–500 bp. Ten percent of each whole-cell lysate was stored as the input, and the rest of the lysate was incubated with 1 μg of primary antibodies against NR5A2 (ab18293, Abcam, Cambridge, MA, USA), DNMT1, H3K27ac (NBP2-54615, Novus), H3K27me3 (NBP2-16840, Novus), mono-methylation of histone H3 at lysine 4 (H3K4me1) (NB21-1021, Novus), and ESR1 (ab32063, Abcam) at 4°C overnight. Then, an additional 2-h pull-down was performed at 4°C using protein-A beads (Thermo Fisher Scientific). Primers designed to encompass approximately 150 bp around the target GnRH genomic regions were used to detect the enrichment of histone deacetylases (HDACs) using qPCR.
Quantitative polymerase chain reaction (qPCR) assay
The DNA templates were detected using a Fast Universal SYBR Green Real-time PCR Master Mix (Roche) under the following conditions: 95°C for 2 min, 50 cycles of 95°C for 5 s, 60°C for 10 s, 72°C for 30 s, and 72°C for 10 min. β-Actin was used as the loading control. The primers used in this study are listed in Table S3.
Statistical analysis
All statistical analyses were performed using SPSS version 20 (IBM, Armonk, NY, USA). For qPCR data, the 2ΔΔct method was used to calculate the expression, enrichment, or probable DNA content. Student’s t test was used to evaluate differences between groups. A p value of less than 0.05 was considered to indicate a significant difference.
Acknowledgments
This project was supported by the National Natural Science Foundation of China (31602600), the Hospital Level Project of Shanghai Children’s Hospital (2018YQN002), the Cultivation Plan for Excellent Young Talents of Shanghai Children’s Hospital (2017YYQ05), Key R&D and Promotion Projects of Henan Province (tackling key scientific and technical problems; 202102310443), and the Suzhou Science and Technology Development Plan Project (SZD0976). We are grateful for the assistance from Shanghai Genefund Biotech Co. Ltd. for generation and regular analysis of the high-throughput sequencing data. We are also grateful for assistance from Coweldgen Scientific Co. Ltd. with the CRIPSR-Cas9 service and bisulfite assay.
Author contributions
Y.S., H.Z., S.Z., and L.Z. collaborated to conduct all experiments and analyze the raw data. Y.S. designed the project and drafted the manuscript. L.C. and Y.H. helped perform the experiments and analyze data. S.Z. and J.L. revised and submitted the manuscript. H.Z., S.Z., Y.H., and J.L. supported this project with their funds.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.006.
Contributor Information
Jun Li, Email: lijun19801005@163.com.
Shasha Zhou, Email: zhoushasha@sjtu.edu.cn.
Supplemental information
References
- 1.Abreu A.P., Kaiser U.B. Pubertal development and regulation. Lancet Diabetes Endocrinol. 2016;4:254–264. doi: 10.1016/S2213-8587(15)00418-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Roux N., Genin E., Carel J.C., Matsuda F., Chaussain J.L., Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc. Natl. Acad. Sci. USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C., Roepke T.A., Kelly M.J., Rønnekleiv O.K. Kisspeptin depolarizes gonadotropin-releasing hormone neurons through activation of TRPC-like cationic channels. J. Neurosci. 2008;28:4423–4434. doi: 10.1523/JNEUROSCI.5352-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wyatt A.K., Zavodna M., Viljoen J.L., Stanton J.A., Gemmell N.J., Jasoni C.L. Changes in methylation patterns of kiss1 and kiss1r gene promoters across puberty. Genet. Epigenet. 2013;5:51–62. doi: 10.4137/GEG.S12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kohli R.M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurian J.R., Louis S., Keen K.L., Wolfe A., Terasawa E., Levine J.E. The Methylcytosine Dioxygenase Ten-Eleven Translocase-2 (tet2) Enables Elevated GnRH Gene Expression and Maintenance of Male Reproductive Function. Endocrinology. 2016;157:3588–3603. doi: 10.1210/en.2016-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jönsson M.E., Ludvik Brattås P., Gustafsson C., Petri R., Yudovich D., Pircs K., Verschuere S., Madsen S., Hansson J., Larsson J., et al. Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat. Commun. 2019;10:3182. doi: 10.1038/s41467-019-11150-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz W.A., Steinhoff C., Florl A.R. Methylation of endogenous human retroelements in health and disease. Curr. Top. Microbiol. Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 9.Ishak C.A., Classon M., De Carvalho D.D. Deregulation of Retroelements as an Emerging Therapeutic Opportunity in Cancer. Trends Cancer. 2018;4:583–597. doi: 10.1016/j.trecan.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Shen Y., Zhou S., Zhao X., Li H., Sun J. Characterization of Genome-Wide DNA Methylation and Hydroxymethylation in Mouse Arcuate Nucleus of Hypothalamus During Puberty Process. Front. Genet. 2020;11:626536. doi: 10.3389/fgene.2020.626536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanasaki H., Oride A., Mijiddorj T., Sukhbaatar U., Kyo S. How is GnRH regulated in GnRH-producing neurons? Studies using GT1-7 cells as a GnRH-producing cell model. Gen. Comp. Endocrinol. 2017;247:138–142. doi: 10.1016/j.ygcen.2017.01.025. [DOI] [PubMed] [Google Scholar]
- 12.Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W., et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 13.Zhao S.G., Chen W.S., Li H., Foye A., Zhang M., Sjöström M., Aggarwal R., Playdle D., Liao A., Alumkal J.J., et al. The DNA methylation landscape of advanced prostate cancer. Nat. Genet. 2020;52:778–789. doi: 10.1038/s41588-020-0648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 15.Markoff A. DNA Press; 2005. Analytical Tools for DNA, Genes and Genomes. [Google Scholar]
- 16.Estécio M.R., Gallegos J., Dekmezian M., Lu Y., Liang S., Issa J.P. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol. Cancer Res. 2012;10:1332–1342. doi: 10.1158/1541-7786.MCR-12-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim H.S., Yumkham S., Choi J.H., Son G.H., Kim K., Ryu S.H., Suh P.G. Serotonin stimulates GnRH secretion through the c-Src-PLC gamma1 pathway in GT1-7 hypothalamic cells. J. Endocrinol. 2006;190:581–591. doi: 10.1677/joe.1.06727. [DOI] [PubMed] [Google Scholar]
- 18.Bhattarai J.P., Roa J., Herbison A.E., Han S.K. Serotonin acts through 5-HT1 and 5-HT2 receptors to exert biphasic actions on GnRH neuron excitability in the mouse. Endocrinology. 2014;155:513–524. doi: 10.1210/en.2013-1692. [DOI] [PubMed] [Google Scholar]
- 19.Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D., et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48(D1):D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crespo B., Gómez A., Mazón M.J., Carrillo M., Zanuy S. Isolation and characterization of Ff1 and Gsdf family genes in European sea bass and identification of early gonadal markers of precocious puberty in males. Gen. Comp. Endocrinol. 2013;191:155–167. doi: 10.1016/j.ygcen.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Faienza M.F., Chiarito M., Baldinotti F., Canale D., Savino C., Paradies G., Corica D., Romeo C., Tyutyusheva N., Caligo M.A., et al. NR5A1 Gene Variants: Variable Phenotypes, New Variants, Different Outcomes. Sex Dev. 2019;13:258–263. doi: 10.1159/000507411. [DOI] [PubMed] [Google Scholar]
- 22.Gao T., He B., Liu S., Zhu H., Tan K., Qian J. EnhancerAtlas: a resource for enhancer annotation and analysis in 105 human cell/tissue types. Bioinformatics. 2016;32:3543–3551. doi: 10.1093/bioinformatics/btw495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han B.Y., Seah M.K.Y., Brooks I.R., Quek D.H.P., Huxley D.R., Foo C.S., Lee L.T., Wollmann H., Guo H., Messerschmidt D.M., Guccione E. Global translation during early development depends on the essential transcription factor PRDM10. Nat. Commun. 2020;11:3603. doi: 10.1038/s41467-020-17304-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J.A., Kim K.C. Expression patterns of PRDM10 during mouse embryonic development. BMB Rep. 2010;43:29–33. doi: 10.5483/bmbrep.2010.43.1.029. [DOI] [PubMed] [Google Scholar]
- 25.Montenegro-Venegas C., Tortosa E., Rosso S., Peretti D., Bollati F., Bisbal M., Jausoro I., Avila J., Cáceres A., Gonzalez-Billault C. MAP1B regulates axonal development by modulating Rho-GTPase Rac1 activity. Mol. Biol. Cell. 2010;21:3518–3528. doi: 10.1091/mbc.E09-08-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tortosa E., Montenegro-Venegas C., Benoist M., Härtel S., González-Billault C., Esteban J.A., Avila J. Microtubule-associated protein 1B (MAP1B) is required for dendritic spine development and synaptic maturation. J. Biol. Chem. 2011;286:40638–40648. doi: 10.1074/jbc.M111.271320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomikawa J., Uenoyama Y., Ozawa M., Fukanuma T., Takase K., Goto T., Abe H., Ieda N., Minabe S., Deura C., et al. Epigenetic regulation of Kiss1 gene expression mediating estrogen-positive feedback action in the mouse brain. Proc. Natl. Acad. Sci. USA. 2012;109:E1294–E1301. doi: 10.1073/pnas.1114245109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goto T., Tomikawa J., Ikegami K., Minabe S., Abe H., Fukanuma T., Imamura T., Takase K., Sanbo M., Tomita K., et al. Identification of hypothalamic arcuate nucleus-specific enhancer region of Kiss1 gene in mice. Mol. Endocrinol. 