Abstract
Temozolomide (TMZ) resistance is an important cause of clinical treatment failure and poor prognosis in gliomas. Increasing evidence indicates that cancer-derived exosomes contribute to chemoresistance; however, the specific contribution of glioma-derived exosomes remains unclear. The aim of this study was to explore the role and underlying mechanisms of exosomal macrophage migration inhibitory factor (MIF) on TMZ resistance in gliomas. We first demonstrated that MIF was upregulated in the exosomes of TMZ-resistant cells, engendering the transfer of TMZ resistance to sensitive cells. Our results indicated that exosomal MIF conferred TMZ resistance to sensitive cells through the enhancement of cell proliferation and the repression of cell apoptosis upon TMZ exposure. MIF knockdown enhanced TMZ sensitivity in resistant glioma cells by upregulating Metalloproteinase Inhibitor 3 (TIMP3) and subsequently suppressing the PI3K/AKT signaling pathway. Additionally, exosomal MIF promoted tumor growth and TMZ resistance of glioma cells in vivo, while IOS-1 (MIF inhibitor) promotes glioma TMZ sensitive in vivo. Taken together, our study demonstrated that exosome-mediated transfer of MIF enhanced TMZ resistance in glioma through downregulating TIMP3 and further activating the PI3K/AKT signaling pathway, highlighting a prognostic biomarker and promising therapeutic target for TMZ treatment in gliomas.
Keywords: glioma, temozolomide resistance, exosomes, MIF, TIMP3
Graphical abstract
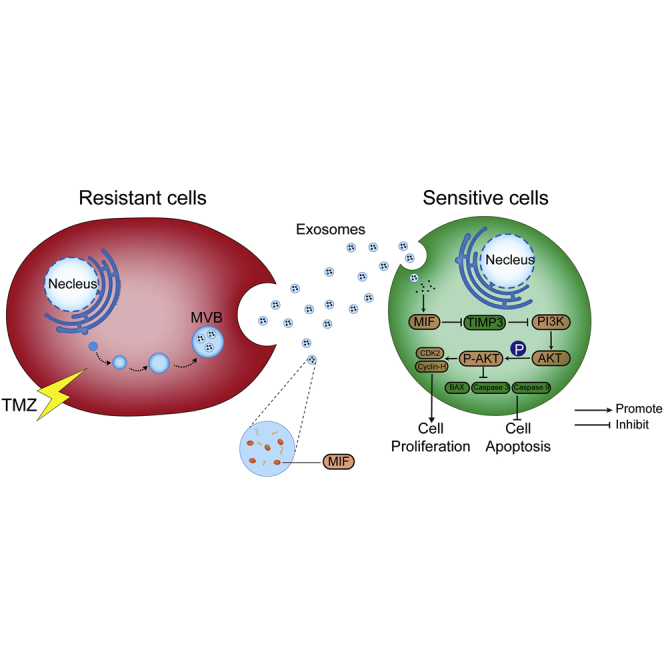
Exosomal MIF derived from TMZ-resistant cells can transfer chemoresistance character to sensitive glioma cells by downregulating TIMP3, activating the PI3K/AKT signaling pathway. ISO-1 (MIF inhibitor) is verified to be a promising treatment for TMZ resistance gliomas in animal models because it could enhance the TMZ sensitivity by inhibiting MIF.
Introduction
Glioma is the most common intracerebral tumor with high morbidity and mortality.1 The current standard glioma treatments include: surgical resection, radiotherapy, and chemotherapy.2 Despite these approaches, most of the high-grade glioma patients suffered tumor recurrence, and the majority of them showed temozolomide (TMZ) resistance.3 Although TMZ is still the standard first-line chemotherapy, drug resistance significantly limits the overall prognosis of glioma patients.4,5 Therefore, it is crucial to research the molecular mechanisms of TMZ resistance and further explore feasible therapy targets.
Exosomes, which can be secreted by a variety of cells, are the 40–150 nm size extracellular vesicle (EVS) with lipid bilayer membranes.6, 7, 8 Tumor-derived exosomes are rich in bioactive molecules, including proteins, nucleic acids, lipids, and various molecules.9 Accumulating evidence has shown that exosomes released by highly malignant cancer cells can be transferred to low grade cancer cells and regulate their growth, metastasis, and chemoresistance.10, 11, 12, 13, 14, 15, 16 Recently, ever more attention has been paid to their potential mechanisms in cancer chemoresistance. For example, exosomes of glioblastoma cells containing long noncoding RNA SBF2-AS1 can induce TMZ resistance in recipient cells.17 Additionally, reactive astrocyte-derived exosomes confer TMZ resistance phenotype to glioma cells via transfer of O6-methylguanine-DNA methyltransferase (MGMT) mRNA.18 Hence, exosomes have emerged as a new model for research into tumor chemoresistance.
Macrophage migration inhibitory factor (MIF; also known as L-dopachrome tautomerase) is a proinflammatory multifunctional cytokine that was originally discovered in 1966.19 Previous studies have shown that MIF overexpressed in many tumors.20,21 Recent evidence shows that MIF is involved in tumor chemoresistance.22,23 As a secretory cytokine, exosomal MIF was highly expressed in lung cancer patients’ plasma.24 Another study found that MIF was highly expressed in pancreatic ductal adenocarcinomas (PDAC) derived exosomes, and MIF blockade prevented liver pre-metastatic niche formation and metastasis.24,25 However, whether exosomal MIF contributes to TMZ resistance in glioma remains to be elucidated. Here, we aim to explore whether MIF could be transmitted by exosomes and involved in TMZ resistance of glioma.
We have dedicated this study to explore the molecular therapy targets of chemoresistance in glioma in our previous studies.26, 27, 28 In this study, we found that MIF was upregulated in U87TR and U251TR (TMZ-resistant) cells, and our in vitro and in vivo results showed that exosomal MIF derived from TMZ-resistant cells can transfer chemoresistance character to sensitive glioma cells by downregulating TIMP3, further activating the PI3K/AKT signaling pathway. Applying ISO-1 (MIF inhibitor) is verified to be a promising treatment strategy for TMZ resistance in glioma animal models, because it could enhance the TMZ sensitivity by inhibiting MIF.
Results
MIF upregulation is associated with poor prognosis in gliomas
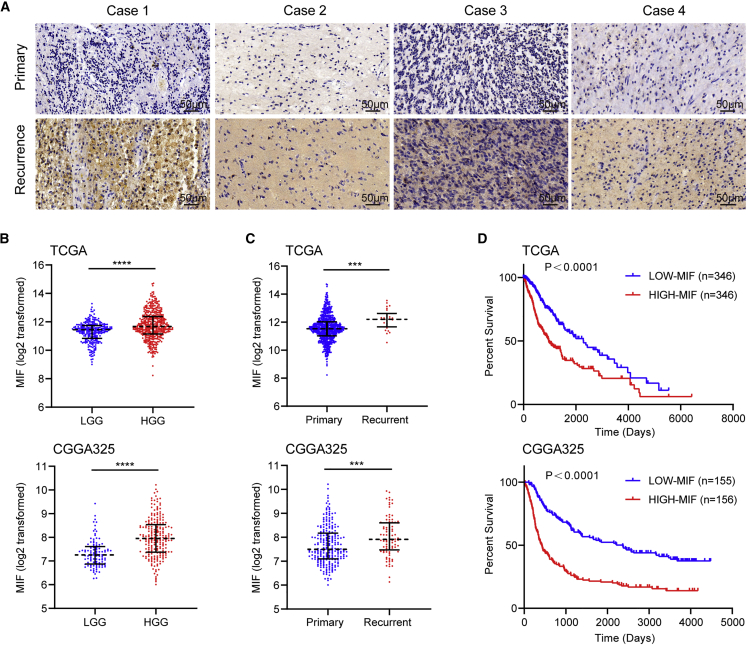
In this research, we demonstrated that MIF was upregulated in higher grade gliomas and recurrent patients by immunohistochemistry (IHC) staining (Figure 1A). Next, we analyzed the mRNA expression data of gliomas in The Cancer Genome Atlas (TCGA) and The Chinese Glioma Genome Atlas (CGGA) cohorts, revealing that MIF was highly expressed in high grade gliomas and recurrent patients. These findings were in accord with our study (Figures 1B and 1C). Survival curves of glioma samples from TCGA and CGGA analyses showed that patients with high MIF expression levels had a worse prognosis (Figure 1D). These results indicated that high expression of MIF was associated with poor prognosis in glioma patients.
Figure 1.
MIF upregulation is associated with poor prognosis in gliomas
(A) MIF was upregulated in 4 typical recurrent glioma patients by IHC staining (scale bars, 50 μm). (B–D) TCGA and CGGA database analysis showed that MIF expression level was positively correlated with glioma grade (B) and recurrence (C), but negatively correlated with prognosis (D). ∗∗∗p<0.001, ∗∗∗∗p<0.0001.
