Summary
The circadian system is comprised three components: a network of core clock cells in the brain that keeps time, input pathways that entrain clock cells to the environment, and output pathways that use this information to ensure appropriate timing of physiological and behavioral processes throughout the day. Core clock cells can be divided into molecularly distinct populations that likely make unique functional contributions. Here we clarify the role of the dorsal neuron 1 (DN1) population of clock neurons in the transmission of circadian information by the Drosophila core clock network. Using an intersectional genetic approach that allowed us to selectively and comprehensively target DN1 cells, we show that suppressing DN1 neuronal activity alters the magnitude of daily activity and sleep without affecting overt rhythmicity. This suggests that DN1 cells are dispensable for both the generation of circadian information and the propagation of this information across output circuits.
Subject areas: Biological sciences, Neuroscience, Behavioral neuroscience, Sensory neuroscience
Graphical abstract
Highlights
-
•
Intersectional genetic approach targets DN1 cells comprehensively and selectively
-
•
DN1p silencing alters distribution and amount of activity and sleep across the day
-
•
DN1p cell firing is neither necessary nor sufficient for circadian activity rhythms
-
•
DN1a silencing subtly alters total activity and sleep but leaves rhythmicity intact
Biological sciences; Neuroscience; Behavioral neuroscience; Sensory neuroscience
Introduction
Animals exhibit daily rhythms in behavioral and physiological processes under control of an endogenous circadian timing system. In Drosophila, ∼150 neurons track time of day via the presence of molecular circadian clocks. These neurons, known as clock cells, can be divided into several distinct populations based on anatomical and functional properties and include the large- and small-ventrolateral neurons (lLNvs and sLNvs, respectively), the dorsolateral neurons (LNds), the lateral posterior neurons, and three groups of dorsal neurons (DN1s, DN2s, and DN3s). Together, these clock cell populations form a core clock network that generates circadian rhythms of behavior through connections with downstream neurons of the so-called output pathways (Dubowy and Sehgal, 2017; Hermann-Luibl and Helfrich-Förster, 2015). Much remains unknown about the cellular mechanisms through which circadian information is coordinated among different clock cells and transmitted across output circuits to produce coherent behavioral rhythms. For example, it is unclear whether multiple parallel output circuits emanate from distinct groups of clock cells, or instead whether circadian information is first consolidated among specific clock cell populations that serve as output nodes within the clock network. Knowledge of mammalian circadian circuits, which share many characteristics with those of flies, is similarly incomplete (Hastings et al., 2019).
Using genetic tools available in Drosophila that allow for the targeted manipulation of defined cell types (Venken et al., 2011), investigators have begun to delineate the contribution of specific populations of clock cells to rest:activity rhythms. Initial studies identified an essential contribution of two groups of lateral neurons, the sLNvs and the LNds, which have come to be considered central pacemakers within the clock network (Grima et al., 2004; Picot et al., 2007; Renn et al., 1999; Stoleru et al., 2004). More recently, the role of DN1 clock neurons has been assessed, facilitated by the development of specific genetic drivers that label these clock cells.
A major question that has emerged from these studies is whether DN1 cells comprise essential components of the core circadian oscillator, or whether they instead serve predominantly output functions. In support of the latter possibility, DN1s have been described as driven oscillators, incapable of maintaining free-running molecular or electrical activity cycles in the absence of input from other clock cells, in particular neuropeptidergic signals from sLNvs and LNds (Klarsfeld et al., 2004; Liang et al., 2017; Yoshii et al., 2009). Indeed, the pace of DN1 molecular clock cycling is dictated by the speed of cycling in lateral clock cells (Chatterjee et al., 2018; Stoleru et al., 2005; Yao et al., 2016; Zhang et al., 2010a). Also consistent with an output function for DN1 cells is the fact that they are synaptically and functionally connected to multiple groups of non-clock output cells (Barber et al., 2021; 2016; Cavanaugh et al., 2014; Guo et al., 2018; Jin et al., 2021; Lamaze et al., 2018; Tabuchi et al., 2018; Zhang et al., 2021a, 2021b). Together, these results suggest a hierarchical organization, with DN1s lying downstream of the lateral clock neurons and serving as intermediaries that propagate pacemaker-derived circadian information to output regions that control behaviors. Other evidence, however, suggests that the DN1s play a more intrinsic role within the clock network. DN1s have been shown to form reciprocal connections with lateral clock neurons, and in this capacity may directly control pacemaker function (Collins et al, 2012, 2014; Díaz et al., 2019; Fernandez et al., 2020; Fujiwara et al., 2018; Guo et al., 2016). In line with this idea, rest:activity rhythms become less coherent when DN1 clocks are desynchronized from those in lateral neurons (Yao et al., 2016).
Further confounding an understanding of DN1 function is the fact that the ∼17 DN1 cells per brain hemisphere can be subdivided into multiple anatomically discrete populations, which may make unique contributions to the regulation of sleep and circadian rhythms. Most broadly, this involves separation into anterior and posterior populations that have distinct developmental origins. The 2 anterior DN1 (DN1a) cells are present from early larval stages, while the ∼15 posterior DN1 (DN1p) neurons, which express the glass transcription factor, are not fully developed until adulthood (Klarsfeld et al., 2004; Liu et al., 2015; Shafer et al., 2006). DN1p neurons have been further subdivided based on differential expression of the blue light photoreceptor CRY, the glutamate transporter VGlut, and also based on distinct axonal projection patterns (Benito et al., 2008; Chatterjee et al., 2018; Guo et al, 2016, 2018; Lamaze et al., 2018). Notably, these molecular and anatomical distinctions have been associated with functional differences, as they define subpopulations that appear to make distinct contributions to locomotor and sleep outputs. For example, it was reported that a group of CRY- and VGlut- DN1p cells regulate locomotor activity in the evening, while CRY+ and VGlut+ cells contribute to morning activity (Chatterjee et al., 2018). Distinct DN1 cell populations also appear to differentially regulate sleep, with some subsets promoting and others inhibiting sleep (Guo et al., 2018; Kunst et al., 2014; Lamaze et al., 2018). The capacity of DN1 cells to regulate these parameters may also depend on environmental conditions, such as light and temperature, as well as time of day (Zhang et al., 2010b).
An unequivocal assessment of the role of DN1 cells in circadian rhythm regulation has been hindered by the use of genetic tools that either lack specificity or fail to target the entire DN1 population. We therefore devised an intersectional genetic approach that allowed us to selectively and comprehensively target DN1 clock cells with effector genes that result in electric silencing. We performed immunohistochemical analysis to confirm that our approach restricted expression to the DN1 cells and was comprehensive among these cells. We then conducted locomotor activity monitoring under constant environmental conditions, to assess the strength of endogenously driven circadian rhythms, and in the presence of daily light cues (but with temperature held constant), to assess entrained behavior. We demonstrate that suppressing DN1 neuronal activity alters the distribution and magnitude of daily activity and sleep behavior but does not impact their rhythmicity. Thus, at least under constant temperature conditions, DN1s appear dispensable for the generation and transmission of circadian information in the control of locomotor activity and sleep.
Results
The search for a comprehensive and selective DN1 driver
To clarify their role in the production of circadian rest:activity rhythms, we determined the consequences of electrically silencing DN1 cells. We used the GAL4-UAS system to drive ectopic expression in DN1 cells of an eGFP-tagged Kir2.1 potassium channel (Kir2.1eGFP), which hyperpolarizes Drosophila clock cells and strongly suppresses neuronal activity, effectively preventing communication with downstream targets (Depetris-Chauvin et al., 2011; Wu et al., 2008). We reasoned that if DN1 cells serve an obligatory function in propagating circadian information from the clock network to cells of the output circuit, then such a manipulation should eliminate behavioral rhythmicity.
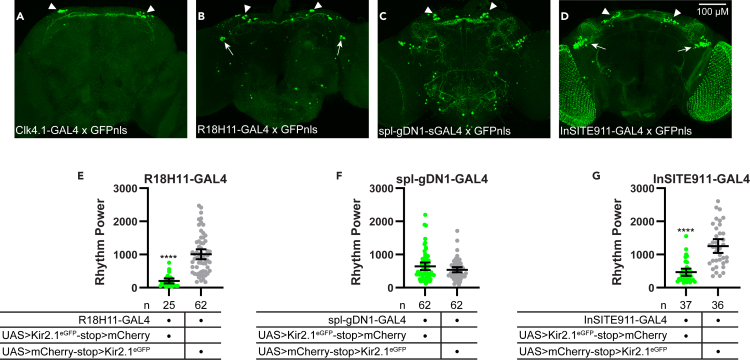
In initial experiments, we focused on the DN1p subset and used a collection of existing GAL4 driver lines to target these cells, including Clk4.1-GAL4, R18H11-GAL4, and a recently developed split GAL4 driver (spl-gDN1-GAL4) (Guo et al., 2018; 2017; 2016; Kunst et al., 2014; Lamaze et al., 2018; Zhang et al., 2010a, 2010b). Consistent with previous reports, we found that these GAL4 lines are expressed in partially overlapping subsets of DN1p cells and are also present to varying extents in non-DN1 neurons (Figures 1A–1C).
Figure 1.
Variable effects of Kir2.1eGFP-mediated neuronal silencing using multiple established DN1p GAL4 drivers
(A–D) Representative maximum-projection confocal images of brains in which GAL4 lines were used to drive expression of a nuclear-localized GFP (GFPnls). Brains were stained for GFP immunofluorescence (green). Arrowheads indicate GFP+ DN1 cells. Arrows in (B) indicate GFP+ LNd clock cells in the R18H11-GAL4 line. Arrows in (D) indicate GFP+ non-clock cells in the vicinity of the LNds in the InSITE911-GAL4 line.
(E–H) Rest:activity rhythm power over 6 days in DD is displayed for the genotypes listed. Lines are means ±95% confidence intervals. Dots represent individual flies. Experimental lines (GAL4 x UAS>Kir2.1eGFP-stop>mCherry) are in green, and control lines (GAL4 x UAS>mCherry-stop>Kir2.1eGFP) are in gray. ∗∗∗∗p < 0.0001, t test. ns listed are number of flies that survived throughout the behavioral monitoring.
Having confirmed the anatomical distribution of these commonly used drivers, we next monitored locomotor activity under constant environmental conditions to assess the strength of free-running circadian rhythms in the face of DN1p cell silencing. Surprisingly, the effect of GAL4-mediated expression of Kir2.1eGFP varied according to the driver used. With Clk4.1-GAL4, we observed complete developmental lethality with no adult eclosion, preventing behavioral assessment of these flies. This is likely due to non-neuronal expression of the Clk4.1-GAL4 driver. We also noted significant lethality with R18H11-GAL4. Although a substantial number of flies eclosed, many died during the course of behavioral monitoring such that only 40.3% of flies loaded for locomotor activity analysis survived throughout the duration of the experiment. Interestingly, these surviving flies exhibited profoundly decreased rest:activity rhythm strength compared to controls (Figure 1E). In contrast, there was no lethality associated with spl-gDN1-GAL4-mediated expression of Kir2.1eGFP and no effect of this manipulation on free-running behavioral rhythms (Figure 1F).
The discrepant behavioral effects associated with these GAL4 drivers could feasibly derive from their expression in different subsets of DN1p cells that make distinct contributions to the regulation of circadian rhythms. (To our knowledge, the overlap between the R18H11- and spl-gDN1-GAL4 lines has not been directly tested, although both label 5–6 DN1p cells). Thus, the reduction in free-running rhythmicity that results from driving Kir2.1eGFP in R18H11-GAL4-expressing cells could represent a necessary contribution of one or more DN1p cells labeled by this line but not by spl-gDN1-GAL4. Alternatively, the R18H11-GAL4 phenotype could occur as a result of Kir2.1eGFP effects in non-DN1 cells that are not also labeled by spl-gDN1-GAL4. To resolve this discrepancy, we sought a GAL4 line that would label the DN1 population in its entirety while minimizing non-DN1 expression.
Sustained free-running rest:activity rhythms following selective Kir2.1eGFP-mediated silencing of DN1p cells
To comprehensively label the DN1p population, we turned to InSITE911-GAL4, which is active in all DN1p cells in addition to a few other groups of neurons, including photoreceptor cells and a cluster of non-clock cells in the vicinity of LNd clock neurons (Cavanaugh et al., 2014) (Figure 1D). Notably, Kir2.1eGFP-mediated silencing of InSITE911-GAL4-expressing cells significantly decreased rest:activity rhythm strength (Figure 1G) without affecting fly survival. This result is consistent with the effects observed with R18H11-GAL4, bolstering the idea that DN1p cell activity is necessary for robust rest:activity rhythms. However, as is the case with R18H11-GAL4, the effect on locomotor activity rhythms produced by InSITE911-GAL4-driven Kir2.1eGFP cannot be unequivocally ascribed to DN1p cells due to the lack of specificity of the InSITE911-GAL4 driver. We therefore adopted an intersectional strategy to narrow expression of Kir2.1eGFP selectively to the DN1ps.
