Abstract
Cisplatin-based chemotherapy remains the standard care for non-small cell lung cancer (NSCLC) patients. Relapse after chemotherapy-induced dormancy affects the overall survival of patients. The evolution of cancer cells under chemotherapy stress is regulated by transcription factors (TFs) with binding sites initially buried deep within inaccessible chromatin. The transcription machinery and dynamic epigenetic alterations during the process of dormancy-reactivation of lung cancer cells after chemotherapy need to be investigated. Here, we investigated the chromatin accessibility of lung cancer cells after cisplatin treatment, using an assay for transposase-accessible chromatin sequencing (ATAC-seq). We observed that global chromatin accessibility was extensively improved. Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST) v.2 was used to elucidate TF-target interaction during the process of dormancy and reactivation. Enhancer regions and motifs specific to key TFs including JUN, MYC, SMAD3, E2F1, SP1, CTCF, SMAD4, STAT3, NFKB1, and KLF4 were enriched in differential loci ATAC-seq peaks of dormant and reactivated cancer cells induced by chemotherapy. The findings suggest that these key TFs regulated gene expressions during the process of dormancy and reactivation of cancer cells through altering promoter accessibility of target genes. Our study helps advance understanding of how cancer cells adapt to the stress induced by chemotherapy through TF binding motif accessibility.
Keywords: NSCLC, chemotherapy, dormancy and reactivation, chromatin accessibility, transcription factors
Graphical abstract
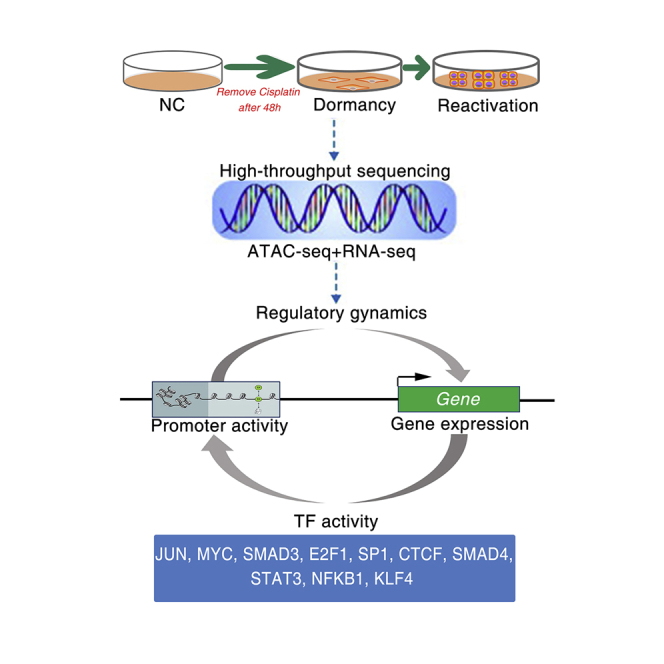
In this study, we profiled chromatin accessibility and transcriptome in cisplatin-responsive dormant and reactivated cells. We observed features of the chromatin landscape of lung cancer cells after chemotherapy. Our integrative analyses revealed that adaptive cellular response is often achieved by transcription factor binding motif accessibility.
Introduction
Lung cancer is the leading cause of cancer-related deaths around the world. The standard treatments for non-small cell lung cancer (NSCLC) are surgery, cisplatin-based chemotherapy, and targeted therapy. Cisplatin binds to DNA to form covalent platinum-DNA adducts and leads to cancer cell death by inhibiting DNA and RNA synthesis.1 Despite decades of application of cisplatin as one of the most potent chemotherapy drugs for lung cancer patients, resistance to cisplatin eventually develops and has been a major clinical obstacle.2
Dormancy is considered to be cellular quiescence. Tumor dormancy, which is a clinically undetectable state of cancer for a long period, is essential for cancer cells to become resistant to cancer therapy and to evade immune destruction.3 Dormant cancer cells cause tumor recurrence and metastatic relapse when they awaken.4,5 Therefore, a better understanding of the biological characteristics of cancer cell dormancy can help to develop potential therapeutic strategies to deal with the residual disease.6
In many eukaryotic organisms, DNA is tightly packed and organized with the formation of nucleosomes.7 When DNA is actively transcribed into RNA, the DNA will be opened and loosened from the nucleosome complex.8 The accessibility of the DNA is important for the activation of genes.9 Many factors, such as the chromatin structure, the position of the nucleosomes, and histone modifications, affect the accessibility of DNA to transcriptional regulatory factors and regulate gene expression.10 Chromatin remodeling may be achieved by several epigenetic mechanisms that involve histone modifications, DNA methylation, and nucleosome remodeling.10 Genome-wide access to chromatin or access to transcription factor (TF) binding has been identified as the most relevant genomic feature associated with gene activity at a particular site.11
In this study, we profiled chromatin accessibility and transcriptome in cisplatin-responsive dormant and reactivated cells. We observed features of the chromatin landscape of lung cancer cells after chemotherapy. As described below, our integrative analyses of assay for transposase-accessible chromatin sequencing (ATAC-seq) and RNA sequencing (RNA-seq) revealed that adaptive cellular response is often achieved by TF binding motif accessibility.
