Abstract
N-acetyltransferase 10 (NAT10) is the key enzyme for N4-acetylcytidine (ac4C) modification of mRNA, which participates in various cellular processes and is related to many diseases. Here, we explore the relationships among osteoblast differentiation, NAT10, and ac4C, and we found that NAT0 expression and the ac4C level of total RNA were decreased in the bone tissues of bilateral ovariectomized (OVX) mice and osteoporosis patients. Adenoviruses overexpressing NAT10 reversed bone loss, and Remodelin, an NAT10 inhibitor, enhanced the loss of bone mass in OVX mice. Moreover, bone marrow-derived mesenchymal stem cells (BMSCs) with low-level ac4C modification formed fewer calcium nodules in vitro with NAT10 silencing, whereas BMSCs with high-level ac4C modification formed more calcium nodules with NAT10 overexpression. Moreover, we demonstrated that the ac4C level of runt-related transcription factor 2 (RUNX2) mRNA was increased after BMSCs were cultured in osteogenic medium (OM) and decreased after NAT10 silencing. The RUNX2 mRNA half-life and protein expression decreased after silencing NAT10 in BMSCs. Therefore, NAT10-based ac4C modification promotes the osteogenic differentiation of BMSCs by regulating the RUNX2 ac4C level. Because abnormal levels of NAT10 are probably one of the mechanisms responsible for osteoporosis, NAT10 is a new potential therapeutic target for this disease.
Keywords: ac4C, NAT10, bone marrow-derived mesenchymal stem cells, osteoblast differentiation, osteoporosis
Graphical abstract
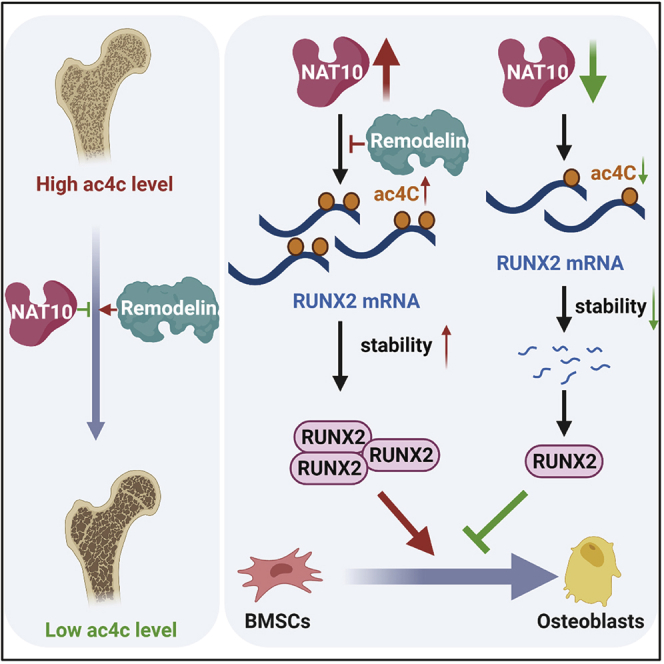
NAT10 catalyzes the addition of ac4C to RUNX2 mRNA, which increases the half-life of RUNX2 mRNA and promotes osteogenic differentiation of BMSCs. Moreover, overexpression of NAT10 with adenovirus reversed the bone loss in OVX mice. These results indicate that NAT10 may be a new potential therapeutic target for osteoporosis.
Introduction
N-acetyltransferase 10 (NAT10), a member of the general control non-repressible 5 (GCN5)-related N-acetyltransferase (GNAT) family, is a lysine acetyltransferase (KAT) that acetylates RNA,1, 2, 3, 4 p53,5 Che-1,6 and α-tubulin.7 It has been reported that autoacetylated NAT10 activates ribosomal RNA (rRNA) transcription, and that Sirt1 deacetylates NAT10 to overcome glucose starvation by inhibiting rRNA biogenesis. Moreover, deacetylated NAT10 fails to acetylate the autophagy regulator Che-1, leading to the autophagy.6,8 NAT10 functions with THUMP domain containing 1 (THUMPD1) to acetylate transfer RNA (tRNA) for tRNA stability.9 In addition, NAT10 plays an important role in p53 activation by acetylating p53 upon DNA damage and stabilizes microtubules by acetylating α-tubulin.5,7 More importantly, NAT10 is related to many diseases, such as Hutchinson-Gilford progeria syndrome (HGPS),10 hepatocellular carcinoma (HCC),11 and breast cancer.12 It is worth noting that the GNAT family regulates bone metabolic diseases. For example, N-acetyltransferase 1 (NAT1) is highly expressed in luminal breast cancer (LBC), promotes osteoclast differentiation, and induces LBC bone metastasis, and single-nucleotide polymorphisms (SNPs) of N-acetyltransferase 2 (NAT2) are related to the osteosarcoma progression and metastasis.13
N4-acetylcytidine (ac4C) is a conservative chemical modification of tRNA, rRNA, and mRNA.1, 2, 3 Addition of ac4C is catalyzed by NAT10, and ac4C on tRNAs or rRNA increases the fidelity or accuracy of protein translation.14,15 Recently, ac4C was found to be widely present on mRNA wobble sites, which helped to maintain mRNA stability and promote translation efficiency. Moreover, changes in NAT10−/− HeLa cell gene expression compared with parental HeLa cells are not associated with the known trans-acting roles of NAT10 (such as 18S rRNA and tRNAser/leu acetylation).3 ac4C is the first acetylation modification found on mRNA and is related to the development and prognosis of many diseases.
Bone marrow-derived mesenchymal stem cells (BMSCs), which possess the ability of multidirectional differentiation, have been widely used in stem cell therapy.16,17 More importantly, the osteogenic differentiation ability of BMSCs contributes to bone formation.18 Abnormal osteogenic differentiation of BMSCs leads to various diseases. The differentiation fate of BMSCs is regulated by a variety of factors, such as signaling pathway regulation,19 transcription factor activation,20 and epigenetic modification.21 Several stimulatory transcription factors, such as β-catenin, runt-related transcription factor 2 (RUNX2), special AT-rich sequence binding protein 2 (SATB2), osterix (OSX), smads, and CCAAT/enhancer-binding proteins (C/EBPs), play an important role in osteoblast differentiation. Moreover, RUNX2 is a master transcription factor for osteoblast differentiation and skeletal development controlled by several signaling pathways, such as the BMP, Wnt, and Notch signaling pathways. RUNX2 is able to induce the expression of many osteoblast markers.22 In addition, epigenetic modifications, such as histone modification, DNA methylation, and RNA modification, determine the fate of BMSCs and the pathogenetic mechanisms of osteoporosis by regulating the gene expression profiles.23,24 It is necessary and important to know whether ac4C modification of mRNA, a type of epigenetic modifications, affects the osteoblast differentiation of BMSCs.
We aimed to explore the relationship among NAT10, ac4C, osteoblast differentiation, and osteoporosis. As a result, we found that the addition of ac4C to mRNA catalyzed by NAT10 prevents osteoporosis and promotes osteoblast differentiation of BMSCs in vitro through ac4C-based posttranscriptional regulation of RUNX2, an important transcription factor for osteogenesis.
Results
NAT10 expression and ac4C modifications are downregulated in osteoporosis
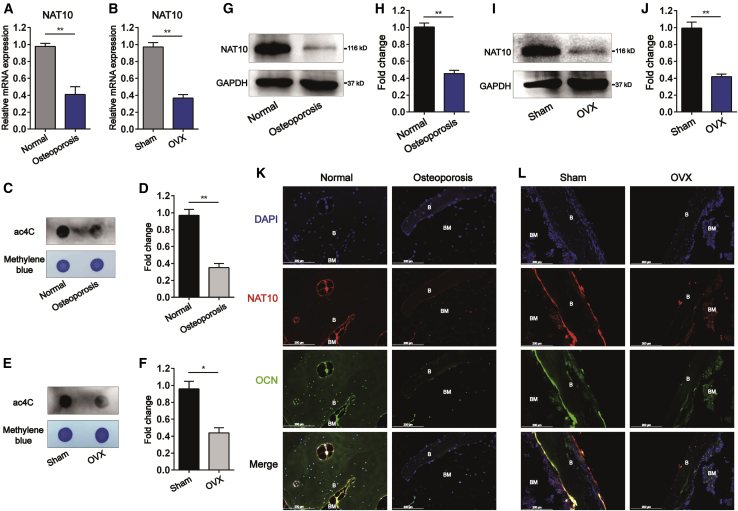
Osteoporosis is a systemic skeletal disease in which bone mineral density is decreased.25 Bone is a dynamic organ that maintains the balance of bone mass by osteoblasts and osteoclasts.26 BMSCs, the progenitor cells of osteoblasts, play an important role in the process of bone remodeling.27 To assess expression of NAT10 and the content of ac4C modifications in osteoporotic bone tissue, we performed quantitative polymerase chain reaction (qPCR) and ac4C dot blot among the total RNA of femurs from patients with osteoporosis, patients with developmental dysplasia, bilateral ovariectomized (OVX) mice, and sham mice. First, we found that mRNA expression of NAT10 (Figures 1A and 1B) and the ac4C content in total RNA (Figures 1C–1F) in the femurs of osteoporosis patients and OVX mice were significantly decreased compared with those in the corresponding control groups. Meanwhile, protein expression levels of NAT10 were also decreased in femurs (Figures 1G–1J). Next, we used immunofluorescence to detect whether NAT10 expression in osteoblasts was downregulated during osteoporosis. We found that OCN+ osteoblasts significantly reduced NAT10 expression in the femoral sections of osteoporosis patients and OVX mice (Figures 1K and 1L). These data indicate that NAT10 may be involved in the pathological process of osteoporosis.
