Abstract
The study of prions and the discovery of candidate therapeutics for prion disease have been facilitated by the ability of prions to replicate in cultured cells. Paradigms in which prion proteins from different species are expressed in cells with low or no expression of endogenous prion protein (PrP) have expanded the range of prion strains that can be propagated. In these systems, cells stably expressing a PrP of interest are typically generated via coexpression of a selectable marker and treatment with an antibiotic. Here, we report the unexpected discovery that the aminoglycoside G418 (Geneticin) interferes with the ability of stably transfected cultured cells to become infected with prions. In G418-resistant lines of N2a or CAD5 cells, the presence of G418 reduced levels of protease-resistant PrP following challenge with the RML or 22L strains of mouse prions. G418 also interfered with the infection of cells expressing hamster PrP with the 263K strain of hamster prions. Interestingly, G418 had minimal to no effect on protease-resistant PrP levels in cells with established prion infection, arguing that G418 selectively interferes with de novo prion infection. As G418 treatment had no discernible effect on cellular PrP levels or its localization, this suggests that G418 may specifically target prion assemblies or processes involved in the earliest stages of prion infection.
Keywords: prion, cell culture, protein misfolding, protein aggregation, neurodegeneration, antibiotics
Abbreviations: DAPI, diamidino-2-phenylindole; GPI, glycosylphosphatidylinositol; HaPrP, hamster PrP; IRES, internal ribosome entry site; MoPrP, mouse PrP; NCAM, neural cell adhesion molecule; PBS, phosphate-buffered saline; PK, proteinase K; PrP, prion protein; RT-QuIC, real-time quaking-induced conversion; TBS, Tris buffered saline; ThT, Thioflavin T
Prion diseases are a group of fatal neurodegenerative disorders caused by the accumulation of toxic protein aggregates in the brain (1, 2). Examples of prion diseases include chronic wasting disease in cervids, scrapie in sheep, “mad cow” disease, and Creutzfeldt–Jakob disease in humans. Prions, which were originally defined as proteinaceous infectious particles (3), arise from the structural rearrangement of the host-encoded prion protein (PrP) (4). In its native state, the cellular prion protein (PrPC) has a predominantly α-helical structure and is attached to the outer leaflet of the cell membrane of neurons and other central nervous system cells via a glycosylphosphatidylinositol (GPI) anchor (5, 6). PrPC has been implicated in a wide range of functions, specifically in regard to myelin maintenance in the peripheral nervous system and regulation of neural cell adhesion molecule (NCAM) polysialylation (7, 8). During disease, PrPC undergoes a profound conformational rearrangement to form β-sheet-rich aggregates that are protease-resistant, neurotoxic, and infectious (9). This conformer is termed PrPSc, and its infectious nature allows it to template the conversion of PrPC into additional copies of PrPSc. Therefore, PrPSc functions as a self-propagating protein assembly, allowing for the accumulation and spread of PrPSc within the brain. This templated conversion process is believed to be the central event in the pathogenesis of prion disease.
Animal bioassays are currently the gold standard for assessing prion transmissibility and studying the pathogenesis of prion disease (10, 11). However, animal experiments are often long and costly, and therefore cellular models that can recapitulate the templated conversion of PrPC to PrPSc have provided a cost-efficient paradigm for investigating the biology associated with prion disease (12). Several different lines of immortalized cells expressing murine PrPC have been identified that can become infected with mouse prions (13, 14, 15, 16, 17, 18, 19). Perhaps the most important use for these cellular models is the platform they provide for identifying small molecule inhibitors of prion replication (20). A high-throughput screen conducted in prion-infected N2a neuroblastoma cells yielded several 2-aminothiazole compounds, including IND24, which could reduce levels of proteinase K (PK)-resistant PrP (PrPres) in cells and significantly increase the survival of prion-infected mice (21, 22). However, while these compounds displayed activity against mouse prions in cultured cells and mice, they were ineffective at inhibiting the replication of human prions in transgenic mice expressing human PrP (22, 23, 24, 25). Thus, antiprion compounds need to be identified and validated using a paradigm capable of replicating the exact type of prions that the drug is intended to treat.
The availability of cellular paradigms for replicating nonmouse prions is comparatively limited. Very few PrPC-expressing cell lines from nonmouse species that can become infected with species-matched prions have been identified (20, 26, 27, 28). A more fruitful approach has been to take a cell line that does not express detectable levels of PrPC and then engineer it to express PrPC from the desired species. For instance, rabbit RK13 cells become susceptible to chronic wasting disease and scrapie prions upon expression of elk or sheep PrPC, respectively (29, 30, 31). Similar paradigms have been developed using cells derived from PrP knockout mice or gene-edited PrPC-null (PrP−/−) cell lines as a starting point (32, 33, 34, 35, 36). In each of these cases, selection of stably transfected cells expressing the desired PrPC is typically accomplished by coexpression of an antibiotic resistance gene and subsequent antibiotic treatment. A commonly used antibiotic for selection of stably transfected cells is G418 (37), also known as Geneticin, which is an aminoglycoside originally identified in the bacterium Micromonospora rhodorangea. G418 binds to the ribosome and inhibits translational elongation in both prokaryotic and eukaryotic cells, thus interfering with protein synthesis and cell growth (38). Resistance to G418 is conferred via expression of phosphotransferase enzymes such as neomycin phosphotransferase II (39).
During our attempts to study prion propagation using gene-edited PrP−/− cells that had been stably transfected with vectors encoding PrPC, we made the surprising discovery that G418 interferes with de novo prion infection in cultured cells. In this study, we investigate the inhibitory role of G418 on prion infection using different cell lines and prion strains. Our findings suggest that to ensure optimal results, strategies to avoid or limit the use of G418 during prion infection experiments should be implemented.
Results
G418 inhibits de novo prion infection in cultured cells
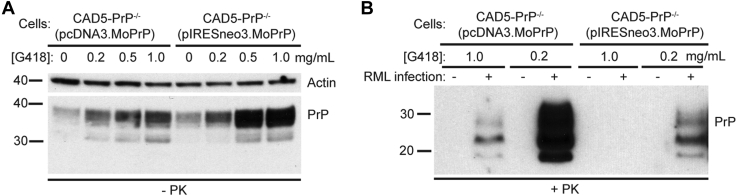
Murine CAD5 cells, which are derived from the catecholaminergic line Cath.a (40, 41) and express mouse PrP (MoPrP), are capable of replicating many different mouse prion strains, including RML, Me7, and 22L (16, 22, 42). We recently generated CAD5-PrP−/− cells lacking endogenous MoPrP and found that, following stable expression of hamster PrP (HaPrP), they can become infected with several strains of hamster prions (34). In parallel, we generated polyclonal pools of CAD5-PrP−/− cells stably expressing MoPrP [CAD5-PrP−/−(MoPrP) cells] using two different vectors (pcDNA3 and pIRESneo3), both of which confer resistance to the selectable agent G418. G418 is typically used at concentrations of 1.0 and 0.2 mg/ml for selection and maintenance of stably transfected cell lines, respectively. For both lines of CAD5-PrP−/−(MoPrP) cells, maintenance of the cells in higher concentrations of G418 resulted in higher levels of PrPC expression suggesting that, in the absence of selective agent, the composition of these polyclonal pools of cells drifts toward cells with lower levels of PrPC expression (Fig. 1A). When CAD5-PrP−/−(MoPrP) cells were challenged with RML prions in the presence of 0.2 mg/ml G418, successful prion infection was observed, as indicated by the presence of PrPres in cellular lysates (Fig. 1B). Paradoxically, when 1.0 mg/ml G418 was used in an attempt to increase PrPC expression levels and thus maximize PrPres production (43), the efficiency of RML prion infection in CAD5-PrP−/−(MoPrP) cells was substantially reduced (Fig. 1B). This led us to hypothesize that G418 may interfere with de novo prion infection in cultured cells.
Figure 1.
Increasing levels of G418 promote expression of plasmid-encoded mouse PrP in PrP−/−cells yet lead to lower PrPreslevels following RML infection.A, immunoblot of PrPC levels in lysates from uninfected CAD5-PrP−/−(pcDNA3.MoPrP) and CAD5-PrP−/−(pIRESneo3.MoPrP) cells cultured in the presence of the indicated concentrations of G418 for three passages. B, immunoblot of proteinase K (PK)-resistant PrP (PrPres) levels in CAD5-PrP−/−(pcDNA3.MoPrP) and CAD5-PrP−/−(pIRESneo3.MoPrP) cells after three passages following infection with mouse RML prions in the presence of the indicated concentrations of G418. PrPres was detected using HuM-P, whereas PrPC was detected with the antibody HuM-D13. In panel A, the blot was reprobed with an actin antibody to assess equal protein loading. Molecular weight markers indicate kDa.