Results
Cisplatin-induced dormancy and reactivation showed distinct gene expression profile
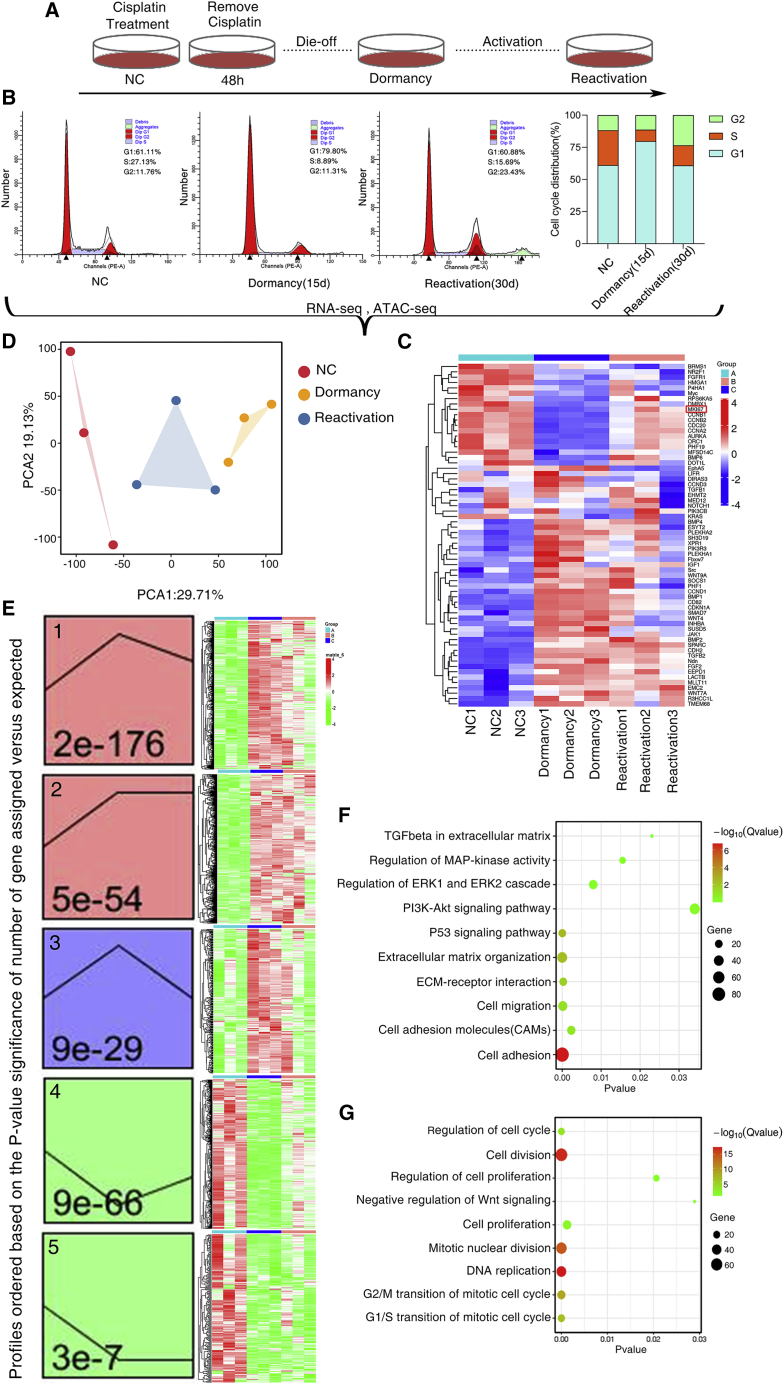
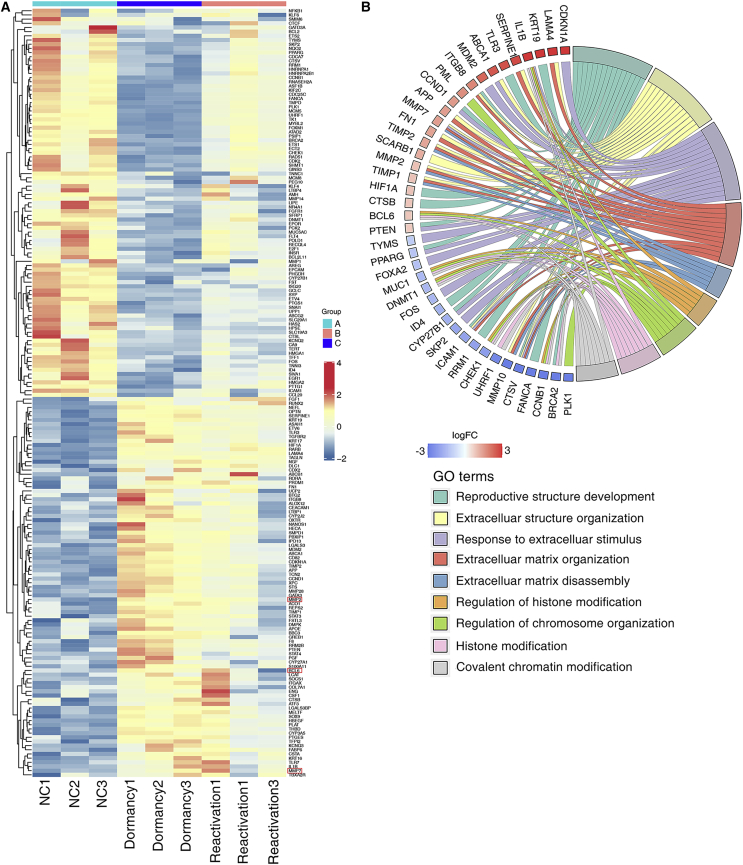
Conventional chemotherapy initially kills most cancer cells; however, the residual resistant cancer cells eventually survive and cause metastatic relapse. We first developed a short-term single-dose cisplatin cell system that mimics the process of reversible dormancy and reactivation after treatment with cisplatin12 (Figure 1A). The NSCLC cells were treated with cisplatin for 2 days at 10 ng/μL. Most cancer cells died, and the remaining cells entered into an inactive stage called the quiescent stage. We defined cellular dormancy as a state in which cells did not proliferate. The cell cycle analysis revealed that remained cells were arrested in G0/G1 phase 15 days after cisplatin withdrawal. About 30 days after withdrawal of cisplatin, the dormant cancer cells underwent outgrowth and progressed into S and G2/M phase (Figure 1B). RNA-seq-based analysis confirmed the cellular states of dormancy and reactivation, in which Ki67 was shown to be reduced and recovered in dormant and reactivated states (Figure 1C). Principal-component analysis showed consistency between three replicates, and the gene expression patterns could easily discriminate between different cell status (Figure 1D). Short Time-series Expression Miner (STEM) analysis of differential expression genes (DEGs) revealed the dynamic mRNA expressions during the process of dormancy and reactivation, which were classified into 5 significant patterns (Figure 1E, p < 0.05). In patterns 1, 2, and 3, mRNA expressions gradually increased during the entry into cellular dormancy. In patterns 4 and 5, mRNA expressions gradually decreased during the entry into cellular dormancy. During the process of shift from dormancy to reactivation, some mRNA expressions maintained stable (patterns 2, 5) and some mRNA expressions were back to the control level (patterns 1, 3, 4). Gene Ontology (GO) enrichment of the patterns indicated that patterns 1, 2, and 3 included the genes related to cell adhesion, transforming growth factor β (TGF-β) in extracellular matrix (ECM), and mitogen-activated protein kinase (MAPK) regulation (Figure 1F). The patterns 4 and 5 whose mRNA expressions increased in the reactivated phase included genes related to DNA replication, cell cycle phase transition et al. (Figure 1G).