Highly expressed MIF promotes TMZ resistance in glioma cells
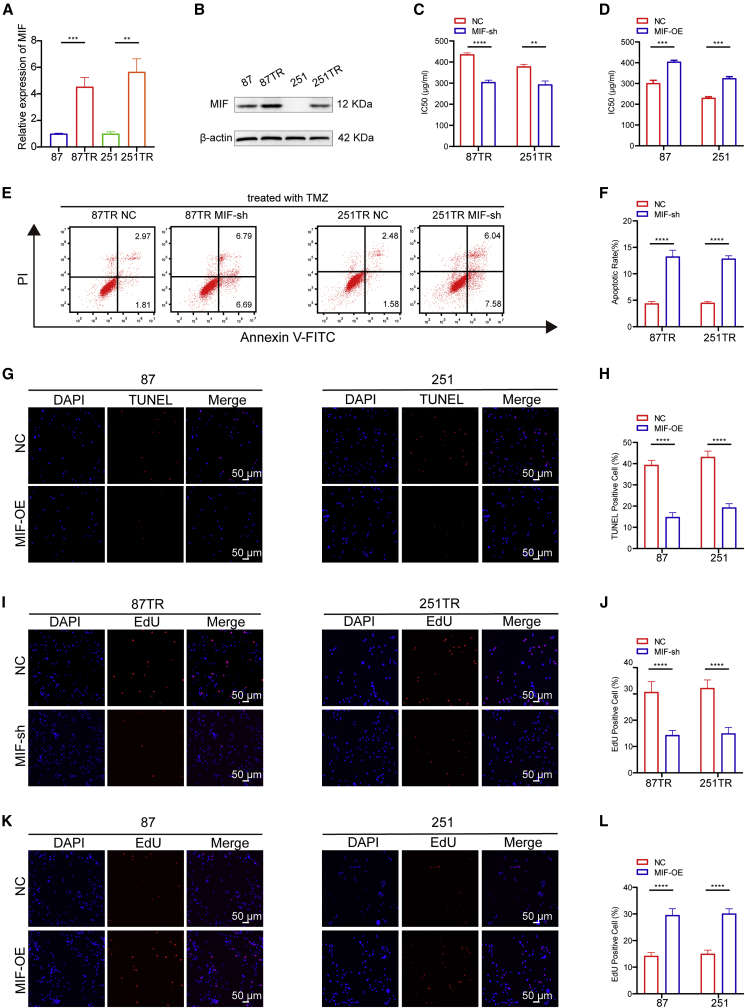
Our group had successfully established two TMZ-resistant glioma cell lines through repetitive exposure to increasing TMZ concentrations in vitro,29 named U87TR and U251TR (TMZ-resistant) cells, which had a higher IC50 (half maximal inhibitory concentration) value than U87TS and U251TS (TMZ-sensitive) cells (Figure S1A).26, 27, 28, 29 qRT-PCR and western blot results found that TR cells had a higher MIF expression level than TS cells (Figures 2A and 2B). To assess the role of MIF in TMZ resistance of glioma, we transfected TR cells with MIF short hairpin RNA (shRNA) lentiviral vectors to knock down MIF expression and confirmed its efficiency by qRT-PCR (Figure S1B). Meanwhile, MIF overexpressed (OE) lentiviral vectors were transfected into TS cells and its overexpression efficiency was confirmed by qRT-PCR assay (Figure S1C). Cell counting kit-8 (CCK-8) assay further showed that knockdown of MIF significantly decreased IC50 values in TR cells (Figure 2C). In contrast, overexpressed MIF significantly increased IC50 values in TS cells (Figure 2D). These results demonstrated that highly expressed MIF promotes TMZ chemoresistance in glioma cells. Furthermore, the flow cytometry (FCM) showed that loss of MIF significantly increased cell apoptosis upon TMZ (50 μg/mL) treatment in TR cells (Figures 2E and 2F). Whereas TUNEL assay showed that overexpression of MIF decreased apoptosis in TS cells (Figures 2G and 2H). The percentage of the EdU-positive cells were significantly decreased in TR-MIF-sh groups than in TR-NC (negative control) groups. On the other hand, the percentage of the EdU-positive cells was significantly increased in TS-MIF-OE groups than in TS-NC groups (Figures 2I–2L). These data suggest that MIF confers TMZ chemoresistance, enhances cell proliferation, and inhibits cell apoptosis in TMZ-resistant cells.
Figure 2.
Highly expressed MIF promotes TMZ resistance in glioma cells
(A and B) qRT-PCR and western blot showed that the expression of MIF in U87TR/251TR cells was significantly upregulated compared with U87/251 cells. (C and D) CCK-8 showed that the IC50 values were significantly deceased after MIF knockdown in TR cells. In contrast, overexpressed MIF significantly increased the IC50 values in TS cells. (E and F) The flow cytometry (FCM) showed that loss of MIF in TR cells significantly increased cell apoptosis upon TMZ treatment for 48 h (50 μg/mL). (G and H) On the contrary, TUNEL assay showed that overexpression of MIF may cause apoptosis rates to decrease in TS cells. (I–L) The percentage of the EdU-positive cells were significantly decreased in TR-MIF-sh groups than in TR-NC groups. On the other hand, the percentage of the EdU-positive cells were significantly increased in TS-MIF-OE groups than in TS-NC groups. ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001.
Exosomal MIF transfers from TR cells to TS cells
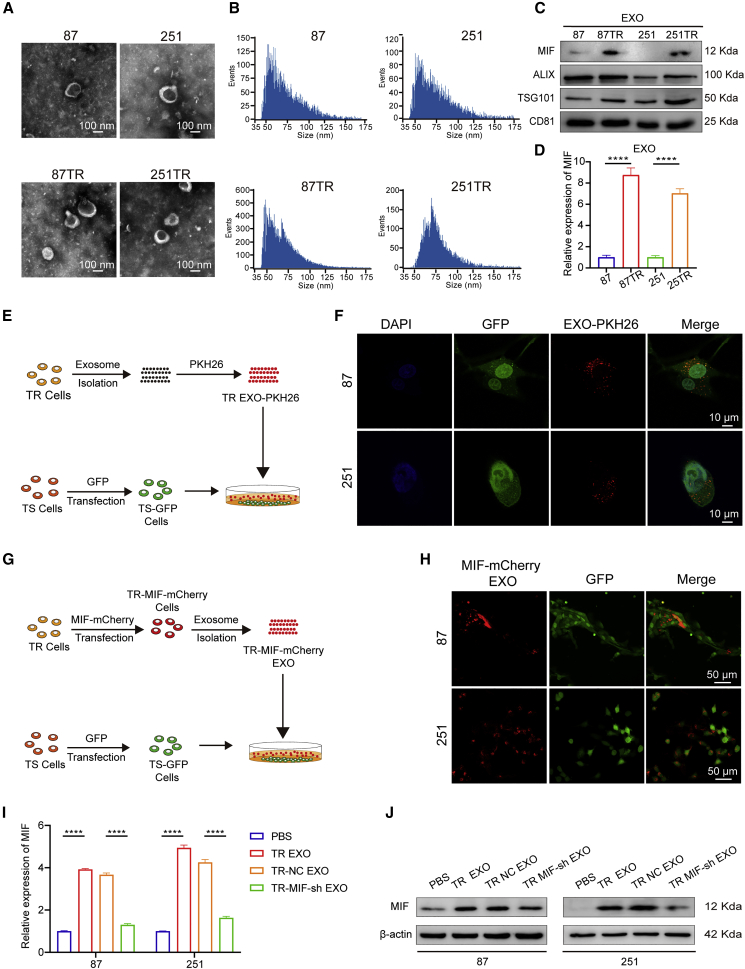
We performed the differential expressed genes (DEGs) analysis of MIF in TCGA database, the cellular component (CC) analysis indicated that the DEGs were enriched in extracellular exosome (Figure S1A). In addition, enzyme-linked immunosorbent assay showed that TR cells secrete more MIF than TS cells (Figure S1B). Then, exosomes were isolated from TR and TS cells’ supernatant. The exosomes derived from glioma cells presented as 40–150 nm round shape vesicles with bilayer membranes, which were confirmed by high-resolution transmission electron microscopy (TEM; Figure 3A). The nano-flow cytometer (nFCM) further identified that the similar predominant size of these vesicles. In addition, TR cells secreted more exosomes than TS cells (Figure 3B). The exosome markers like ALIX, CD81, and TSG101 were detected in these vesicles (Figure 3C). qRT-PCR and western blot showed that the exosomes secreted by TR cells (TR EXO) expressed more MIF than those from TS cells (Figures 3C and 3D).
Figure 3.
Exosomal MIF transfers from TR cells to TMZ-sensitive cells
(A) The representative micrograph of round-shaped vesicles derived from TMZ-resistant and TMZ-sensitive cells by TEM (scale bars, 100 nm). (B) The nFCM further identified that the similar predominant size of these vesicles. (C) Western blot analysis showed the presence of MIF, ALIX, TSG101, and CD81 in exosomes. (D) qRT-PCR showed that the exosomes secreted by TR cells (TR EXO) expressed more MIF than those from TS cells. (E and F) Schematic diagram: TR exosomes were labeled with PKH26 and co-incubated with TS-GFP cells. Confocal photography found that exosomes could be successfully ingested by TS cells (scale bars, 10 μm). (G and H) Schematic diagram: MIF in TR cells was labeled with mCherry, and then exosomes were extracted and added into the supernatant of TS-GFP cells. After 24 h of coculture, we observed strong red fluorescence in the cytoplasm of TS-GFP cells (scale bars, 50 μm). (I and J) qRT-PCR and western blot were used to verify the expression level of MIF in TS cells after they were co-incubated with PBS, TR EXO, TR-NC EXO, or TR-MIF-sh EXO separately. ∗∗∗∗p<0.0001.