Figure 1.
NAT10 expression and the ac4C content of total RNA are decreased in osteoporosis bone tissues
(A and B) mRNA expression of NAT10 in femurs from osteoporosis patients and OVX mice was decreased compared with expression in their corresponding controls, as determined by quantitative real-time PCR, n = 6. (C–F) Decreases in the ac4C content in total RNAs extracted from femurs of osteoporosis patients and OVX mice compared with in the corresponding control determined by ac4C dot blot analysis. (D) Densitometry quantitation of (C). (F) Densitometry quantitation of (E), n = 6. (G–J) The protein level of NAT10 in femurs from osteoporosis patients and OVX mice was decreased compared with expression in their corresponding controls, as determined by western blotting. (H) Densitometry quantitation of (G). (J) Densitometry quantitation of (I), n = 6. (K and L) The expression of NAT10 in the femurs of osteoporosis patients and OVX mice was decreased relative to that in their control groups, as determined by immunofluorescence staining (scale bars: 200 μm), n = 6. All data are expressed as the means ± SD. ∗p < 0.05, ∗∗p < 0.01 (n = 3 independent experiments).
Overexpression of NAT10 promotes bone formation in OVX
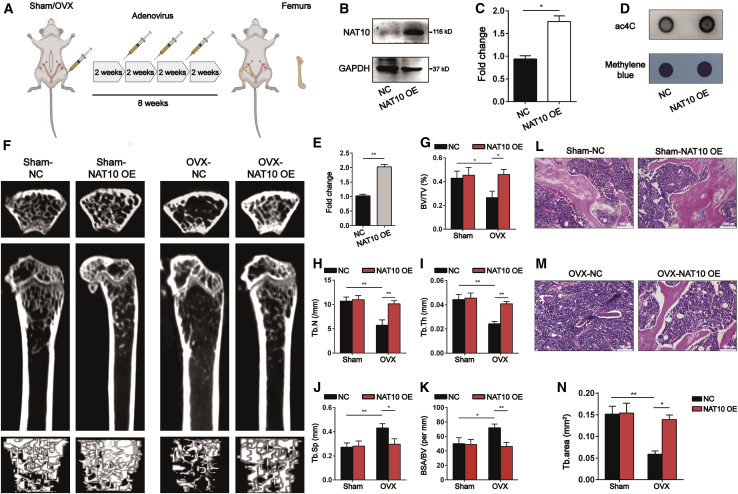
To test whether NAT10 is a regulator of bone formation, we used adenoviruses overexpressing NAT10 and injected them into C57BL/6 mice intramuscularly as shown in the illustration (Figure 2A). The protein levels of NAT10 were significantly increased in bone tissues of NAT10-overexpressing OVX mice (Figures 2B and 2C). Dot blot analysis showed that the ac4C content was also enhanced in the bone tissues of OVX mice overexpressing NAT10 (Figures 2D and 2E). Microcomputed tomography (μCT) analysis demonstrated that overexpression of NAT10 in OVX mice prevented trabecular bone loss. However, the sham mice overexpressing NAT10 had no significant changes in trabecular bone loss (Figure 2F). Quantitative measurement of bone microstructural parameters showed that trabecular bone volume fraction (BV/TV), trabecular number (Tb.N), and trabecular thickness (Tb.Th) were decreased, and that trabecular separation (Tb.Sp) and BSA/BV were increased in OVX mice, but these parameters in OVX mice were all reversed by overexpressing NAT10. However, these parameters did not change significantly in the sham mice overexpressing NAT10 (Figures 2G–2K). Hematoxylin and eosin (H&E) staining also showed that OVX mice overexpressing NAT10 reversed the reduction in bone formation, and sham mice overexpressing NAT10 did not have a change in bone formation (Figures 2L–2N). These results show that NAT10 is a protective factor against osteoporosis.
Figure 2.
Overexpression of NAT10 prevents bone loss in OVX mice
(A) Illustration of adenoviruses overexpressing NAT10 or empty vector for OVX mice or sham mice, n = 6. (B and C) Confirmation of the efficiency of adenoviruses overexpressing NAT10 by western blotting. (C) Quantification of band intensities (B), n = 6. (D) The ac4C content of total RNA was increased in adenoviruses overexpressing NAT10 group compared with empty vector adenoviruses group, as determined by dot blot, n = 6. (E) Densitometry quantitation of (D), n = 6. (F) Representative μCT images of four groups of mice, n = 6. (G–K) Quantification of bone microstructural parameters of BV/TV (G), Tb.N (H), Tb.Th (I), Tb.Sp (J), and BSA/BV (K), n = 6. (L and M) OVX mice treated with adenoviruses overexpressing NAT10 demonstrated aggravated bone loss, but the bone mass of sham mice treated with Remodelin had no difference from sham mice treated with empty vector adenoviruses (scale bars: 100 μm), as determined by H&E staining, n = 6. (N) Quantification of the trabecular area from four groups of mice, n = 6. All data are expressed as the means ± SD. ∗p < 0.05, ∗∗p < 0.01 (n = 3 independent experiments).
Remodelin, an effective and specific inhibitor of NAT10, negatively regulates bone mass and density in OVX mice
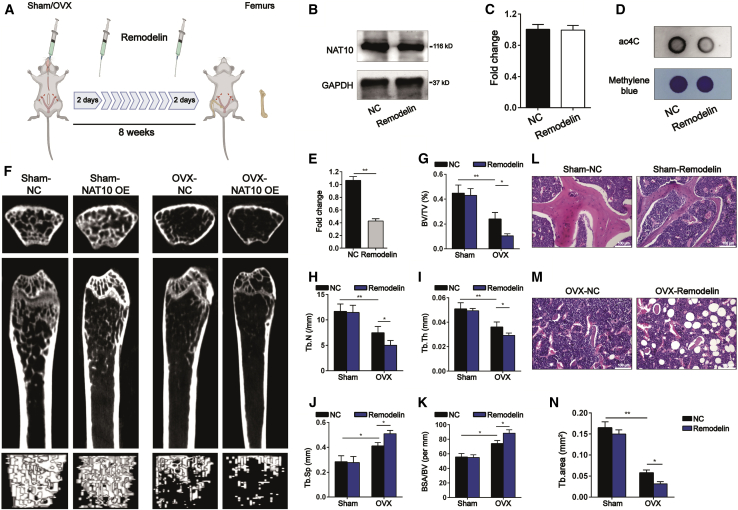
Remodelin is a novel and effective inhibitor of the acetyl-transferase protein NAT10.10 To explore whether the inhibition of NAT10 activity affected bone formation, we treated the OVX mice and sham mice with a Remodelin solution (100 mg/kg) by oral gavage every 2 days as shown in the illustration (Figure 3A). Although protein expression of NAT10 was unchanged after treatment with Remodelin (Figures 3B and 3C), dot blot analysis showed that the ac4C content in OVX mice treated with Remodelin was significantly decreased compared with that in the control group (Figures 3D and 3E). μCT demonstrated that Remodelin aggravated trabecular bone loss in OVX mice, while the bone mass of mice in the sham group remained unchanged (Figure 3F). Remodelin decreased the bone microstructural parameters BV/TV, Tb.N, and Tb.Th and increased Tb.Sp and BSA/BV in OVX mice, whereas these parameters were unchanged in sham mice after treatment with Remodelin (Figures 3G–3K). H&E staining also showed that Remodelin increased bone loss in OVX mice, but not in the sham mice (Figures 3L–3N). Thus, Remodelin appears to aggravate bone loss in OVX mice.
Figure 3.