Figure 1.
Cisplatin-induced dormancy and reactivation showed distinct gene expression profile
A549 cells showed reversible states of dormancy and reactivation after 2-day treatment with cisplatin. (A) Schematic diagram of the cisplatin treatment experiment. (B) Cell cycle analysis of dormant and reactivated cancer cells induced by cisplatin. (C) The differential gene expression profiles measured by RNA-seq. (D) Principal-component analysis showed that the three groups of cells had globally different gene expression profiles. (E) Short Time-series Expression Miner (STEM) analysis of differential expression genes (DEGs) revealed the dynamic mRNA expressions during the process of dormancy and reactivation, which were classified into 5 significant patterns. The p values are shown in the lower left corner of the profile boxes. (F) GO enrichment of the patterns indicated that differentially expressed genes in dormant cancer cells enriched in the genes related to cell adhesion, TGF-β in extracellular matrix, and MAPK regulation. (G) GO enrichment of the patterns indicated that differentially expressed genes in reactivated cancer cells enriched in the genes related to DNA replication and cell cycle phase transition.
Identifying transcription factors key to cisplatin-induced dormancy and reactivation transition
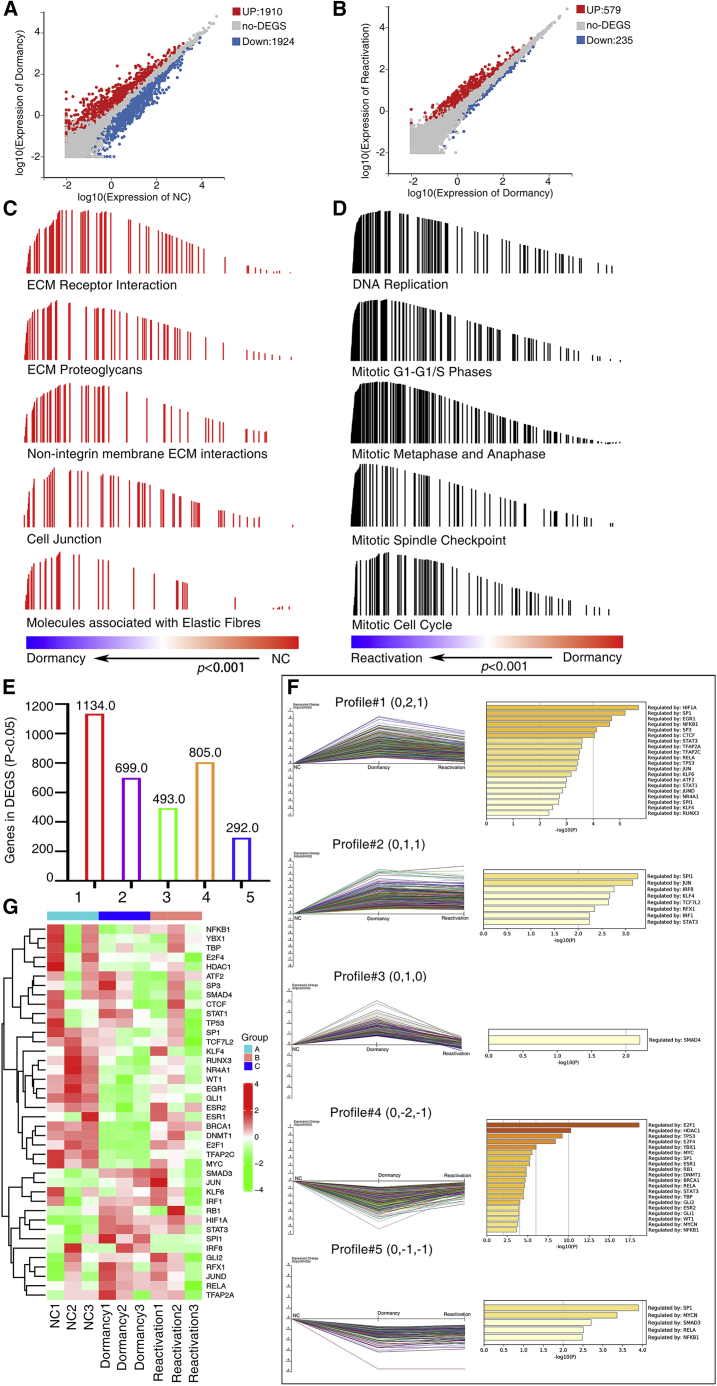
We then performed detailed analysis to determine the genes that are critical to cisplatin-induced dormancy and reactivation transition. We found that 1,910 genes were upregulated and 1,924 genes were downregulated in dormant status, whereas 579 genes were upregulated and 235 genes were downregulated in reactivated status (Figures 2A and 2B). Pathway analyses using the Gene Set Enrichment Analysis (GSEA) program revealed that, out of a total of 1,432 gene sets (c2.cp.v7.0.symbols.gmt), 972 gene sets were significantly upregulated in dormant lung cancer cells compared with intact control cells (Table S1). Four hundred ninety-three gene sets were significantly activated during the process from dormancy to reactivation (Table S2). The sets of genes that were mostly enriched in dormant cancer cells included ECM receptor interaction, ECM proteoglycans, and cell junction (Figure 2C). The sets of genes that were mostly enriched in reactivated cancer cells included DNA replication and cell cycle regulation (Figure 2D). This indicates that lung cancer cells surviving chemotherapy are characteristic of regulating ECM remodeling at the dormant phase and regulating cell cycle at the reactivated phase.