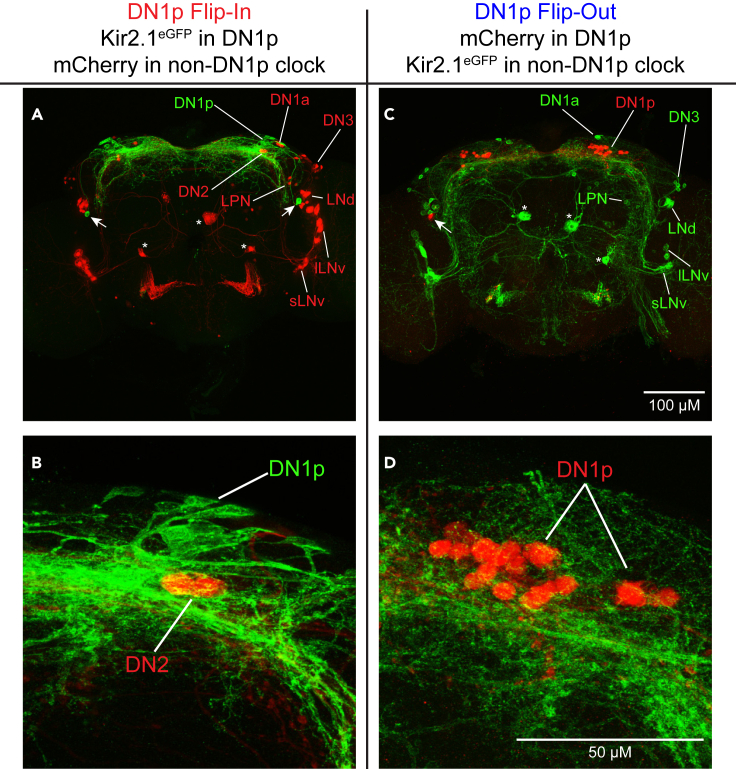
This approach involved the use effector lines that allowed for GAL4-driven Kir2.1eGFP to be either flipped into or out of a population of interest (Watanabe et al., 2017). To target Kir2.1eGFP specifically to the DN1p subset of InSITE911-expressing cells, we drove UAS>mCherry-stop>Kir2.1eGFP (“>” indicates a flippase (FLP)-sensitive FRT site) with Clk856-GAL4, which is active in most clock neurons in the brain (Gummadova et al., 2009). We paired this with the orthogonal QF-QUAS system, using an InSITE911-QF line to dictate expression of QUAS-FLP. In the presence of FLP, the intervening mCherry-stop cassette is excised, permitting GAL4-driven expression of Kir2.1eGFP. Immunostaining confirmed the effectiveness of this approach, which results in Kir2.1eGFP expression in all DN1p cells and virtually nowhere else in the brain, with the occasional exception of a 1–2 additional cells in the InSITE911 cluster adjacent to the LNds (Figures 2A and 2B). In these animals, which we call DN1p Flip-In flies, the entire DN1p neuronal subset should be electrically silenced via ectopic Kir2.1eGFP expression, with no effect elsewhere in the brain.
Figure 2.
An intersectional genetic strategy to selectively label DN1p cells
(A) Representative maximum projection confocal image of a DN1p Flip-In brain co-stained for Kir2.1eGFP (green) and mCherry (red). The major clock cell populations are labeled. In addition to the DN1p cells, Kir2.1eGFP is also present in two cells near the LNd clock neurons (arrows). mCherry is present in most non-DN1p clock cells and is additionally expressed in 3 medial non-clock cells (asterisks).
(B) Magnified image of the DN1 cells from the brain in (A). Kir2.1eGFP labels all DN1p cells. The 2 mCherry+ DN2 cells are also visible in this projection.
(C) Representative maximum projection confocal image of a DN1p Flip-Out brain co-stained for Kir2.1eGFP (green) and mCherry (red).
(D) Magnified image of the DN1 cells from the brain in (C). Expression of Kir2.1eGFP and mCherry in this brain are the converse of that observed in the DN1p Flip-In brain. See also Figure S1 for a demonstration of PER staining within labeled DN1p cells.
We also took a complementary approach to generate DN1p Flip-Out flies, in which a Kir2.1eGFP-stop cassette is excised from cells in which InSITE911-QF drives FLP expression. In these flies, Kir2.1eGFP is present throughout the clock network with the exception of DN1p cells, which instead express an inert mCherry reporter (Figures 2C and 2D). Importantly, we counted 15.0 ± 0.47 mCherry-expressing DN1p cells per hemisphere in DN1p Flip-Out brains, including 97.8 ± 1.5% of PER+ DN1p cells (n = 9 brains), demonstrating that the intersectional strategy targets the DN1ps comprehensively (Figure S1). Together, DN1p Flip-In and Flip-Out flies enabled us to test both the necessity and sufficiency of DN1p neuronal activity in the generation of locomotor rest:activity rhythms.
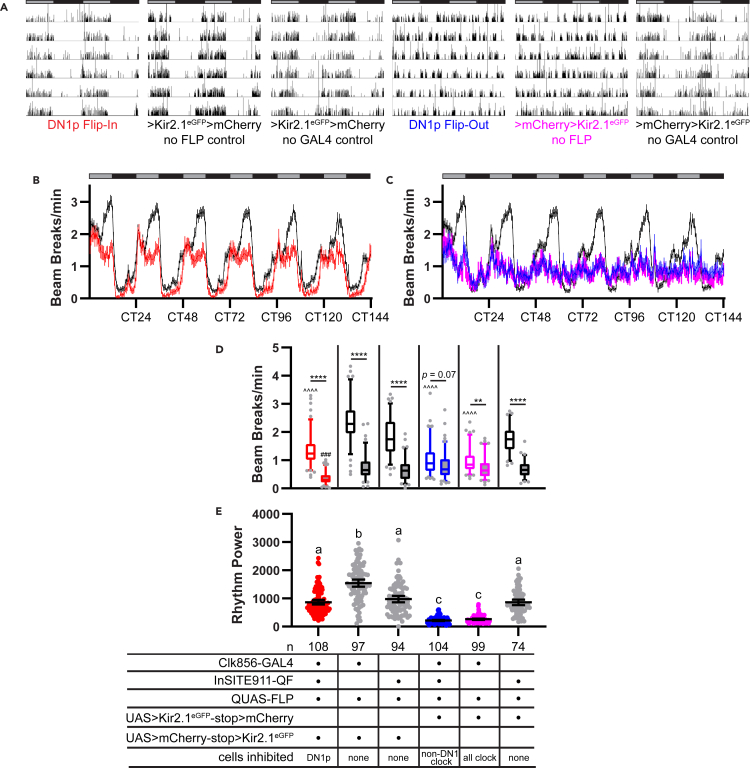
Notably, under constant dark (DD) conditions, DN1p Flip-In flies, in which DN1p cells selectively expressed Kir2.1eGFP, maintained robust daily oscillations in activity levels (Figures 3A and 3B), with higher levels of activity in the subjective day as compared to the subjective night (Figure 3D). Because of this, rest:activity period and rhythm strength of DN1p Flip-In flies was generally indistinguishable from control flies lacking Kir2.1eGFP expression (Table 1; Figure 3E). We note that DN1p Flip-In flies did show significantly reduced rest:activity rhythm strength compared to 1 of the 3 control lines, which expressed mCherry in all clock neurons due to a lack of InSITE911-QF to drive FLP expression. However, this control line exhibited extremely strong rest:activity rhythms that were significantly different from all other flies tested, including other controls. Thus, based on the lack of difference between the other two controls and DN1p Flip-In flies, we conclude that selective DN1p silencing is without effect on rest:activity rhythm strength. These results furthermore suggest that the reduction in rest:activity rhythm strength we observed with R18H11- and InSITE911-GAL4 represents nonspecific effects of Kir2.1eGFP in non-DN1p cells.
Figure 3.
Sustained free-running rest:activity rhythms following selective and comprehensive expression of Kir2.1eGFP in DN1p cells
(A) Representative single-fly activity records are shown for the genotypes listed. Activity in infrared beam breaks/min is plotted for each minute over 6 d in DD conditions. Data are double plotted, with 48 hr on each line and the second 24 hr replotted at the start of the next line. Gray and black bars above each plot represent subjective day and night, respectively.
(B) Mean number of DAM beam breaks/min (±95% confidence interval) is plotted over 6 days in DD conditions for DN1p Flip-In flies (red) compared with control flies (black).
(C) Mean DAM beam breaks/min is plotted as in (B) for DN1p Flip-Out flies (blue), flies in which all Clk856+ clock neurons are silenced with Kir2.1eGFP (pink), and control flies (black). The same control data are graphed in (B) and (C) and consist of a combination of 3 control lines lacking Kir2.1eGFP expression (see STAR Methods). Gray and black bars indicate subjective day and night, respectively.
(D) Box and whisker plots of mean DAM beam breaks/min over the 12-hr subjective day (open boxes) and 12-hr subjective night (filled boxes) are shown for the indicated genotypes. Box extends from 25th to 75th percentiles; whiskers extend to 5th and 95th percentiles. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001, within genotype comparison between subjective day and night activity; ˆˆˆˆp < 0.0001 compared to subjective day activity of all 3 control lines; ##p < 0.01 compared with subjective night activity of all three control lines, Tukey’s multiple comparisons test following two-way ANOVA.
(E) Rest:activity rhythm power over 6 days in DD is displayed for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. Different letters indicate significant difference (p < 0.05), Tukey’s multiple comparisons test following one-way ANOVA. The same letter indicates no significant different (p > 0.05).
Table 1.
Effect of DN1p manipulations on free-running circadian rhythms
| Group | n | % Rhythmic | Period | Power | Amplitude |
|---|---|---|---|---|---|
| DN1p Flip-In | 108 | 95.37 | 23.64 ± 0.03 | 865.8 ± 43.0 | 2.22 ± 0.07∗ |
| >mCherry>Kir2.1eGFP no FLP control | 97 | 97.94 | 23.48 ± 0.02 | 1544.3 ± 64.6 | 3.41 ± 0.09 |
| >mCherry>Kir2.1eGFP no GAL4 control | 94 | 97.87 | 23.60 ± 0.03 | 977.08 ± 56.3 | 2.71 ± 0.08 |
| DN1p Flip-Out | 104 | 2.88 | NA | 216.2 ± 10.9∗∗∗∗ | NA |
| >Kir2.1eGFP>mCherry no FLP | 99 | 1.01 | NA | 260.5 ± 12.7∗∗∗∗ | NA |
| >Kir2.1eGFP>mCherry no GAL4 control | 73 | 96.77 | 23.57 ± 0.04 | 867.9 ± 49.3 | 2.56 ± 0.07 |
Numbers given for period, power and amplitude are means ± SEM.
∗p < 0.05, ∗∗∗∗p < 0.0001 compared to all 3 control lines, Tukey’s multiple comparisons test following one-way ANOVA.
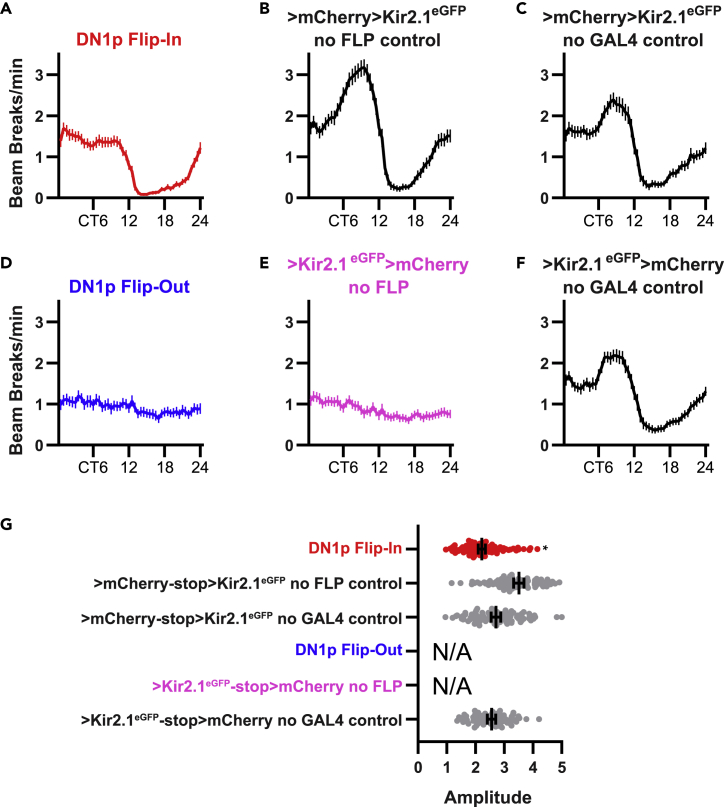
Despite the presence of normal strength circadian rest:activity rhythms, there were several important effects of DN1p neuronal silencing on the overall amount and distribution of activity across the 24-hr day. First, DN1p Flip-In flies were hypoactive compared to controls during both the subjective day and night (Figure 3D). However, as both daytime and nighttime activity were suppressed, peak to trough rhythm amplitude was not drastically altered (Table 1; Figures 4A–4G). DN1p Flip-In flies also lacked the prominent peak of activity present during the late subjective day that was present in control flies (Figures 3B and 4A–4G). As a result, in comparison to control lines, which exhibited highest activity levels in the late evening (Figures 4B, 4C, and 4F), activity levels of DN1p Flip-In flies remained relatively flat throughout most of the subjective day, if anything peaking during the early morning (Figure 4A). We conclude that though DN1p neuronal firing is dispensable for the maintenance of free-running rest:activity rhythms, these cells do appear to generally promote activity, especially during the second half of the subjective day.