Accumulating evidence has shown that MIF can be packaged into exosomes derived from tumor cells and promotes tumor progression.24,25 Besides, several studies have demonstrated that exosomal RNA derived from resistant cells transfer chemoresistance characteristics to sensitive cells.30, 31, 32 Combining literature and our results, we speculated that MIF could transfer TMZ-resistant character via exosomes. To visualize exosome transfer, we labeled TR EXO with PKH26 (TR EXO-PKH26). TS cells transfected with lentivirus vectors carrying green fluorescent protein (TS-GFP) were incubated with the TR EXO-PKH26 for 3 h (Figure 3E). The confocal microscope captured the PKH26-labeled exosomes that were internalized by the recipient TS cells (Figure 3F). Then we validated whether the transfer of MIF occurs via exosomes. Exosomes, which were isolated from TR cells that transfected with the MIF-mCherry lentiviral activation vectors (TR MIF-mCherry), were added into culture medium of TS-GFP cells (Figure 3G). After 24 h of coculture, we observed strong red fluorescence in the cytoplasm of TS-GFP cells (Figure 3H). Furthermore, after co-incubation with TR EXO or TR-MIF-sh EXO separately, the cellular level of MIF was corresponding upregulated or downregulated in recipient cells (Figures 3I and 3J). These data demonstrated that MIF could be directly transferred from TR cells to sensitive cells through exosomes.
Exosomal MIF confers TMZ resistance via promotion of proliferation and inhibition of apoptosis in recipient cells
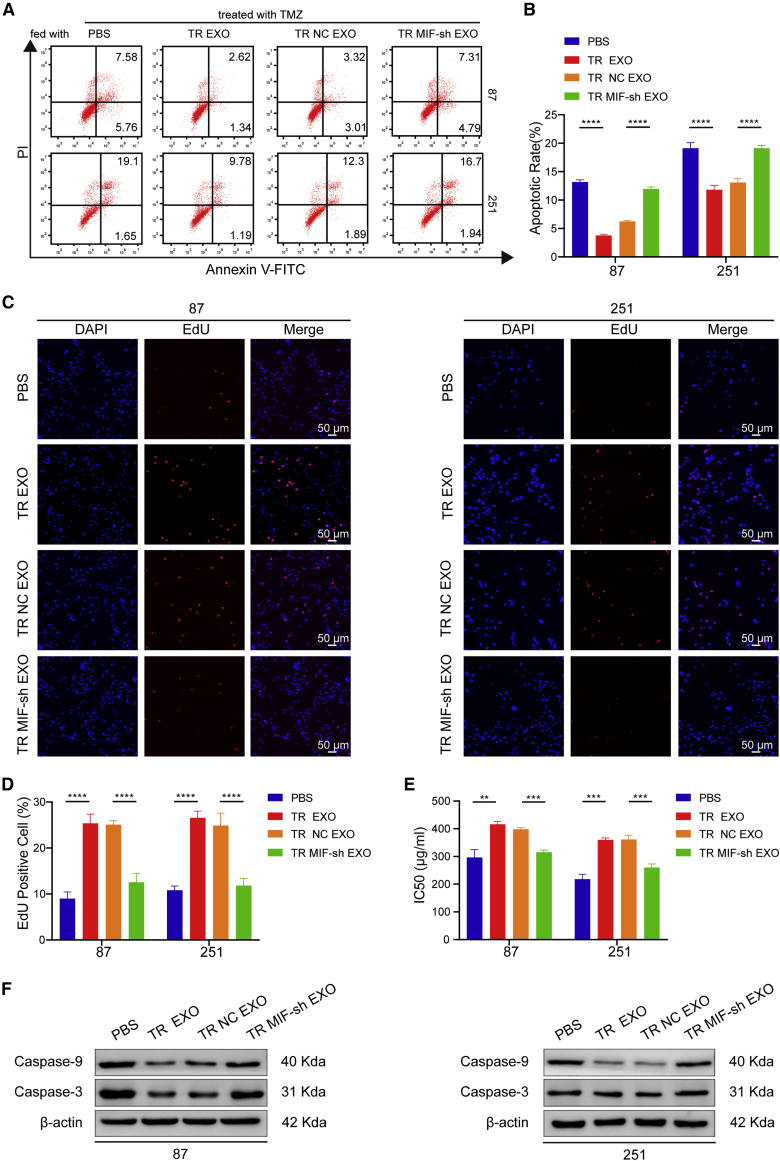
We next investigated whether exosomal MIF could confer TMZ resistance phenotype to recipient TS glioma cells. TS cells were incubated with PBS, TR EXO, TR-NC EXO, TR-MIF-sh EXO for 24 h, and then followed with TMZ treatment for 48 h. The FCM results showed that the apoptosis rates in TR EXO and TR-NC EXO groups were significantly lower than in PBS and TR-MIF-sh EXO groups in TS cells (Figures 4A and 4B). Similarly, EdU assay demonstrated that TR EXO and TR-NC EXO groups had higher cell proliferation than PBS and TR-MIF-sh EXO groups (Figures 4C and 4D). Meanwhile, TR EXO and TR-NC EXO significantly enhance the TMZ-resistant character after co-culture with TS cells (Figure 4E). In addition, TR EXO and TR-NC EXO obviously decrease the apoptosis-related protein (caspase-3 and caspase-9) expression level in recipient TS cells (Figure 4F). Together, these results suggested that exosomal MIF conferred TMZ resistance to recipient TS cells by promoting cell proliferation and inhibiting cell apoptosis.
Figure 4.
Exosomal MIF confers TMZ resistance via promotion of proliferation and inhibition of apoptosis in recipient cells
(A and B) The FCM results showed that the apoptosis rates in TR EXO and TR-NC EXO groups were significantly lower than in PBS and TR-MIF-sh EXO groups (TMZ = 50 μg/mL). (C) EdU assay demonstrated that TR EXO and TR-NC EXO groups had a higher cell proliferation rate than PBS and TR-MIF-sh EXO groups (scale bars, 50 μm). (D) EdU statistics. (E) CCK-8 showed that TR EXO and TR-NC EXO significantly enhanced the TMZ resistant characteristic in TS cells. (F) Western blot for caspase-3 and caspase-9 expression in the different treated glioma cells. ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001.
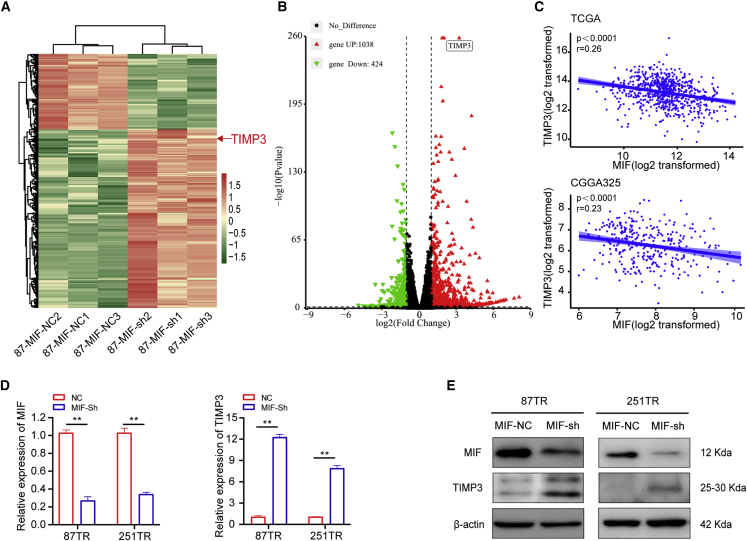
TIMP3 is a downstream target of MIF in glioma cells
To ascertain whether the latent mechanism of MIF induces TMZ resistance in glioma cells, we performed RNA sequencing (RNA-seq) analysis to predict the possible target genes. Tissue inhibitors of metalloproteinases-3 (TIMP3) was the most top significantly upregulated genes in U87 MIF-sh cells, and it was predicted to be the highly possible downstream target of MIF in glioma cells (Figures 5A and 5B). TIMP3, a member of the TIMP family, is a 24-kDa secreted glycoprotein and can suppress cell proliferation and enhance chemosensitivity by activating apoptosis-related signal pathways.33, 34, 35, 36, 37 In addition, the analysis of TCGA and CGGA cohorts showed significant negative correlation between MIF and TIMP3 (Figure 5C). Further experiments verified that TIMP3 was upregulated at both mRNA and proteins levels after knockdown of MIF in TR cells (Figures 5D and 5E). These results indicated that TIMP3 served as a target gene of MIF in glioma cells.