Figure 2.
Identifying transcription factors key to cisplatin-induced dormancy and reactivation transition
RNA-seq was performed to identify gene expression profiles. (A and B) Scatterplots of RNA-seq data comparing mRNA abundance between dormant cancer cells and untreated control cells (A) and between reactivated cancer cells and dormant cancer cells (B). The cutoff values were fold change of ≥1.5 or ≤0.5 for up- or downregulated genes. (C) Pathway analyses using the Gene Set Enrichment Analysis (GSEA) program revealed that the sets of genes that were mostly enriched in dormant cancer cells included ECM receptor interaction, ECM proteoglycans, and cell junction et al. (D) The sets of genes that were mostly enriched in reactivated cancer cells included DNA replication and cell cycle regulation. (E) Five patterns of gene expression based on the STEM analysis were shown. On the top of each profile are the values for relative gene expressions, presented in log2 (V(i)/(V(0)). (F) Transcription factor-target interaction during the process of dormancy and reactivation was elucidated by TRRUST v.2. (G) Gene expression profiles of key transcription factors.
Based on the STEM analysis, 1,134 genes, 699 genes, 493 genes, 805 genes, and 292 DEGs were observed respectively among these 5 patterns (Figure 2E). These DEGs in these 5 patterns showed similar expression patterns. Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining (TRRUST) v.2 was used to elucidate TF-target interaction during the process of dormancy and reactivation (https://www.grnpedia.org/trrust). We found that DEGs in these patterns were regulated by different TFs. For example, HIF1A, SP1, EGR1 et al. play roles in pattern 1, and SMAD4 plays a role in pattern 3; SP1, MYCN, SMAD3 et al. play roles in pattern 5; E2F1, HDAC1, and TP53 play regulatory roles in pattern 4; and SPI1, JUN, IRF8 et al. play roles in pattern 2 (Figure 2F).We defined TFs HIF1A, SMAD4, STAT3 et al., which function in the switch into dormant status, and TFs SP1, NFKB1, E2F1 et al., which function in the switch from dormant status to reactivated status (Figure 2F). The expression profiles of these TFs showed dynamic correspondence during the process of dormancy and reactivation of cancer cells, suggesting that TFs may play crucial roles in this process (Figure 2G).
Cisplatin affects chromatin accessibility
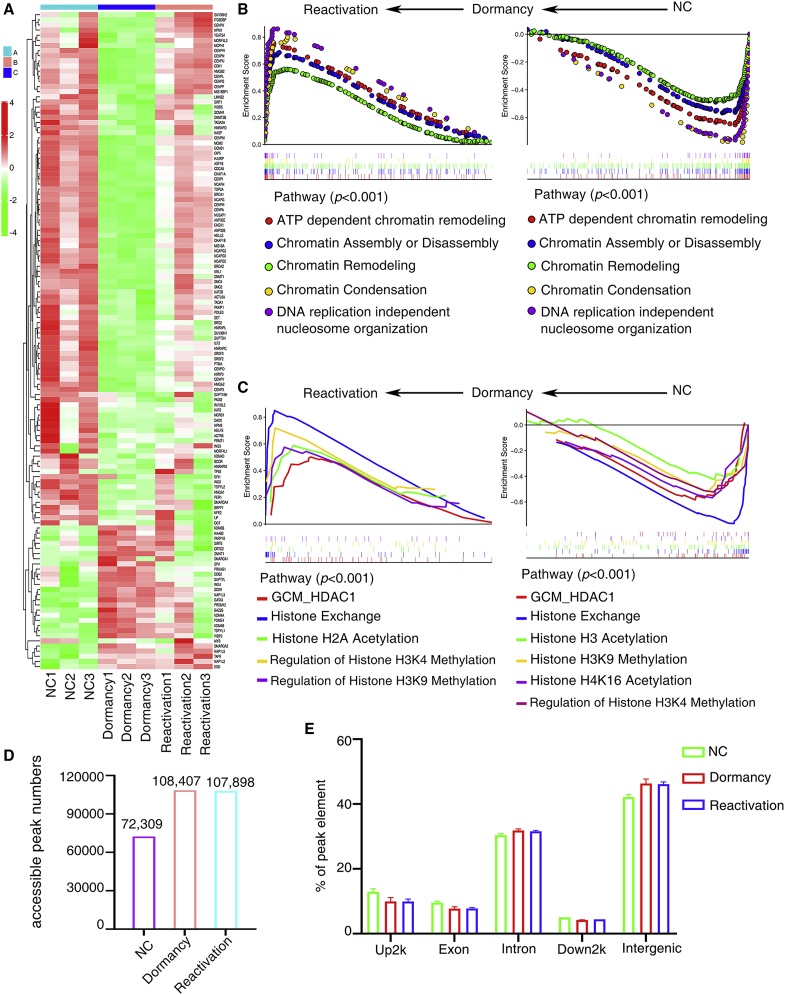
Chromatin remodeling and histone modification are important mechanisms regulating eukaryotic gene expression. Cisplatin covalently modifies DNA and inhibits DNA and RNA synthesis. We found that chromatin-remodeling complexes and histone-modifying enzymes showed differential expression in cisplatin-induced dormant and reactivated cancer cells (Figure 3A). Pathway analyses using the GSEA program revealed that chromatin remodeling, chromatin H2A acetylation, H3K4 methylation, and H3K9 methylation were the significantly upregulated gene sets in reactivated cancer cells compared with dormant cancer cells. These gene sets were downregulated in dormant cancer cells compared with untreated cancer cells, suggesting that chromatin modifications act as potential regulatory factors during the process of cisplatin-induced dormancy and reactivation of cancer cells (Figures 3B and 3C).