Figure 4.
Neuronal silencing of DN1p cells suppresses the prominent evening activity peak
(A–F) Mean DAM bream breaks/min, averaged into 30 min bins over 6 DD d (±95% confidence interval), are shown for the indicated genotypes.
(G) Peak to trough rhythm amplitude is shown for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. ∗p < 0.05 compared with all 3 control lines; Tukey’s multiple comparisons test following one-way ANOVA. Peak to trough amplitude was not determined for DN1p Flip-Out and >Kir2.1eGFP-stop>mCherry no FLP flies because the overwhelming majority of these flies exhibited arrhythmic behavior.
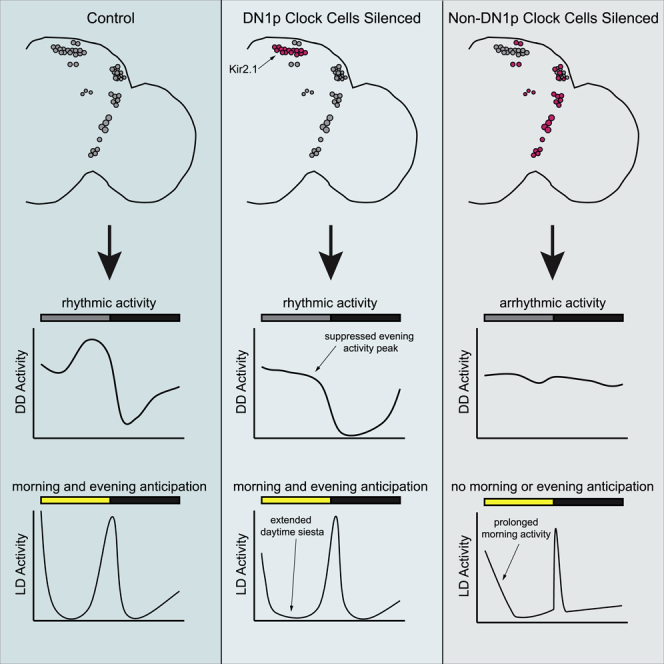
In contrast to the relatively subtle effects associated with selective silencing of DN1p cells, we observed drastic degradation of rest:activity rhythm strength in DN1p Flip-Out flies, in which only DN1p cells should be active among clock neurons. The vast majority of these flies showed arrhythmic activity patterns (Table 1; Figure 3A), and after ∼1 day in constant darkness, locomotor activity of DN1p Flip-Out flies failed to oscillate (Figure 3C). This drastically curtailed subjective day/night differences in activity levels (Figure 3D), substantially reducing the power of rest:activity rhythms (Figure 3E). In fact, in these measures, DN1p Flip-Out flies performed equivalently to flies in which all clock neurons, including DN1ps, were electrically silenced (Figures 3A, 3C, 3D, and 3E), indicating that DN1p neuronal firing is not sufficient for circadian rhythms of behavior when neuronal communication is prevented in the rest of the clock network.
DN1p flip-in flies exhibit normal morning and evening anticipatory activity under light-entrained conditions
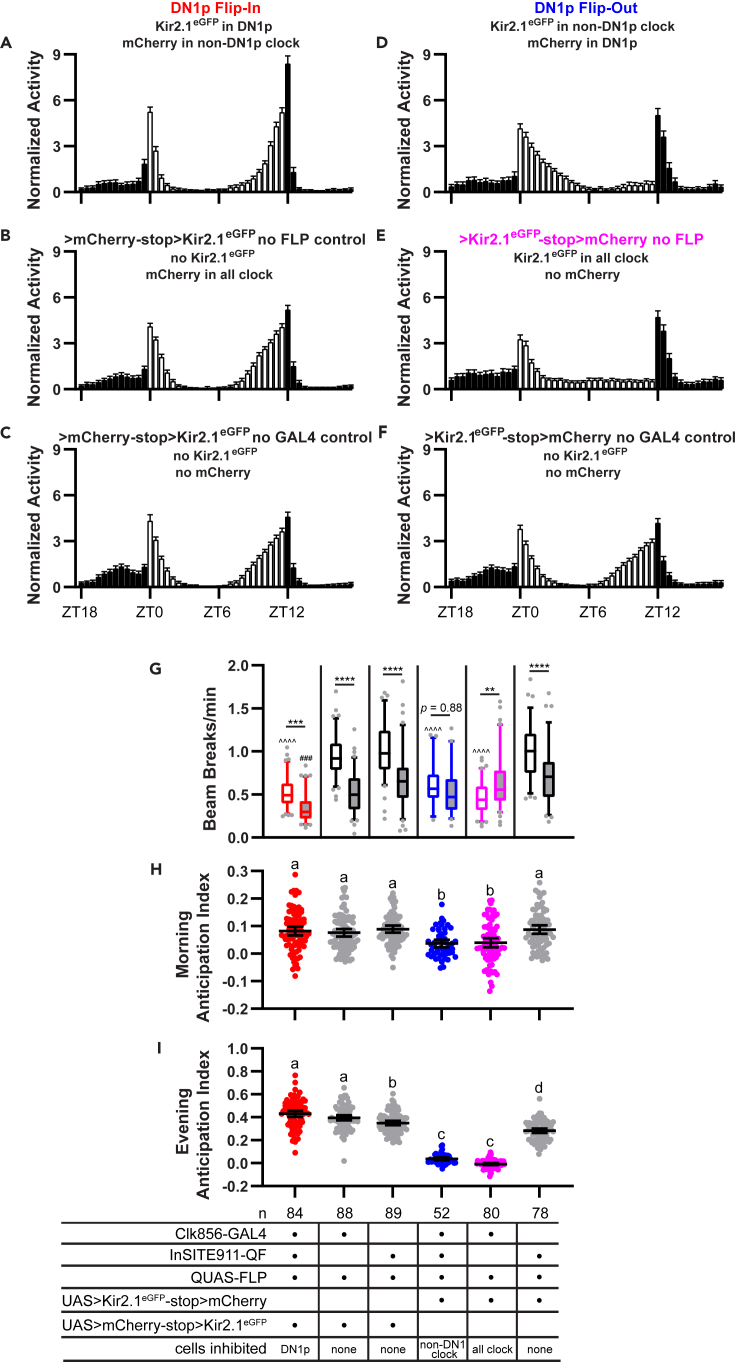
We also assessed the effect of DN1p silencing on light-entrained behavioral rhythms. Under normal light-dark (LD) cycles, flies exhibit crepuscular activity patterns, with morning and evening bouts of activity interspersed by two major sleep periods: a daytime siesta, and a consolidated period of nighttime sleep. Notably, morning and evening activity gradually ramp up preceding the lights-on and lights-off transitions, and this anticipatory activity occurs under control of the endogenous circadian clock (Dubowy and Sehgal, 2017). As expected, control flies exhibited both morning and evening anticipation (Figures 5B, 5C, 5F, 5H, and 5I), although under our experimental conditions, the evening peak was much more prominent than the morning peak. Importantly, we found no effect of Kir2.1eGFP expression in DN1p cells (in DN1p Flip-In flies) on the magnitude of morning or evening anticipation (Figures 5A, 5H, and 5I), again suggesting that DN1p cell activity is not required for circadian regulation of rest:activity rhythms. Nevertheless, as in DD, activity levels were lower across the day and night (Figures 5G and S2A). In particular, DN1p Flip-In flies appeared to have an extended mid-day siesta period compared with controls.
Figure 5.
Kir2.1eGFP expression in DN1p cells does not affect anticipatory locomotor activity in light-entrained conditions
(A–F) LD eduction plots are displayed for the indicated genotypes. Data show mean normalized DAM beam breaks/min (±95% confidence interval) in 30 min bins throughout the 24-hr day (see STAR Methods for explanation of normalization and averaging procedure). White and black bars represent activity during the light and dark periods, respectively. See also Figure S2 for non-normalized activity plots over the 5 d LD monitoring period.
(G) Box and whisker plots of mean DAM beam breaks/min over the 12-hr light period (open boxes) and 12-hr dark period (filled boxes) are shown for the indicated genotypes. Box extends from 25th to 75th percentiles; whiskers extend to 5th and 95th percentiles. ∗∗p < 0.01, ∗∗∗p < 0.001,∗∗∗∗p < 0.0001, within genotype comparison between light and dark period activity; ˆˆˆˆp < 0.0001 compared to light period activity of all 3 control lines; ###p < 0.001 compared with dark period activity of all three control lines; Tukey’s multiple comparisons test following two-way ANOVA.
(H and I) Morning and evening anticipation indices are plotted for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. Different letters indicate significant difference (p < 0.05), one-way ANOVA followed by Tukey’s multiple comparisons test. The same letter indicates no significant different (p > 0.05).
In contrast, DN1p Flip-Out flies had significantly reduced morning and evening anticipation compared to control lines, and in fact performed equally poorly on these measures as flies in which all clock neurons were silenced (Figures 5H and 5I). We note that because morning anticipation was subtle in control lines under our conditions, the magnitude of the effect on morning anticipation was small, making definitive conclusions difficult for this parameter. DN1p Flip-Out flies also lost the typical day-night differences in activity levels (Figure 5G). Interestingly, compared with flies in which all clock cells expressed Kir2.1eGFP, DN1p Flip-Out flies had an extended period of activity following the lights-on transition (Figures 5D and 5E), indicating that a function of these neurons in LD is to drive early morning activity. Nevertheless, the overall persistence of anticipatory activity following DN1p neuronal silencing, and its absence when DN1p cells alone retain neuronal firing, suggest that DN1ps are neither necessary nor sufficient for circadian rhythms under light-entrained conditions.
Kir2.1eGFP expression in DN1p cells increases sleep duration
DN1ps have recently been ascribed a sleep-regulatory function. There are conflicting reports as to whether these cells promote or inhibit sleep; however, this is likely due to the fact that distinct DN1p subpopulations may differentially regulate sleep and that the contribution of a single subpopulation may differ according to time of day or neuronal activity pattern (Guo et al, 2016, 2017, 2018; Kunst et al., 2014; Lamaze et al., 2018; Tabuchi et al., 2018). In fact, using a conceptually very similar intersectional genetic approach to that which we have applied here, Guo et al. showed that there are both sleep-promoting and sleep-suppressing subsets of DN1p neurons. Notably, however, the combined impact of the entire DN1p population on sleep has not been studied due to a lack of appropriately comprehensive drivers. We therefore assessed the timing and duration of sleep in both DN1p Flip-In and Flip-Out flies.