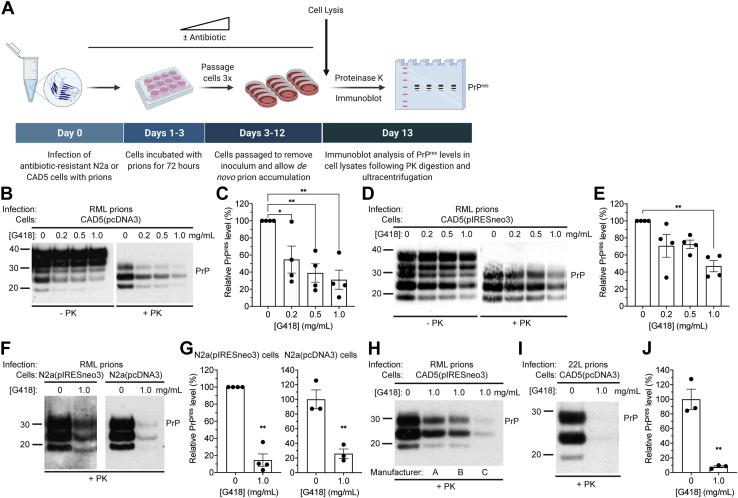
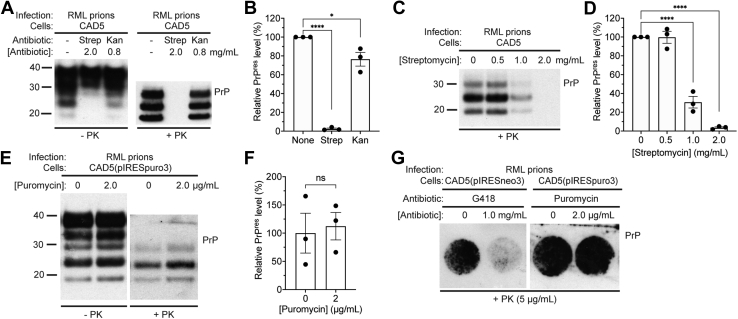
To test the effect of G418 on RML prion infection, we stably transfected wild-type CAD5 cells with two different empty vectors that confer G418 resistance to generate CAD5(pcDNA3) and CAD5(pIRESneo3) cells. These G418-resistant cells, which express endogenous levels of MoPrP, were then treated with RML prions in the presence of increasing concentrations of G418 (Fig. 2A). Following prion exposure, cells were passaged three times to allow for complete removal of the inoculum by dilution and the emergence of de novo RML prion-infected cells. In CAD5(pcDNA3) cells, increasing concentrations of G418 resulted in a dose-dependent reduction in PrPres levels obtained following challenge with RML prions (Fig. 2, B and C). Similar results were obtained when CAD5(pIRESneo3) cells were used (Fig. 2, D and E). To analyze whether this effect was specific to CAD5 cells, we generated G418-resistant murine N2a neuroblastoma cells by stably transfecting them with either pcDNA3 or pIRESneo3 and then challenged them with RML prions in the presence or absence of G418. N2a cells are widely used for studying prion propagation but are susceptible to a more limited range of mouse prion strains than CAD5 cells (14, 16). As with CAD5 cells, the presence of G418 significantly decreased PrPres levels in N2a(pcDNA3) and N2a(pIRESneo3) cells following RML prion infection (Fig. 2, F and G). Preparations of G418 obtained from three different manufacturers interfered with RML prion infection in CAD5(pIRESneo3) cells (Fig. 2H). Moreover, this effect was not specific to the RML strain since G418 also hindered infection of CAD5(pcDNA3) cells with the 22L strain of mouse prions (Fig. 2, I and J). Thus, G418 obstructs de novo prion infection in cells challenged with mouse prion strains.
Figure 2.
G418 inhibits de novo prion infection in cultured cells expressing endogenous mouse PrP.A, schematic of the experimental workflow for infection of antibiotic-resistant cell lines with prions. All infection experiments were analyzed following three passages. B, immunoblots of total PrP (−PK) and PrPres (+PK) levels in CAD5(pcDNA3) cells following infection with RML prions in the presence of the indicated concentrations of G418. C, quantification of PrPres levels in CAD5(pcDNA3) cells infected with RML prions in the presence of increasing concentrations of G418. n = 4 independent biological replicates (data is mean ± SEM). ∗p = 0.034 for 0.2 mg/ml, ∗∗p = 0.0055 for 0.5 mg/ml, and ∗∗p = 0.0023 for 1.0 mg/ml G418 by one-way ANOVA followed by Dunnett’s multiple comparisons test. D, immunoblots of total PrP and PrPres levels in CAD5(pIRESneo3) cells following infection with RML prions in the presence of the indicated concentrations of G418. E, quantification of PrPres levels in CAD5(pIRESneo3) cells infected with RML prions in the presence of G418. n = 4 independent biological replicates (data is mean ± SEM). ∗∗p = 0.0039 compared with infections performed in the absence of G418 by one-way ANOVA followed by Dunnett’s multiple comparisons test. F, immunoblots of PrPres levels in N2a(pIRESneo3) and N2a(pcDNA3) cells following infection with RML prions in the absence or presence of 1 mg/ml G418. G, quantification of PrPres levels (data is mean ± SEM) in N2a(pIRESneo3) and N2a(pcDNA3) cells infected with RML prions in the presence or absence of G418 (n = 4 or 3 independent biological replicates, respectively). For N2a(pIRESneo3) cells, ∗∗p = 0.0013 by a one sample t test. For N2a(pcDNA3) cells, ∗∗p = 0.0067 by a two-tailed unpaired t test. H, immunoblot of PrPres levels in CAD5(pIRESneo3) cells following infection with RML prions in the absence or presence of 1 mg/ml G418 sourced from three different manufacturers. I, immunoblot of PrPres levels in CAD5(pcDNA3) cells following infection with 22L prions in the absence or presence of 1 mg/ml G418. J, quantification of PrPres levels (data is mean ± SEM) in CAD5(pcDNA3) cells infected with 22L prions in the presence or absence of G418 (n = 3 independent biological replicates). ∗∗p = 0.0027 by a two-tailed unpaired t test. In B, D, F, H, and I, undigested PrP blots were probed with the antibody HuM-D13, whereas blots of PrPres were probed with HuM-P. Molecular weight markers indicate kDa.
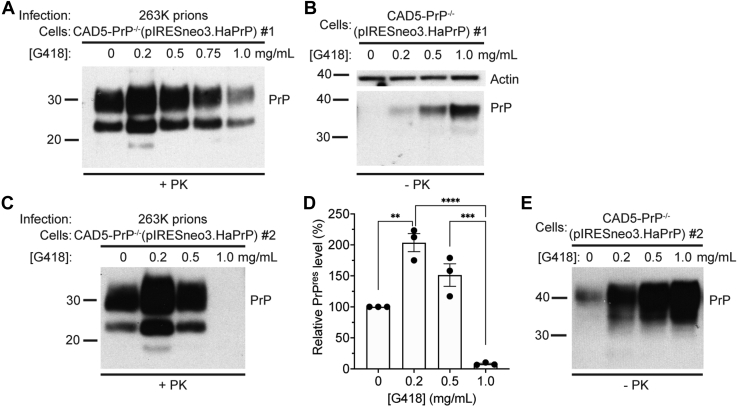
We next asked whether G418 interferes with infection of cultured cells with a nonmouse prion strain, the 263K strain of hamster prions. For this purpose, we used CAD5-PrP−/− cells that had been transfected with a vector that encodes HaPrP and confers resistance to G418 (pIRESneo3.HaPrP). We expanded these cells in the presence of G418 to generate a polyclonal pool of stably transfected cells [CAD5-PrP−/−(HaPrP) cells] (34). Exposure of these cells to 263K prions in the presence of a high concentration of G418 hindered prion infection compared with cells that were treated with a low concentration of G418 (Fig. 3A). Infection of cells in the presence of 0.2 mg/ml G418 resulted in higher levels of PrPres than in cells that were infected in the absence of G418. We speculate that this is because, similar to what we observed in polyclonal CAD5-PrP−/−(MoPrP) cells (Fig. 1A), higher PrPC levels were found in CAD5-PrP−/−(HaPrP) cells maintained in the presence of G418 (Fig. 3B). We repeated these experiments using an independent polyclonal line of CAD5-PrP−/−(HaPrP) cells and obtained similar results (Fig. 3, C–E). Thus, G418 interferes with the infection of cultured cells with mouse or hamster prion strains, regardless of whether PrP is expressed endogenously or via a vector-based heterologous promoter.
Figure 3.
G418 inhibits infection of cultured PrP−/−cells expressing hamster PrP with hamster prions.A, immunoblot of PrPres levels in CAD5-PrP−/−(pIRESneo3.HaPrP) cells (line #1) after three passages following infection with hamster 263K prions in the presence of the indicated concentrations of G418. B, immunoblot of PrPC levels in uninfected CAD5-PrP−/−(pIRESneo3.HaPrP) cells (line #1) cultured in the presence of the indicated concentrations of G418 for three passages. C, immunoblot of PrPres levels in an independent line of CAD5-PrP−/−(pIRESneo3.HaPrP) cells (line #2) after three passages following infection with hamster 263K prions in the presence of the indicated concentrations of G418. D, quantification of PrPres levels in CAD5-PrP−/−(pIRESneo3.HaPrP) cells (line #2) infected with 263K prions in the presence of increasing concentrations of G418. n = 3 independent biological replicates (data is mean ± SEM). ∗∗p = 0.0011, ∗∗∗p = 0.00011, and ∗∗∗∗p = 0.000011 by one-way ANOVA followed by Tukey’s multiple comparisons test. E, immunoblot of PrPC levels in uninfected CAD5-PrP−/−(pIRESneo3.HaPrP) cells (line #2) cultured in the presence of the indicated concentrations of G418 for three passages. PrPres was detected using HuM-P whereas PrPC was detected with the antibody HuM-D13. In panel B, the blot was reprobed with an actin antibody to assess equal protein loading. Molecular weight markers indicate kDa.