Figure 3.
Cisplatin affects chromatin accessibility
(A) Gene expression profiles of genes related to chromatin remodeling complexes and histone-modifying enzymes. (B and C) GSEA analysis of dormant and reactivated cancer cells. (D) Accessible peaks in dormant and reactivated cancer cells evaluated by ATAC-seq. (E) Location of ATAC peaks.
To integratively analyze the chromatin accessibility profiles during the process of cisplatin-responsive dormancy and reactivation, we performed ATAC-seq to generate a profile of chromatin accessibility in dormant and reactivated A549 cells. Three independent chemotherapy treatments were performed. We sequenced 3 samples per group for hg38 species with ATAC-seq, on average generating 112,428,090 raw reads and 112,366,083 clean reads after filtering dirty reads, including low-quality reads, N reads, and adaptor sequences. We obtained an average of 86% mapping rates. We identified a significant number of differentially accessible chromatin regions between cisplatin-treated groups and control group by deepTools. More regions were opened in dormant cancer cells and reactivated cancer cells. ATAC-seq profiles revealed the peak location and peak length in dormant and reactivated lung cancer cells. The differentially accessible peaks (false discovery rate [FDR] < 0.05) were identified between dormant cancer cells and control untreated cancer cells. On average, we identified 108,407 accessible peaks in dormant cells and 107,898 accessible peaks in reactivated cells, which possessed more open chromatin regions than untreated control cells (peak number = 72,309) (Figure 3D). A large proportion of ATAC peaks are located in intergenic regions (Figure 3E).
Accessible chromatin overlapped extensively with putative cis-regulatory sequences
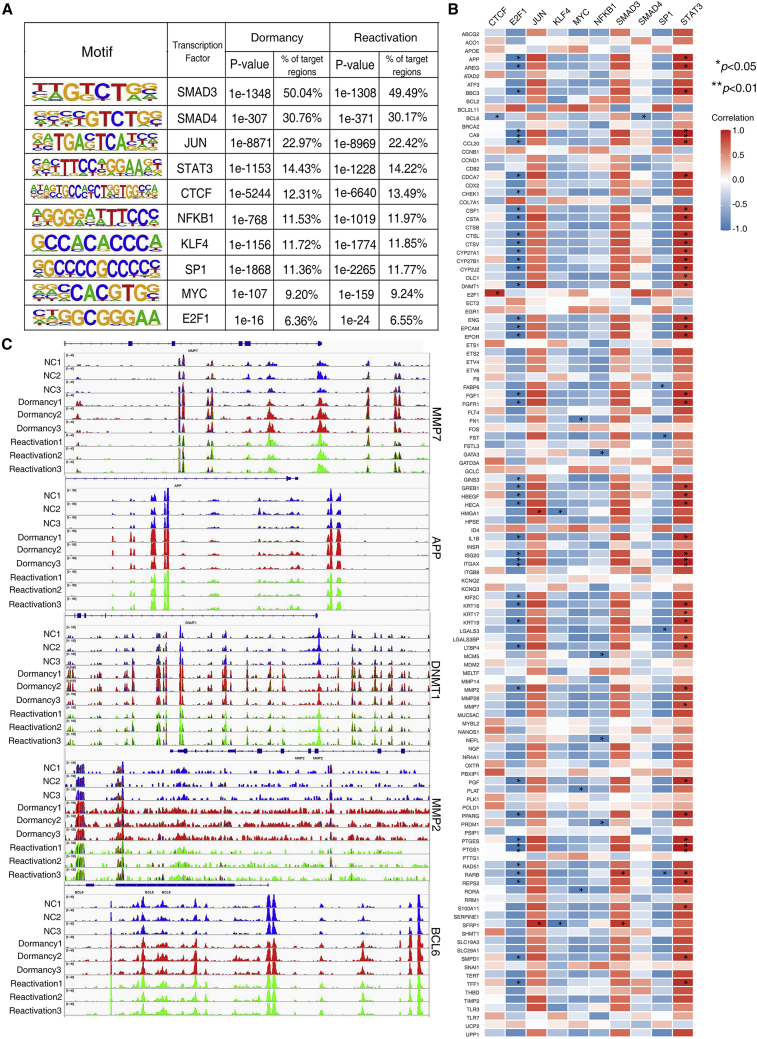
To define the differential TF activity and explore the relationship between TF binding sites (TFBSs) and chromatin accessibility, we scanned for motifs for TFs within open regions in chromosomes with Hypergeometric Optimization of Motif EnRichment (HOMER). This showed the significantly enriched motifs for TFs such as JUN, MYC, SMAD3, E2F1, SP1, CTCF, SMAD4, STAT3, NFKB1, and KLF4 in dormant cells and reactivated cells (Figure 4A). We then explored the relationship between TF expression and chromatin accessibility of their target genes. Pearson’s correlation analysis revealed that the promoter accessibility of their target genes was correlated with the expression of TFs (Figure 4B). For example, the promoter accessibility of MMP2 and MMP7 is positively correlated with the expression of STAT3, and the promoter accessibility of BCL6 is negatively correlated with the expression of SMAD4. The Integrative Genomics Viewer (IGV) was used to analyze the genomic profiles of specific differentially accessible regions. We identified regions of significant interest containing a putative enhancer region and predicted binding sites for these TFs in 2 kb upstream of transcription start site (TSS) (Figure 4C).