Remodelin, an inhibitor of NAT10, aggravates bone loss in OVX mice
(A) Illustration of Remodelin treatment or solution alone for OVX mice or sham mice, n = 6. (B and C) The protein level of NAT10 was decreased in Remodelin treatments group compared with the solution alone group, as determined by western blotting, (C) Quantification of band intensities (B), n = 6. (D) The ac4C level of total RNA was decreased in the Remodelin treatments group compared with the solution alone group, as determined by western blotting, n = 6. (E) Densitometry quantitation of (D), n = 6. (F) Representative μCT images of four groups of mice, n = 6. (G–K) Quantification of bone microstructural parameters of BV/TV (G), Tb.N (H), Tb.Th (I), Tb.Sp (J), and BSA/BV (K), n = 6. (L and M) OVX mice treated with Remodelin demonstrated aggravated bone loss, but the bone mass of sham mice treated with Remodelin had no difference from sham mice treated with solution alone (scale bars: 100 μm), as determined by H&E staining, n = 6. (N) Quantification of the trabecular area from four groups of mice, n = 6. All data are expressed as the means ± SD. ∗p < 0.05, ∗∗p < 0.01 (n = 3 independent experiments).
The ac4C contents of total RNA and NAT10 expression are elevated during osteogenic differentiation of BMSCs compared with proliferation
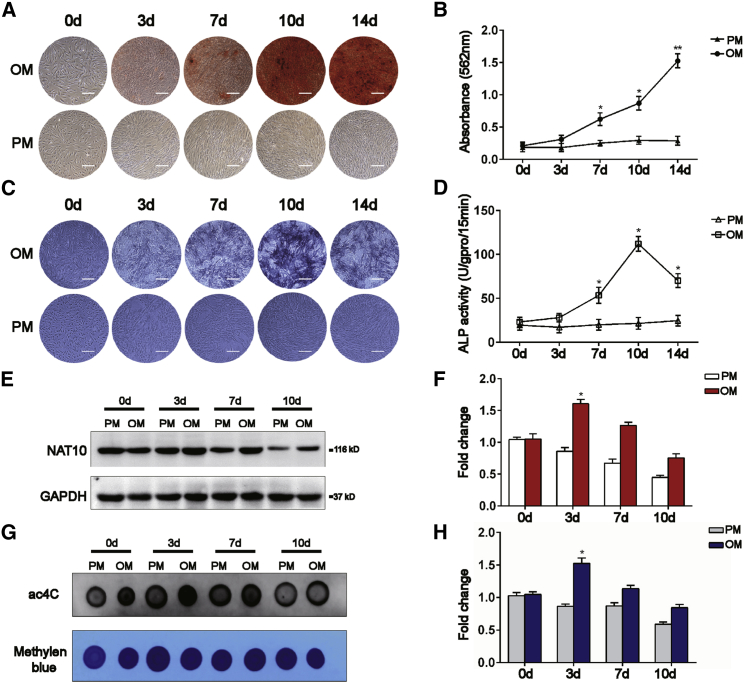
The above results indicate that NAT10 regulates bone formation in vivo, but the specific mechanism is unclear. The immunofluorescence results (Figures 1K and 1L) showed that osteoblasts from the femurs of osteoporosis patients and OVX mice expressed a low level of NAT10, and the ac4C content of total RNA decreased (Figures 1G–1J). We hypothesized that NAT10 regulated the osteoblast differentiation of BMSCs through ac4C modification. We isolated BMSCs from healthy donor bone marrow and identified their phenotypes by flow cytometry as previously described. Previous results showed that isolated BMSCs were positive for CD29, CD44, and CD105 and negative for CD14 and CD45. Moreover, the isolated BMSCs could differentiate into osteoblasts, adipocytes, and chondrocytes in vitro.28 As we found before, alizarin red S (ARS) staining revealed increased calcium nodule formation; staining for alkaline phosphatase (ALP) showed it to be elevated in BMSCs cultured in osteogenic medium (OM), but not in those cultured in proliferation medium (PM) (Figures 4A–4D). Moreover, the protein level of NAT10 and the ac4C level of total RNA were increased at the early stage of osteogenic differentiation of BMSCs (Figures 4E–4H). These results indicate that increased expression of NAT10 at the early stage may be related to the osteogenic differentiation of BMSCs.
Figure 4.
The ac4C content of total RNA and NAT10 expression are elevated when BMSCs are cultured in OM rather than PM
(A and B) BMSCs cultured in OM formed more calcium nodules than those cultured in PM from days 7 to 14, as determined by ARS staining (scale bar: 250 μm), n = 6. (C and D) BMSCs cultured in OM synthesized more ALP than BMSCs cultured in PM from days 7 to 14. During osteoblast differentiation, ALP staining peaked on day 10, as determined by ALP staining (scale bar: 250 μm), n = 6. (E) The protein level of NAT10 was increased in BMSCs cultured in OM compared with PM, n = 6. (F) Quantification of band intensities, n = 6. (G) The ac4C level of BMSCs cultured in OM increased compared with PM, as determined by dot blot analysis, n = 6. (H) Densitometry quantitation of (G), n = 6. All data are presented as the means ± SD. ∗p < 0.05, ∗∗p < 0.01 (n = 3 independent experiments).
NAT10 positively regulates the osteogenic differentiation of BMSCs in vitro
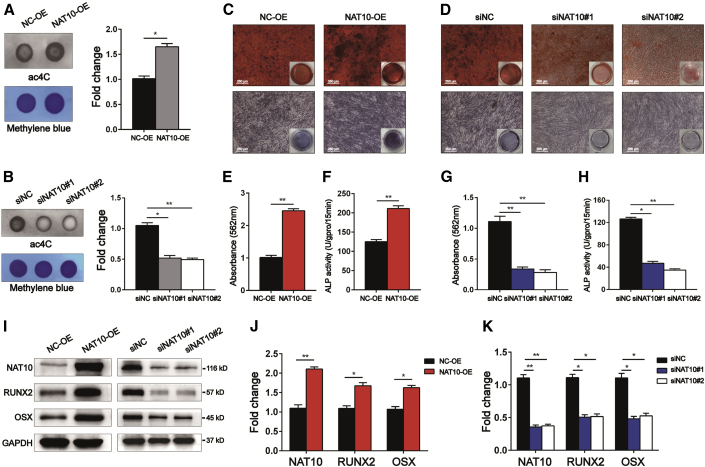
We explored whether the expression of NAT10 affects the osteogenic differentiation potential of BMSCs. BMSCs were infected with lentiviruses overexpressing NAT10 or short interfering RNA (siRNA) (siNAT10). The efficiency of NAT10 overexpression or NAT10 silencing in BMSCs was confirmed by western blotting on the 14th day of osteogenic differentiation. Dot blot analysis showed that compared with that of the corresponding control group, the ac4C content of total RNA was increased after NAT10 overexpression and decreased after silencing NAT10 in BMSCs on the 14th day of osteogenic differentiation (Figures 5A and 5B). Remodelin also decreased the ac4C content of total RNA (Figures S1A and S1B). ARS staining and ALP activity assays were used to detect the osteogenic differentiation ability of BMSCs. ARS staining results demonstrated that BMSCs overexpressing NAT10 formed more calcium nodules, and BMSCs with silenced NAT10 or Remodelin formed fewer calcium nodules. The ALP activity assay results were consistent with the results of ARS staining (Figures 5C–5H and S1C–S1E). Furthermore, we detected expression of osteogenic differentiation markers, such as RUNX2 and OSX. The results showed that overexpressing NAT10 led to an increase in the expression of RUNX2 and OSX, and silencing NAT10 or treatment with Remodelin led to a decrease in RUNX2 and OSX expression in BMSCs on the 14th day of osteogenic differentiation (Figures 5I–5K, S1F, and S1G). The above results suggest that NAT10 may promote the osteogenic differentiation of BMSCs by upregulating ac4C modification.
Figure 5.
NAT10 positively regulates the osteogenic differentiation of BMSCs in vitro
(A and B) The ac4C level of total RNA increased in BMSCs infected with lentiviruses overexpressing NAT10 and decreased in BMSCs infected with siNAT10, as determined by dot blot analysis, n = 6. (C) ARS staining showed that overexpressing NAT10 caused more calcium nodule formation (upper panels), and ALP staining increased after overexpressing NAT10 (lower panels) (scale bars: 250 μm), n = 6. (D) NAT10 knockdown by siRNA decreased calcium nodule formation (upper panels) and ALP staining (lower panels) (scale bars: 250 μm), n = 6. (E and G) ARS staining was quantified as the absorbance at 562 nm, n = 6. (F and H) ALP activity was determined as units per gram of protein per 15 min, n = 6. (I) Protein levels of the markers of osteoblast differentiation, RUNX2, and Osterix were increased after overexpressing NAT10 during osteogenic induction (left), and knocking down NAT10 had the opposite effect (right), n = 6. (J and K) Quantification of band intensities, n = 6. All data are presented as the means ± SD. ∗p < 0.05, ∗∗p < 0.01 (n = 3 independent experiments).