G418 does not affect prion propagation in cells with established prion infection
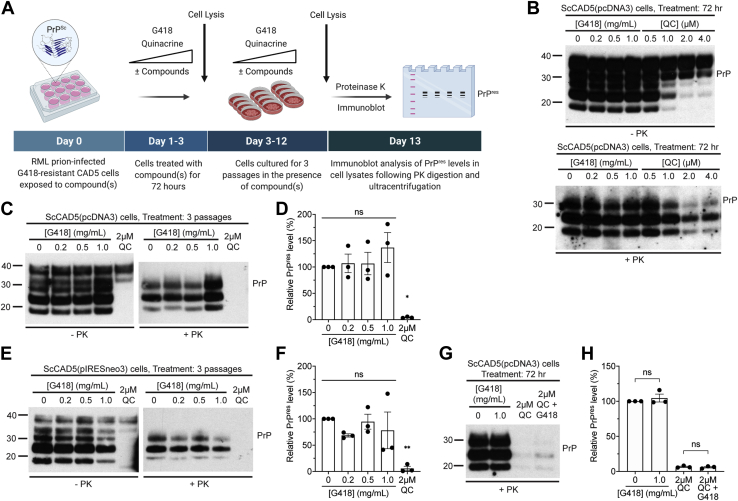
Having demonstrated that the presence of G418 at the time of prion infection reduces the accumulation of PrPres in both CAD5 and N2a cells, we next asked whether G418 is able to modulate established prion infection in cultured cells. G418-resistant CAD5(pcDNA3) and CAD5(pIRESneo3) cells were infected with RML prions in the absence of G418 to generate ScCAD5(pcDNA3) and ScCAD5(pIRESneo3) cells and then treated with increasing concentrations of G418 (Fig. 4A). As a positive control, cells were treated with quinacrine, a small molecule known to reduce PrPres levels in RML-infected N2a and CAD5 cells (22, 44). After a single 72-h treatment, G418 treatment did not reduce levels of PrPres in ScCAD5(pcDNA3) cells, whereas quinacrine reduced PrPres levels in a dose-dependent fashion (Fig. 4B). The treatment duration was then extended to encompass three passages (12–15 days of treatment) in the presence of varying doses of G418. After three passages, RML infection was cured in quinacrine-treated cells, whereas G418-treated ScCAD5(pcDNA3) and ScCAD5(pIRESneo3) cells showed no consistent reduction in PrPres levels (Fig. 4, C–F). To check whether G418 may only be effective at reducing prion infection in cells with lower levels of PrPres, as may occur during the initial stages of de novo prion infection, we simultaneously treated ScCAD5(pcDNA3) cells for 72 h with both quinacrine (to lower PrPres levels) and G418. We observed no additional reduction of PrPres in cells treated with both quinacrine and G418 (Fig. 4, G and H), even when a lower concentration of quinacrine was used (Fig. S1). Collectively, these results suggest that G418 does not have a major effect on PrPres levels in cells with established prion infection.
Figure 4.
Treatment of RML prion-infected cells with G418 has minimal effects on prion propagation.A, schematic of the experimental workflow for treatment of prion-infected G418-resistant cells with G418 or quinacrine (QC). PrPres levels in cell lysates were analyzed following treatment for either 72 h or three passages. B, immunoblots of total PrP (−PK) and PrPres (+PK) levels in RML prion-infected CAD5(pcDNA3) cells [ScCAD5(pcDNA3)] treated with the indicated concentrations of G418 or QC for 72 h. C, immunoblots of total PrP and PrPres levels in ScCAD5(pcDNA3) cells following three passages in the presence of the indicated concentrations of G418 or 2 μM QC. D, quantification of PrPres levels in ScCAD5(pcDNA3) cells following three passages in the presence of increasing concentrations of G418 or 2 μM QC. n = 3 independent biological replicates (data is mean ± SEM). ∗p = 0.011 compared with untreated cells by one-way ANOVA followed by Dunnett’s multiple comparisons test; ns, not significant. E, immunoblots of total PrP and PrPres levels in ScCAD5(pIRESneo3) cells following three passages in the presence of the indicated concentrations of G418 or 2 μM QC. F, quantification of PrPres levels in ScCAD5(pIRESneo3) cells following three passages in the presence of increasing concentrations of G418 or 2 μM QC. n = 3 independent biological replicates (data is mean ± SEM). ∗∗p = 0.0093 compared to untreated cells by one-way ANOVA followed by Dunnett’s multiple comparisons test; ns, not significant. G, immunoblot of PrPres levels in ScCAD5(pcDNA3) cells treated with the indicated concentrations of G418, 2 μM QC, or 2 μM QC + 1.0 mg/ml G418 for 72 h. H, quantification of PrPres levels in ScCAD5(pcDNA3) cells following treatment with G418, QC, or QC + G418 for 72 h. n = 3 independent biological replicates (data is mean ± SEM). PrPres levels were not significantly different (p = 0.99) in cells treated with 2 μM QC or 2 μM QC + 1.0 mg/ml G418 as determined by one-way ANOVA followed by Tukey’s multiple comparisons test. In B, C, E, and G undigested PrP blots were probed with the antibody HuM-D13, whereas blots of PrPres were probed with HuM-P. Molecular weight markers indicate kDa.
Streptomycin, but not puromycin, inhibits de novo prion infection
G418 belongs to a larger family of aminoglycoside antibiotics that inhibit protein synthesis in predominantly Gram-negative bacteria. Unlike G418, other commonly used aminoglycosides like streptomycin and kanamycin are only effective against the bacterial ribosome. We wanted to see if these antibiotics, which are harmless to mammalian cells, could also inhibit de novo prion replication. Therefore, wild-type CAD5 cells were challenged with RML prions in the presence of concentrations of kanamycin or streptomycin that are equimolar to 1.0 mg/ml (1.4 mM) G418. Whereas PrPres levels were decreased by ∼25% in cells cultured in the presence of kanamycin compared with untreated cells, prion infection was essentially blocked in cells cultured in the presence of streptomycin (Fig. 5, A and B). This indicates that other aminoglycoside antibiotics can hinder de novo prion infection in cultured cells to varying degrees. As the effect of streptomycin was so pronounced and given that streptomycin at 0.1 mg/ml is commonly added to cell culture medium to discourage bacterial growth, we assessed the effect of culturing cells in lower streptomycin concentrations on RML prion infection. While PrPres levels in CAD5 cells treated with 1.0 mg/ml streptomycin were decreased by ∼70%, PrPres levels were not significantly altered when 0.5 mg/ml streptomycin was used (Fig. 5, C and D).
Figure 5.
The effects of aminoglycoside and aminonucleoside antibiotics on de novo infection of cultured cells with RML prions.A, immunoblots of total PrP (−PK) and PrPres (+PK) levels in CAD5 cells after three passages following challenge with RML prions in the presence of either 2.0 mg/ml streptomycin (Strep) or 0.8 mg/ml kanamycin (Kan). B, quantification of PrPres levels (data is mean ± SEM) in CAD5 cells infected with RML prions in the presence or absence of 2.0 mg/ml streptomycin or 0.8 mg/ml kanamycin (n = 3 independent biological replicates). ∗p = 0.014 and ∗∗∗∗p < 0.0001 compared with untreated cells by one-way ANOVA followed by Dunnett’s multiple comparisons test. C, immunoblot of PrPres levels in CAD5 cells at passage three following challenge with RML prions in the presence of the indicated concentrations of streptomycin. Streptomycin is commonly added to cell media at a concentration of 0.1 mg/ml to discourage microbial growth. D, quantification of PrPres levels (data is mean ± SEM) in CAD5 cells infected with RML prions in the presence of the indicated concentrations of streptomycin (n = 3 independent biological replicates). ∗∗∗∗p < 0.0001 compared with untreated cells by one-way ANOVA followed by Dunnett’s multiple comparisons test. E, immunoblots of total PrP and PrPres levels in CAD5(pIRESpuro3) cells at passage three following challenge with RML prions in the absence or presence of 2 μg/ml puromycin. F, quantification of PrPres levels in CAD5(pIRESpuro3) cells infected with RML prions in the presence or absence of puromycin (n = 3 independent biological replicates, data is mean ± SEM). No significant difference (ns) in PrPres levels was observed, as assessed by an unpaired two-tailed t test (p = 0.79). G, cell immunoblots of CAD5(pIRESneo3) cells and CAD5(pIRESpuro3) cells after three passages following infection with RML prions and treatment with the indicated concentrations of G418 or puromycin. In A and B, undigested PrP blots were probed with the antibody HuM-D13, whereas blots of PrPres were probed with HuM-P. Molecular weight markers indicate kDa.
Puromycin, an aminonucleoside antibiotic commonly used as a selectable marker, also inhibits protein synthesis. Unlike the aminoglycosides, puromycin disrupts protein synthesis by causing premature chain termination. We wanted to see if the effects of the aminoglycoside antibiotics were recapitulated by a structurally distinct but somewhat functionally similar antibiotic such as puromycin. CAD5 cells stably transfected with an empty vector encoding puromycin resistance (pIRESpuro3) were used in this experiment. Puromycin-resistant CAD5(pIRESpuro3) cells were challenged with RML prions in the presence of either 0 or 2 μg/ml puromycin, the latter being a common concentration used for selecting puromycin-resistant cell lines. Following three passages, cell lysates were analyzed for the presence of PrPres. We found that puromycin had no effect on the accumulation of PrPres (Fig. 5, E and F). To further analyze how G418 and puromycin affect the accumulation of PrPres immediately following prion infection, cell blots (45) were generated on RML-challenged CAD5(pIRESneo3) or CAD5(pIRESpuro3) cells that had been infected in the absence or presence of G418 or puromycin, respectively. G418 substantially reduced the proportion of PrPres-positive cells, whereas puromycin had no effect. (Fig. 5G). Thus, aminoglycoside, but not aminonucleoside, antibiotics interfere with de novo prion infection in cultured cells.