Figure 4.
Accessible chromatin overlapped extensively with putative cis-regulatory sequences
(A) Motifs for transcription factors within open regions in chromosomes were identified with HOMER. (B) The correlation between the expression of transcription factors and the promoter accessibility of their target genes was evaluated by Pearson’s correlation analysis. (C) The genomic profiles of specific differentially accessible regions of transcription factor target genes visualized by Integrative Genomics Viewer (IGV).
We then tried to explore the relationship of TF binding profiles, which were derived from ATAC-seq peaks, and the target gene expression level. We found that expression of target genes of TFs JUN, MYC, SMAD3, E2F1, SP1, CTCF, SMAD4, STAT3, NFKB1, and KLF4 overlapped with the expression profiles in dormant and reactivated states (Figure 5A). The expression profiles of the target genes of these TFs showed dynamic correspondence with the promoter accessibility. For example, in dormant cancer cells, STAT3 upregulates MMP2 and MMP7 through increasing their promoter accessibility. SMAD4 downregulates BCL6 through decreasing BCL6 promoter accessibility. The upregulated target genes were used to identify enriched GO categories. GO categories related to ECM remodeling and chromosome organization were the most enriched categories (Figure 5B). This indicated that TFs controlled the adapted alteration of lung cancer cells with chemotherapy.
Figure 5.
The relationship of TF binding profiles
(A) Heatmap of key transcription factors and their target genes. (B) GO enrichment of the patterns indicated that target genes were enriched in key molecular functions.
Discussion
Here we provide a genome-wide survey of accessible chromatin in dormant and reactivated states of lung cancer cells induced by chemotherapy. Combined with the gene expression profiles, we found that ECM remodeling and cell cycle regulation dominate the process of dormancy and reactivation. The expression of TFs JUN, MYC, SMAD3, E2F1, SP1, CTCF, SMAD4, STAT3, NFKB1, and KLF4 affects transcriptional outcome of their target genes by regulating chromatin accessibility of gene promoter. Based on GO analysis of the upregulated target genes of enriched TFs, we found that the target genes of these TFs were mainly involved in the biological processes in the dormant and reactivated cells.
Lung cancer recurrence, despite surgery and chemotherapy, may occur at variable times after primary tumor removal and, finally, death due to metastases.13 Mathematical modeling of clinical data and experiments in mouse models suggest an extended period of dormancy at premetastatic sites.14 Newly uncovered mechanisms that govern cancer dormancy and reactivation include tumor evolution, stem cell signaling, and micro-environmental niches.14,15 In this study, we first analyzed the transcriptomic profiles of dormant and reactivated lung cancer cells induced by cisplatin. We found the key TFs governing the shift of cellular dormancy and reactivation, which control the entry of cancer cells into dormant states and control the reentry of dormant cancer cells into the cell cycle. The target genes of these TFs displayed corresponding expression during the process of dormancy and reactivation, indicating that these TFs can determine cell fate. These master TFs regulate expression of each specific gene with the involvement of chromatin remodeling. It is obvious that the regulation of these TFs is of crucial importance for the gene expression profiles during the reversible process of dormancy and reactivation. Within eukaryotes, specific genes are expressed under conditions in which RNA polymerase and TFs access the cis-regulatory elements.16 Transcription is a complex process that involves sequence-specific factors along with RNA polymerase transcriptional machinery and chromatin-remodeling factors that control the opening of chromatin.17 Epigenetic reprogramming is considered to have the potential to simultaneously regulate multiple genes.18 Recent studies in high-throughput technologies highlighted the importance of simultaneous activation of genes rather than alterations in a single gene. “Open” and “closed” chromatins represent active and repressed states of individual genes. Chromatin remodeling events can change the transcriptional state of cells, resulting in a higher possibility of allowing the transcription of genes in cancer growth and therapy resistance.15,19 In this study, we used ATAC-seq to detect the changes in chromatin access and gene expression during the progression of lung cancer after cisplatin therapy. ATAC-seq is an effective method to reveal chromatin accessibility at a genome-wide level, and it can identify H3K4, H3K36, and H3K79 trimethylation in the open chromatin region.20 This technology linked with RNA-seq can furnish high resolution of the possible functional interactions.21 Recent evidence has indicated that epigenetic mechanisms, such as abnormal DNA methylation, may regulate the long-time commitment of disseminated tumor cells to stand still while preserving growth potential.22 The expression of the anti-angiogenic genes TIMP3 and CDH1 is raised during dormancy and is reduced during the transition to active growth via changes in histone modification and DNA methylation.23 The analysis of chromatin remodeling led to the identification of epigenetic modification in multiple TFs that determine the process of dormancy and reactivation.
In this study, we report the analysis that lung cancer cells that survived chemotherapy displayed ECM remodeling during the dormant phase and cell cycle transit during the activation phase. Sp1-like factors and Krüppel-like factors (KLFs) are important components of the cellular transcriptional machinery, regulating multiple cancerous biological processes such as cell growth, differentiation, angiogenesis, apoptosis, and stem cell reprogramming.24 ECM and its related cell-cell adhesion exert a direct effect on cellular quiescence.25 KLFs belong to the relatively large family of Sp1-like TFs.26 KLF4 is enriched in the quiescent cortical cells of the thymus epithelium and regulates the major EMC protein laminin α.27,28 Sp1 family members and E2F cooperatively activate exit from and the progression of the cell cycle.29 In this study, the integrated computational analysis identified multiple TFs, in particular the Sp1 family and its interacting proteins such as smad3 and smad4.