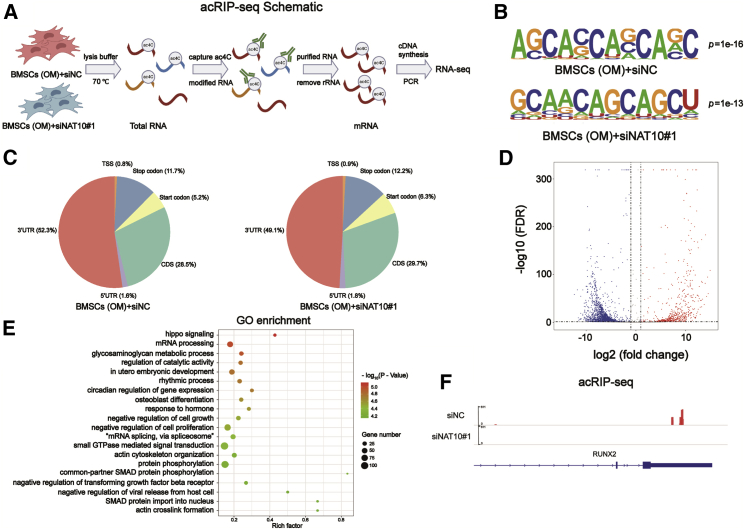
Identification of NAT10 targets through acetylated RNA immunoprecipitation and sequencing (acRIP-seq)
As we found above, expression of NAT10 and ac4C modification of total RNA were increased during osteogenic differentiation of BMSCs in vitro. Moreover, silencing NAT10 weakened ac4C modification in BMSCs. To explore the possible role of ac4C in the osteogenesis of BMSCs, we performed acRIP-seq between BMSCs cultured with short interfering RNA negative control (siNC) and siNAT10 during osteogenic differentiation in vitro (Figure 6A). Consistent with a previous article,28 most ac4C peaks contained distinct CXX repeats (Figure 6B), four obligate cytidines separated by two nonobligate nucleotides. However, unlike a previous article, our results showed that most ac4C peaks were enriched in 3′ UTR noncoding sequences (Figure 6C). According to the volcano map, the number of hypoacetylated genes was decreased after silencing NAT10 (Figure 6D). Gene Ontology (GO) enrichment analysis showed that these differentially acetylated genes (Data S1) were enriched in signaling pathways, including Hippo signaling (p = 7.71E−06), mRNA processing (p = 9.94E−06), osteoblast differentiation (p = 2.35E−05), and so on (Figure 6E; Data S2). The above results indicated that the osteoblast differentiation pathway might be regulated by NAT10, and that NAT10 might have facilitated osteoblast differentiation of BMSCs through the ac4C mechanism. We found four genes closely related to osteogenesis in the osteoblast differentiation pathway, RUNX2, collagen type I alpha 1 chain (COL1A1), tenascin C (TNC), and smad family member 3 (SMAD3). Among these genes, RUNX2 was the master gene for osteoblast differentiation, which determined the fate of BMSCs. The ac4C peak existed on the 3′ UTR of RUNX2 mRNA and disappeared after silencing NAT10 (Figure 6F). These data suggest that NAT10 may regulate the osteogenic differentiation ability through the ac4C modification of these genes.
Figure 6.
Identification of NAT10 targets through acRIP-seq
(A) Schematic illustration of acRIP-seq performed in two healthy donor BMSCs, which were divided into two groups shown in the picture (two biological repeats per group). (B) Enriched motifs of ac4C peaks within two groups of BMSCs. (C) Pie charts show the percentage of ac4C peak distribution in the acetylated transcripts. (D) Volcano map of differentially expressed ac4C gene peaks in BMSCs transfected with siNAT10 versus siNC. (E) GO enrichment analysis of the differentially expressed ac4C gene peaks. (F) The acRIP-seq assay showed the ac4C peaks on RUNX2 mRNA between the two groups. ac4C sites (red) were indicated in the 3′ UTR. p = 2.04E-09.
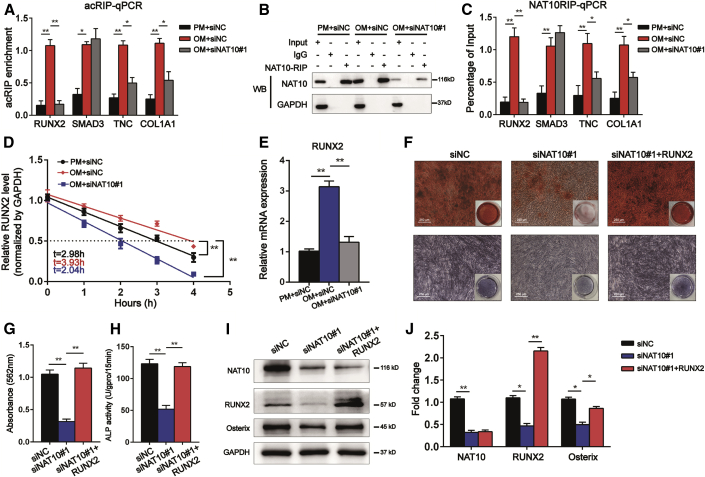
RUNX2 is a target mRNA of NAT10 and a mediator of osteogenic differentiation of the NAT10/ac4C modification axis
Because we found that the ac4C content of total RNA was upregulated after osteogenic induction, we hypothesized that the ac4C level of the genes in the above osteoblast differentiation signaling pathway was also increased in BMSCs cultured in OM compared with PM. Then we performed acRIP-qPCR and NAT10 RNA immunoprecipitation (NAT10RIP)-qPCR among BMSCs in PM with siNC, BMSCs in OM with siNC, and BMSCs in OM with siNAT10. The ac4C pull-down assay and qPCR analysis showed that the ac4C levels of RUNX2, SMAD3, TNC, and COL1A1 in BMSCs cultured with OM were increased compared with the levels in the PM, while the ac4C levels of RUNX2 were downregulated most significantly after silencing NAT10 (Figure 7A). Similarly, the NAT10 pull-down assay and qPCR analysis demonstrated that NAT10 binds to RUNX2 mRNA in BMSCs after osteogenic induction. After silencing NAT10 in BMSCs cultured with OM, the percentage of NAT10 binding to RUNX2 mRNA decreased most significantly (Figures 7B and 7C). Because RUNX2 is an important transcription factor that promotes osteoblast differentiation, we presumed that NAT10 enhanced the osteoblast differentiation ability of BMSCs by increasing the ac4C mRNA level of RUNX2. Considering that ac4C promotes mRNA expression by increasing the stability of mRNA,3 we tested the half-life of RUNX2. The half-life of RUNX2 in BMSCs cultured in OM increased compared with that in BMSCs cultured in PM, while the half-life of RUNX2 decreased after silencing NAT10 (Figure 7D). The mRNA and protein levels of RUNX2 were significantly decreased after silencing NAT10 (Figures 5I and 7E). Moreover, overexpressing NAT10 increased the half-life of RUNX2, and Remodelin reversed this effect of NAT10 (Figure S2A). The mRNA expression of RUNX2 was increased after overexpressing NAT10, and Remodelin reversed this increase (Figure S2B). Nevertheless, the half-life of Osterix did not change after silencing NAT10, but the mRNA expression was decreased (Figures S2C and S2D).
Figure 7.
RUNX2 is a target mRNA of NAT10 and a mediator of osteogenic differentiation of the NAT10/ac4C modification axis
(A) acRIP-qPCR analysis of alterations in the ac4C levels of RUNX2, SMAD3, TNC, and COL1A1 in BMSCs cultured in PM, OM, and OM along with siNAT10, n = 6. (B) Immunoblotting of NAT10 after NAT10RIP assay with cell lysate among three groups of BMSCs. GAPDH blotting confirmed immunoprecipitation (IP) specificity, n = 6. (C) Quantitative real-time PCR was performed after NAT10RIP was performed, n = 6. (D) The decay rate of mRNA and qPCR analysis of RUNX2 at the indicated times after actinomycin D (5 μg/mL) treatment among three groups of BMSCs, n = 6. (E) RUNX2 expression was quantified by qPCR after treatment with siNAT10, n = 6. (F) ARS staining (upper panels) and ALP staining (lower panels) assays showed that the weakened osteogenic differentiation ability of BMSCs cultured in siNAT10 was rescued by overexpressing RUNX2 (scale bar: 250 μm), n = 6. (G) ARS staining was quantified as the absorbance at 562 nm, n = 6. (H) ALP activity was determined as units per gram of protein per 15 min, n = 6. (I) RUNX2 overexpression rescued the decreased protein level of Osterix, n = 6. (J) Quantification of band intensities, n = 6. All data are presented as the means ± SD. ∗p < 0.05, ∗∗p < 0.01 (n = 3 independent experiments).