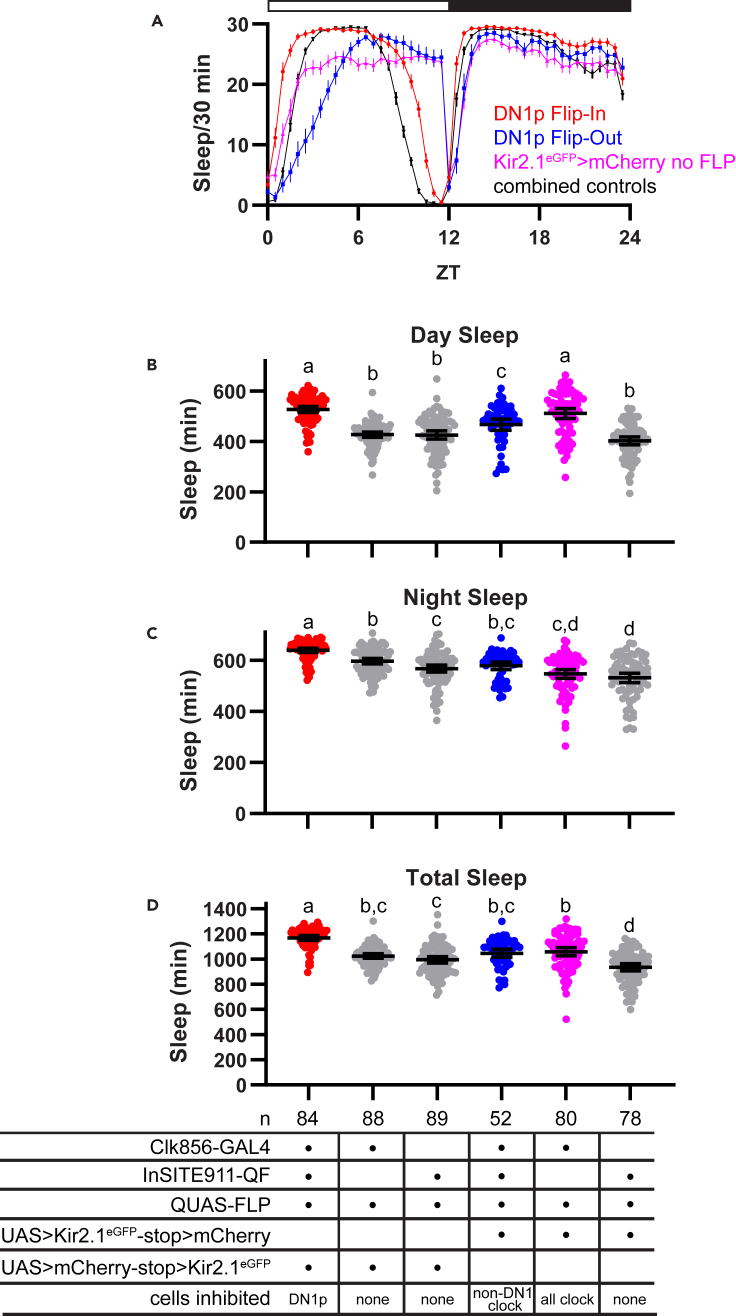
We found that selective Kir2.1eGFP-mediated inhibition of DN1p cells (in DN1p Flip-In flies) significantly increased sleep during both the day and night (Figures 6A–6D). Thus, although clearly the DN1p group is made up of functionally distinct subpopulations that differentially regulate sleep (Guo et al., 2018; Lamaze et al., 2018), our results suggest that the net impact of DN1p cell firing under normal conditions is to suppress sleep. We also recorded increased daytime sleep in DN1p Flip-Out flies, and this was associated with an apparent shift in the daytime siesta later into the light phase such that flies did not reach maximal daytime sleep until ∼ZT6 (ZT stands for zeitgeber time, with ZT0 corresponding to the time of lights-on) but then slept at near maximal levels until the lights-off transition (Figure 6A). In contrast, although DN1p Flip-In flies exhibited increased sleep across the duration of the typical siesta period, sleep levels began to decline around ZT6 such that these flies were largely awake at the end of the light period, at a time when DN1p Flip-Out flies remained asleep (Figure 6A). These results suggest that maintenance of neuronal activity in DN1p cells prevents early morning sleep (a conclusion that is consistent with the fact that selective DN1p inhibition drastically increases sleep at this time), and furthermore that neuronal activity in non-DN1p clock cells is responsible for driving late evening arousal.
Figure 6.
Kir2.1eGFP expression in DN1p cells increases sleep duration
(A) Mean sleep/30 min (±95% confidence interval) over the course of a 24-hr day is plotted for DN1p Flip-In flies (red), DN1p Flip-Out flies (blue), flies in which all Clk856-GAL4+ clock cells are silenced (pink), and control flies (black). Control plot consists of data from a combination of 3 control lines lacking Kir2.1eGFP expression (see STAR Methods). Data are averaged over 5 days in LD conditions. White and black bars indicate the light and dark periods, respectively.
(B–D) Total daytime, nighttime, or 24-hr sleep is plotted for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. Different letters indicate significant difference (p < 0.05); Tukey’s multiple comparisons test following one-way ANOVA. The same letter indicates no significant different (p > 0.05). See also Figure S3 for effects of DN1p silencing on sleep in DD conditions.
Notably, although sleep was increased throughout the day and night in DN1p Flip-In flies, they retained normal cycles of sleep and wakefulness, with consolidated periods of sleep during the midday siesta and at night, interspersed by periods of activity, which occurred primarily around lighting transitions. The retention of such cycles, which we note has also been observed with previous DN1p manipulations (Guo et al., 2018; Jin et al., 2021; Kunst et al., 2014), underscores our conclusion that DN1p cells are dispensable for circadian behavioral rhythms. We observed similar effects on sleep in free-running conditions. Thus, in DD, DN1p Flip-In flies had elevated sleep during both the subjective day and night compared with control lines but maintained sleep-wake rhythms, with relatively more sleep during the subjective night than the subjective day (Figures S3A–S3D). In contrast, sleep in DN1p Flip-Out flies was flat across the day and night, mimicking the effect produced by silencing all clock neurons (Figures S3A–S3D).
DN1a cells make minimal contributions to rest:activity rhythms under free-running and entrained conditions
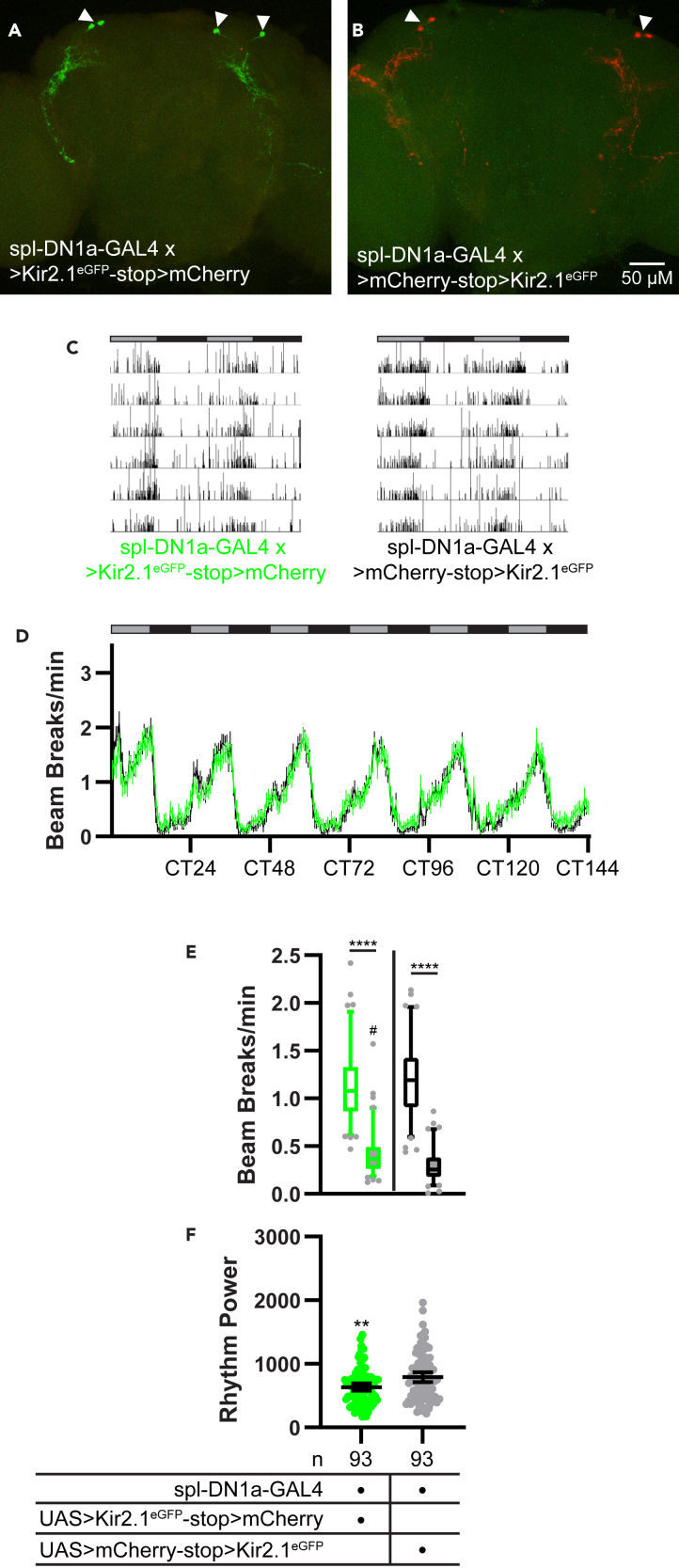
Owing to a lack of specific drivers, the role of DN1a clock cells in the production of circadian rest:activity rhythms has not been assessed in adult flies. Recently, however, split GAL4 lines have been developed that label groups of clock neurons more selectively, including the DN1a cells (Sekiguchi et al., 2020), enabling a more precise determination of their function. We therefore targeted Kir2.1eGFP to DN1a cells using a split-GAL4 line (which we call spl-DN1a-GAL4) that labels the DN1a cells and no other neurons in the brain (Sekiguchi et al., 2020) (Figures 7A and 7B) and monitored locomotor activity in these flies under both free-running and light-entrained conditioned.
Figure 7.
Minor effects of DN1a silencing on free-running rest:activity rhythms
(A and B) Representative maximum projection confocal images of brains from flies in which spl-DN1a-GAL4 was used to drive UAS>Kir2.1eGFP-stop>mCherry (A) or UAS>mCherry-stop>Kir2.1eGFP (B). Brains were co-stained for Kir2.1eGFP (green) and mCherry (red) immunofluorescence. Arrowheads indicate DN1a cells. Note that expression was restricted to 2 DN1a cells per hemisphere with this split GAL4 line.
(C) Representative single-fly activity records are shown for the genotypes listed. Activity in infrared beam breaks/min is plotted for each minute over 6 d in DD conditions.
(D) Mean number of DAM beam breaks/min (±95% confidence interval) is plotted over 6 days in DD conditions for flies with spl-DN1-GAL4-driven Kir2.1eGFP (green) and control flies with spl-DN1a-GAL4-driven mCherry (black). Gray and black bars indicate subjective day and night, respectively.
(E) Box and whisker plots of mean DAM beam breaks/min over the 12-hr subjective day (open boxes) and 12-hr subjective night (filled boxes) are shown for the indicated genotypes. Box extends from 25th to 75th percentiles; whiskers extend to 5th and 95th percentiles. ∗∗∗∗p < 0.0001, within genotype comparison between subjective day and night activity; #p < 0.05 compared with subjective night activity of control flies; Tukey’s multiple comparisons test following two-way ANOVA.
(F) Rest:activity rhythm power over 6 days in DD is plotted for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. ∗∗p < 0.01, t test.
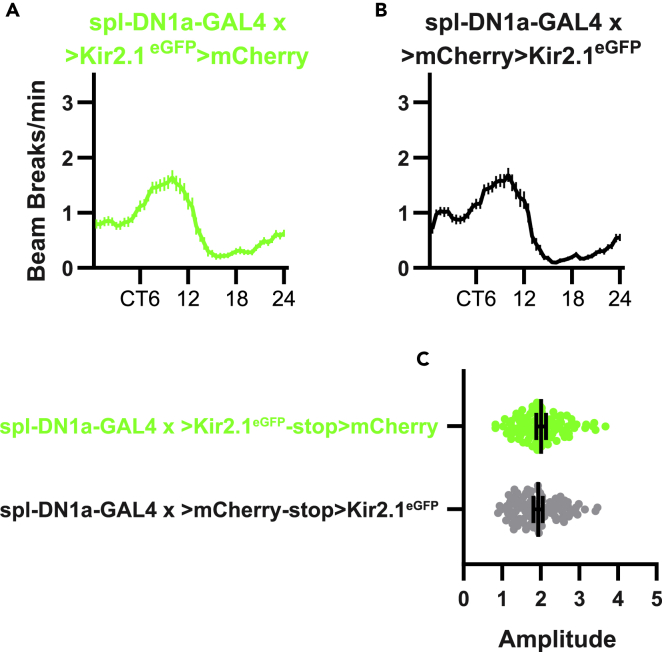
Kir2.1eGFP-mediated silencing of DN1a cells had only subtle effects on locomotor behavior under DD conditions. DN1a-silenced flies retained prominent oscillations in activity and rest (Figures 7C and 7D); however, they were slightly hypoactive compared with controls during the subjective day and slightly hyperactive during the subjective night (Figure 7E). The cumulative effect of these changes was to subtly flatten rest:activity oscillations, which caused a minor reduction in overall rest:activity rhythm strength (Table 2; Figure 7F). Despite this change in rhythm strength, we saw no effects on peak-to-trough rhythm amplitude (Table 2; Figures 8A–8C). These data suggest that DN1a cells make a limited contribution to rest:activity rhythms under DD conditions.