In conclusion, here we tried to apply a new technology, integration of ATAC-seq and RNA-seq, to study differences in gene expression profile during chemotherapy-induced dormancy and reactivation. We identified chemotherapy-responsive accessible chromatin regions and motifs at differential chromatin states via ATAC-seq. We found some crucial TFs that could control the adaptive cellular response after chemotherapy, and this provides insights into therapeutic strategies to deal with the residual disease after chemotherapy.
Materials and methods
Cell culture and treatment
Human lung adenocarcinoma cell line A549 obtained from ATCC was cultured in RPMI-1640 medium (HyClone Life Sciences, USA) supplemented with penicillin G (100 U/mL), streptomycin (100 mg/mL), and 10% fetal calf serum. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. Cisplatin (P4394) was purchased from Sigma-Aldrich (St. Louis, MO, USA). A549 cells were treated with cisplatin at 10 ng/μL for 48 h.
Cell cycle analysis
Cells were treated with cisplatin for 48 h. Cells were harvested at different times and fixed with cold 70% ethanol. Cells were washed twice with cold PBS and incubated for 30 min with PI/RNase solution (C1052, Beyotime, China). Samples were analyzed on a FACScan flow cytometer (Becton Dickinson, California, USA).
RNA-seq
RNA-seq analysis was performed with the BGISEQ-500 platform (BGI, Shenzhen, China). Total RNA was isolated, and a cDNA library was constructed. High-quality clean reads were aligned to the human reference genome with Bowtie2. The expression levels for each of the genes were normalized as fragments per kilobase of exon model per million mapped reads (FPKM) by expectation maximization (RSEM). The cutoff values were fold change of ≥1.5 or ≤0.5 for up- or downregulated genes. The RNA-seq data presented in this study can be found in the NCBI Sequence Read Archive (SRA) database. The accession number is SRA PRJNA730205.
STEM analysis
STEM software (version 1.3.13) was used to identify the dynamic gene expression clusters in dormant and reactivated cancer cells induced by cisplatin.30 The statistically enriched genes with similar expression patterns were assessed according to the default parameter.
ATAC-seq
ATAC-seq analysis was performed by BGI-Shenzhen (https://en.genomics.cn). Cells were harvested and resuspended in cold lysis buffer, followed by spinning down by centrifuge. Crude nuclei were prepared and immediately continued to the transposition reaction. After transposition, purified DNA was amplified. qPCR-based methods were used to quantify ATAC-seq libraries. The libraries were sequenced with Illumina high-throughput sequencing instruments. Raw reads were filtered to remove low-quality or adaptor sequences by SOAPnuke. Cleaned reads were mapped to the reference genome of hg38 with Bowtie2 (version 2.2.5). We used MACS2 (version 2.1.2) to call peaks (open chromatin regions). IDR (v.2.0.4) was applied to measure the reproducibility of findings identified from replicate samples. The promoter accessibility was presented by normalized reads counted by featureCounts. The different enrichment peaks from different samples were plotted by IGV. The gene element annotation of peak or different enrichment peak from different samples was carried out by bedtools intersect mode with overlap 50% as well. To identify the position of nucleosomes, broad peaks called by MACS2 were analyzed with NucleoATAC (v.0.3.4). The ATAC-seq data presented in this study can be found in the NCBI SRA database. The accession number is SRA PRJNA731334.
TF-target interaction analysis
TRRUST was used to elucidate the TF-target interaction.31 A cutoff for −log10(p value) was presented to select significant potential TF-target interactions. Values of the −log10 bigger than 1.3 were considered to be statistically significant.
Motif analysis
De novo motif discovery from ATAC-seq was achieved by HOMER. The motif with the most significant p value predicted by HOMER was selected as key TF.
Acknowledgments
This work was supported by National Natural Science Foundation, China (grant numbers 81972198, 81773147, 81472695); Strategic Priority Research Program of Central South University (ZLXD2017004); and Key Research and Development Program of Hunan (2019SK2253).
Author contributions
L.W., J.T., and J. Xiang designed and conceived the experiments. Q.P., N.Y., Y.X., J. Xu, A.C., and J.Y. performed the experiments and analyzed the data. L.W., J.T., and J. Xiang wrote the paper.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.07.019.