Figure 5.
TIMP3 is a downstream target of MIF in glioma cells
(A and B) After knocking down MIF, sequencing analysis revealed that TIMP3 was significantly upregulated. (C) TCGA and CGGA database analysis further confirmed the negative correlation between MIF and TIMP3. (D and E) qRT-PCR and western blot verified that TIMP3 was upregulated at both mRNA and proteins levels after knocking down MIF in TR cells. ∗∗p<0.01.
The PI3K/AKT signaling pathway is involved in MIF-regulated TIMP3 in TMZ resistance
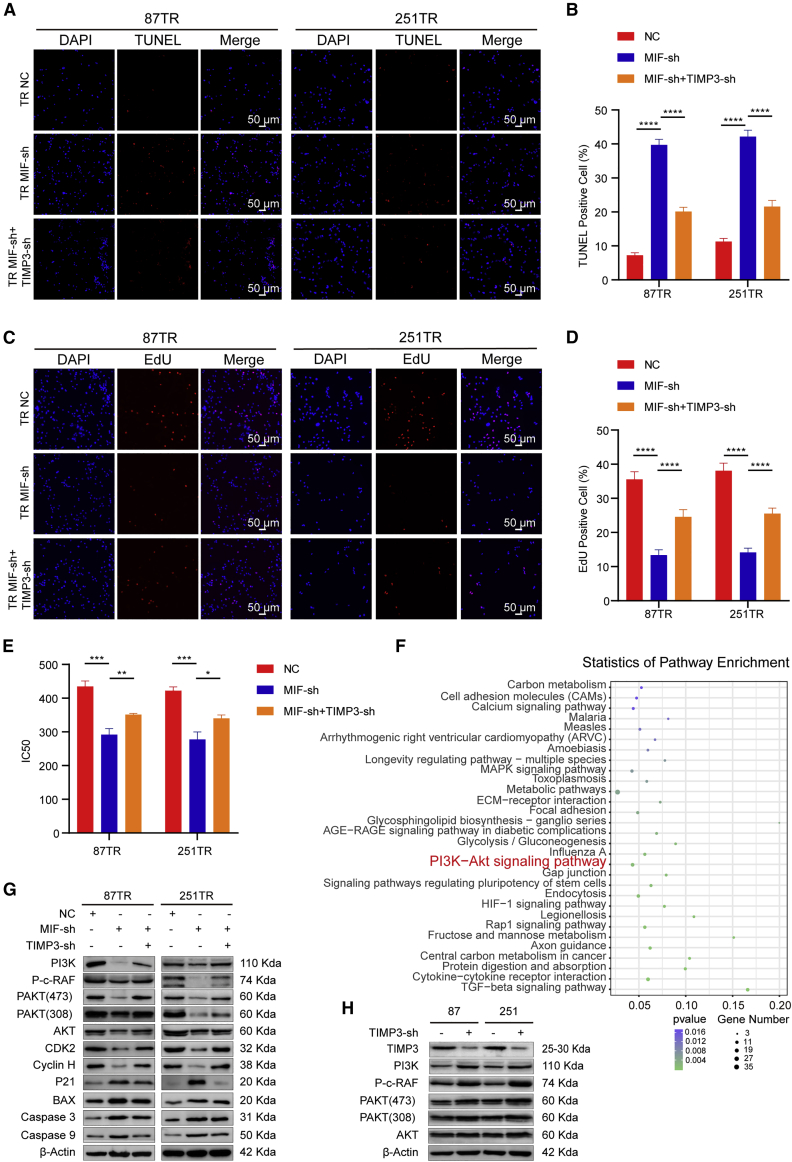
To further explore whether TIMP3 was involved in TMZ resistance mediated by MIF, we co-transfected TIMP3 shRNA lentiviral vectors into the TR-MIF-sh cells. TUNEL assay showed that TIMP3 knockdown can markedly inhibit cell apoptosis in TR-MIF-sh cells (Figures 6A and 6B). In addition, the EdU assay revealed that the percentage of the proliferated cells was increased after the knockdown of TIMP3 in the TR-MIF-sh cells (Figures 6C and 6D). The rescue experiments exhibited that knockdown of TIMP3 could reverse the decrease in IC50 values caused by the knocking down of MIF (Figure 6E). To identify the potential mechanism of MIF/TIMP3 axis to induce TMZ resistance in glioma cells, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of the DEGs. In statistics of these top 30 KEGG pathways (Figure 6F), the PI3K/Akt signaling pathway was proved to play a pivotal role in cancer progression and chemoresistance, and applying PI3K inhibitor may be a promising therapy strategy.38, 39, 40, 41, 42 Based on this evidence, we hypothesized that the MIF/TIMP3 axis induced TMZ resistance via regulating the PI3K/Akt signaling pathway. Western blot assay showed that knockdown of MIF expression suppressed the activation of the PI3K/Akt signaling pathway and significantly decreased the expression level of downstream target genes, such as PI3K, AKT, PAKT, Phospho-c-Raf, Cyclin H, and CDK2, which are related to cell proliferation. On the other hand, knockdown of MIF increased the expression level of downstream target genes, such as P21, an antioncogene; the others including BAX, caspase-3, and caspase-9, which are related to cell apoptosis. Furthermore, knockdown of TIMP3 in TR-MIF-sh cells can reverse the changes of these proteins (Figure 6G). For the sake of verifying whether MIF/TIMP3 signaling could direct modulate PI3K/AKT, western blot analysis was done. TIMP3 was knocked down in TS cells because TIMP3 is highly expressed in TS cells (Figure S3A). Western blot showed that knockdown of TIMP3 activated the PI3K/Akt signaling pathway and significantly increased the expression level of downstream target genes, such as PI3K, AKT, PAKT, and Phospho-c-Raf (Figures 6H). Together, these results suggested that MIF regulated chemoresistance via TIMP3/PI3K/Akt axis.
Figure 6.
The PI3K/AKT signaling pathway is involved in MIF-regulating TIMP3 in TMZ resistance
(A and B) TUNEL analysis of TR-NC, MIF-Sh, MIF-Sh + TIMP3-Sh, after TMZ treatment for 48 h (50 μg/mL) (scale bars, 50 μm). (C) Cell proliferation in different groups (scale bars, 50 μm). (D) EdU statistics. (E) CCK-8 assay showed that knockdown of TIMP3 could reverse the decrease in IC50 values caused by knockdown of MIF. (F) KEGG pathway analyses of the DEGs showed that PI3K/Akt signaling pathway might play a pivotal role in gliomas progression. (G) The protein levels of PI3K/AKT signaling pathway-related genes in TR-NC, TR-MIF-sh, and TR-MIF-sh+TIMP3-sh transfected cells. (H) The protein levels of PI3K/AKT signaling pathway-related genes in TS-NC and TS-TIMP3-sh transfected cells. ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001.
Exosomal MIF induces TMZ resistance in vivo
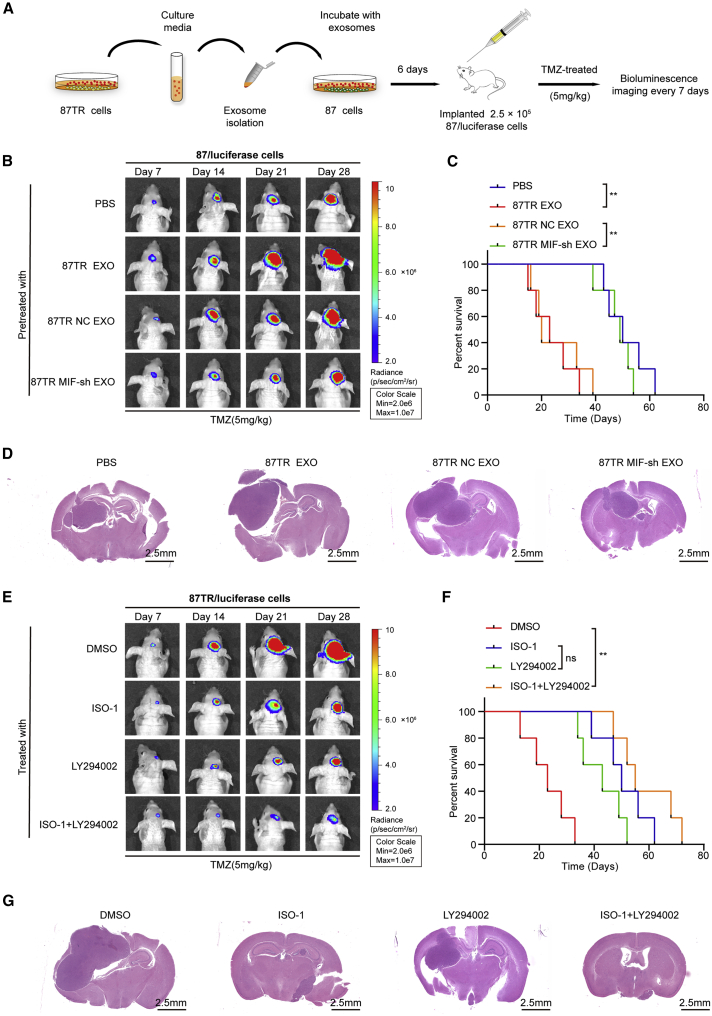
To investigate whether exosomal MIF regulated its target genes and induced TMZ resistance in vivo, we orthotopically implanted mice with 2.5 × 105 U87/luciferase cells following pre-treatment with PBS or exosomes derived from different cells (U87TR EXO, U87TR-NC EXO, or U87TR-MIF-sh EXO) for 6 days (Figure 7A). Bioluminescence imaging results indicated that, compared to the negative PBS control group, U87TR EXO pretreatment prominently promoted TMZ resistance (Figures 7B and 7D) and reduced the overall survival of tumor-bearing mice (Figure 7C). Moreover, in contrast to U87TR-MIF-sh EXO, TR-NC EXO pretreatment also promoted TMZ resistance (Figures 7B and 7D) and reduced the overall survival (Figure 7C). Together, these results suggested that exosomal MIF promoted TMZ resistance in vivo. To further assess the antitumor effect of combined ISO-1 and LY294002 (PI3K inhibitor) treatment in vivo, we treated mice harboring U87TR/luciferase cells with DMSO, ISO-1, LY294002, or ISO-1+LY294002. The results revealed that combined ISO-1 and LY294002 treatment prominently inhibited tumor growth and improved the overall survival of tumor-bearing mice in comparison with the control group (Figures 7E–7G). Together, these results strongly revealed that exosomal MIF conferred TMZ resistance in glioma in vivo and that MIF could be a potent therapy target for overcoming chemoresistance.