Finally, we explored whether overexpressing RUNX2 reversed the NAT10 inhibitory function of BMSC osteoblast differentiation. After silencing NAT10 by siRNA in BMSCs, RUNX2 lentivirus was used to overexpress RUNX2. ARS and ALP staining showed that BMSCs formed fewer calcium nodules after treatment with siNAT10, but the inhibitory function of NAT10 was reversed by overexpressing RUNX2. The results of the ALP staining assay were consistent with the results of ARS staining (Figures 7F–7H). Moreover, the western blotting results showed that overexpressing NAT10 rescued the loss of OSX, a marker of mature osteoblasts (Figures 7I and 7J). In conclusion, NAT10 directly controls the RUNX2 expression through ac4C modification to regulate the osteoblast differentiation ability of BMSCs in vitro and may be regarded as a new potential therapeutic target for osteoporosis in vitro.
Discussion
First, we found that the expression of NAT10 and the ac4C modification of total RNA in osteoporosis patients and OVX mice were decreased compared with those in the corresponding control. Immunofluorescence showed that osteoblasts expressed low levels of NAT10 in femoral tissue sections of osteoporosis patients and OVX mice. Second, overexpression of NAT10 with adenovirus reversed bone loss in OVX mice. OVX mice treated with Remodelin, an NAT10 inhibitor, exhibited aggravated bone loss. However, sham mice treated with Remodelin did not exhibit osteoporosis. A previous article reported that NAT10 knockout leads to embryonic lethality in mice, NAT10 haploinsufficient mice have no significant changes in bone mass compared with WT mice, and Remodelin does not cause changes in the mineral density and bone mineral content in mice with HGPS.10 Our results show that Remodelin aggravated bone loss in OVX mice but had no influence on sham mice. These results demonstrate that NAT10 is an antiosteoporosis molecule. However, further studies are needed to confirm the specific mechanism by which NAT10 regulates osteoporosis in vivo. It is of great interest to investigate whether NAT10 haploinsufficient mice with OVX exhibit aggravated bone loss.
The imbalance of bone homeostasis is the main reason for osteoporosis.26 Osteoblasts, which originate from BMSCs, facilitate bone formation and osteoclasts, originating from hemopoietic stem cells, and lead to bone resorption.27,29 Although overexpression of NAT10 with adenovirus reversed bone loss in OVX mice, it was not certain whether NAT10 had an effect on the differentiation of osteoblasts and osteoclasts. Hence we cultured human CD14+ monocytes with RANKL and macrophage colony-stimulating factor (M-CSF) in vitro, which led to osteoclast differentiation. After overexpressing or silencing NAT10, the number of TRAP+ osteoclasts did not change (data not shown). Because NAT10 did not affect the process of osteoclast differentiation, it is quite possible that NAT10 regulates the osteoblast differentiation of BMSCs. As mentioned above, NAT10 is one of the members of the GNAT superfamily, which can be able to transfer an acyl moiety to many substrates. It has been reported that GCN5 expression in bone sections from OVX mice was decreased, and that the protein levels of GCN5 in BMSCs from OVX mice were significantly decreased.30, 31, 32 Overexpression of GCN5 accelerated the osteoblast differentiation of BMSCs, and knockdown of GCN5 inhibited the osteoblast differentiation of BMSCs in vitro.31,32 Another study found that GCN5 regulated expression of the Hox gene and was essential for mouse skeletal development.33 As a result, it is likely that NAT10 also regulates the osteoblast differentiation of BMSCs.
Next, we verified that the osteogenic differentiation ability of BMSCs treated with lentiviruses overexpressing NAT10 was enhanced in vitro, and the osteogenic differentiation ability of BMSCs after silencing NAT10 or treatment with Remodelin was weakened. However, the specific mechanism by which NAT10 regulates the osteogenic differentiation of BMSCs remains unknown. As a previous article reported, NAT10 could catalyze the addition of ac4C modifications on mRNA, which promoted the stability and translation of mRNA. The ac4C modification of total RNA was enhanced in BMSCs treated with OM. Considering that 18S rRNA and tRNaser/leu also undergo ac4C modifications, these RNA modifications can have a global impact on translation. Regardless, the functions of 18S rRNA and tRNaser/leu in NAT0−/− HeLa cells did not change.3 Nevertheless, more experiments need to be performed to verify these functions in BMSCs. Therefore, the influence of NAT10 on osteoblast differentiation may be caused by the ac4C modification of mRNA.
To determine the targets regulated by NAT10 through ac4C modification, we performed acRIP-seq to compare the differences in ac4C-modified mRNA between BMSCs cultured in OM after treatment with siNC or siNAT10. We found that the ac4C peaks of several osteogenic differentiation pathway genes were decreased in BMSCs after silencing NAT10. RUNX2 is a transcription factor that plays an important role in osteoblast differentiation and skeletal morphogenesis, which directs the differentiation of BMSCs to preosteoblasts. The expression of RUNX2 is increased in preosteoblasts, reaches its peak expression in immature osteoblasts, and decreases in mature osteoblasts. RUNX2 activates the osteoblast differentiation process. The expression of ALP and OCN was absent in Runx2−/− mice.34 Several signaling pathways, including the BMP and Wnt, phosphatidylinositol 3-kinase (PI3K)/AKT, and Notch signaling pathways, regulate RUNX2 expression.35, 36, 37 During the process of osteoblast differentiation, RUNX2 is regulated at the transcriptional, posttranscriptional, translational, and posttranslational levels. Before this study, it was not known whether the ac4C in RUNX2 mRNA exists in BMSCs or regulates the osteoblast differentiation process. Thus, we identified a new posttranscriptional mode, ac4C mRNA, to regulate the osteoblast differentiation of BMSCs by affecting RUNX2 mRNA stability and protein expression.
acRIP-qPCR was performed to verify the acRIP-seq results. As expected, we found that the mRNA ac4C modification of RUNX2 was increased in BMSCs after osteogenic induction and decreased after NAT10 was knocked down. Consistent with acRIP-qPCR, the results of NAT10RIP-qPCR showed that NAT10 bound to RUNX2 mRNA during BMSC osteoblast differentiation. After knocking down NAT10, less NAT10 bound to RUNX2 mRNA. Therefore, these results indicate that NAT10 binds to RUNX2 mRNA after the osteogenic differentiation of BMSCs and promotes ac4C modification of RUNX2 mRNA. In addition, the mRNA half-life of RUNX2-enriched ac4C was prolonged after osteogenic induction and expression of RUNX2 was increased, thereby promoting the osteogenic differentiation of BMSCs. Moreover, silencing or overexpressing NAT10 could decrease or increase the half-life and the expression of RUNX2, respectively. Although the expression of Osterix was decreased after silencing NAT10, the half-life of Osterix did not change. These results indicated that NAT10 does not regulate expression of Osterix through ac4C modification. Finally, overexpression of RUNX2 reversed the weakened osteogenic differentiation ability of BMSCs treated with siNAT10. These results confirm that NAT10 promotes BMSC osteoblast differentiation directly by increasing the ac4C level on RUNX2 mRNA and improving the protein level of RUNX2. Although NAT10, a writer of ac4C, promotes the osteogenic differentiation of BMSCs, it is unknown whether erasers and readers exist to regulate the function of ac4C. Further studies need to be carried out to explore how the erasers and readers of ac4C regulate the osteogenic differentiation of BMSCs. Furthermore, it is unknown whether NAT10 regulates osteoblast differentiation through RUNX2 in vivo. More animal studies should be performed to demonstrate the specific mechanisms of NAT10 in OVX mice.
Common drug therapies for osteoporosis include calcium, vitamin D, selective estrogen-receptor modulator, denosumab, romosozumab, bisphosphonates, and the PTH analog teriparatide, among others.38, 39, 40, 41, 42 Despite significant progress in preventing and treating osteoporosis with such drugs, new therapies are needed; for example, stem cell therapy may be the best alternative for the treatment of osteoporosis in the future. Preclinical studies have demonstrated that genetically modified BMSCs are suitable for osteoporosis treatment.43 Our study provides a new mechanism for the stem cell therapy of osteoporosis.
In summary, we discovered that NAT10 promotes the osteoblast differentiation of BMSCs and is an antiosteoporosis factor. Expression of endogenous NAT10 and the ac4C level in the bone tissues of osteoporosis patients or OVX mice were reduced. Overexpression of NAT10 with adenovirus reversed the bone loss in OVX mice, and overexpression of NAT10 with lentivirus strengthened the osteogenic differentiation ability of BMSCs in vitro. Our study shows that NAT10 catalyzes the addition of ac4C to RUNX2 mRNA, which increases the half-life of RUNX2 mRNA and increases protein expression of RUNX2 during osteogenic differentiation in BMSCs. These results indicate that NAT10 may be a biomarker of bone formation and a new potential therapeutic target for osteoporosis.
Materials and methods
Osteoporosis models and treatments
This study was approved by the ethics committee of the Eighth Affiliated Hospital, Sun Yat-sen University, Shenzhen, China. All animals were purchased from the Laboratory Animal Center of Sun Yat-Sen University (n = 6 per group). Eight-week-old C57BL/6 female mice were subjected to either OVX or a sham operation (sham).