G418 does not interfere with PrPC expression or localization
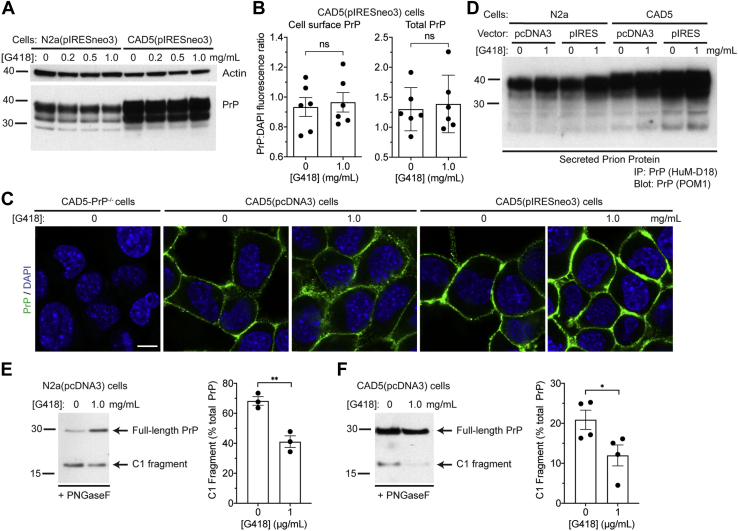
To probe the mechanism by which G418 might interfere with de novo prion infection, we tested its effect on PrPC levels in G418-resistant CAD5 and N2a cells. Both lines were exposed to increasing concentrations of G418 for 72 h. No apparent change in PrPC levels within cell lysates was detected in either line (Fig. 6A). To quantify cell surface and total PrPC levels, cells were treated for 72 h in microplates, and then fluorescence was measured using a plate reader. When using an antibody that binds to residues 95 to 105 of PrP (HuM-D13), we found that both cell-surface and total PrPC expression levels were similar following treatment with G418 (Fig. 6B). Next, we investigated whether the localization of PrPC was altered by G418. CAD5(pcDNA3) and CAD5(pIRESneo3) cells were treated with G418 or left untreated and then fixed, immunolabeled for PrP, and visualized using confocal microscopy. The levels and distribution of PrPC at the cell surface were not appreciably altered by G418 treatment (Fig. 6C). As expected, no PrP staining was observed in CAD5-PrP−/− cells.
Figure 6.
G418 treatment does not interfere with PrPCexpression or its cellular localization.A, immunoblot of PrPC levels in N2a(pIRESneo3) and CAD5(pIRESneo3) cells that were treated with the indicated concentrations of G418 for 72 h. PrP was detected using the antibody HuM-D13. The blot was reprobed with an actin antibody to assess equal protein loading. B, quantification of full-length cell-surface and total PrPC levels in CAD5(pIRESneo3) cells treated with the indicated G418 concentrations for 72 h using a microplate assay (data is mean ± SEM). Cells were stained with DAPI and the anti-PrP antibody HuM-D13, and then the PrP:DAPI fluorescence ratio was calculated. n = 6 independent biological replicates. No significant differences (ns) in cell-surface (p = 0.73) or total (p = 0.73) PrPC levels were observed between untreated and G418-treated cells, as assessed by unpaired two-tailed t tests. C, immunofluorescence images of PrPC in non-permeabilized CAD5-PrP−/−, CAD5(pcDNA3), and CAD5(pIRESneo3) cells that were left untreated or treated with 1 mg/ml G418 for 72 h. Cells were stained with DAPI (blue) and the anti-PrP antibody POM1 (green). Scale bar = 10 μM (applies to all images). D, immunoblot of secreted PrP levels in either N2a or CAD5 cells stably transfected with the pcDNA3 or pIRESneo3 vectors. Cells were either treated with 1 mg/ml G418 for 24 h or left untreated. Secreted PrP in the conditioned medium was isolated by immunoprecipitation with HuM-D18 and then detected using the antibody POM1. E and F, assessment of PrPC cleavage in G418-treated cells. Immunoblots for PrP in PNGaseF-treated lysates as well as quantification of C1 fragment levels in N2a(pcDNA3) (E) and CAD5(pcDNA3) (F) cells that were treated with the indicated concentrations of G418 for 72 h (n = 3 or 4 independent biological replicates, respectively; data is mean ± SEM). ∗∗p = 0.0049 and ∗p = 0.047 by unpaired, two-tailed t tests. Full-length PrP and its C1 cleavage fragment were detected using the antibody HuM-D18. Molecular weight markers indicate kDa.
G418 and other aminoglycosides have been reported to partially influence the activity of phospholipases, resulting in increased shedding of GPI-anchored proteins into the media (46). Since PrPC that has been shed into the media might bind to and sequester PrPSc seeds at the time of infection, thereby reducing the ability of PrPSc to interact with membrane-bound PrPC, we tested whether G418 modulates PrPC shedding. Four different lines of G418-resistant CAD5 or N2a cells were treated with G418, and the conditioned media were collected 24 h later and then analyzed for PrP following immunoprecipitation. Levels of secreted PrPC were only slightly increased in the G418-treated compared with the untreated samples (Fig. 6D), suggesting that G418 is unlikely to interfere with prion infection via an increase in PrPC shedding. Finally, we tested whether proteolytic processing of PrPC is affected by G418 treatment. A proportion of PrPC normally undergoes endoproteolytic cleavage in the vicinity of residues 110/111 to produce a C1 fragment, which is believed to be unable to undergo conversion into PrPSc (47, 48, 49). In G418-resistant N2a and CAD5 cells, C1 cleavage of PrPC was moderately but significantly reduced following treatment with G418 for 72 h (Fig. 6, E and F).
Effects of antibiotics on the biophysical properties of recombinant PrP
To investigate whether G418 or puromycin might interact directly with PrPC, we generated recombinant MoPrP (recPrP) and performed differential scanning fluorimetry, a technique capable of detecting shifts in PrP thermal stability upon binding of small molecules (50, 51). In the presence of a tenfold molar excess of either puromycin, G418, or quinacrine, a ratio commonly used in protein thermal shift experiments (52), the melting temperature of recPrP remained unchanged, suggesting that none of these compounds likely act by binding to PrPC and modulating its stability (Fig. 7A). Next, we tested whether G418 modulates the aggregation kinetics of recPrP using a Thioflavin T (ThT) fluorescence assay. A tenfold molar excess of G418 did not alter the lag phase for shaking-induced recPrP aggregation nor the shape of the aggregation curve, suggesting that G418 does not influence the spontaneous polymerization of recPrP (Fig. 7, B and C). An important caveat to this finding is that recPrP aggregates formed spontaneously in vitro are not highly infectious and are structurally distinct from PrPres generated in a brain or cellular environment (53). Therefore, these results do not exclude the possibility that G418 may affect the spontaneous polymerization of membrane-anchored and glycosylated PrPC in vivo. Finally, we tested the ability of puromycin and G418 to modulate the prion-induced templating of recPrP into aggregates using real-time quaking-induced conversion (RT-QuIC) (54). Both G418 and puromycin modestly increased the lag phase for RT-QuIC reactions when using RML prions as the seed (Fig. 7, D and E). Given that G418, but not puromycin, affects de novo prion infection, it seems unlikely that this small shift in lag phase is sufficient to explain the effects observed in cultured cells.
Figure 7.
G418 does not affect the stability or the aggregation of recombinant PrP.A, melting temperatures (mean ± SEM) of recombinant MoPrP (residues 23–230), as determined by differential scanning fluorimetry, in the presence or absence of a tenfold molar excess of either G418, puromycin, or quinacrine. n = 4 independent biological replicates; ns, not significant by one-way ANOVA (p = 0.69). B, ThT fluorescence curves for the spontaneous aggregation of recombinant MoPrP in the absence (black) or presence (red) of a tenfold molar excess of G418. Each data point represents the mean ± SEM of six independent biological replicates. C, calculated lag phases for the spontaneous aggregation of recombinant MoPrP in the absence (black) or presence (red) of a tenfold molar excess of G418. n = 6 independent biological replicates; ns, not significant by an unpaired two-tailed t test (p = 0.062). D, RT-QuIC analysis of RML-seeded recombinant MoPrP in the absence (black) or presence of a tenfold molar excess of either G418 (red) or puromycin (blue). Each data point represents the mean ThT fluorescence ± SEM of four independent biological replicates. E, lag phases (mean ± SEM) for RML-seeded RT-QuIC reactions using recombinant MoPrP in the absence (black) or presence of a tenfold molar excess of either G418 (red) or puromycin (blue). n = 4 independent biological replicates; ∗p = 0.015 compared with control reactions by one-way ANOVA followed by Dunnett’s multiple comparisons test.