Figure 7.
Exosomal MIF induces TMZ resistance in vivo
(A) Schematic of the in vivo model: BALB/c mice were orthotopically injected intracranially with 2.5 × 105 U87/luciferase cells that were pre-incubated with PBS or 40 μg/mL exosomes from U87 TR, U87TR-NC, or U87TR-MIF-sh cells for 6 days (n = 5 per group). (B) Representative pseudocolor bioluminescence images of orthotopic tumors. (C) Kaplan-Meier survival curve of mice with intracranial xenografts derived from indicated exosomes-treated U87/luciferase cells. (D) Representative H&E staining for tumor cytostructures (scale bars, 2.5 mm). PBS: U87/luciferase cells pretreated with PBS, TR EXO: U87/luciferase cells pretreated with U87 TR cells-derived exosomes, TR NC EXO: U87/luciferase cells pretreated with the exosomes derived from U87TR-NC cells, TR MIF-sh EXO: U87/luciferase cells pretreated with the exosomes derived from U87TR-MIF-sh cells. (E–G) U87TR/luciferase cells were orthotopically implanted into the brain of BALB/c mice. DMSO, ISO, LY294002, or ISO+LY294002 (n = 5 per group) were injected (i.p.) into the mice twice per week for 28 days. (E) Representative pseudocolor bioluminescence images of orthotopic tumors. (F) Kaplan-Meier survival curve of mice with different treatment. (G) Representative H&E staining for tumor cytostructures (scale bars, 2.5 mm). ∗∗p<0.01.
Discussion
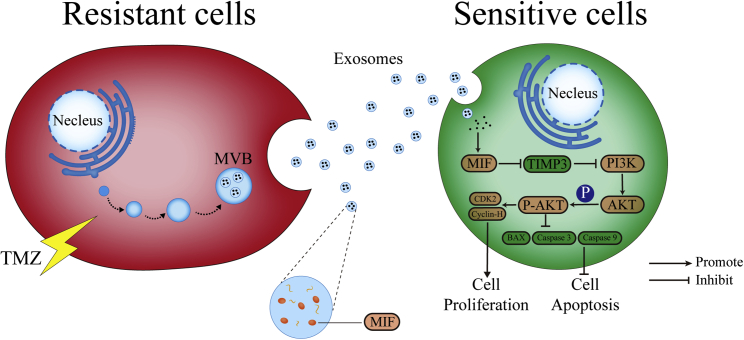
In this study, there are several major findings. First, MIF was overexpressed in TMZ-resistant glioma cells and was confirmed to play a significant role in TMZ chemoresistance. Second, TR-cell-derived exosomes were full enriched MIF and transferred TMZ resistance characteristic between glioma cells via cell apoptosis suppression and proliferation promotion. Third, MIF inhibitor ISO-1 could enhance the TMZ sensitivity in vivo. Therefore, our results exhibited that a novel mechanism that TR cells may transfer chemoresistance characteristic via exosomal MIF delivering (Figure 8).
Figure 8.
Schematic diagram of the potential roles of exosomal MIF in glioma TMZ resistance
The TR cells-secreted exosomes containing MIF can be taken up by the TS cells. Exosomal MIF suppresses apoptosis by downregulating TIMP3 and activating PI3K/Akt signaling pathway, resulting in enhanced cell proliferation, suppressed cell apoptosis, and promoted TMZ resistance of the recipient TS cells.
Emerging evidence showed that exosomes played a significant role in chemoresistance. Chen et al.43 found that exosomal UCA1 lncRNA enhanced cell proliferation and impaired the gefitinib-induced cell apoptosis. Yu et al.18 reported that exosomal MGMT mRNA, which transfers TMZ resistance to the recipient glioma cells, leading to TMZ resistance in these cells and recurrence.
In this study, we focused on MIF, which is a well-known gene but lack of sufficient alertness in the past, especially in drug resistance. Previous studies demonstrated that MIF expression was highly positively related to WHO grade and chemoresistance tumors.20, 21, 22, 23 As a secretory cytokine, exosomal MIF was found to be highly expressed in lung cancer and to take part in the development of pancreatic ductal adenocarcinomas liver metastasis.24,25 Our study showed that MIF was highly expressed in both TR cells and their exosomes. TR cell-derived exosomes transferred TMZ resistance characteristics between glioma cells via cell apoptosis suppression and proliferation promotion both in vivo and vitro, and this coincided with previous literature.
Jackson et al.44 reported that many mechanisms of preventing TIMP3 expression in diverse human cancers support that TIMP3 acts as a tumor suppressor. Han et al.37 reported that TIMP3 overexpression suppressed cell proliferation and enhanced cisplatin sensitivity by activating apoptosis-related signaling pathways. But, so far, the relationship between MIF and TIMP3 has not been reported yet. During knockdown of MIF in glioma cells, TIMP3 was obtained significantly upregulated by RNA-seq analysis in our study. Our further research demonstrated that TIMP3 could counteract the TMZ resistance character caused by MIF via suppressing cell proliferation and enhancing cell apoptosis.
A former study showed that MIF can lead to chemoresistance via enhancing tumor proliferation via the AKT signaling pathway, but IOS-1 can reverse the chemoresistance caused by MIF and promote chemotherapeutic sensitivity.45 Cheng et al.46 found that ISO-1 exerts anti-cancer effects on pancreatic cancer cells by inhibiting proliferation, migration, and invasion in vitro, and suppressed tumor growth in vivo. Considering the aim of our research was to verify a novel molecular mechanism, we demonstrated that ISO-1 could enhance TMZ sensitivity by suppressing the PI3K/AKT signaling pathway in vivo. On the other hand, according to our in vivo results, we have also found suboptimal efficacy of LY294002, the PI3K inhibitor monotherapy, which indicates that MIF is likely to act via other additional unknown mechanisms apart from PI3K/AKT pathway. According to statistics of pathway enrichment of our RNA-seq data, downregulating of MIF resulting in several pathway changed besides the PI3K-Akt signaling pathway, such as TGF-beta signaling pathway. Other features (e.g., metabolic pathways, calcium signaling) appear to be affected as well, while further research is needed to demonstrate whether these pathways are involved in TMZ resistance mediated by MIF. Combined ISO-1 (MIF inhibitor) and LY294002 (PI3K inhibitor) treatment has the most obvious anti-tumor effect, demonstrating that MIF may target the PI3K-Akt signaling pathways during the TMZ resistance process. However, the metabolic pathways also attracted our attention. When we knocked down MIF in GBM cells, the cell proliferation was significantly inhibited and the cell apoptosis was also promoted under TMZ treatment, which indicated that MIF may influence tumor cell growth and apoptosis process. As to the metabolic pathways, we believe that its differential enrichment is related to the intracellular energy uptake and exocrine process in GBM cells. It will be a good research focus in our further study. No matter how, our results identified that applying ISO-1 was a promising treatment strategy for TMZ resistance gliomas in animal models.
In future research, we will focus on the inducement and upstream mechanisms of MIF upregulation both in TR cells and TR cell-derived exosomes. Meanwhile, small molecule inhibitor of MIF, which is suitable for glioma patients, is needed to be further developed to improve the prognosis in TMZ-resistant patients.
Conclusions
In conclusion, our results demonstrate that MIF is delivered by packing into exosomes and enhances TMZ resistance in recipient glioma cells by inhibiting TIMP3, subsequently stimulating the PI3K/AKT signaling pathway. MIF inhibitor is a promising treatment for TMZ resistance, and it may bring hope in the future for TMZ-resistant patients. Taken together, our findings highlight a novel molecular mechanism for TMZ resistance in glioma and provide a new target for TMZ resistance treatment.