Table 2.
Effect of DN1a manipulations on free-running circadian rhythms
| Group | n | % Rhythmic | Period | Power | Amplitude |
|---|---|---|---|---|---|
| spDN1a-GAL4 x >Kir2.1eGFP>mCherry | 93 | 96.77 | 23.73 ± 0.04 | 634.0 ± 29.3∗∗ | 1.94 ± 0.06 |
| spDN1a-GAL4 x >mCherry>Kir2.1eGFP | 93 | 100 | 23.68 ± 0.03 | 791.9 ± 38.7 | 2.01 ± 0.06 |
Numbers given for period, power and amplitude are means ± SEM.
∗∗p < 0.01, t test.
Figure 8.
Kir2.1eGFP expression in DN1a cells does not affect the distribution of activity across the subjective day
(A and B) Mean DAM bream breaks/min, averaged into 30 min bins over 6 DD d (±95% confidence interval), are shown for the indicated genotypes.
(C) Peak to trough rhythm amplitude is shown for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. There was no effect of DN1a silencing on rhythm amplitude.
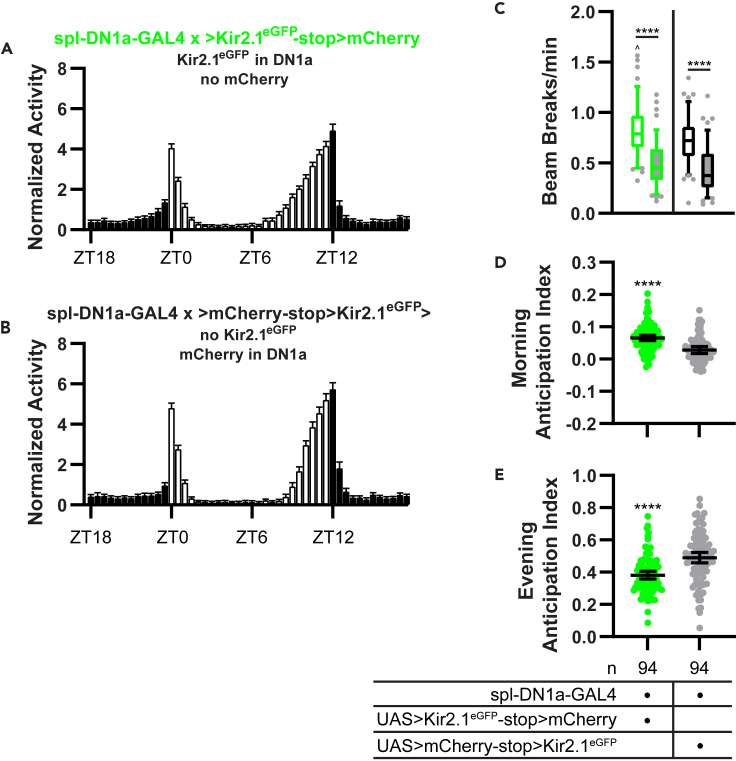
We also observed subtle changes in behavior under LD conditions. DN1a-silenced flies exhibited slightly exaggerated morning anticipatory activity compared with controls (Figures 9A, 9B, and 9D), although we note that morning anticipation was generally low in these control flies compared to those from other experiments. DN1a-silenced flies also exhibited prolonged evening anticipatory activity, beginning earlier in the day than control flies. This resulted in a reduced evening anticipation index (which is calculated based on the slope of the activity increase prior to lights-off), although anticipatory activity reached a similar maximum in both groups (Figures 9A, 9B, 9E, and S4A). This prolonged evening anticipation was likely responsible for the overall increased daytime activity in DN1a-silenced flies (Figure 9C).
Figure 9.
Sustained anticipatory locomotor activity in light-entrained conditions following selective Kir2.1eGFP expression in DN1a cells
(A and B) LD eduction plots are displayed for the indicated genotypes. Data show mean normalized DAM beam breaks/min (±95% confidence interval) in 30 min bins throughout the 24-hr day (see STAR Methods for explanation of normalization and averaging procedure). White and black bars represent activity during the light and dark periods, respectively. See also Figure S4 for non-normalized activity plots over the 5 d LD monitoring period.
(C) Box and whisker plots of mean DAM beam breaks/min over the 12-hr light period (open boxes) and 12-hr dark period (filled boxes) are shown for the indicated genotypes. Box extends from 25th to 75th percentiles; whiskers extend to 5th and 95th percentiles. ∗∗∗∗p < 0.0001, within genotype difference between light and dark period activity; ˆp < 0.05 compared with light period activity of control flies, Tukey’s multiple comparisons test following two-way ANOVA.
(D and E) Morning and evening anticipation indices are graphed for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. ∗∗∗∗p < 0.0001, t test. Morning and evening anticipation remain intact in the face of DN1a silencing, although morning anticipation is slightly enhanced and evening anticipation slightly dampened compared with controls. For (C–E), experimental flies with spl-DN1a-GAL4-driven expression of Kir2.1eGFP are shown in green and control files with spl-DN1a-GAL4-driven expression of mCherry are shown in black/gray.
Kir2.1eGFP expression in DN1a cells decreases sleep duration
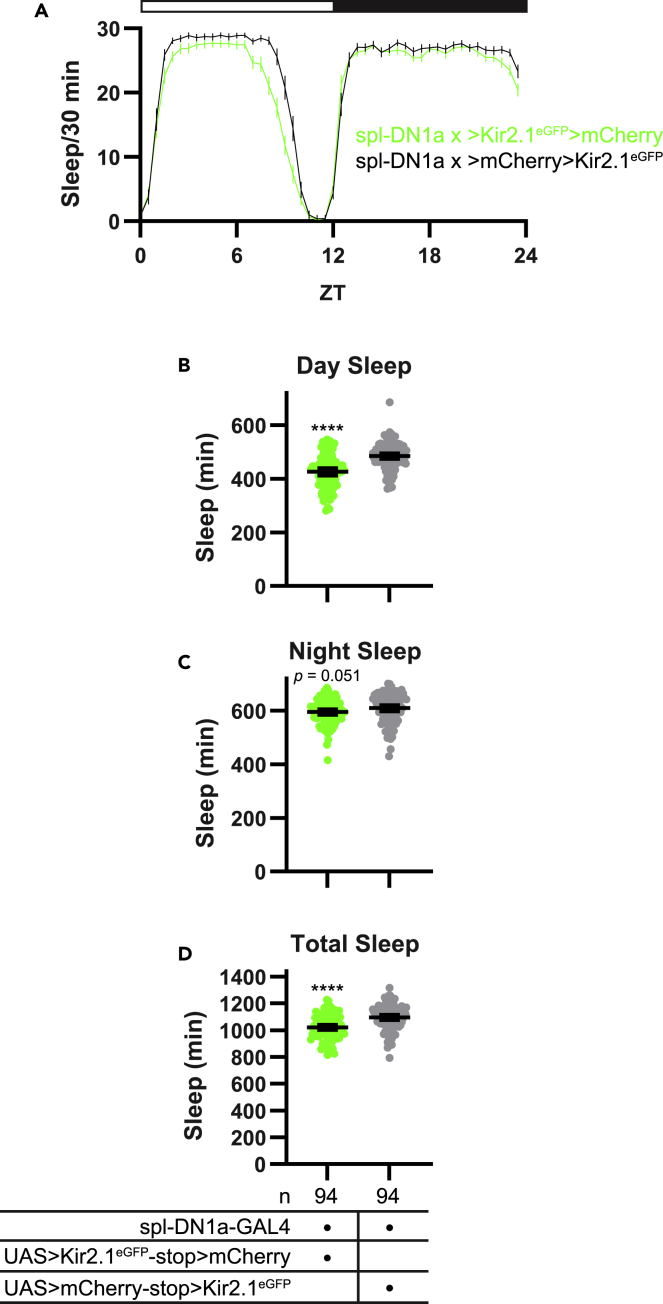
The increased daytime activity produced by Kir2.1eGFP expression in DN1a cells was associated with a corresponding decrease in daytime sleep, especially toward the end of the light period (Figures 10A and 10B). We also observed a reduction in sleep toward the end of the night, although this reduction narrowly failed to reach statistical significance when sleep was quantified across the entire dark period (Figures 10A and 10C). Under DD conditions, the effect of DN1a silencing was also to reduce sleep, although here the reduction was quite small and limited to the subjective night (Figures S5A–S5D). We note that these effects on sleep are opposite those we observed with DN1p manipulations, suggesting that these two clock cell groups provide counteracting influences on sleep behavior.
Figure 10.
Kir2.1eGFP expression in DN1a cells decreases sleep duration
(A) Mean sleep/30 min (±95% confidence interval) over the course of a 24-hr day is plotted for flies with spl-DN1a-GAL4-driven expression of Kir2.1eGFP (green) and control flies with spl-DN1a-GAL4 driven mCherry (black). Data are averaged over 5 days in LD conditions. White and black bars indicate the light and dark periods, respectively.
(B–D) Total daytime, nighttime, or 24-hr sleep is plotted for the indicated genotypes. Lines are means ±95% confidence intervals. Dots represent individual flies. ∗∗∗∗p < 0.0001, t test. See also Figure S5 for effects of DN1a silencing on sleep in DD conditions.
Discussion
The central circadian clock is comprised a network of interconnected populations of clock cells that can be distinguished based on molecular and anatomical properties. Although each clock cell contains a semi-autonomous molecular clock, the generation of coherent circadian outputs is an emergent property of the clock network that depends critically on intercellular communication (Dubowy and Sehgal, 2017; Hastings et al., 2019; Hermann-Luibl and Helfrich-Förster, 2015). A current focus of circadian research is therefore to delineate the circuit-level interactions through which circadian information is consolidated across clock cells and to define the pathways through which cells of the clock network access downstream brain regions to modulate behavioral processes.
In flies, this effort has been facilitated by the use of binary expression systems (such as GAL4-UAS) that enable the selective manipulation of molecularly defined neuronal populations. Because of this, progress in our understanding of the role of the different clock cells has often followed the development of drivers that label specific clock cell groups. For example, the discovery that the LNvs uniquely express the pigment-dispersing factor (Pdf) neuropeptide led not only to the identification of the central function of Pdf in clock network synchronization but also to the generation of highly selective Pdf-GAL4 lines that could be used for cell-specific LNv manipulations (Helfrich-Förster, 1995; Renn et al., 1999). These lines have proven instrumental in defining the essential pacemaker function of the LNvs within the clock network.
More recently, identification of DN1-selective drivers has led to an investigation into their contribution to circadian behavior. These studies have offered conflicting conclusions regarding the exact role of DN1 clock cells. For example, while molecular clock cycling in the Clk4.1-GAL4-expressing DN1p cells is neither necessary nor sufficient for free-running rhythms in DD conditions (Bulthuis et al., 2019; Schlichting et al., 2019; Zhang et al., 2010b), manipulations of these same cells that alter expression or function of Pdf receptor or the narrow abdomen ion channel can decrease DD rhythm strength (Flourakis et al., 2015; Seluzicki et al., 2014; Zhang et al., 2010a). Furthermore, some experiments in which the molecular clock in DN1p cells has been made to cycle at a different pace than other clock cells have implicated the DN1s in the determination of ensemble circadian period (Yao et al., 2016), while others have emphasized an output role important for setting circadian phase but not for determining period or overall rhythmicity (Chatterjee et al., 2018).
One complication in interpreting the results of these studies is the fact that the Clk4.1-GAL4 line labels only a subset of DN1p cells. This lack of comprehensiveness, which is also characteristic of other commonly used DN1p GAL4 lines, precludes an unequivocal determination of the role of DN1p cells due to the possibility that redundant contributions from the remaining, unaffected cells could provide sufficient function, as is known to occur for both the sLNv and LNd clock cells (Bulthuis et al., 2019; Helfrich-Förster, 1998). In addition to a lack of comprehensiveness, common DN1p-targeting GAL4 lines also exhibit expression outside of DN1p cells, which could result in non-specific effects of manipulations using these lines. These differences in expression could account for the variable behavioral phenotypes we and others have observed using different DN1p drivers. It could also underlie the substantial lethality we found with manipulations of Clk4.1-GAL4- or R18H11-GAL4-expressing cells, which may derive from non-DN1p expression that must be taken into account when assessing the effects of manipulations using these drivers.