Exposure to G418 does not lead to the selection of drug-resistant prions
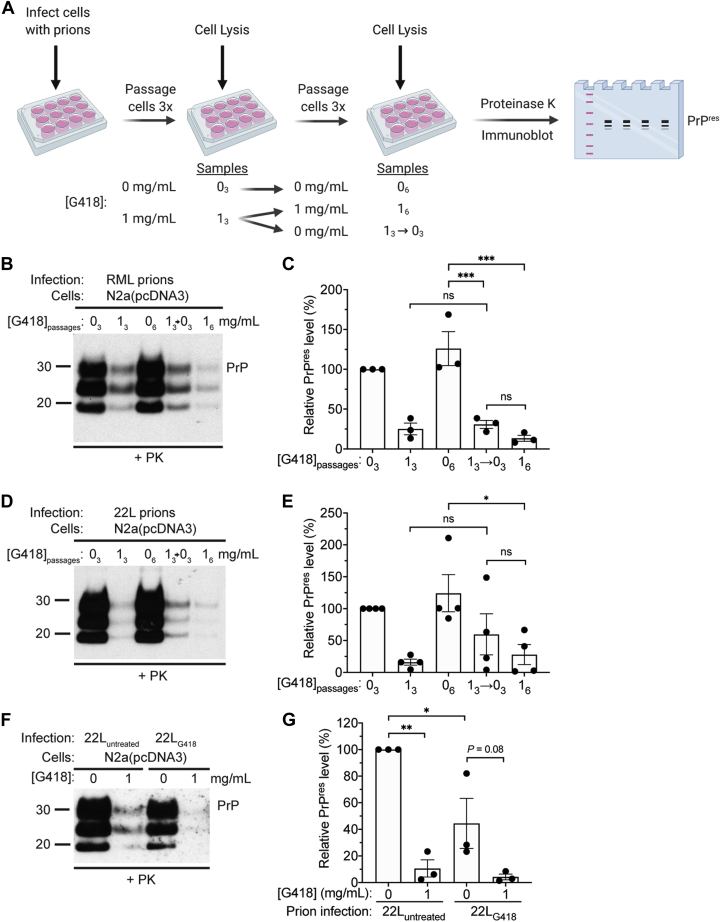
To determine whether the decrease in PrPres levels that occurs when cells are infected in the presence of G418 is reversible, we compared PrPres levels in RML prion-challenged N2a(pcDNA3) cells following six passages in the presence or absence of G418 to cells in which G418 treatment was applied for three passages and then removed for the remaining three passages (Fig. 8A). As expected, PrPres levels were strikingly reduced in cells exposed to G418 compared with untreated cells following either three or six passages (Fig. 8, B and C). However, there was no difference in PrPres levels between cells exposed to G418 for three or six passages following infection and cells exposed to G418 for three passages followed by removal of the G418 for an additional three passages. Similar results were obtained when we repeated this experiment using the 22L strain as the source of prions (Fig. 8, D and E). Two possible explanations for the lack of PrPres recovery following cessation of G418 treatment are: 1) the additional three passages following removal of G418 are insufficient to allow recovery of PrPres to normal levels; 2) infection in the presence of G418 leads to the selection and propagation of a G418-resistant prion strain that accumulates to lower steady-state levels than the parental strain.
Figure 8.
G418 treatment does not lead to the selection of G418-resistant prions.A, schematic of the cellular infection experiment. B, immunoblot of PrPres levels in RML prion-challenged N2a(pcDNA3) cells following three passages in the absence (03) or presence (13) of 1.0 mg/ml G418, six passages in the absence (06) or presence (16) of 1.0 mg/ml G418, or three passages in the presence of 1.0 mg/ml G418 followed by three passages in the absence of G418 (13→03). C, quantification of PrPres levels in N2a(pcDNA3) cells challenged with RML prions in the presence or absence of 1.0 mg/ml G418 and cultured for the indicated the number of passage (n = 3 independent biological replicates, data is mean ± SEM). ∗∗∗p < 0.001 by one-way ANOVA followed by Tukey’s multiple comparisons test; ns, not significant. D, immunoblot of PrPres levels in 22L prion-challenged N2a(pcDNA3) cells following three passages in the absence or presence of 1.0 mg/ml G418, six passages in the absence or presence of 1.0 mg/ml G418, or three passages in the presence of 1.0 mg/ml G418 followed by three passages in the absence of G418. E, quantification of PrPres levels in N2a(pcDNA3) cells challenged with 22L prions in the presence or absence of 1.0 mg/ml G418 and cultured for the indicated the number of passage (n = 4 independent biological replicates, data is mean ± SEM). ∗p < 0.05 by one-way ANOVA followed by Tukey’s multiple comparisons test. F, immunoblot of PrPres levels in N2a(pcDNA3) cells infected with prions from N2a(pcDNA3) cells (passage 6) that were infected with 22L prions either in the absence (22Luntreated) or presence (22LG418) of 1.0 mg/ml G418. Cells were passaged three times in the absence or presence of 1.0 mg/ml G418. G, quantification of PrPres levels in N2a(pcDNA3) cells infected with either 22Luntreated or 22LG418 prions in the presence or absence of 1.0 mg/ml G418 (n = 3 independent biological replicates, data is mean ± SEM). ∗∗p < 0.01, ∗p < 0.05 by one-way ANOVA followed by Tukey’s multiple comparisons test. Molecular weight markers indicate kDa.
To evaluate whether G418 treatment at the time of prion infection leads to the propagation of a drug-resistant prion strain, we infected N2a(pcDNA3) cells with 22L prions in either the presence or absence of 1.0 mg/ml G418 and then passaged them six times. As expected, PrPres levels were much lower in the cells exposed to G418 (Fig. 8, D and E). Cellular homogenates from 22L-infected untreated (22Luntreated) or G418-treated (22LG418) cells were then used to infect naïve N2a(pcDNA3) cells, again in the presence or absence of G418. As predicted, G418 treatment reduced PrPres levels in cells that were infected with 22Luntreated prions (Fig. 8, F and G). Due to the difference in PrPres levels between 22Luntreated and 22LG418 at the time of infection (Fig. 8, D and E), prion accumulation in untreated cells was lower when 22LG418 prions were applied (Fig. 8, F and G). When 22LG418 prions were applied to cells, PrPres levels were still substantially lower in cells infected in the presence of G418, although this did not reach statistical significance (Fig. 8, F and G). If G418 treatment had led to the selection and propagation of a G418-resistant prion strain, we would have expected to see similar PrPres levels in G418-treated and untreated cells following infection with 22LG418 prions. Thus, G418 treatment is unlikely to result in the emergence of drug-resistant prions.
Discussion
In this study, we investigated the inhibitory role of G418 and other aminoglycosides when attempting to infect antibiotic-resistant cultured cells with prions. We show that G418 inhibits de novo prion replication in a dose-dependent manner when present at the time of infection and during initial cellular passaging. This phenomenon was reproduced using distinct plasmid vectors, different murine cell lines, and two unique strains of mouse prions. Moreover, G418 also interfered with the infection of cells with hamster prions. This indicates that G418 hinders de novo prion infection in a fundamental way that applies across different PrP species and prion strains. We also found that another aminoglycoside antibiotic, streptomycin, can significantly inhibit the ability of cells to become infected with prions, although the inhibitory effect of streptomycin was only observed at concentrations exceeding those that would normally be used for discouraging microbial growth in cultured cells. Thus, the antibiotic composition of growth medium used when infecting cultured cells with prions needs to be carefully considered to maximize the efficiency of prion infection.
Infection of cultured PrP−/− cells engineered to express PrP alleles of interest with prions constitutes a powerful paradigm for dissecting the molecular determinants of prion replication. Several strategies can be envisioned to eliminate or reduce the negative effects of G418 in such prion infection experiments. First, lower concentrations of G418 can be used to minimize the antiprion activity of G418 while still promoting the maintenance of PrPC levels within a heterogeneous pool of cells. We found that PrPC levels in polyclonal CAD5-PrP−/− cell lines stably expressing MoPrP or HaPrP were directly correlated with the concentration of G418 present in the media. For both RML and 263K prion strains, infection of cells in the presence of 0.2 mg/ml G418 permitted prion replication despite the presence of lower PrPC levels. Second, monoclonal lines of stably PrP-transfected PrP−/− cells can be created to circumvent the genetic drift issue and remove the necessity for continuous presence of G418 in the culture medium. Finally, it may be possible to use a different selectable marker. In our experiments, puromycin had no effect on the ability of CAD5 cells with endogenous MoPrP expression to become infected with RML prions, although we do not know whether this is true for all prion strains. Additional caution should be exercised as puromycin treatment can lead to the emergence of puromycin-resistant subclones in aneuploid cell lines, even in cells that have not been transfected with a puromycin-resistance gene (55).
Other antibiotics have been documented to influence prion propagation in cells or animals. Treatment of prion-infected cells with the polyene antifungal agent amphotericin B reduces levels of PrPres (56), and hamsters or mice treated with amphotericin B exhibit moderate survival extensions following prion infection (57, 58). Tetracycline and doxycycline have also been shown to decrease PrPres levels in vitro and could extend the survival of prion-challenged animals (59, 60). However, neither amphotericin B nor doxycycline has proven to be effective at treating prion disease in humans (61, 62, 63). Despite its ability to hinder de novo prion infection in cultured cells, it is highly unlikely that G418 will be a viable treatment option for prion diseases. G418 inhibits protein synthesis in eukaryotic cells by blocking translational elongation and thus would be expected to have toxic effects following prolonged exposure in humans. It is possible, however, that other aminoglycosides or G418 derivatives might exist that could prove beneficial for treating prion disease.