NAT10 overexpression adenovirus (NAT10-OE) and empty vector adenovirus (NC-OE) were constructed by OBiO Technology (Shanghai, China). Eight-week-old female C57BL/6 mice were divided into four groups (n = 6 per group). Two groups underwent OVX, and the others underwent a sham operation. After the surgery, 25 μl of NAT10-carrying adenovirus (1 × 1010 plaque-forming units [PFUs]/mL) or NC-OE was intramuscularly injected into the lateral thigh every 2 weeks for 4 consecutive injections. The first treatment and the operations were performed at the same time. Two months later, the four groups of mice were sacrificed, and femurs were collected for further study.
Remodelin was dissolved in a solution (200 μL volume) of 20% DMSO, 65% (45% 2-hydroxypropyl-b-cyclodextrin) solution (H5784; Sigma), and 15% Tween 80 (HY-Y1891; MCE). The mice were treated with solution alone or Remodelin solution by oral gavage at 100 mg/kg every 2 days for 2 months after receiving OVX or a sham operation. The first treatment and the operations were performed at the same time. Two months later, these mice were sacrificed, and femurs were collected for further study.
μCT analysis
The collected femurs were fixed in 4% paraformaldehyde (PFA) for 48 h. High-resolution μCT was performed using an Inveon MM system (Siemens). Images were acquired at a resolution of 8.82 μm/pixel, with a voltage of 80 kV, a current of 500 μA, and an exposure time of 1,500 ms in each of the 360 rotational steps. The trabecular bone parameters of the distal femurs, including the BV/TV, Tb.N, Tb.Th, Tb.Sp, and bone surface area/bone volume (BSA/BV), were analyzed according to the guidelines. Three-dimensional and two-dimensional bone structure image slices were reconstructed.
Human bone samples
This study was approved by the ethics committee of the Eighth Affiliated Hospital, Sun Yat-sen University, Shenzhen, China. Bone samples were obtained from six female patients diagnosed with postmenopausal osteoporosis who had fractures of the neck, as well as from six female control subjects with developmental dysplasia of the hip during the surgery. The patient information is presented in Table S1. The samples were fixed in 4% posttranscriptional PFA at room temperature for 3 days and decalcified in 10% ethylenediaminetetraacetic acid (EDTA), which was exchanged every 2 days at room temperature for 2 weeks. Samples were washed with phosphate-buffered saline (PBS), soaked with 30% sucrose in PBS at 4°C overnight, and embedded in paraffin. Sections (5-mm thickness) were used in the following study.
Quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen) and reverse transcribed using the PrimeScript RT Reagent Kit (TaKaRa Bio). Then quantitative real-time PCR was performed using SYBR Green Pro Taq (AG). Relative mRNA expression was determined using the 2−ΔΔCT method and was normalized to GAPDH expression. Primers are listed in Table S2.
Western blot analysis
In short, cells were washed with cold PBS three times and lysed with radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors for 30 min on ice. The total protein concentration was quantified by a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Samples were separated by SDS/polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride membranes (Millipore). Then the membranes were blocked with 5% fat-free milk dissolved in TBST (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, and 0.05% Tween 20) at room temperature for 1 h and incubated with primary antibodies against GAPDH (1:1,000, CW0100M; CWBIO), NAT10 (1:1,000, 13365-1-AP; Proteintech), RUNX2 (1:1,000 dilution, sc-390351; Santa Cruz), and Osterix (1:1,000, ab209484; Abcam) overnight at 4°C. After that, the membranes were incubated with the appropriate secondary antibody, either horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:3,000; Beyotime) or anti-mouse IgG (1:3,000; Beyotime), at room temperature for 1 h and visualized with chemiluminescent HRP substrate (Millipore, Billerica, MA, USA). ImageJ software was used to analyze the band intensities.
ac4C dot blot
Total RNA was heated to 75°C for 5 min, placed on ice for 1 min, and loaded onto Hybond-N+ membranes. Membranes were crosslinked with 150 mJ/cm2 in a UV254 nm Stratalinker 2400 (Stratagene). Then membranes were blocked with 5% nonfat milk in 0.1% Tween 20 PBS (PBST) for 1 h at room temperature and incubated with an anti-ac4C antibody (ab252215; Abcam) in PBST (1:1,000) at 4°C overnight. The membranes were then washed three times with 0.1% PBST, probed with a HRP-conjugated secondary anti-rabbit IgG in PBST (1:1,000) at room temperature for 1 h, and washed three times with 0.1% PBST. Chemiluminescent HRP substrate (Millipore, Billerica, MA, USA) was used to visualize the dots.
H&E staining
The femur sections were stained with H&E to evaluate the bone formation ability. First, the sections were deparaffinized at 65°C for 1 h, hydrated, and incubated with hematoxylin for 5 min. Then the sections were cleared in 1% HCl and 70% alcohol and stained with eosin for 2 min. ImagePro software was used to measure the trabecular area within the selected area.
Immunofluorescence staining
The sections were deparaffinized at 65°C for 1 h and treated with 100% xylene for 5 min twice, 100% ethanol for 5 min, 95% ethanol for 5 min, 85% ethanol for 5 min, and PBS for 2 min twice. Pepsin was used for antigen retrieval at 37°C for 20 min. Blocking was performed with goat serum at room temperature for 20 min. Anti-NAT10 (1:50 dilution, 13365-1-AP; Proteintech) and anti-osteocalcin (1:50 dilution, sc-365797; Santa Cruz) were used at 4°C overnight. The secondary antibodies used were anti-rabbit IgG (1:500, 4416; Cell Signaling Technology) and anti-mouse IgG (1:500, 4409; Cell Signaling Technology). The nuclei were stained with DAPI. An LSM 5 Exciter confocal imaging system (Carl Zeiss) was used to obtain images.
Isolation and culture of BMSCs
BMSCs were isolated and cultured as previously described.44 In brief, BMSCs were isolated from the bone marrow of healthy donors by density gradient centrifugation. Then BMSCs were resuspended in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Sijiqing, Hangzhou, China), seeded in flasks, and cultured in an incubator at 37°C and 5% CO2. The medium was removed after 48 h, and fresh DMEM was added, which was changed every 3 days. When BMSCs reached 80%–90% confluence, 0.25% trypsin containing 0.53 mM EDTA was used to digest the cells, and BMSCs were divided and reseeded in two new flasks at passage 1. The BMSCs were expanded to passages 3–5 for further study.
Osteogenic differentiation assay
As previously described, passage 3 BMSCs were seeded in 12-well plates at a density of 0.5 × 105 cells/well in PM (DMEM with 10% FBS).45 After 1 day, the cells had adhered to the wells, and the PM was changed with osteogenic medium consisting of DMEM with 10% FBS, 100 IU/mL penicillin, 100 IU/mL streptomycin, 0.1 μM dexamethasone, 10 mM β-glycerol phosphate, and 50 μM ascorbic acid (Sigma-Aldrich, St. Louis, MO, USA). The osteogenic differentiation medium was changed every 3 days.
ALP staining and activity assay
BMSCs cultured in osteogenic differentiation medium were fixed in 4% PFA on day 7 and stained with a BCIP/NBT ALP kit (Beyotime) for ALP staining according to the manufacturer’s instructions. An ALP assay kit (Nanjing Jiancheng Biotech, China) was used to test the ALP activity of BMSCs treated with osteogenic differentiation medium on day 7. The total protein concentration was tested by a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). The ALP activity was normalized to the total protein content and reported as units per gram of protein per 15 min.
ARS staining and quantification
BMSCs were fixed in 4% PFA at room temperature for 30 min and stained with 1% ARS (pH 4.3) for 15 min after culturing in OM on day 14. To remove nonspecific staining, we washed BMSCs three times with PBS, observed them under a microscope, and photographed them. For ARS quantification, 10% cetylpyridinium chloride monohydrate (Sigma-Aldrich) was used to destain the cells for 1 h at room temperature. After that, 200 μL of liquid was transferred to a 96-well plate, and spectrophotometric absorbance was measured at 562 nm.