To circumvent these issues, we devised an intersectional genetic strategy that labeled the DN1p cells completely and with negligible non-DN1 expression, and paired this with a relatively straightforward genetic manipulation, of neuronal silencing through expression of the Kir2.1 potassium channel. This approach, which minimized the potential for redundant or non-specific effects, provided evidence arguing against a necessary contribution of DN1p clock cells to either free-running or light-entrained rest:activity rhythms. We note that such a conclusion, based on negative results, would not have been possible with existing DN1p GAL4 lines. Recent studies have demonstrated that DN1p neurons are a heterogeneous group that can be further divided into multiple subpopulations that likely make distinct contributions to behavior (Chatterjee et al., 2018; Guo et al., 2018; Lamaze et al., 2018; Ma et al., 2021). These findings indicate that a thorough understanding of the role of DN1p neurons in circadian control will require a fine-grained analysis of different DN1p subpopulations. However, although it is undoubtedly of interest to define the smallest functional units of behavioral control, our experiments emphasize the utility of also taking a comprehensive approach, in particular for loss-of-function manipulations that test for cellular necessity.
Our results have important implications for our understanding of the organizational logic of the clock cell network. First, as the DN1ps in our DN1p Flip-In flies ostensibly lack communication with target cells, the persistence of behavioral rhythms argues against a necessary function of DN1ps in transmitting circadian information to downstream components of the circadian output circuits, as least with regard to control of locomotor activity rhythms. In other words, the clock network must be able to access output circuitry through alternate or parallel pathways that do not involve the DN1ps. In addition, our results suggest that feedback from DN1p cells to other clock neuron populations (Collins et al, 2012, 2014; Díaz et al., 2019; Fernandez et al., 2020; Guo et al., 2016) is unnecessary for those other cells to maintain rhythmicity. These conclusions are consistent with the recent proposal that lateral clock neurons access motor control centers in the ellipsoid body through intervening dopaminergic neurons and also potentially through a direct action of Pdf on ellipsoid body cells (Liang et al., 2019; Pírez et al., 2013). They are further supported by studies of glass mutant flies, which lack DN1p cells in addition to eye photoreceptors but have essentially normal rest:activity rhythms under DD conditions (Helfrich-Förster et al., 2001; Klarsfeld et al., 2004).
In previous work, we identified specific populations of cells in the pars intercerebralis (PI) region of the brain that serve as critical components of the circadian output circuitry regulating rest activity rhythms, and furthermore suggested that clock inputs to these PI cells could be provided by DN1p neurons (Cavanaugh et al., 2014). In support of this model, we also demonstrated that DN1p axons make close anatomical contact with PI output cells. Recently, this connectivity has been functionally confirmed through calcium imaging of PI cells during genetic activation of DN1p neurons (Barber et al., 2021; Jin et al., 2021). However, given our current results that indicate that rest:activity rhythms persist in the face of Kir2.1eGFP-mediated silencing of DN1p cells, it is clear that this DN1p to PI connection is not strictly necessary for circadian regulation of behavior. This could be either because other groups of clock neurons, including LNd cells, directly regulate PI output cells (Barber et al., 2016), thus allowing for circadian control of PI cells even in the absence of DN1p activity, or because circuits accessing non-PI output centers that do not require DN1p inputs are sufficient to drive behavioral rhythmicity.
Although DN1p Flip-In flies retained robust circadian rhythms, we did observe changes in the overall distribution and amount of daily activity and sleep. In both LD and DD, we found that silencing DN1p cells decreased locomotor activity and concomitantly increased sleep duration. The most prominent effect of DN1p silencing under LD conditions occurred in the early morning, following the lights-on transition. Thus, DN1p Flip-In flies exhibited an extended daytime siesta that began hours earlier than in control flies. Conversely, DN1p Flip-Out flies, in which all clock cells except DN1ps were made to express Kir2.1eGFP, displayed an extended period of activity following lights-on. Together, these results highlight an important role for the DN1ps in driving early morning activity. As neuronal activity of DN1ps is high at this time (Flourakis et al., 2015), we suggest that they provide important excitatory drive onto downstream activity-promoting cells. Importantly, the impact of DN1p silencing on sleep and activity that we observed indicates that the DN1ps do regulate locomotor and sleep outputs under the conditions in which we conducted our behavioral monitoring, despite the fact that circadian oscillations in these behavioral outputs persist in DN1p Flip-In flies.
To determine whether any behavioral effects of manipulations using these flies could result from expression of effector genes in non-central nervous system tissues, we also conducted a gross assessment of GAL4 activity outside the brain (data not shown). Though both Clk856-GAL4 and InSITE911-GAL4 individually exhibited minimal activity outside the brain (Clk856 in processes as well as a few intrinsic cells of the ventral nerve cord; InSITE911 in cells of the ejaculatory duct of the reproductive tract), these were in non-overlapping regions such that use of our intersectional approach produced no observable expression outside the brain. Thus, it is unlikely that the behavioral effects we observed in our DN1p Flip-In flies occur due to off-target impacts of Kir2.1eGFP expression in non-DN1p cells.
In contrast to the DN1ps, comparatively few studies have assessed the function of DN1a clock cells in adult flies. Like DN1ps, DN1a cells show circadian oscillations of neuronal activity (Alpert et al., 2020), and furthermore are thought to form reciprocal connections with lateral clock neurons (Fujiwara et al., 2018). In line with our findings of only minor effects of DN1a silencing on circadian modulation of activity and sleep, manipulations of CCHamide, which, among clock neurons, is selectively expressed by the DN1as, subtly alter morning activity and siesta length under LD conditions, with little impact on DD rhythmicity (Fujiwara et al., 2018). DN1a cells were also recently implicated in mediating the effects of cold temperature on sleep. In particular, suppression of neurotransmitter release in DN1a cells via expression of tetanus toxin was found to increase morning sleep, partially occluding the normal effect of cold temperature to suppress early morning activity (Alpert et al., 2020). In contrast, we found that Kir2.1eGFP-induced silencing of DN1a cells increased sleep, especially in the late evening. It is difficult to account for the differential effects produced by electrical silencing in our study compared with those observed by Alpert et al. using tetanus toxin expression; however, they could be due to the use of different split GAL4 and effector lines to target and manipulate the DN1a cells. Importantly, we tested for activity of our spl-DN1a-GAL4 line outside the brain and found no additional staining apart from very minimal expression in processes of the ventral nerve cord, indicating that the phenotypes we observed were likely due to manipulation of DN1a cells.
DN1 cells exhibit properties consistent with a role in circadian control of behavior, including circadian oscillations of neuronal activity (Alpert et al., 2020; Flourakis et al., 2015) and functional connections with both output cells (Barber et al., 2016; Cavanaugh et al., 2014; Jin et al., 2021; Tabuchi et al., 2018; Zhang et al., 2021b) and clock cells (Collins et al, 2012, 2014; Díaz et al., 2019; Fernandez et al., 2020; Guo et al., 2016). Nevertheless, our findings indicate a lack of necessity of DN1 cell activity in the generation of circadian rest:activity rhythms, raising the question of what role, if any, the DN1s play in circadian regulation. We suggest several possibilities. First, they could provide for parallel (but redundant) control of output circuits regulating rest:activity rhythms. If this were the case, then rest:activity rhythms would be expected to persist following DN1 silencing, due to the presence of alternate output circuitry. Second, they might be important only under certain environmental conditions, perhaps serving to integrate environmental stimuli such as light and temperature to regulate core clock pacemaking activity. Consistent with this possibility, both DN1a and DN1p cells have been found to be sensitive to acute temperature changes (Alpert et al., 2020; Yadlapalli et al., 2018), and temperature cycles act as potent zeitgebers for entraining DN1 molecular clocks (Yoshii et al., 2010). Finally, it is possible that DN1s are coupled to behavioral outputs other than rest and activity. In this regard, it is of interest that DN1 cells have previously been implicated in regulating various clock-controlled outputs, including temperature preference, male sex drive, courtship, oogenesis, and memory (Chen et al., 2017; Fujii and Amrein, 2010; Head et al., 2015; Yoshii et al., 2010; Zhang et al., 2021a, 2021b). Future studies aimed at testing the consequences of DN1 silencing using our comprehensive intersectional driver under more natural environmental conditions (for example, in the presence of ramping temperature cycles), or using different behavioral end points, should help to differentiate among these various possibilities.
Limitations of the study
A limitation of this study is our use of constitutive genetic manipulations. Because neuronal silencing was present throughout development, we cannot rule out the possibility of compensatory adaptations obscuring a more prominent role of DN1 cells in circadian regulation. This issue could be circumvented with adult-specific manipulations, for example using the TARGET system to restrict GAL4 activity to within a desired temporal window (McGuire et al., 2004), although this would be difficult given the already complex genetic makeup of our flies inherent in our intersectional approach. A related issue is that we did not assess developmental expression of our transgenes. Given that adult expression patterns often differ substantially from those observed developmentally, it is possible that Kir2.1eGFP was expressed more broadly during development compared with the highly selective adult pattern, thereby affecting a less specific group of cells. This concern is at least partially mitigated by the relatively subtle phenotypes we observed in adult flies, arguing against widespread developmental effects.
Furthermore, as we have not directly confirmed Kir2.1eGFP-induced silencing, it is possible that some cells retain neuronal activity even though our genetic manipulations result in comprehensive labeling of either the DN1p or DN1a population with Kir2.1eGFP. In addition, as it was recently demonstrated that neuropeptide release can occur in Drosophila neurons in the absence of neuronal activity (Klose et al., 2021), it is possible that DN1 signaling could persist even in the face of complete silencing. If either of these possibilities were to hold true, the general lack of effect associated with Kir2.1eGFP expression in DN1 cell could have resulted from incomplete prevention of DN1 signaling.
Finally, it is possible that DN1 cells do make a contribution to the generation or maintenance of locomotor activity rhythms, but that the effects are not apparent until multiple cycles under free-running conditions, for example, as has been observed for some parameters of rhythmicity following manipulations of Pdf-expressing sLNv clock cells (Renn et al., 1999). As our behavioral observations were limited to ∼1 week in DD, our experiments could have missed slower acting effects.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFP | Invitrogen | Cat# A-11122; RRID:AB_221569 |
| Guinea Pig anti-PER UP1140 | Gift of Amita Sehgal | N/A |
| Chicken anti-GFP | Invitrogen | Cat# A10262; RRID:AB_2534023 |
| Rabbit anti-dsRed | TaKaRa | Cat# 632496; RRID:AB_10013483 |
| FITC donkey anti-rabbit | Jackson | Cat# 711-095-152; RRID:AB_2315776 |
| Cy5 donkey anti-guinea pig | Jackson | Cat# 706-175-148; RRID:AB_2340462 |
| Cy3 donkey anti-rabbit | Jackson | Cat# 711-165-152; RRID:AB_2307443 |
| AlexaFluor 488 donkey anti-chicken | Jackson | Cat# 703-545-155; RRID:AB_2340375 |
| Experimental models: Organisms/strains | ||
| D. melanogaster: R18H11-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_48832 |
| D. melanogaster: InSITE911-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_63890 |
| D. melanogaster: QUAS-FLP | Bloomington Drosophila Stock Center | RRID:BDSC_30127 |
| D. melanogaster: UAS-GFP.nls | Bloomington Drosophila Stock Center | RRID:BDSC_4775 |
| D. melanogaster: tub>GAL80> | Bloomington Drosophila Stock Center | RRID:BDSC_38881 |
| D. melanogaster: 10XUAS-IVS-myr::tdTomato | Bloomington Drosophila Stock Center | RRID:BDSC_32221 |
| D. melanogaster: Clk4.1-GAL4 | Gift of Paul Hardin | RRID:BDSC_36316 |
| D. melanogaster: spl-gDN1-GAL4 | Gift of Gerry Rubin | SS00781 |
| D. melanogaster: InSITE911-QF | Gift of Liqun Luo | N/A |
| D. melanogaster: Clk856-GAL4 | Gift of Orie Shafer | FBtp0069616 |
| D. melanogaster: UAS>Kir2.1eGFP-stop>mCherry | Gift of David Anderson | N/A |
| D. melanogaster: UAS>mCherry-stop>Kir2.1eGFP | Gift of David Anderson | N/A |
| D. melanogaster: R43D05-p65.AD | Bloomington Drosophila Stock Center | RRID:BDSC_69883 |
| D. melanogaster: R93B11-DBD | Bloomington Drosophila Stock Center | RRID:BDSC_70370 |
| Software and algorithms | ||
| GraphPad Prism 9 | GraphPad | RRID:SCR_002798 |
| ClockLab | Actimetrics | RRID:SCR_014309 |
| Sleep and Circadian Analysis MATLAB Program (SCAMP) v2 | Chris Vecsey | N/A |
| Photoshop 2021 | Adobe | RRID:SCR_014199 |
| ImageJ | https://imagej.nih.gov/ij/index.html | RRID:SCR_003070 |
Resource availability
Lead contact
Further information and requests for resources, reagents and raw datasets should be directed to and will be fulfilled by the lead contact, Daniel Cavanaugh (dcavanaugh1@luc.edu).