2015;29:121–129. doi: 10.1210/me.2014-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richardson S.R., Doucet A.J., Kopera H.C., Moldovan J.B., Garcia-Perez J.L., Moran J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0061-2014. MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogiwara I., Miya M., Ohshima K., Okada N. Retropositional parasitism of SINEs on LINEs: identification of SINEs and LINEs in elasmobranchs. Mol. Biol. Evol. 1999;16:1238–1250. doi: 10.1093/oxfordjournals.molbev.a026214. [DOI] [PubMed] [Google Scholar]
- 31.Huang D., Wang X., Tang Z., Yuan Y., Xu Y., He J., Jiang X., Peng S.A., Li L., Butelli E., et al. Subfunctionalization of the Ruby2-Ruby1 gene cluster during the domestication of citrus. Nat. Plants. 2018;4:930–941. doi: 10.1038/s41477-018-0287-6. [DOI] [PubMed] [Google Scholar]
- 32.Varshney D., Vavrova-Anderson J., Oler A.J., Cowling V.H., Cairns B.R., White R.J. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015;6:6569. doi: 10.1038/ncomms7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh C.P., Chaillet J.R., Bestor T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- 34.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sardina J.L., Collombet S., Tian T.V., Gómez A., Di Stefano B., Berenguer C., Brumbaugh J., Stadhouders R., Segura-Morales C., Gut M., et al. Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem Cell. 2018;23:727–741.e9. doi: 10.1016/j.stem.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Li J., Wu X., Zhou Y., Lee M., Guo L., Han W., Mo W., Cao W.M., Sun D., Xie R., Huang Y. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res. 2018;46:2883–2900. doi: 10.1093/nar/gky063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu M.H., Li X.F., McCausland B., Li S.Y., Gresham R., Kinsey-Jones J.S., Gardiner J.V., Sam A.H., Bloom S.R., Poston L., et al. Relative Importance of the Arcuate and Anteroventral Periventricular Kisspeptin Neurons in Control of Puberty and Reproductive Function in Female Rats. Endocrinology. 2015;156:2619–2631. doi: 10.1210/en.2014-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenwood M., Bordieri L., Greenwood M.P., Rosso Melo M., Colombari D.S., Colombari E., Paton J.F.R., Murphy D. Transcription factor CREB3L1 regulates vasopressin gene expression in the rat hypothalamus. J. Neurosci. 2014;34:3810–3820. doi: 10.1523/JNEUROSCI.4343-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews S. Babraham Bioinformatics; 2013. FastQC A Quality Control tool for High Throughput Sequence Data. [Google Scholar]
- 42.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazee A.C., Pertea G., Jaffe A.E., Langmead B., Salzberg S.L., Leek J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015;33:243–246. doi: 10.1038/nbt.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krismer K., Guo Y., Gifford D.K. IDR2D identifies reproducible genomic interactions. Nucleic Acids Res. 2020;48:e31. doi: 10.1093/nar/gkaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- 50.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202-8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao Z., Zhang Y., Yuan G.C., Orkin S.H., Waxman D.J. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13:R16. doi: 10.1186/gb-2012-13-3-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Agarwal P., Singh D., Raisuddin S., Kumar R. Amelioration of ochratoxin-A induced cytotoxicity by prophylactic treatment of N-Acetyl-L-Tryptophan in human embryonic kidney cells. Toxicology. 2020;429:152324. doi: 10.1016/j.tox.2019.152324. [DOI] [PubMed] [Google Scholar]
- 53.Roy D., Angelini N.L., Belsham D.D. Estrogen directly respresses gonadotropin-releasing hormone (GnRH) gene expression in estrogen receptor-alpha (ERalpha)- and ERbeta-expressing GT1-7 GnRH neurons. Endocrinology. 1999;140:5045–5053. doi: 10.1210/endo.140.11.7117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.