Supplemental information
References
- 1.Mymryk J.S., Zaniewski E., Archer T.K. Cisplatin inhibits chromatin remodeling, transcription factor binding, and transcription from the mouse mammary tumor virus promoter in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:2076–2080. doi: 10.1073/pnas.92.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen S.H., Chang J.Y. New Insights into Mechanisms of Cisplatin Resistance: From Tumor Cell to Microenvironment. Int. J. Mol. Sci. 2019;20:4136. doi: 10.3390/ijms20174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahanban-Esfahlan R., Seidi K., Manjili M.H., Jahanban-Esfahlan A., Javaheri T., Zare P. Tumor Cell Dormancy: Threat or Opportunity in the Fight against Cancer. Cancers (Basel) 2019;11:1207. doi: 10.3390/cancers11081207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Recasens A., Munoz L. Targeting Cancer Cell Dormancy. Trends Pharmacol. Sci. 2019;40:128–141. doi: 10.1016/j.tips.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 5.Marx V. How to pull the blanket off dormant cancer cells. Nat. Methods. 2018;15:249–252. doi: 10.1038/nmeth.4640. [DOI] [PubMed] [Google Scholar]
- 6.Yadav A.S., Pandey P.R., Butti R., Radharani N.N.V., Roy S., Bhalara S.R., Gorain M., Kundu G.C., Kumar D. The Biology and Therapeutic Implications of Tumor Dormancy and Reactivation. Front. Oncol. 2018;8:72. doi: 10.3389/fonc.2018.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ordu O., Lusser A., Dekker N.H. Recent insights from in vitro single-molecule studies into nucleosome structure and dynamics. Biophys. Rev. 2016;8(Suppl 1):33–49. doi: 10.1007/s12551-016-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miskimen K.L.S., Chan E.R., Haines J.L. Assay for Transposase-Accessible Chromatin Using Sequencing (ATAC-seq) Data Analysis. Curr. Protoc. Hum. Genet. 2017;92:20.4.1–20.4.13. doi: 10.1002/cphg.32. [DOI] [PubMed] [Google Scholar]
- 9.Chereji R.V., Eriksson P.R., Ocampo J., Prajapati H.K., Clark D.J. Accessibility of promoter DNA is not the primary determinant of chromatin-mediated gene regulation. Genome Res. 2019;29:1985–1995. doi: 10.1101/gr.249326.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciechomska I.A., Jayaprakash C., Maleszewska M., Kaminska B. Histone Modifying Enzymes and Chromatin Modifiers in Glioma Pathobiology and Therapy Responses. Adv. Exp. Med. Biol. 2020;1202:259–279. doi: 10.1007/978-3-030-30651-9_13. [DOI] [PubMed] [Google Scholar]
- 11.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S., Kennedy M., Payne S., Kennedy K., Seewaldt V.L., Pizzo S.V., Bachelder R.E. Model of tumor dormancy/recurrence after short-term chemotherapy. PLoS ONE. 2014;9:e98021. doi: 10.1371/journal.pone.0098021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelsey C.R., Fornili M., Ambrogi F., Higgins K., Boyd J.A., Biganzoli E., Demicheli R. Metastasis dynamics for non-small-cell lung cancer: effect of patient and tumor-related factors. Clin. Lung Cancer. 2013;14:425–432. doi: 10.1016/j.cllc.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Giancotti F.G. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braadland P.R., Urbanucci A. Chromatin reprogramming as an adaptation mechanism in advanced prostate cancer. Endocr. Relat. Cancer. 2019;26:R211–R235. doi: 10.1530/ERC-18-0579. [DOI] [PubMed] [Google Scholar]
- 16.Kadonaga J.T. Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell. 2004;116:247–257. doi: 10.1016/s0092-8674(03)01078-x. [DOI] [PubMed] [Google Scholar]
- 17.Hager G.L., McNally J.G., Misteli T. Transcription dynamics. Mol. Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asano N., Takeshima H., Yamashita S., Takamatsu H., Hattori N., Kubo T., Yoshida A., Kobayashi E., Nakayama R., Matsumoto M., et al. Epigenetic reprogramming underlies efficacy of DNA demethylation therapy in osteosarcomas. Sci. Rep. 2019;9:20360. doi: 10.1038/s41598-019-56883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sur I., Taipale J. The role of enhancers in cancer. Nat. Rev. Cancer. 2016;16:483–493. doi: 10.1038/nrc.2016.62. [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Lowe E.K., Cuomo C., Voronov D., Arnone M.I. Using ATAC-seq and RNA-seq to increase resolution in GRN connectivity. Methods Cell Biol. 2019;151:115–126. doi: 10.1016/bs.mcb.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Bedi U., Mishra V.K., Wasilewski D., Scheel C., Johnsen S.A. Epigenetic plasticity: a central regulator of epithelial-to-mesenchymal transition in cancer. Oncotarget. 2014;5:2016–2029. doi: 10.18632/oncotarget.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyu T., Jia N., Wang J., Yan X., Yu Y., Lu Z., Bast R.C., Jr., Hua K., Feng W. Expression and epigenetic regulation of angiogenesis-related factors during dormancy and recurrent growth of ovarian carcinoma. Epigenetics. 2013;8:1330–1346. doi: 10.4161/epi.26675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vellingiri B., Iyer M., Devi Subramaniam M., Jayaramayya K., Siama Z., Giridharan B., Narayanasamy A., Abdal Dayem A., Cho S.G. Understanding the Role of the Transcription Factor Sp1 in Ovarian Cancer: from Theory to Practice. Int. J. Mol. Sci. 2020;21:1153. doi: 10.3390/ijms21031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho I.J., Lui P.P., Obajdin J., Riccio F., Stroukov W., Willis T.L., Spagnoli F., Watt F.M. Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence. Stem Cell Reports. 2019;12:1190–1200. doi: 10.1016/j.stemcr.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nandan M.O., Yang V.W. The role of Krüppel-like factors in the reprogramming of somatic cells to induced pluripotent stem cells. Histol. Histopathol. 2009;24:1343–1355. doi: 10.14670/hh-24.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panigada M., Porcellini S., Sutti F., Doneda L., Pozzoli O., Consalez G.G., Guttinger M., Grassi F. GKLF in thymus epithelium as a developmentally regulated element of thymocyte-stroma cross-talk. Mech. Dev. 1999;81:103–113. doi: 10.1016/s0925-4773(98)00237-8. [DOI] [PubMed] [Google Scholar]
- 28.Miller K.A., Eklund E.A., Peddinghaus M.L., Cao Z., Fernandes N., Turk P.W., Thimmapaya B., Weitzman S.A. Kruppel-like factor 4 regulates laminin alpha 3A expression in mammary epithelial cells. J. Biol. Chem. 2001;276:42863–42868. doi: 10.1074/jbc.M108130200. [DOI] [PubMed] [Google Scholar]
- 29.Lin S.Y., Black A.R., Kostic D., Pajovic S., Hoover C.N., Azizkhan J.C. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol. Cell. Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst J., Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han H., Cho J.W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E., et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.