Materials availability
All fly stocks used in this study are available from the lead contact.
Experimental model and subject details
Fly lines
∼7d-old male flies were used for all experiments. Flies were raised on cornmeal-molasses medium and were entrained to a 12:12 LD cycle at 25°C for ≥ 5 d prior to behavioral or immunohistochemical experiments. The following fly stocks were used: R18H11-GAL4 (RRID:BDSC_48832), InSITE911-GAL4 (RRID:BDSC_63890) (Gohl et al., 2011), QUAS-FLP (RRID:BDSC_30127), UAS-GFP.nls (RRID:BDSC_4775), tub>GAL80> (RRID:BDSC_38881), and 10XUAS-IVS-myr::tdTomato (RRID:BDSC_32221) were ordered from the Bloomington Drosophila Stock Center. Clk4.1-GAL4 (RRID:BDSC_36316) (Zhang et al., 2010a, 2010b) was a gift from Paul Hardin. spl-gDN1-GAL4 (SS00781) (Guo et al., 2017) was a gift from Gerry Rubin. InSITE911-QF flies were made by swapping in a QF element to replace the GAL4 in the InSITE911-GAL4 line (generated by Xiaojing Gao and Liqun Luo). Clk856-GAL4 (Gummadova et al., 2009) was a gift from Orie Shafer. UAS>Kir2.1eGFP-stop>mCherry and UAS>mCherry-stop>Kir2.1eGFP (Watanabe et al., 2017) were gifts from David Anderson. The spl-DN1a-GAL4 line was made from a combination of R43D05-p65.AD (RRID:BDSC_69883) and R93B11-DBD (RRID:BDSC_70370) (Sekiguchi et al., 2020).
Method details
Free-running rest:activity rhythm analysis
Following entrainment, individual ∼7 d old male flies were loaded into glass tubes containing 5% sucrose and 2% agar for locomotor activity analysis with the Drosophila Activity Monitoring (DAM) System (Trikinetics, Waltham MA). Monitoring with DAM2 monitors was conducted at 25°C in DD conditions and data were acquired every minute. For each individual fly, rest:activity rhythm period and strength (power) were determined for 1 min bins over days 2-7 of DD with ClockLab software (Actimetrics, Wilmette IL) using chi-square periodogram analysis. Rhythm power was calculated as the amplitude of the periodogram line at the dominant period minus the chi-square significance line (at a significance of p < 0.01). Flies that died during the course of behavioral monitoring were identified via visual inspection of activity records and removed from analysis. All flies that survived through the end of the one-week monitoring period were included in mean rest:activity power determination. Because rhythm strength cannot be negative, flies with a calculated power < 0 were assigned a power of 0 for subsequent analysis. Presence or absence of behavioral rhythmicity was determined with ClockLab software based on two criteria (Ewer et al., 1992; Shafer and Taghert, 2009): 1) rhythm power (calculated with 30 min bins over d 2-7 of DD) had to be ≥ 10, and 2) the width of the major periodogram peak had to be ≥ 1.5 hrs. Only rhythmic flies were included in assessment of free-running period.
To determine peak to trough rhythm amplitude, we first averaged single fly activity data into 30 min bins and then averaged these across days 2-7 of DD. Peak to trough amplitude was determined for each fly as the absolute difference in activity level between the bins with highest and lowest activity.
Morning and evening anticipation analysis
Flies were entrained and loaded for DAMS monitoring as described above. DAMS monitoring was conducted at 25°C in 12:12 LD conditions, at a light intensity of ∼700 lux. Flies were transferred into behavioral monitoring incubators at the day-night transition (ZT12) and analysis was performed on 5 consecutive days of recording, beginning with the lights-on transition (ZT0) on d 2. This eliminated the first ∼12 hours of data, which allowed for acclimation to the monitoring conditions. Activity counts were acquired in 1 min bins. These data were subsequently averaged into 30 min bins and normalized on a fly-by-fly basis by dividing each data point by the average amount of activity per min for that fly over the 5 d of analysis. These normalized data were used to create LD eduction plots and to calculate morning and evening anticipation indices. Eduction plots (Figures 5A–5F, 9A and 9B) combine the 5 d of data into a single average day. Morning and evening anticipation indices were quantified for each individual fly and consisted of the slope of the least-squares linear regression line fitted to the 12 data points (representing 6 hr of data) preceding the lights-on (morning anticipation) or lights-off (evening anticipation) transition (Fernandez et al., 2020).
Sleep analysis
The same locomotor activity data collected for DD and LD circadian analyses were used to assess mean daily sleep and activity. Sleep and activity levels were calculated with version 2 of the Sleep and Circadian Analysis MATLAB Program (SCAMP) (Donelson et al., 2012). The stdur (Total Sleep Duration) and atcounts (Total Activity Counts) functions were used to determine average total sleep and activity, and the s30 function (Minutes of Sleep/30 Mins) was used to plot average sleep duration across the 24-hr day in 30 min bins (Figures 6A, 10A, S3A, and S5A). For these analyses, data from individual flies were first averaged across 6 d (for DD experiments) or 5d (for LD experiments), beginning at the start of d 2, before being combined to calculate genotype averages. Sleep was defined as ≥ 5 consecutive min of inactivity, as determined by a lack of DAM beam breaks (Hendricks et al., 2000).
Immunohistochemistry
Adult (∼7d old) male fly brains were dissected in phosphate-buffered saline with 0.1% Triton-X (PBST) and fixed in 4% formaldehyde for 20-35 min. Brains were rinsed 3 X 15 min with PBST, blocked for 60 min in 5% normal donkey serum in PBST (NDST), and incubated for 24 hrs at RT in primary antibodies diluted in NDST. Brains were then rinsed 3 X 15 min in PBST, incubated for 24 hrs in secondary antibodies diluted in NDST, rinsed 3 X 15 min in PBST, cleared for 5 min in 50% glycerol in PBST, and mounted in Vectashield. To label GFP.nls (Figures 1A–1D), the primary antibody was rabbit anti-GFP 1:1000 (Invitrogen A-11122) and the secondary antibody was FITC donkey anti-rabbit 1:1000 (Jackson 711-095-152). For simultaneous labeling of Kir2.1eGFP, mCherry and PER (Figures 2A–2D, 7A, 7B, and S1), the primary antibodies were chicken anti-GFP 1:1000 (Invitrogen A10262), rabbit anti-dsRed 1:1000 (Takara 632496) and guinea pig anti-PER (UP1140) 1:1000, and the secondary antibodies were Alexa Fluor488 donkey anti-chicken 1:1000 (Jackson 703-545-155), Cy3 donkey anti-rabbit 1:1000 (Jackson 711-165-152) and Cy5 donkey anti-guinea pig 1:1000 (Jackson 706-175-148). Immunolabeled brains were visualized with a Fluoview 1000 (Olympus) or LSM880 (Zeiss) confocal microscope. Images from different channels were combined in Adobe Photoshop.
ImageJ was used to count the number of mCherry+ and PER+ DN1p cells and determine the extent of overlap between these two markers in DN1p Flip-Out fly brains. In addition to DN1ps, PER was also present in nearby DN1a and DN2 clock cells, as well as glial cells. It was occasionally difficult to differentiate PER staining in DN1p cells from that of nearby glia; however, glial PER label tended to be restricted to a smaller area and to have an absence of staining at the cell center. Nevertheless, it is possible that in some cases, we misclassified a glial cell as a DN1 neuron.
Gross assessment of non-neuronal GAL4 expression
We imaged endogenous fluorescence of a myr-tdTomato reporter in acutely dissected, unfixed tissue from male flies. We dissected out all part of the fly, including the brain, ventral nerve cord, digestive tract, reproductive tract, thorax, abdomen, legs, wings, eyes and proboscis, mounted them in Vectashield and immediately visualized with a Fluoview 1000 (Olympus) confocal microscope. We acquired images in both the red channel (to determine specific signal) and green channel (to account for non-specific autofluorescence). For Clk856-GAL4, InSITE911-GAL4, spl-DN1a-GAL4 lines, we directly drove expression of 10XUAS-IVS-myr::tdTomato. For our DN1p intersectional approach, we monitored for tdTomato expression in flies of the genotype Clk856-GAL4/10XUAS-IVS-myr::tdTomato; InSITE911-QF,QUAS-FLP/tub>GAL80>, in which tdTomato is limited to cells expressing both Clk856-GAL4 and InSITE911-QF. We also imaged control flies containing 10XUAS-IVS-myr::tdTomato alone to account for leaky tdTomato expression. We only considered a tissue to be positive for GAL4 activity if we observed specific signal that was absent in control flies and could be differentiated from autofluorescence. In some tissues, such as the rectal ampulla, autofluorescence was extremely high, precluding unequivocal determination of specific signal.
Quantification and statistical analysis
To ensure reproducibility, we conducted 2-4 independent experimental replicates for each behavioral analysis, and data from all flies of a given genotype that survived the duration of the experiment were pooled. For experiments depicted in Figures 1, 7, 8, 9, and 10, experimental flies consisted of different GAL4 lines crossed to a UAS>Kir2.1eGFP-stop>mCherry effector line. In the absence of FLP, these flies express Kir2.1eGFP in all GAL4-expressing cells. Controls for these experiments consisted of the same GAL4 lines driving expression of UAS>mCherry-stop>Kir2.1eGFP, which in the absence of FLP will express mCherry in GAL4-expressing cells. For the experiments depicted in Figures 2, 3, 4, 5, and 6, we compared several genotypes that expressed Kir2.1eGFP in different subsets of cells. Controls for these experiments lacked Kir2.1eGFP expression, either because of a lack of a GAL4 to drive expression, or a lack of FLP in flies expressing UAS>mCherry-stop>Kir2.1eGFP. Statistical analyses were performed with GraphPad Prism 9.0.0 software (La Jolla, CA). Circadian rhythm period, power, amplitude, morning and evening anticipation, sleep and activity amounts were analyzed using a t-test (for experiments with only 2 groups run simultaneously) or a one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test (for experiments with 3 or more groups). Total morning and evening activity were analyzed using a two-way ANOVA (with genotype and time of day as factors) followed by Tukey’s multiple comparisons test. For all statistical tests, p < 0.05 was considered significant. For experiments with multiple control groups, effects were only deemed significant if experimental lines were significantly different from all control lines.
Acknowledgments
We thank Drs. David Anderson, Paul Hardin, Gerry Rubin, and Orie Shafer for fly stocks, Dr. Amita Sehgal for antibodies, and Dr. Chris Vecsey for making available the SCAMP analysis program. This work was supported by the National Institute of General Medical Sciences, www.nigms.nih.gov/, Grant R15GM128170 to D.J.C.
Author contributions
Conceptualization, supervision, writing – original draft, and funding acquisition, D.J.C.; methodology, visualization preparation, investigation, and writing – review & editing, E.A.N, T.S., A.N., S.S., and D.J.C.; formal analysis, E.A.N, T.S., A.N., and D.J.C.
Declaration of interests
The authors declare no competing interests.