The effects of G418 and other aminoglycosides on de novo prion infection in antibiotic-resistant cell lines are likely independent of their ability to disrupt protein synthesis since puromycin, which inhibits translation by an orthogonal method, had no discernible effect, whereas streptomycin, which has no effect on mammalian ribosomes, significantly hindered cellular prion infection. The differential activities of equimolar amounts of G418, streptomycin, and kanamycin in RML-challenged CAD5 cells (∼70%, ∼95%, and ∼25% reduction in PrPres levels, respectively) further suggest that there is an element of specificity to the response. It is conceivable that the action of G418 and streptomycin may be related to their ability to permit read-through of premature termination codons and normal STOP codons (64, 65), possibly leading to enhanced production of a protein that interferes with de novo prion infection. Indeed, G418-induced translational errors have been shown to modulate the production of the yeast prion [URE3] (66). However, the precise mechanism by which G418 hinders de novo prion infection in cultured cells remains enigmatic. At the present time, we know more about the cellular mechanisms relevant to prion infection that remain unaffected by G418 treatment. G418 treatment had no discernible effects on PrPC levels, PrPC cell surface localization, the thermal stability of recPrP, or the spontaneous formation of recPrP aggregates. Collectively, these results suggest that G418 is unlikely to act by binding to PrPC and/or by influencing its biogenesis. A moderate decrease in the extent of PrP C1 cleavage was observed following G418 treatment. However, if anything, this should result in an increased amount of PrPres since the PrP C1 fragment has been proposed to be a dominant-negative inhibitor of prion replication (47). We also saw no evidence that G418 treatment alters strain properties in PrPSc, including the glycoform ratio in PrPres and its molecular weight. Moreover, G418 did not lead to the emergence of a novel drug-resistant prion strain, as has been observed following treatment with other antiprion small molecules (22, 67, 68, 69). Finally, it is unlikely that G418 treatment resulted in the emergence of protease-sensitive prions (70, 71). It has been shown that PK-resistant and PK-sensitive preparations possess similar levels of infectivity and lead to identical amount of PrPres being produced upon injection into hamsters (71). Infecting cells with homogenates from cells that were previously infected with 22L prions in the presence of G418 resulted in lower levels of PrPres than when using homogenates from cells that were not exposed to G418 (Fig. 8G). This suggests that G418 treatment during de novo cellular prion infection leads to a reduction in PrPres levels and a corresponding decrease in prion infectivity, implying that G418 treatment does not increase levels of PK-sensitive prions.
Despite having a pronounced inhibitory effect on de novo prion infection in cultured cells, G418 had little to no effect on PrPres levels in cells with established prion infection. While there is no shortage of small molecules capable of reducing or eliminating PrPres in cultured cells infected with mouse prions (20, 72), to the best of our knowledge, G418 is the only drug that selectively exerts its influence during the initial phases of cellular infection. We envision two scenarios that may explain the selective effect of G418 on de novo cellular prion infection compared with established cellular prion infection:
G418 may specifically interfere with prion replication by blocking a PrPSc species that is selectively present in the prion inocula
To generate the source of prions used for the cellular prion infection experiments, we homogenized cells using a bead beater apparatus. During this process, smaller PrPSc species may be generated, which are known to be more infectious (73), and it is conceivable that G418 may selectively act on smaller PrPSc assemblies. Consistent with this theory, the presence of G418 significantly delayed the seeded formation of recPrP aggregates in an RT-QuIC assay, potentially suggesting that G418 might partially interfere with an interaction between PrPC and PrPSc. However, these results should be interpreted with caution since puromycin, which had no effect in a cellular prion infection assay, also slightly delayed the seeded formation of recPrP aggregates. A candidate PrPSc species that may be selectively present in prion inoculum preparations is nonanchored PrPSc. In persistently infected cells, it has been determined that cell–cell contact, possibly via tunneling nanotubes, or mother-to-daughter transmission during cell division are the primary mechanisms responsible for the spreading and accumulation of prion infection (74, 75, 76). While PrPSc can also be released into the medium of cultured cells, perhaps via exosomes, infection via cell medium is not as efficient (74, 77). Prion replication via cell–cell contact or vertical transmission would both involve an interaction between membrane-anchored PrPC and PrPSc, whereas transmission through the cell medium, which is more akin to the procedure used to infect cells de novo, is more likely to involve PrPSc species that have been released from the membrane. G418 may be better at blocking prion replication when PrPSc is not constrained to the plasma membrane.
G418 may sensitize cells to a toxic PrP species that is only present in the initial stages of de novo cellular prion infection
Cytopathic effects have not been commonly documented in prion-infected immortalized cell lines, suggesting that cells with established prion infection do not produce highly toxic PrP species. However, it has been hypothesized that neurotoxic PrP species catalyzed by an interaction between PrPSc and PrPC are responsible for neuronal dysfunction and death in mice and cultured primary neurons (78, 79, 80). Such toxic species may be transiently generated during the initial stages of cellular prion infection, possibly due to a specific type of PrPSc-PrPC interaction that does not occur in chronically infected cells. The toxicity of this PrP species may be enhanced by G418, and this could lead to a selective elimination of cells with high levels of PrPres. Interestingly, cells expressing a neurotoxic PrP mutant lacking the hydrophobic tract (PrPΔ105–125) are more rapidly eliminated following G418 treatment than cells expressing wild-type PrP (81, 82), which may be related to the formation of pores in the membrane and/or induction of spontaneous ion currents (83, 84). Thus, G418 may be having a similar effect on cells during de novo prion infection.
Our findings reveal that the negative effects of G418 and other antibiotics need to be considered when trying to establish accurate and efficient cellular models of prion replication. The cellular determinants of prion susceptibility in cell culture remain largely unknown, and therefore it is prudent to be diligent about the factors present during the infection and propagation phases of cellular prion infection. On the other hand, molecules such as G418 may also provide a unique opportunity to dissect early events during prion infection. New insight into this process may yield novel strategies for blocking the earliest stages of prion replication during prion disease.
Experimental procedures
Chemicals
G418 sulfate powder (Geneticin) was purchased from either Thermo Fisher Scientific (#11811031), BioShop (#GEN418.5), or FroggaBio (#400-111P-5G). Puromycin dihydrochloride (#PUR333.100), kanamycin sulfate (#KAN201.10), and streptomycin sulfate (#STP101.25) were obtained from BioShop. All cell culture reagents were purchased from Thermo Fisher Scientific unless otherwise indicated.
Cell culture
Mouse Neuro2a (N2a) neuroblastoma cells (ATCC# CCL-131) were cultured in DMEM medium (Thermo Fisher #11965118) containing 10% (v/v) FBS (Thermo Fisher #12483020), 1× GlutaMAX (Thermo Fisher #35050061), and 0.2× penicillin-streptomycin (Thermo Fisher #15140122). Mouse CAD5 catecholaminergic and CAD5-PrP−/− (clone D6) cells (34) were cultured in Opti-MEM medium (Thermo Fisher #31985088) containing 5 to 10% (v/v) FBS, 1× GlutaMAX, and 0.2× penicillin-streptomycin. All cell lines were kept at 37 °C in a 5% CO2 incubator. CAD5-PrP−/− cells were generated previously using CRISPR/Cas9 gene editing technology (34, 85). N2a cells were passaged at 1:10 dilution every 2 to 3 days using 0.25% trypsin-EDTA solution (Thermo Fisher #25200056). CAD5 cells were passaged at 1:5 dilution every 4 to 5 days using an enzyme-free cell dissociation reagent (Millipore #S-014-B).
Generation of stable cell lines
The open reading frames of either mouse or Syrian hamster PrP were inserted into the vectors pcDNA3 (Thermo Fisher), pIRESneo3 (Clontech), or pIRESpuro3 (Clontech). pcDNA3 and pIRESneo3 both confer resistance to G418, whereas pIRESpuro3 confers resistance to puromycin. In pIRESneo3 and pIRESpuro3, expression of the antibiotic resistance gene is coupled to expression of a gene of interest through use of an internal ribosome entry site (IRES). In pcDNA3, expression of the gene of interest and the antibiotic resistance gene are controlled by distinct promoters. For cloning into pcDNA3, the BamHI and XbaI restriction sites were used, whereas NheI and BamHI sites were used for the pIRESpuro3 and pIRESneo3 plasmids. For generation of stably transfected cells, CAD5, CAD5-PrP−/−, or N2a cells were plated in a 6-well plate at a density of 5 to 7 × 105 cells/well. These cells were then transfected with either empty vector or a PrP-encoding plasmid using Lipofectamine-2000 at a 2 μg plasmid DNA:4 μl Lipofectamine ratio in Opti-MEM medium. The next day, the cells were passaged into the appropriate medium containing either 1 mg/ml G418 or 2 μg/ml puromycin in 10 cm tissue culture dishes. The selection was conducted over 2 to 3 weeks, after which G418-resistant cells were maintained in 0.2 mg/ml G418. All stable cell lines constitute polyclonal pools of cells; limiting dilution subcloning was not performed.
Prion strains
Mouse RML and 22L prion inocula were obtained from RML- or 22L-infected CAD5 cells (34). The cells were grown to confluency, scraped in 1 ml PBS per 10 cm dish, and then homogenized using a Minilys bead homogenizer and CK14 homogenization tubes (Bertin). Benzonase (50 units/ml; EMD Millipore #70746-4) was added into the scraped culture, and then the culture was homogenized for three cycles of 30 s at maximum speed, with 5 min of incubation on ice between each cycle. Cell homogenates were then stored at −80 °C. Syrian hamster 263K prion inoculum was prepared in a similar way from 263K-infected CAD5-PrP−/− cells stably expressing HaPrP (34).