Lentivirus construction and infection
The lentivirus infection protocol was described previously.46 In brief, lentiviruses overexpressing NAT10 (OE-NAT10; 1 × 108 transfection units [TUs]/mL) and a vector control (OE-NC, 1 × 108 TUs/mL) were constructed by OBiO Technology (Shanghai, China). Lentiviruses and polybrene (5 μg/mL; Sigma) were added to the medium and incubated with BMSCs for 24 h at a multiplicity of infection (MOI) of 50. After 24 h, the medium was changed. The infected cells were used for further study.
siRNA transfection
Passage 3 BMSCs were seeded in 12-well plates at a density of 0.5 × 105 cells/well. When BMSCs reached 60%–70% confluence, siRNA (GenePharma, Shanghai, China) was added to the medium with Lipofectamine RNA iMAX reagent (MAN0007825; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The sequences of the siRNAs used are listed in Table S3.
acRIP-seq
acRIP-seq was conducted in two groups of BMSCs (two biological repeats per group), including BMSCs cultured in OM with siNC and OM with siNAT10 on day 7. Total RNA was collected using the method described above. Total RNA was heated to 70°C for 6 min. Then EDTA was used to stop the reaction. The fragmented RNA was purified and collected by a Zymo RNA Clean and Concentrator-25 kit (R1017; Zymo Research). An anti-ac4C antibody, Dynabeads Protein G (10004D; Invitrogen), and purified RNA were incubated at 4°C for 6 h. The immunoprecipitated RNA was collected according to the manufacturer’s instructions. The library was constructed using an EpiTM Mini LongRNA-seq Kit (E1802; Epibiotek) according to the manufacturer’s protocol. Quality control of the library was conducted with a Bioptic Qsep100 Analyzer (Bioptic). NovaSeq, a high-throughput sequencing platform, was used for sequencing by Epibiotek (Guangzhou, China).
acRIP-qPCR and NAT10RIP-qPCR
acRIP and NAT10RIP were conducted by an RNA Immunoprecipitation Kit (P0101; GENESEED). In brief, cells were washed with PBS twice and collected by centrifugation at 1,000 × g for 5 min. Then 1 mL of RIP lysis buffer was added to the cells on ice for 10 min. Then 100 μL of lysis buffer was stored at −80°C. An anti-ac4C antibody (1:50, ab252215; Abcam), anti-NAT10 (1:50 dilution, 13365-1-AP; Proteintech), normal rabbit IgG (1:50, 2729S; Cell Signaling Technology), or normal mouse IgG (1:50, 12-371; Merck Millipore) was mixed with protein A/G beads at 4°C for 2 h. Then 450 μL of lysis buffer was incubated with the beads above at 4°C for 2 h. The beads were washed with buffer, and RNA was extracted. The RNA was analyzed by quantitative real-time PCR.
RNA stability assay
BMSCs were seeded in 12-well plates and treated with actinomycin D (5 mg/mL; catalog #HY-17559; Sigma) for 0, 1, 2, 3 and 4 h. Total RNA was collected using the method described above for quantitative real-time PCR analysis. The mRNA half-life was estimated by linear regression analysis. All data are expressed as the mean ± SD.
Statistical analysis
All experiments were repeated three times independently, and each experiment contained BMSCs from six different donors. All data are expressed as the mean ± SEM. SPSS 18.0 software was used to analyze the data. Two-group comparisons were performed with the Student’s t test. Multiple-group comparisons (one independent variable) were performed with one-way analysis of variance (ANOVA), and comparison between the groups was performed with proper post hoc analyses. The comparisons of more than two groups (two independent variables) were performed with two-way ANOVA. A p value <0.05 was considered statistically significant.
Acknowledgments
We thank American Journal Experts for providing the English language editing service for this manuscript. We also thank Yuanqi Feng and Fengxin Gao for valuable comments on acRIP-seq data analysis. All graphical abstracts were created with BioRender.com. This study was supported by the National Natural Science Foundation of China (81971518), the Key Realm R&D Programme of Guangdong Province (2019B020236001), the Shenzhen Key Medical Discipline Construction Fund (ZDSYS20190902092851024), the Natural Science Foundation of Guangdong Province (2018A030313232), and the Health Welfare Fund Project of Futian District (FTWS2020078).
Author contributions
H.S., M.M., and W.Y. designed this study. W.Y., Y.W., and H.L. performed the experiments. R.M., Y.J., and Y.L. participated in the animal experiments. Y.L. and X.S. helped with the cell experiments. R.M. and X.S. were in charge of the isolation and culture of BMSCs. R.M., Y.L., and M.M. helped with the statistical analysis. W.Y., Y.W., and H.L. wrote this manuscript. W.L. helped with the revision. All authors reviewed and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.06.022.
Contributor Information
M.J. Ma, Email: mamj8@mail.sysu.edu.cn.
H.Y. Shen, Email: shenhuiy@mail.sysu.edu.cn.
Supplemental information
References
- 1.Kawai G., Hashizume T., Miyazawa T., McCloskey J.A., Yokoyama S. Conformational characteristics of 4-acetylcytidine found in tRNA. Nucleic Acids Symp. Ser. 1989;1989:61–62. [PubMed] [Google Scholar]
- 2.Bruenger E., Kowalak J.A., Kuchino Y., McCloskey J.A., Mizushima H., Stetter K.O., Crain P.F. 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum. FASEB J. 1993;7:196–200. doi: 10.1096/fasebj.7.1.8422966. [DOI] [PubMed] [Google Scholar]
- 3.Arango D., Sturgill D., Alhusaini N., Dillman A.A., Sweet T.J., Hanson G., Hosogane M., Sinclair W.R., Nanan K.K., Mandler M.D., et al. Acetylation of Cytidine in mRNA Promotes Translation Efficiency. Cell. 2018;175:1872–1886.e1824. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito S., Horikawa S., Suzuki T., Kawauchi H., Tanaka Y., Suzuki T., Suzuki T. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA) J. Biol. Chem. 2014;289:35724–35730. doi: 10.1074/jbc.C114.602698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Tan Y., Zhang C., Zhang Y., Zhang L., Ren P., Deng H., Luo J., Ke Y., Du X. NAT10 regulates p53 activation through acetylating p53 at K120 and ubiquitinating Mdm2. EMBO Rep. 2016;17:349–366. doi: 10.15252/embr.201540505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X., Cai S., Zhang C., Liu Z., Luo J., Xing B., Du X. Deacetylation of NAT10 by Sirt1 promotes the transition from rRNA biogenesis to autophagy upon energy stress. Nucleic Acids Res. 2018;46:9601–9616. doi: 10.1093/nar/gky777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen Q., Zheng X., McNutt M.A., Guang L., Sun Y., Wang J., Gong Y., Hou L., Zhang B. NAT10, a nucleolar protein, localizes to the midbody and regulates cytokinesis and acetylation of microtubules. Exp. Cell Res. 2009;315:1653–1667. doi: 10.1016/j.yexcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Cai S., Liu X., Zhang C., Xing B., Du X. Autoacetylation of NAT10 is critical for its function in rRNA transcription activation. Biochem. Biophys. Res. Commun. 2017;483:624–629. doi: 10.1016/j.bbrc.2016.12.092. [DOI] [PubMed] [Google Scholar]
- 9.Sharma S., Langhendries J.L., Watzinger P., Kötter P., Entian K.D., Lafontaine D.L. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242–2258. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balmus G., Larrieu D., Barros A.C., Collins C., Abrudan M., Demir M., Geisler N.J., Lelliott C.J., White J.K., Karp N.A., et al. Sanger Mouse Genetics Project Targeting of NAT10 enhances healthspan in a mouse model of human accelerated aging syndrome. Nat. Commun. 2018;9:1700. doi: 10.1038/s41467-018-03770-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tschida B.R., Temiz N.A., Kuka T.P., Lee L.A., Riordan J.D., Tierrablanca C.A., Hullsiek R., Wagner S., Hudson W.A., Linden M.A., et al. Sleeping Beauty Insertional Mutagenesis in Mice Identifies Drivers of Steatosis-Associated Hepatic Tumors. Cancer Res. 2017;77:6576–6588. doi: 10.1158/0008-5472.CAN-17-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H.Y., Liu Y.Y., Yang F., Zhang L., Zhang F.L., Hu X., Shao Z.M., Li D.Q. Acetylation of MORC2 by NAT10 regulates cell-cycle checkpoint control and resistance to DNA-damaging chemotherapy and radiotherapy in breast cancer. Nucleic Acids Res. 2020;48:3638–3656. doi: 10.1093/nar/gkaa130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Z., Yuan L., Jiang Z., Wang D. Associations of polymorphisms in NAT2 gene with risk and metastasis of osteosarcoma in young Chinese population. OncoTargets Ther. 2015;8:2675–2680. doi: 10.2147/OTT.S92275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumbhar B.V., Kamble A.D., Sonawane K.D. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at “wobble” 34th position in the anticodon loop of tRNA. Cell Biochem. Biophys. 2013;66:797–816. doi: 10.1007/s12013-013-9525-8. [DOI] [PubMed] [Google Scholar]