Materials and methods
Clinical samples and ethical approval
Glioma paraffin sections were obtained from glioma patients who underwent surgery in the First Affiliated Hospital of Shantou University Medical College. 4 typical recurrent glioma samples were selected. The project protocol was approved by the Ethics Committee of The First Affiliated Hospital of Shantou University Medical College and written informed consents were obtained from all patients enrolled in this study.
IHC staining
IHC staining was performed according to standard procedures. The paraffin-embedded glioma specimens were deparaffinized and dehydrated by a graded ethanol series following antigen retrieval by boiling citric acid buffer. Then the nonspecific antigens were blocked by 5% BSA. The permeabilization of sections was performed by incubating in 0.01% Triton X-100. These sections were incubated with the primary antibodies overnight at 4°C and then biotinylated secondary antibody (1:500 dilutions, Santa Cruz Biotechnology, USA) for 2 h at room temperature. The following primary antibodies were applied: MIF (Novus Biologicals, USA). The samples were then counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and observed by microscope (DM2500, Leica).
Cell lines
The human glioma cell lines U87 and U251 were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China), which was authenticated and tested for mycoplasma contamination. Our laboratory established and maintained two TR cell lines (U87TR and U251TR) as described previously.29 All cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; Hyclone, Logan, UT, USA), penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37°C in 5% CO2 humidified air incubator (Thermo Scientific, Waltham, MA, USA).
Cell transfection
For lentiviral transfection, cells were seeded at 50% density confluence in 6-well plates and cultured in DMEM with 10% FBS overnight. Then cells were cultured in a 1 mL mixture of OPTI-MEM (Invitrogen, USA) and polybrene (5 μg/mL, Genechem, Shanghai, China). Control shRNA lentiviral vectors, MIF shRNA lentiviral vectors, TIMP3 shRNA lentiviral vectors, control lentiviral activation vectors, and MIF-mCherry lentiviral activation vectors were added in the culture medium, respectively. All the lentiviral vectors were obtained from Genechem (Genechem, Shanghai, China). After 24 h of transfection, cells were cultured in DMEM with 10% FBS. Transfected cells were selected by puromycin (8 μg/mL, sc-108071, Santa Cruz) for 1 week. The target gene expression level was detected by qRT-PCR or western blot.
Enzyme-linked immunosorbent assay
The culture medium of glioma cells was used to determine secreted MIF levels using a MIF enzyme-linked immunosorbent assay kit (R&D Systems) according to the manufacturer’s instructions. The absorbance at 562 nm was detected by Ultra Multifunctional Microplate Reader (Tecan, Switzerland), and the values were calculated by Graphpad Prism 8 software.
Isolation and identification of exosomes
Exosomes of FBS were depleted by ultracentrifugation at 120,000 × g for 6 h at 4°C. The glioma cell lines were cultured in the normal medium until 70%–80% confluency, and thereafter, cultured in DMEM supplemented with 10% exosome-depleted FBS for 48 h. Exosomes of glioma cells were obtained according to the differential centrifugation as previously described.47 Briefly, 40 mL of conditioned medium was centrifuged at 300 × g for 10 min, 2,000 × g for 10 min and then 10,000 × g for 30 min at 4°C to remove cell debris. Next, exosomes were pelleted by ultracentrifugation at 100,000 × g for 70 min, resuspended in 100 μL PBS, and stored at −80°C. Exosomes were visualized by high-resolution TEM and confirmed by the expression levels of CD81, TSG101, and Alix, which are specific proteins of exosomes. The concentration and size distribution of exosomes were detected by nFCM as previously described,48 and then exosomes were ready for RNA/protein extraction or cell treatment.
TEM
Exosomes were suspended in 100 μL PBS and were fixed with 5% glutaraldehyde at incubation temperature and then maintained at 4°C until TEM analysis. According to the TEM sample preparation procedure, we placed a drop of exosome sample on a carbon-coated copper grid and immersed it in 2% phosphotungstic acid solution (pH 7.0) for 30 s. The preparations were observed with a transmission electron microscope (Tecnai G2 Spirit Bio TWIN; FEI, USA).
RNA isolation, reverse transcription, and quantitative real-time PCR
Total RNA from cells or exosomes was obtained by RNeasy Mini kit (QIAGEN, USA) according to the manufacturer’s protocol. The yield and quality of the RNA were measured through the absorbance at 260 and 280 nm. The cDNA was amplified by qRT-PCR using SYBR GREEN PCR Master Mix (Takara Bio, Shiga, Japan) on a 7900 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). GAPDH was used as an internal reference. The quantitative PCR primers are listed in Table S1. The relative quantification values of genes were calculated by the ΔΔCt method with at least 3 independent experiments.
Exosome uptake
The glioma cell derived exosomes were labeled with PKH26 Red Fluorescent Cell Linker Kit (Sigma-Aldrich, USA) according to the manufacturer’s protocol. Briefly, exosomes were resuspended by a mixture of 1 μL PKH26 and 500 μL of Diluent C and incubated for 5 min. For binding excess PKH26, the equal volume of 10% bovine serum albumin (BSA) was added and incubated for 1 min. Then the labeled exosomes were obtained by ultracentrifugation at 120,000 × g for 2 h at 4°C. And the exosomes were resuspended in complete medium and incubated with glioma cells (4 × 105) for 3 h at 37°C. Then the cells were fixed with 4% paraformaldehyde (PFA) and stained by DAPI. Cellular internalization of PKH26-labeled exosomes in glioma cells were observed by a confocal microscopy (Leica, Germany).
Protein extraction and western blot analysis
Total proteins were extracted from cells using Whole Cell Lysis Assay (KeyGEN BioTECH, China) on ice and quantified by bicinchoninic acid protein assay kit (Beyotime, Shanghai, China). The western blot was performed according to standard procedures. The following primary antibodies were applied: MIF (Novus Biologicals, USA), TIMP3, PI3K, CD81, TSG101, ALIX (Abcam, USA), caspase-3, caspase-9, P-c-Raf, AKT, P-Akt (Ser473), P-Akt (Thr308), β-actin (Cell Signaling Technology, USA), BAX (Bioss ANTIBODIES, USA), P21 (Proteintech, USA), CDK2 (ImmunoWay, USA), and Cyclin H (ZenBio Science, China). The antibody information is listed in Table S2. After primary antibody incubation, the membranes were incubated with HRP-labeled goat anti-rabbit or goat anti-mouse secondary antibodies (Cell Signaling Technology, USA). The bands were captured by chemiluminescence (Biosciences, Foster City, CA, USA) through ECL detection system. Finally, the protein expression was analyzed by the software of ImageJ.
TMZ chemosensitivity and cell viability assay
The viability of cells was detected by the CCK-8 (KeyGEN BioTECH, China) according to the manufacturer’s instructions. Different glioma cells were seeded in 96-well plates and treated with TMZ at different concentrations (0 μg/mL, 100 μg/mL, 200 μg/mL, 300 μg/mL, 400 μg/mL, 500 μg/mL) for 48 h. Then the cells were incubated with fresh medium containing 10% CCK-8 solution for 2 h, the absorbance at 450 nm was detected by Ultra Multifunctional Microplate Reader (Tecan, Switzerland), and the IC50 values of cells were calculated by Graphpad Prism 8 software. For cell viability assay, glioma cells were treated with 50 μg/mL TMZ for 24 h, 48 h, 72 h, and 96 h, and then the absorbance at 450 nm was measured after incubating with a mixture of fresh medium and 10% CCK-8 solution for 2 h.
EdU staining assay
Glioma cell proliferation was detected by Cell-Light EdU DNA cell proliferation kit (RiboBio, Guangzhou, China) according to the manufacturer’s protocol. Cells were seeded in a cover glass of 6-well plates and incubated with 10 μM EdU reagent for 2 h at 37°C. Then the cells were fixed with 4% formaldehyde for 30 min. Followed by washing, cells were incubated with Apollo for 30 min, and staining nucleus by Hoechst 33342. The EdU-positive cells were visualized by a confocal microscopy (Leica, Germany). The ratio of cell proliferation was calculated with Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA).
Flow cytometric analysis of cell apoptosis
The apoptosis rate of glioma cells induced by TMZ (50 μg/mL) was detected by Annexin-V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (KeyGEN BioTECH, China) according to the manufacturer’s instructions. Then the apoptotic rate was analyzed by fluorescence-activated cell sorting (FACS) cytometry (BD Biosciences, Franklin, NJ, USA).
TUNEL assay
TUNEL assay was performed by the one-step TUNEL apoptosis assay kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions. Glioma cells were fixed in 4% paraformaldehyde for 30 min, and PBS containing 0.1% Triton X-100 was added for 2 min in the ice bath. Then cells were incubated with TUNEL reaction mixture for 1 h at 37°C in a humidity chamber. After incubation, cells were washed by PBS twice and the nuclei were stained with DAPI. Finally, cell smears were placed on coverslips with anti-fade fluorescence mounting medium. Images were captured using a confocal microscopy (Leica, Germany).