Cellular prion infections
For all experiments involving prion-infected cells, penicillin-streptomycin was omitted from the medium. Cells to be infected with prions were plated 24 h prior to infection in a 12-well dish at 1.5 × 105 cells/well or 3 × 105 cells/well for N2a and CAD5 cells, respectively. Prion-containing cellular homogenates were quantified using a bicinchoninic acid (BCA) assay, and then 100 μg of each homogenate was diluted in the appropriate growth medium and added onto cells, which were 60 to 70% confluent at the time of infection. Aminoglycoside or puromycin antibiotics were added to the diluted cellular homogenate at the desired concentration at the time of infection to prepare the final inoculum. Cells were exposed to the inoculum for 72 h and then passaged in the presence or absence of antibiotics for three passages. At this point, the cells were split off in 6-well plates or 6-cm dishes and lysed as described below. To generate the ScCAD5 cells, CAD5 cells stably transfected with empty vector were infected with RML prions and then passaged a minimum of 3 to 6 times in the absence of G418.
Cell lysis and immunoblotting
Cells were washed twice in PBS and then lysed in lysis buffer [50 mM Tris-HCl pH 8, 150 mM NaCl, 0.5% (v/v) NP40 and 0.5% (w/v) sodium deoxycholate] containing Halt protease inhibitor cocktail (Thermo Fisher #87786). The lysates were then incubated on ice for 15 min, with 30 s of vortexing every 5 min. The samples were then centrifuged at 1000g for 5 min to remove debris. The lysates were quantified using a BCA assay, and then the desired amount of protein was prepared in 1× Bolt LDS loading buffer (Thermo Fisher #B0007) containing 2.5% (v/v) β-mercaptoethanol. Samples were then boiled and loaded onto 10% or 4–12% Bolt Bis-Tris gels (Thermo Fisher) for SDS-PAGE. Following gel electrophoresis, proteins were transferred onto Immobilon-P membranes using Tris-Glycine transfer buffer (137 mM Glycine, 100 mM Tris-HCl pH 8). Membranes were then incubated in blocking buffer [Tris buffered saline (TBS) with 5% skim milk and 0.05% (v/v) Tween-20] for 1 h. The blocked membranes were then incubated in primary antibodies overnight at 4 °C. The next day, the membranes were washed three times with TBST [TBS +0.05% (v/v) Tween-20] for 10 min each. Secondary antibodies conjugated with horseradish peroxidase (BioRad) were then added to the membranes in blocking buffer on a rocker for 1 h at room temperature. The blots were again washed three times with TBST, for 10 min each. Following these washes, the blots were developed using Western Lightning ECL Pro (PerkinElmer), and exposed to HyBlot CL x-ray film. PrP antibodies used in this study were either recombinant humanized Fabs, including HuM-D18 (1:5000 dilution), HuM-D13 (1:5000 dilution), and HuM-P (1:10,000 dilution) (86, 87), or the mouse monoclonal antibody POM1 (1:2000 dilution; MilliporeSigma #MABN2285) (88). Actin blots were conducted on the previously probed blots following inactivation of horseradish peroxidase using 0.05% (w/v) sodium azide. Following washes with TBST, the blots were probed with the Actin 20–33 antibody (1:10,000 dilution; Sigma Aldrich #A5060) in blocking buffer. Immunoblots were scanned and then densitometry was performed using ImageJ.
Enzymatic digestions
For experiments involving prion-infected cells, protease inhibitor was omitted from the cell lysis buffer. Cell lysates were quantified using a BCA assay and volumes adjusted with lysis buffer to achieve identical protein concentrations. Then, 0.5 to 1 mg of lysate was digested with proteinase K (PK) (Thermo Fisher Scientific #EO0491) at 50 μg/ml for 1 h at 37 °C with shaking at 600 rpm. The PK:protein ratio was kept at 1:50 to allow for rigorous digestion of PrPC. The reactions were stopped by the addition of PMSF to a final concentration of 2 mM, and the samples were then mixed with sarkosyl (Sigma Aldrich #61747) to a final concentration of 2% (v/v). The digested samples were ultracentrifuged at 100,000g for 1 h at 4 °C using a TLA-55 rotor (Beckman Coulter). The supernatant was discarded, and the pellet was resuspended in 30 μl of 1× Bolt LDS loading buffer containing 2.5% (v/v) β-mercaptoethanol. The samples were boiled for 10 min and then analyzed by immunoblotting. To analyze PrP C1 cleavage, cell lysates were deglycosylated for 4 h at 37 °C using PNGase F (New England BioLabs #P0704S) according to the manufacturer’s protocol. To quantify the PrP-C1 fragment, levels of C1 were expressed as a percentage of total PrP signal (C1 fragment + full-length PrP).
Cell blotting
Cell blotting experiments were done as previously described (45). Cells were plated at 6 × 105 cells/well in a 6-well dish, which contained 2 to 3 circular coverslips of 1 cm diameter, and then infected and passaged in the presence or absence or antibiotics as described above. The cells were grown to confluency and then an appropriately sized nitrocellulose membrane was activated in sterile dH2O and soaked in lysis buffer. The coverslips were removed and placed onto a filter paper soaked in lysis buffer. The soaked membrane was pressed against the coverslips firmly for 30 s. The coverslips were carefully removed from the membrane and discarded. The membrane was air dried and then stored at −20 °C until further use. The membranes were activated in lysis buffer and then subjected to PK digestion (5 μg/ml) in lysis buffer for 2 h at room temperature. The PK was inactivated with 2 mM PMSF and then the membrane was washed three times with dH2O followed by treatment with 4 M guanidine hydrochloride for 10 min to denature PrPSc. The membrane was washed three times in dH2O and was then further processed using the immunoblotting protocol described above.
Immunofluorescence
For quantitative immunofluorescence, CAD5 cells stably transfected with empty pIRESneo3 vector were plated at 12,000 cells/well in a 96-well plate. One day after plating, the cells were exposed to either 0 or 1 mg/ml G418 in quadruplicates. After 48 h, the cells were fixed with 4% (v/v) paraformaldehyde for 15 min at room temperature. The cells were then washed 1× with phosphate-buffered saline (PBS) and then either permeabilized with 0.1% (v/v) Triton X-100 in PBS for 10 min to detect total PrP or left untreated for the detection of cell-surface PrP. The cells were washed again with 1× PBS and then blocked for 1 h with 2% (v/v) goat serum (in 1× PBS). The cells were then incubated with the anti-PrP antibody HuM-D13 at a dilution of 1:500 overnight at 4 °C. The next day, the cells were washed 2× and then incubated with a goat anti-human AlexaFluor 488-conjugated secondary antibody (Thermo Fisher) at 1:500 in blocking solution (2% goat serum/PBS) for 1.5 h at room temperature. The cells were then washed 2× in PBS and then incubated with 4′,6-diamidino-2-phenylindole (DAPI) at 1:1000 dilution in PBS for 10 min. At this point, the 96-well plate was analyzed for AlexaFluor 488 (PrPC) fluorescence (excitation: 488 ± 7 nm; emission: 535 ± 15 nm) and DAPI fluorescence (excitation: 405 ± 10 nm; emission: 460 ± 15 nm) using the BMG CLARIOstar plate reader set at 1500 gain. Cellular PrPC expression levels were calculated using the PrPC/DAPI signal ratio to provide a quantitative readout. For qualitative immunofluorescence, 1 × 105 cells were plated in 24-well plates with #1.5 glass-like coverslip bottoms (Cellvis #P24-1.5P), which were then subjected to treatment with 0 or 1 mg/ml G418 for 48 h. Cells were fixed with 4% (v/v) paraformaldehyde for 15 min, blocked, and then PrPC was detected using the antibody POM1 at a dilution of 1:500. Cells were washed and stained with DAPI as described above and then imaged using a Zeiss LSM880 confocal microscope.
Analysis of secreted PrPC
CAD5 or N2a cells stably transfected with empty pcDNA3 or pIRESneo3 vectors were plated in a 6-well plate at 5 × 105 cells/well in growth media lacking G418. One day later, the cells were treated with Opti-MEM media containing 0 or 1 mg/ml G418 for 24 h, and then the conditioned media was collected. The conditioned medium was centrifuged at 1500g to remove cellular material. Secreted PrP was immunoprecipitated from the conditioned medium using the antibody HuM-D18 as follows. KappaSelect resin (Cytiva #17545801) was used as the resin for the conjugation of HuM-D18. The resin was washed 3× in PBS and then conjugated with 5 μg of HuM-D18 antibody overnight at 4 °C with end-over-end rotation. The following day, the conjugated resin was washed 3× and then added to the conditioned medium and then left overnight at 4 °C with end-over-end rotation. The next day, the resin was washed 3×, and then eluates were obtained by boiling the resin in 1× Bolt LDS sample buffer for 10 min. The eluates were then analyzed by immunoblotting using the POM1 antibody for detection of PrP.