- 15.Tafforeau L., Zorbas C., Langhendries J.L., Mullineux S.T., Stamatopoulou V., Mullier R., Wacheul L., Lafontaine D.L. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol. Cell. 2013;51:539–551. doi: 10.1016/j.molcel.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Tompkins B.A., Balkan W., Winkler J., Gyöngyösi M., Goliasch G., Fernández-Avilés F., Hare J.M. Preclinical Studies of Stem Cell Therapy for Heart Disease. Circ. Res. 2018;122:1006–1020. doi: 10.1161/CIRCRESAHA.117.312486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antebi B., Pelled G., Gazit D. Stem cell therapy for osteoporosis. Curr. Osteoporos. Rep. 2014;12:41–47. doi: 10.1007/s11914-013-0184-x. [DOI] [PubMed] [Google Scholar]
- 18.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 19.Chen Q., Shou P., Zheng C., Jiang M., Cao G., Yang Q., Cao J., Xie N., Velletri T., Zhang X., et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128–1139. doi: 10.1038/cdd.2015.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almalki S.G., Agrawal D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation. 2016;92:41–51. doi: 10.1016/j.diff.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin B., Yu F., Wang C., Li B., Liu M., Ye L. Epigenetic Control of Mesenchymal Stem Cell Fate Decision via Histone Methyltransferase Ash1l. Stem Cells. 2019;37:115–127. doi: 10.1002/stem.2918. [DOI] [PubMed] [Google Scholar]
- 22.Fakhry M., Hamade E., Badran B., Buchet R., Magne D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J. Stem Cells. 2013;5:136–148. doi: 10.4252/wjsc.v5.i4.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marini F., Cianferotti L., Brandi M.L. Epigenetic Mechanisms in Bone Biology and Osteoporosis: Can They Drive Therapeutic Choices? Int. J. Mol. Sci. 2016;17:1329. doi: 10.3390/ijms17081329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Meurs J.B., Boer C.G., Lopez-Delgado L., Riancho J.A. Role of Epigenomics in Bone and Cartilage Disease. J. Bone Miner. Res. 2019;34:215–230. doi: 10.1002/jbmr.3662. [DOI] [PubMed] [Google Scholar]
- 25.Kanis J.A. Diagnosis of osteoporosis. Osteoporos. Int. 1997;7(Suppl 3):S108–S116. doi: 10.1007/BF03194355. [DOI] [PubMed] [Google Scholar]
- 26.Sotorník I. [Osteoporosis - epidemiology and pathogenesis] Vnitr. Lek. 2016;62(Suppl 6):84–87. [PubMed] [Google Scholar]
- 27.Kohli N., Ho S., Brown S.J., Sawadkar P., Sharma V., Snow M., García-Gareta E. Bone remodelling in vitro: Where are we headed?: -A review on the current understanding of physiological bone remodelling and inflammation and the strategies for testing biomaterials in vitro. Bone. 2018;110:38–46. doi: 10.1016/j.bone.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Larrieu D., Britton S., Demir M., Rodriguez R., Jackson S.P. Chemical inhibition of NAT10 corrects defects of laminopathic cells. Science. 2014;344:527–532. doi: 10.1126/science.1252651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Udagawa N., Takahashi N., Akatsu T., Tanaka H., Sasaki T., Nishihara T., Koga T., Martin T.J., Suda T. Origin of osteoclasts: mature monocytes and macrophages are capable of differentiating into osteoclasts under a suitable microenvironment prepared by bone marrow-derived stromal cells. Proc. Natl. Acad. Sci. USA. 1990;87:7260–7264. doi: 10.1073/pnas.87.18.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jing H., Liao L., Su X., Shuai Y., Zhang X., Deng Z., Jin Y. Declining histone acetyltransferase GCN5 represses BMSC-mediated angiogenesis during osteoporosis. FASEB J. 2017;31:4422–4433. doi: 10.1096/fj.201700118R. [DOI] [PubMed] [Google Scholar]
- 31.Jing H., Su X., Gao B., Shuai Y., Chen J., Deng Z., Liao L., Jin Y. Epigenetic inhibition of Wnt pathway suppresses osteogenic differentiation of BMSCs during osteoporosis. Cell Death Dis. 2018;9:176. doi: 10.1038/s41419-017-0231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang P., Liu Y., Jin C., Zhang M., Tang F., Zhou Y. Histone Acetyltransferase GCN5 Regulates Osteogenic Differentiation of Mesenchymal Stem Cells by Inhibiting NF-κB. J. Bone Miner. Res. 2016;31:391–402. doi: 10.1002/jbmr.2704. [DOI] [PubMed] [Google Scholar]
- 33.Lin W., Zhang Z., Chen C.H., Behringer R.R., Dent S.Y. Proper Gcn5 histone acetyltransferase expression is required for normal anteroposterior patterning of the mouse skeleton. Dev. Growth Differ. 2008;50:321–330. doi: 10.1111/j.1440-169X.2008.01041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R.T., Gao Y.H., Inada M., et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 35.Bian Q., Liu S.F., Huang J.H., Yang Z., Tang D.Z., Zhou Q., Ning Y., Zhao Y.J., Lu S., Shen Z.Y., Wang Y.J. Oleanolic acid exerts an osteoprotective effect in ovariectomy-induced osteoporotic rats and stimulates the osteoblastic differentiation of bone mesenchymal stem cells in vitro. Menopause. 2012;19:225–233. doi: 10.1097/gme.0b013e3182272ef1. [DOI] [PubMed] [Google Scholar]
- 36.Zhao X.L., Chen J.J., Zhang G.N., Wang Y.C., Si S.Y., Chen L.F., Wang Z. Small molecule T63 suppresses osteoporosis by modulating osteoblast differentiation via BMP and WNT signaling pathways. Sci. Rep. 2017;7:10397. doi: 10.1038/s41598-017-10929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen G.Y., Ren H., Huang J.J., Zhang Z.D., Zhao W.H., Yu X., Shang Q., Qiu T., Zhang Y.Z., Tang J.J., et al. Plastrum Testudinis Extracts Promote BMSC Proliferation and Osteogenic Differentiation by Regulating Let-7f-5p and the TNFR2/PI3K/AKT Signaling Pathway. Cell. Physiol. Biochem. 2018;47:2307–2318. doi: 10.1159/000491541. [DOI] [PubMed] [Google Scholar]
- 38.Tang B.M., Eslick G.D., Nowson C., Smith C., Bensoussan A. Use of calcium or calcium in combination with vitamin D supplementation to prevent fractures and bone loss in people aged 50 years and older: a meta-analysis. Lancet. 2007;370:657–666. doi: 10.1016/S0140-6736(07)61342-7. [DOI] [PubMed] [Google Scholar]
- 39.Riggs B.L., Hartmann L.C. Selective estrogen-receptor modulators – mechanisms of action and application to clinical practice. N. Engl. J. Med. 2003;348:618–629. doi: 10.1056/NEJMra022219. [DOI] [PubMed] [Google Scholar]
- 40.Cosman F., Crittenden D.B., Adachi J.D., Binkley N., Czerwinski E., Ferrari S., Hofbauer L.C., Lau E., Lewiecki E.M., Miyauchi A., et al. Romosozumab Treatment in Postmenopausal Women with Osteoporosis. N. Engl. J. Med. 2016;375:1532–1543. doi: 10.1056/NEJMoa1607948. [DOI] [PubMed] [Google Scholar]
- 41.Kearns A.E., Khosla S., Kostenuik P.J. Receptor activator of nuclear factor kappaB ligand and osteoprotegerin regulation of bone remodeling in health and disease. Endocr. Rev. 2008;29:155–192. doi: 10.1210/er.2007-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller P.D., Hattersley G., Riis B.J., Williams G.C., Lau E., Russo L.A., Alexandersen P., Zerbini C.A., Hu M.Y., Harris A.G., et al. ACTIVE Study Investigators Effect of Abaloparatide vs Placebo on New Vertebral Fractures in Postmenopausal Women With Osteoporosis: A Randomized Clinical Trial. JAMA. 2016;316:722–733. doi: 10.1001/jama.2016.11136. [DOI] [PubMed] [Google Scholar]
- 43.Tang Y., Tang W., Lin Y., Long J., Wang H., Liu L., Tian W. Combination of bone tissue engineering and BMP-2 gene transfection promotes bone healing in osteoporotic rats. Cell Biol. Int. 2008;32:1150–1157. doi: 10.1016/j.cellbi.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 44.Tang S., Xie Z., Wang P., Li J., Wang S., Liu W., Li M., Wu X., Su H., Cen S., et al. LncRNA-OG Promotes the Osteogenic Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells Under the Regulation of hnRNPK. Stem Cells. 2019;37:270–283. doi: 10.1002/stem.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., Wang P., Xie Z., Wang S., Cen S., Li M., Liu W., Tang S., Ye G., Zheng G., et al. TRAF4 positively regulates the osteogenic differentiation of mesenchymal stem cells by acting as an E3 ubiquitin ligase to degrade Smurf2. Cell Death Differ. 2019;26:2652–2666. doi: 10.1038/s41418-019-0328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu W., Wang P., Xie Z., Wang S., Ma M., Li J., Li M., Cen S., Tang S., Zheng G., et al. Abnormal inhibition of osteoclastogenesis by mesenchymal stem cells through the miR-4284/CXCL5 axis in ankylosing spondylitis. Cell Death Dis. 2019;10:188. doi: 10.1038/s41419-019-1448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.