Published: September 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103001.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Alpert M.H., Frank D.D., Kaspi E., Flourakis M., Zaharieva E.E., Allada R., Para A., Gallio M. A circuit encoding absolute cold temperature in Drosophila. Curr. Biol. 2020;30:2275–2288.e5. doi: 10.1016/j.cub.2020.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber A.F., Erion R., Holmes T.C., Sehgal A. Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev. 2016;30:2596–2606. doi: 10.1101/gad.288258.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber A.F., Fong S.Y., Kolesnik A., Fetchko M., Sehgal A. Drosophila clock cells use multiple mechanisms to transmit time-of-day signals in the brain. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2019826118. e2019826118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito J., Houl J.H., Roman G.W., Hardin P.E. The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J. Biol. Rhythms. 2008;23:296–307. doi: 10.1177/0748730408318588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulthuis N., Spontak K.R., Kleeman B., Cavanaugh D.J. Neuronal activity in non-LNv clock cells is required to produce free-running rest:activity rhythms in Drosophila. J. Biol. Rhythms. 2019;34:249–271. doi: 10.1177/0748730419841468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh D.J., Geratowski J.D., Wooltorton J.R.A., Spaethling J.M., Hector C.E., Zheng X., Johnson E.C., Eberwine J.H., Sehgal A. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell. 2014;157:689–701. doi: 10.1016/j.cell.2014.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A., Lamaze A., De J., Mena W., Chélot E., Martin B., Hardin P., Kadener S., Emery P., Rouyer F. Reconfiguration of a multi-oscillator network by light in the Drosophila circadian clock. Curr. Biol. 2018;28:2007–2017. doi: 10.1016/j.cub.2018.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Sitaraman D., Chen N., Jin X., Han C., Chen J., Sun M., Baker B.S., Nitabach M.N., Pan Y. Genetic and neuronal mechanisms governing the sex-specific interaction between sleep and sexual behaviors in Drosophila. Nat. Commun. 2017;8:154. doi: 10.1038/s41467-017-00087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Kane E.A., Reeves D.C., Akabas M.H., Blau J. Balance of activity between LNvs and glutamatergic dorsal clock neurons promotes robust circadian rhythms in Drosophila. Neuron. 2012;74:706–718. doi: 10.1016/j.neuron.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B., Kaplan H.S., Cavey M., Lelito K.R., Bahle A.H., Zhu Z., Macara A.M., Roman G., Shafer O.T., Blau J. Differentially timed extracellular signals synchronize pacemaker neuron clocks. PLoS Biol. 2014;12:e1001959. doi: 10.1371/journal.pbio.1001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A., Berni J., Aranovich E.J., Muraro N.I., Beckwith E.J., Ceriani M.F. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr. Biol. 2011;21:1783–1793. doi: 10.1016/j.cub.2011.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz M.M., Schlichting M., Abruzzi K.C., Long X., Rosbash M. Allatostatin-C/AstC-R2 is a novel pathway to modulate the circadian activity pattern in Drosophila. Curr. Biol. 2019;29:13–22.e3. doi: 10.1016/j.cub.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson N.C., Kim E.Z., Slawson J.B., Vecsey C.G., Huber R., Griffith L.C. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the “tracker” program. PLoS One. 2012;7:e37250. doi: 10.1371/journal.pone.0037250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy C., Sehgal A. Circadian rhythms and sleep in Drosophila melanogaster. Genetics. 2017;205:1373–1397. doi: 10.1534/genetics.115.185157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewer J., Frisch B., Hamblen-Coyle M.J., Rosbash M., Hall J.C. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J. Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M.P., Pettibone H.L., Bogart J.T., Roell C.J., Davey C.E., Pranevicius A., Huynh K.V., Lennox S.M., Kostadinov B.S., Shafer O.T. Sites of circadian clock neuron plasticity mediate sensory integration and entrainment. Curr. Biol. 2020;30:2225–2237.e5. doi: 10.1016/j.cub.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M., Kula-Eversole E., Hutchison A.L., Han T.H., Aranda K., Moose D.L., White K.P., Dinner A.R., Lear B.C., Ren D. A conserved bicycle model for circadian clock control of membrane excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S., Amrein H. Ventral lateral and DN1 clock neurons mediate distinct properties of male sex drive rhythm in Drosophila. Proc. Natl. Acad. Sci. U S A. 2010;107:10590–10595. doi: 10.1073/pnas.0912457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Hermann-Luibl C., Katsura M., Sekiguchi M., Ida T., Helfrich-Forster C., Yoshii T. The CCHamide1 neuropeptide expressed in the anterior dorsal neuron 1 conveys a circadian signal to the ventral lateral neurons in Drosophila melanogaster. Front. Physiol. 2018;9:1276. doi: 10.3389/fphys.2018.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl D.M., Silies M.A., Gao X.J., Bhalerao S., Luongo F.J., Lin C.-C., Potter C.J., Clandinin T.R. A versatile in vivo system for directed dissection of gene expression patterns. Nat. Methods. 2011;8:231–237. doi: 10.1038/nmeth.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B., Chélot E., Xia R., Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Gummadova J.O., Coutts G.A., Glossop N.R.J. Analysis of the Drosophila clock promoter reveals heterogeneity in expression between subgroups of central oscillator cells and identifies a novel enhancer region. J. Biol. Rhythms. 2009;24:353–367. doi: 10.1177/0748730409343890. [DOI] [PubMed] [Google Scholar]
- Guo F., Chen X., Rosbash M. Temporal calcium profiling of specific circadian neurons in freely moving flies. Proc. Natl. Acad. Sci. U S A. 2017;114:E8780–E8787. doi: 10.1073/pnas.1706608114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F., Holla M., Díaz M.M., Rosbash M. A circadian output circuit controls sleep-wake arousal in Drosophila. Neuron. 2018;100:624–635.e4. doi: 10.1016/j.neuron.2018.09.002. [DOI] [PubMed] [Google Scholar]
- Guo F., Yu J., Jung H.J., Abruzzi K.C., Luo W., Griffith L.C., Rosbash M. Circadian neuron feedback controls the Drosophila sleep–activity profile. Nature. 2016;536:292–297. doi: 10.1038/nature19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M.H., Maywood E.S., Brancaccio M. The mammalian circadian timing system and the suprachiasmatic nucleus as its pacemaker. Biology (Basel) 2019;8:13. doi: 10.3390/biology8010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head L.M., Tang X., Hayley S.E., Goda T., Umezaki Y., Chang E.C., Leslie J.R., Fujiwara M., Garrity P.A., Hamada F.N. The influence of light on temperature preference in Drosophila. Curr. Biol. 2015;25:1063–1068. doi: 10.1016/j.cub.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J. Comp. Physiol. A. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- Helfrich-Förster C., Winter C., Hofbauer A., Hall J.C., Stanewsky R. The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron. 2001;30:249–261. doi: 10.1016/s0896-6273(01)00277-x. [DOI] [PubMed] [Google Scholar]
- Hendricks J.C., Finn S.M., Panckeri K.A., Chavkin J., Williams J.A., Sehgal A., Pack A.I. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Hermann-Luibl C., Helfrich-Förster C. Clock network in Drosophila. Curr. Opin. Insect Sci. 2015;7:65–70. doi: 10.1016/j.cois.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Jin X., Tian Y., Zhang Z.C., Gu P., Liu C., Han J. A subset of DN1p neurons integrates thermosensory inputs to promote wakefulness via CNMa signaling. Curr. Biol. 2021;31:2075–2087.e6. doi: 10.1016/j.cub.2021.02.048. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A., Malpel S., Michard-Vanhée C., Picot M., Chélot E., Rouyer F. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. J. Neurosci. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose M.K., Bruchez M.P., Deitcher D.L., Levitan E.S. Temporally and spatially partitioned neuropeptide release from individual clock neurons. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2101818118. e2101818118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M., Hughes M.E., Raccuglia D., Felix M., Li M., Barnett G., Duah J., Nitabach M.N. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol. 2014;24:2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamaze A., Krätschmer P., Chen K.-F., Lowe S., Jepson J.E.C. A wake-promoting circadian output circuit in Drosophila. Curr. Biol. 2018;28:3098–3105.e3. doi: 10.1016/j.cub.2018.07.024. [DOI] [PubMed] [Google Scholar]
- Liang X., Ho M.C.W., Zhang Y., Li Y., Wu M.N., Holy T.E., Taghert P.H. Morning and evening circadian pacemakers independently drive premotor centers via a specific dopamine relay. Neuron. 2019;102:843–857.e4. doi: 10.1016/j.neuron.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Holy T.E., Taghert P.H. A series of suppressive signals within the Drosophila circadian neural circuit generates sequential daily outputs. Neuron. 2017;94:1173–1189.e4. doi: 10.1016/j.neuron.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Mahesh G., Houl J.H., Hardin P.E. Circadian activators are expressed days before they initiate clock function in late pacemaker neurons from Drosophila. J. Neurosci. 2015;35:8662–8671. doi: 10.1523/JNEUROSCI.0250-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Przybylski D., Abruzzi K.C., Schlichting M., Li Q., Long X., Rosbash M. A transcriptomic taxonomy of Drosophila circadian neurons around the clock. Elife. 2021;10:e63056. doi: 10.7554/eLife.63056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S.E., Mao Z., Davis R.L. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Picot M., Cusumano P., Klarsfeld A., Ueda R., Rouyer F. Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 2007;5:e315. doi: 10.1371/journal.pbio.0050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pírez N., Christmann B.L., Griffith L.C. Daily rhythms in locomotor circuits in Drosophila involve PDF. J. Neurophysiol. 2013;110:700–708. doi: 10.1152/jn.00126.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn S.C., Park J.H., Rosbash M., Hall J.C., Taghert P.H. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Schlichting M., Diaz M.M., Xin J., Rosbash M. Neuron-specific knockouts indicate the importance of network communication to Drosophila rhythmicity. Elife. 2019;8:e48301. doi: 10.7554/eLife.48301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi M., Inoue K., Yang T., Luo D.-G., Yoshii T. A catalog of GAL4 drivers for labeling and manipulating circadian clock neurons in Drosophila melanogaster. J. Biol. Rhythms. 2020 doi: 10.1177/0748730419895154. [DOI] [PubMed] [Google Scholar]
- Seluzicki A., Flourakis M., Kula-Eversole E., Zhang L., Kilman V., Allada R. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol. 2014;12:e1001810. doi: 10.1371/journal.pbio.1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer O.T., Helfrich-Förster C., Renn S.C.P., Taghert P.H. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J. Comp. Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer O.T., Taghert P.H. RNA-interference knockdown of Drosophila pigment dispersing factor in neuronal subsets: the anatomical basis of a neuropeptide’s circadian functions. PLoS One. 2009;4:e8298. doi: 10.1371/journal.pone.0008298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J., Rosbash M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Nawathean P., Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Tabuchi M., Monaco J.D., Duan G., Bell B., Liu S., Liu Q., Zhang K., Wu M.N. Clock-generated temporal codes determine synaptic plasticity to control sleep. Cell. 2018;175:1213–1227.e18. doi: 10.1016/j.cell.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J.T., Simpson J.H., Bellen H.J. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Chiu H., Pfeiffer B.D., Wong A.M., Hoopfer E.D., Rubin G.M., Anderson D.J. A circuit node that integrates convergent input from neuromodulatory and social behavior-promoting neurons to control aggression in Drosophila. Neuron. 2017;95:1112–1128.e7. doi: 10.1016/j.neuron.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Cao G., Nitabach M.N. Electrical silencing of PDF neurons advances the phase of non-PDF clock neurons in Drosophila. J. Biol. Rhythms. 2008;23:117–128. doi: 10.1177/0748730407312984. [DOI] [PubMed] [Google Scholar]
- Yadlapalli S., Jiang C., Bahle A., Reddy P., Meyhofer E., Shafer O.T. Circadian clock neurons constantly monitor environmental temperature to set sleep timing. Nature. 2018;555:98–102. doi: 10.1038/nature25740. [DOI] [PubMed] [Google Scholar]
- Yao Z., Bennett A.J., Clem J.L., Shafer O.T. The Drosophila clock neuron network features diverse coupling modes and requires network-wide coherence for robust circadian rhythms. Cell Rep. 2016;17:2873–2881. doi: 10.1016/j.celrep.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Hermann C., Helfrich-Förster C. Cryptochrome-positive and -negative clock neurons in Drosophila entrain differentially to light and temperature. J. Biol. Rhythms. 2010;25:387–398. doi: 10.1177/0748730410381962. [DOI] [PubMed] [Google Scholar]
- Yoshii T., Wulbeck C., Sehadova H., Veleri S., Bichler D., Stanewsky R., Helfrich-Forster C. The neuropeptide pigment-dispersing factor adjusts period and phase of Drosophila’s clock. J. Neurosci. 2009;29:2597–2610. doi: 10.1523/JNEUROSCI.5439-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chung B.Y., Lear B.C., Kilman V.L., Liu Y., Mahesh G., Meissner R.-A., Hardin P.E., Allada R. DN1p circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in <em>Drosophila</em>. Curr. Biol. 2010;20:591–599. doi: 10.1016/j.cub.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Y., Bilodeau-Wentworth D., Hardin P.E., Emery P. Light and temperature control the contribution of specific DN1 neurons to <em>Drosophila</em> circadian behavior. Curr. Biol. 2010;20:600–605. doi: 10.1016/j.cub.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Daubnerova I., Jang Y.-H., Kondo S., Žitňan D., Kim Y.-J. The neuropeptide allatostatin C from clock-associated DN1p neurons generates the circadian rhythm for oogenesis. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2016878118. e2016878118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhou Y., Zhang X., Wang L., Zhong Y. Clock neurons gate memory extinction in Drosophila. Curr. Biol. 2021;31:1337–1343.e4. doi: 10.1016/j.cub.2021.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.