RNA-seq and bioinformatics analysis
Total RNA was isolated from U87 cells transfected with control shRNA or MIF shRNA lentiviral vectors. The RNA integrity of each sample evaluated by the Agilent 2200 TapeStation (Agilent Technologies, USA) was more than 7.0. In brief, rRNAs were depleted by EpicenterRibo-Zero rRNA Removal Kit (Illumina, USA) and then total RNA were fragmented to approximately 200 bp. Subsequently, the RNAs were subjected to cDNA synthesis, adaptor ligation, and enrichment according to the manufacturer’s protocol of NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA). After they were evaluated by Agilent 2200 TapeStation and Qubit2.0 (Life Technologies, USA), the library products were pair-end sequenced (PE150, 2 × 150 bp) using HiSeq3000. After removal of reads with ploy-N, adaptor, and low-quality raw data, the clean reads were aligned to the human reference genome hg19 with default parameters using HISAT2. Then the aligned short reads were converted into read counts of each gene model by HTSeq.
We utilized the publicly available TCGA database from Xena platform (https://xenabrowser.net/) and CGGA mRNaseq_325 database (http://www.cgga.org.cn/) for all examinations of gene expression. Differences of MIF expression in different subtypes and grade were examined by one-way ANOVA, following by Bonferroni correction for the multiple comparison. All patients were divided into MIF-upregulated and MIF-downregulated groups based on MIF expression using median as split points. Kaplan-Meier method and log-rank test were used for survival curves analysis. Correlation between MIF and other genes was determined by Pearson correlation coefficients. The DEGs analysis, gene ontology (GO) enrichment analysis, and KEGG pathway enrichment analysis were performed by R language (version 3.6.1). Gene set enrichment analysis (GSEA) was performed by GSEA software (version 4.1.0). The criteria of DEG were fold change > 2 and adjusted p value < 0.05. p value < 0.05 was also used as the threshold for significant enrichment of the gene sets.
Xenograft studies and treatment experiments
Female nude mice (4–5 weeks old) were purchased from the Laboratory Animal Center of Guangdong (Guangzhou, China) after obtaining approval from Animal Ethical Committee of Zhujiang Hospital. U87 and U87TR cells expressed the luciferase reporter (U87/luciferase or U87TR/luciferase cells) were established for the study in vivo. TMZ, LY294002 (PI3K inhibitor, MedChem Express), and ISO-1 (MIF inhibitor, MedChem Express) were used for treatment. These mice were raised in a specific-pathogen-free facility and randomly divided into 8 groups (n = 5 per group) U87/luciferase cells were incubated with different exosomes (U87TR Exo, U87TR-MIFsh Exo, U87TR-NC Exo) for 6 days. Approximately 2.5 × 105 U87 or U87TR/luciferase cells were stereotactically implanted into the right brain of nude mice. 1 week later, the mice were administrated with TMZ (5 mg/kg), ISO-1 (5 mg/kg), LY294002 (10 mg/kg), DMSO, or PBS by intraperitoneal (i.p.) injection twice weekly respectively according to the grouping. The intracranial tumors were detected weekly using bioluminescence imaging with an i.p. injection of D-luciferin (150 mg/kg).
Statistical analysis
All in vitro experiments were performed in triplicate and presented as mean ± standard deviation (SD) subjected to Student’s t test for pairwise comparison or ANOVA for multivariate analysis. Kaplan-Meier survival analysis was used to evaluate the correlation of different factors with survival rate. All statistical analyses were performed by the Graphpad Prism 8.0 (GraphPad Software, San Diego, CA, USA) and SPSS 19.0 software (SPSS, Chicago, IL, USA) and values of p < 0.05 were considered significant statistically.
Acknowledgments
This study was supported by the Natural Science Foundation of Guangdong Province (2017A030308001), the National Natural Science Foundation of China (81874079, 82073193), Guangdong Medical Science and Technology Research Fund Project, China (A2020535), and Guangdong Provincial Clinical Medical Centre for Neurosurgery (2013B020400005).
Author contributions
H.G. designed the research study, Q.W., B.L., and H.J. performed most of the experiments with assistance from Y.L, J.Z., X.Z., C.L., Q.H., and C.W. W.T. analyzed the data. Y.X. collected the clinical data. Q.W., B.L., and Y.L. wrote and revised the paper. All authors read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2021.08.004.
Supplemental information
References
- 1.Ostrom Q.T., Cioffi G., Gittleman H., Patil N., Waite K., Kruchko C., Barnholtz-Sloan J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro-oncol. 2019;21:v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan A.C., Ashley D.M., López G.Y., Malinzak M., Friedman H.S., Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020;70:299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 3.Munoz J.L., Rodriguez-Cruz V., Walker N.D., Greco S.J., Rameshwar P. Temozolomide resistance and tumor recurrence: Halting the Hedgehog. Cancer Cell Microenviron. 2015;2:e747. doi: 10.14800/ccm.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lefranc F., Sadeghi N., Camby I., Metens T., Dewitte O., Kiss R. Present and potential future issues in glioblastoma treatment. Expert Rev. Anticancer Ther. 2006;6:719–732. doi: 10.1586/14737140.6.5.719. [DOI] [PubMed] [Google Scholar]
- 5.Saito T., Sugiyama K., Takeshima Y., Amatya V.J., Yamasaki F., Takayasu T., Nosaka R., Muragaki Y., Kawamata T., Kurisu K. Prognostic implications of the subcellular localization of survivin in glioblastomas treated with radiotherapy plus concomitant and adjuvant temozolomide. J. Neurosurg. 2018;128:679–684. doi: 10.3171/2016.11.JNS162326. [DOI] [PubMed] [Google Scholar]
- 6.Couzin J. Cell biology: The ins and outs of exosomes. Science. 2005;308:1862–1863. doi: 10.1126/science.308.5730.1862. [DOI] [PubMed] [Google Scholar]
- 7.Jiang L., Gu Y., Du Y., Liu J. Exosomes: Diagnostic Biomarkers and Therapeutic Delivery Vehicles for Cancer. Mol. Pharm. 2019;16:3333–3349. doi: 10.1021/acs.molpharmaceut.9b00409. [DOI] [PubMed] [Google Scholar]
- 8.van Niel G., D’Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 9.Nawaz M., Shah N., Zanetti B.R., Maugeri M., Silvestre R.N., Fatima F., Neder L., Valadi H. Extracellular Vesicles and Matrix Remodeling Enzymes: The Emerging Roles in Extracellular Matrix Remodeling, Progression of Diseases and Tissue Repair. Cells. 2018;7:167. doi: 10.3390/cells7100167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng J., Meng J., Zhu L., Peng Y. Exosomal noncoding RNAs in Glioma: biological functions and potential clinical applications. Mol. Cancer. 2020;19:66. doi: 10.1186/s12943-020-01189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao H., Chung J., Lee K., Balaj L., Min C., Carter B.S., Hochberg F.H., Breakefield X.O., Lee H., Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B., Mao J.H., Wang B.Y., Wang L.X., Wen H.Y., Xu L.J., Fu J.X., Yang H. Exosomal miR-1910-3p promotes proliferation, metastasis, and autophagy of breast cancer cells by targeting MTMR3 and activating the NF-κB signaling pathway. Cancer Lett. 2020;489:87–99. doi: 10.1016/j.canlet.2020.05.038. [DOI] [PubMed] [Google Scholar]
- 13.Ge X., Liu W., Zhao W., Feng S., Duan A., Ji C., Shen K., Liu W., Zhou J., Jiang D. Exosomal Transfer of LCP1 Promotes Osteosarcoma Cell Tumorigenesis and Metastasis by Activating the JAK2/STAT3 Signaling Pathway. Mol. Ther. Nucleic Acids. 2020;21:900–915. doi: 10.1016/j.omtn.2020.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Z., Wang X., Yang Y., Chen W., Zhang K., Teng B., Huang C., Zhao Q., Qiu Z. Hypoxic Tumor-Derived Exosomal Long Noncoding RNA UCA1 Promotes Angiogenesis via miR-96-5p/AMOTL2 in Pancreatic Cancer. Mol. Ther. Nucleic Acids. 2020;22:179–195. doi: 10.1016/j.omtn.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y., Wang S., Lai Q., Fang Y., Wu C., Liu Y., Li Q., Wang X., Gu C., Chen J. Cancer-associated fibroblasts-derived exosomal miR-17-5p promotes colorectal cancer aggressive phenotype by initiating a RUNX3/MYC/TGF-β1 positive feedback loop. Cancer Lett. 2020;491:22–35. doi: 10.1016/j.canlet.2020.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Tong Y., Yang L., Yu C., Zhu W., Zhou X., Xiong Y., Wang W., Ji F., He D., Cao X. Tumor-Secreted Exosomal lncRNA POU3F3 Promotes Cisplatin Resistance in ESCC by Inducing Fibroblast Differentiation into CAFs. Mol. Ther. Oncolytics. 2020;18:1–13. doi: 10.1016/j.omto.2020.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Yin J., Lu C., Wei Y., Zeng A., You Y. Exosomal transfer of long non-coding RNA SBF2-AS1 enhances chemoresistance to temozolomide in glioblastoma. J. Exp. Clin. Cancer Res. 2019;38:166. doi: 10.1186/s13046-019-1139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu T., Wang X., Zhi T., Zhang J., Wang Y., Nie E., Zhou F., You Y., Liu N. Delivery of MGMT mRNA to glioma cells by reactive astrocyte-derived exosomes confers a temozolomide resistance phenotype. Cancer Lett. 2018;433:210–220. doi: 10.1016/j.canlet.2018.06.041. [DOI] [PubMed] [Google Scholar]
- 19.Bloom B.R., Bennett B. Mechanism of a reaction in vitro associated with delayed-type hypersensitivity. Science. 1966;153:80–82. doi: 10.1126/science.153.3731.80. [DOI] [PubMed] [Google Scholar]
- 20.Fukaya R., Ohta S., Yaguchi T., Matsuzaki Y., Sugihara E., Okano H., Saya H., Kawakami Y., Kawase T., Yoshida K., Toda M. MIF Maintains the Tumorigenic Capacity of Brain Tumor-Initiating Cells by Directly Inhibiting p53. Cancer Res. 2016;76:2813–2823. doi: 10.1158/0008-5472.CAN-15-1011. [DOI] [PubMed] [Google Scholar]
- 21.Mittelbronn M., Platten M., Zeiner P., Dombrowski Y., Frank B., Zachskorn C., Harter P.N., Weller M., Wischhusen J. Macrophage migration inhibitory factor (MIF) expression in human malignant gliomas contributes to immune escape and tumour progression. Acta Neuropathol. 2011;122:353–365. doi: 10.1007/s00401-011-0858-3. [DOI] [PubMed] [Google Scholar]
- 22.Parekh A., Das S., Parida S., Das C.K., Dutta D., Mallick S.K., Wu P.H., Kumar B.N.P., Bharti R., Dey G. Multi-nucleated cells use ROS to induce breast cancer chemo-resistance in vitro and in vivo. Oncogene. 2018;37:4546–4561. doi: 10.1038/s41388-018-0272-6. [DOI] [PubMed] [Google Scholar]
- 23.Wu M.Y., Fu J., Xu J., O’Malley B.W., Wu R.C. Steroid receptor coactivator 3 regulates autophagy in breast cancer cells through macrophage migration inhibitory factor. Cell Res. 2012;22:1003–1021. doi: 10.1038/cr.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pan D., Chen J., Feng C., Wu W., Wang Y., Tong J., Zhou D. Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells. Int. J. Mol. Sci. 2019;20:323. doi: 10.3390/ijms20020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu B., Zhou J., Wang C., Chi Y., Wei Q., Fu Z., Lian C., Huang Q., Liao C., Yang Z. LncRNA SOX2OT promotes temozolomide resistance by elevating SOX2 expression via ALKBH5-mediated epigenetic regulation in glioblastoma. Cell Death Dis. 2020;11:384. doi: 10.1038/s41419-020-2540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu N., Liu B., Lian C., Doycheva D.M., Fu Z., Liu Y., Zhou J., He Z., Yang Z., Huang Q. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis. 2018;9:1139. doi: 10.1038/s41419-018-1183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng H., Yang Z., Xu N., Liu B., Fu Z., Lian C., Guo H. Connective tissue growth factor promotes temozolomide resistance in glioblastoma through TGF-β1-dependent activation of Smad/ERK signaling. Cell Death Dis. 2017;8:e2885. doi: 10.1038/cddis.2017.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng H., Xu N., Liu Y., Liu B., Yang Z., Fu Z., Lian C., Guo H. Genomic profiling of long non-coding RNA and mRNA expression associated with acquired temozolomide resistance in glioblastoma cells. Int. J. Oncol. 2017;51:445–455. doi: 10.3892/ijo.2017.4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu X., Liu Y., Li Y., Chen H., Zhang Y., Liu J., Deng S., Zheng Y., Sun X., Wang J. Selective exosome exclusion of miR-375 by glioma cells promotes glioma progression by activating the CTGF-EGFR pathway. J. Exp. Clin. Cancer Res. 2021;40:16. doi: 10.1186/s13046-020-01810-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin J., Zeng A., Zhang Z., Shi Z., Yan W., You Y. Exosomal transfer of miR-1238 contributes to temozolomide-resistance in glioblastoma. EBioMedicine. 2019;42:238–251. doi: 10.1016/j.ebiom.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng A., Wei Z., Yan W., Yin J., Huang X., Zhou X., Li R., Shen F., Wu W., Wang X., You Y. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018;436:10–21. doi: 10.1016/j.canlet.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Su C.W., Lin C.W., Yang W.E., Yang S.F. TIMP-3 as a therapeutic target for cancer. Ther. Adv. Med. Oncol. 2019;11 doi: 10.1177/1758835919864247. 1758835919864247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bodnar M., Szylberg Ł., Kazmierczak W., Marszalek A. Tumor progression driven by pathways activating matrix metalloproteinases and their inhibitors. J. Oral Pathol. Med. 2015;44:437–443. doi: 10.1111/jop.12270. [DOI] [PubMed] [Google Scholar]
- 35.Liang J., Chen M., Hughes D., Chumanevich A.A., Altilia S., Kaza V., Lim C.U., Kiaris H., Mythreye K., Pena M.M. CDK8 Selectively Promotes the Growth of Colon Cancer Metastases in the Liver by Regulating Gene Expression of TIMP3 and Matrix Metalloproteinases. Cancer Res. 2018;78:6594–6606. doi: 10.1158/0008-5472.CAN-18-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garofalo M., Di Leva G., Romano G., Nuovo G., Suh S.S., Ngankeu A., Taccioli C., Pichiorri F., Alder H., Secchiero P. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Han X.G., Mo H.M., Liu X.Q., Li Y., Du L., Qiao H., Fan Q.M., Zhao J., Zhang S.H., Tang T.T. TIMP3 Overexpression Improves the Sensitivity of Osteosarcoma to Cisplatin by Reducing IL-6 Production. Front. Genet. 2018;9:135. doi: 10.3389/fgene.2018.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C., Wu H., Li Y., Shen L., Yu R., Yin H., Sun T., Sun C., Zhou Y., Du Z. SALL4 suppresses PTEN expression to promote glioma cell proliferation via PI3K/AKT signaling pathway. J. Neurooncol. 2017;135:263–272. doi: 10.1007/s11060-017-2589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lv D., Jia F., Hou Y., Sang Y., Alvarez A.A., Zhang W., Gao W.-Q., Hu B., Cheng S.-Y., Ge J. Histone Acetyltransferase KAT6A Upregulates PI3K/AKT Signaling through TRIM24 Binding. Cancer Res. 2017;77:6190–6201. doi: 10.1158/0008-5472.CAN-17-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanhaesebroeck B., Perry M.W.D., Brown J.R., André F., Okkenhaug K. PI3K inhibitors are finally coming of age. Nat. Rev. Drug Discov. 2021 doi: 10.1038/s41573-021-00209-1. Published online June 14, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quail D.F., Bowman R.L., Akkari L., Quick M.L., Schuhmacher A.J., Huse J.T., Holland E.C., Sutton J.C., Joyce J.A. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352:aad3018. doi: 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juric D., Castel P., Griffith M., Griffith O.L., Won H.H., Ellis H., Ebbesen S.H., Ainscough B.J., Ramu A., Iyer G. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. Nature. 2015;518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X., Wang Z., Tong F., Dong X., Wu G., Zhang R. lncRNA UCA1 Promotes Gefitinib Resistance as a ceRNA to Target FOSL2 by Sponging miR-143 in Non-small Cell Lung Cancer. Mol. Ther. Nucleic Acids. 2020;19:643–653. doi: 10.1016/j.omtn.2019.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Jackson H.W., Defamie V., Waterhouse P., Khokha R. TIMPs: versatile extracellular regulators in cancer. Nat. Rev. Cancer. 2017;17:38–53. doi: 10.1038/nrc.2016.115. [DOI] [PubMed] [Google Scholar]
- 45.Russo R., Matrone N., Belli V., Ciardiello D., Valletta M., Esposito S., Pedone P.V., Ciardiello F., Troiani T., Chambery A. Macrophage Migration Inhibitory Factor Is a Molecular Determinant of the Anti-EGFR Monoclonal Antibody Cetuximab Resistance in Human Colorectal Cancer Cells. Cancers (Basel) 2019;11:1430. doi: 10.3390/cancers11101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng B., Wang Q., Song Y., Liu Y., Liu Y., Yang S., Li D., Zhang Y., Zhu C. MIF inhibitor, ISO-1, attenuates human pancreatic cancer cell proliferation, migration and invasion in vitro, and suppresses xenograft tumour growth in vivo. Sci. Rep. 2020;10:6741. doi: 10.1038/s41598-020-63778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 48.Tian Y., Gong M., Hu Y., Liu H., Zhang W., Zhang M., Hu X., Aubert D., Zhu S., Wu L., Yan X. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles. 2019;9:1697028. doi: 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.