Real-time quaking-induced conversion (RT-QuIC)
Recombinant MoPrP(23–230) was generated as described previously for recombinant Syrian hamster PrP (34). ScCAD5 cells were homogenized as described above, and then protein levels in homogenates quantified using the BCA assay. RT-QuIC was carried out as described previously, with conditions optimized for detection using MoPrP as the substrate (89). Briefly, RT-QuIC substrate mixture was prepared as follows: 0.1 mg/ml (4 μM) recombinant MoPrP(23–230), PBS (130 mM NaCl, 10 mM sodium phosphate pH 7.3), 10 μM ThT, and 1 mM EDTA. Prior to addition to the reaction mixture, recombinant MoPrP was centrifuged at 21,000g for 15 min to remove any preexisting aggregates. A tenfold molar excess (40 μM) of either G418 or puromycin was added where indicated. Ninety eight microliters of this reaction mixture was added to each well of a black, clear bottom 96-well plate (Nunc). Then, 2 μl of the seed mixture [PBS containing 0.05% (w/v) SDS, 1× N-2 Supplement (ThermoFisher), and 10 ng of ScCAD5 homogenate] was added to each well. The plate was sealed and then incubated at 42 °C in a BMG CLARIOstar microplate reader. The plate was subjected to alternating cycles of 1 min shaking (700 rpm, double orbital) and 1 min rest. ThT fluorescence (excitation: 444 ± 5 nm; emission: 485 ± 5 nm) was measured every cycle (every 2 min) for up to 38 h total using a gain setting of 1600.
ThT aggregation assay
Recombinant MoPrP(23–230) was dialyzed to pH 7.4 in 10 mM sodium phosphate buffer. Following dialysis, reaction mixture was prepared using 10 mM sodium phosphate pH 7.4, 0.1 mg/ml (4 μM) MoPrP(23–230), 10 μM ThT, and 135 mM NaCl. A tenfold molar excess of G418 (40 μM) was added to the mixture where indicated. One hundred microliters of this mixture was pipetted to the bottom of each well of a black, clear bottom 96 well plate (Nunc). The plate was sealed and then subjected to continuous shaking (700 rpm, double orbital) for up to 80 h at 37 °C in a BMG CLARIOstar microplate reader. ThT readings (excitation: 444 ± 5 nm; emission: 485 ± 5 nm) were taken every 5 min. Lag phases for aggregation were calculated by fitting the kinetic curves to a sigmoidal dose–response (variable slope) model in GraphPad Prism, and then using the equation T50 − [1/(2∗k)], where k is the Hill slope and T50 is the time at which fluorescence is halfway between the baseline and plateau values (90).
Differential scanning fluorimetry
Recombinant MoPrP(23–230) at a concentration of 0.1 mg/ml (4 μM) was incubated with SYPRO Orange according to the manufacturer’s instructions (ThermoFisher #4461146). The temperature was increased in slow 2 °C/min increments using a quantitative PCR machine (Light Cycler 480, Roche), and the fluorescence from the SYPRO orange dye was measured over time. Increased binding of the dye is directly proportional to exposure of hydrophobic residues as the protein becomes denatured. Where indicated, the reaction mixture contained a tenfold molar excess (40 μM) of either quinacrine, puromycin, or G418. Melting temperatures were calculated using Protein Thermal Shift software (Roche).
Statistical analysis
In prion infection experiments comparing the means of two experimental conditions where all replicate samples were run together on the same gel, an unpaired two-tailed t test was used. In prion infection experiments where experimental replicates were run on different gels, samples were normalized to their respective control replicates, which were set at 100%, and then a one sample t test was performed. When greater than two experimental conditions were compared, a one-way ANOVA was used followed by Dunnett’s (when samples were compared with a defined control) or Tukey’s (when all samples were compared with each other) multiple comparisons test. The distribution of the data was assumed to be normal, but this was not formally tested. All statistical analysis was performed using GraphPad Prism software (version 9.0.1) with a significance threshold of p < 0.05.
Data availability
All data are contained within the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors thank Jason Bartz and Ronald Shikiya for providing the 263K hamster prion strain, and David Westaway for providing the 22L mouse prion strain. Experimental schematics were created using BioRender.com.
This work was funded by a grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (#RGPIN-2015-05112).
Author contributions
H. A., G. S.-U., and J. C. W. conceptualization; H. A. and Z. P. data curation; H. A., Z. P., and J. C. W. formal analysis; J. C. W. funding acquisition; H. A., Z. P., M. M., M. E. C. B., and Z. A. M. A.-A. investigation; H. A., Z. P., M. M., M. E. C. B., and Z. A. M. A.-A. methodology; J. C. W. project administration; G. S.-U. and J. C. W. supervision; H. A. validation; H. A. and J. C. W. visualization; H. A. and J. C. W. writing–original draft; H. A., Z. P., M. M., M. E. C. B., Z. A. M. A.-A., G. S.-U., and J. C. W. writing–review and editing.
Funding and additional information
Z. P. and Z. A. M. A.-A. were supported by NSERC undergraduate student research awards.
Edited by Ursula Jakob
Footnotes
Present address for Mohadeseh Mehrabian: Sangamo Therapeutics, Brisbane, California, USA.
Supporting information
References
- 1.Colby D.W., Prusiner S.B. Prions. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheckel C., Aguzzi A. Prions, prionoids and protein misfolding disorders. Nat. Rev. Genet. 2018;19:405–418. doi: 10.1038/s41576-018-0011-4. [DOI] [PubMed] [Google Scholar]
- 3.Prusiner S.B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 4.Oesch B., Westaway D., Wälchli M., McKinley M.P., Kent S.B.H., Aebersold R., Barry R.A., Tempst P., Teplow D.B., Hood L.E., Prusiner S.B., Weissmann C. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 5.Stahl N., Borchelt D.R., Hsiao K., Prusiner S.B. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 1987;51:229–240. doi: 10.1016/0092-8674(87)90150-4. [DOI] [PubMed] [Google Scholar]
- 6.Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., Wuthrich K. NMR structure of the mouse prion protein domain PrP(121-231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 7.Watts J.C., Bourkas M.E.C., Arshad H. The function of the cellular prion protein in health and disease. Acta Neuropathol. 2018;135:159–178. doi: 10.1007/s00401-017-1790-y. [DOI] [PubMed] [Google Scholar]
- 8.Mehrabian M., Hildebrandt H., Schmitt-Ulms G. NCAM1 polysialylation: The prion protein's elusive reason for being? ASN Neuro. 2016;8 doi: 10.1177/1759091416679074. 1759091416679074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan K.-M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R.J., Cohen F.E., Prusiner S.B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watts J.C., Prusiner S.B. Mouse models for studying the formation and propagation of prions. J. Biol. Chem. 2014;289:19841–19849. doi: 10.1074/jbc.R114.550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandner S., Jaunmuktane Z. Prion disease: Experimental models and reality. Acta Neuropathol. 2017;133:197–222. doi: 10.1007/s00401-017-1670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grassmann A., Wolf H., Hofmann J., Graham J., Vorberg I. Cellular aspects of prion replication in vitro. Viruses. 2013;5:374–405. doi: 10.3390/v5010374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Race R.E., Fadness L.H., Chesebro B. Characterization of scrapie infection in mouse neuroblastoma cells. J. Gen. Virol. 1987;68:1391–1399. doi: 10.1099/0022-1317-68-5-1391. [DOI] [PubMed] [Google Scholar]
- 14.Butler D.A., Scott M.R., Bockman J.M., Borchelt D.R., Taraboulos A., Hsiao K.K., Kingsbury D.T., Prusiner S.B. Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J. Virol. 1988;62:1558–1564. doi: 10.1128/jvi.62.5.1558-1564.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schatzl H.M., Laszlo L., Holtzman D.M., Tatzelt J., DeArmond S.J., Weiner R.I., Mobley W.C., Prusiner S.B. A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J. Virol. 1997;71:8821–8831. doi: 10.1128/jvi.71.11.8821-8831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahal S.P., Baker C.A., Demczyk C.A., Smith E.W., Julius C., Weissmann C. Prion strain discrimination in cell culture: The cell panel assay. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20908–20913. doi: 10.1073/pnas.0710054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birkett C.R., Hennion R.M., Bembridge D.A., Clarke M.C., Chree A., Bruce M.E., Bostock C.J. Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. EMBO J. 2001;20:3351–3358. doi: 10.1093/emboj/20.13.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vorberg I., Raines A., Story B., Priola S.A. Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. J. Infect. Dis. 2004;189:431–439. doi: 10.1086/381166. [DOI] [PubMed] [Google Scholar]
- 19.Herbst A., Banser P., Velasquez C.D., Mays C.E., Sim V.L., Westaway D., Aiken J.M., McKenzie D. Infectious prions accumulate to high levels in non proliferative C2C12 myotubes. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krance S.H., Luke R., Shenouda M., Israwi A.R., Colpitts S.J., Darwish L., Strauss M., Watts J.C. Cellular models for discovering prion disease therapeutics: Progress and challenges. J. Neurochem. 2020;153:150–172. doi: 10.1111/jnc.14956. [DOI] [PubMed] [Google Scholar]
- 21.Ghaemmaghami S., May B.C.H., Renslo A.R., Prusiner S.B. Discovery of 2-aminothiazoles as potent antiprion compounds. J. Virol. 2010;84:3408–3412. doi: 10.1128/JVI.02145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry D.B., Lu D., Geva M., Watts J.C., Bhardwaj S., Oehler A., Renslo A.R., DeArmond S.J., Prusiner S.B., Giles K. Drug resistance confounding prion therapeutics. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E4160–E4169. doi: 10.1073/pnas.1317164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu D., Giles K., Li Z., Rao S., Dolghih E., Gever J.R., Geva M., Elepano M.L., Oehler A., Bryant C., Renslo A.R., Jacobson M.P., DeArmond S.J., Silber B.M., Prusiner S.B. Biaryl amides and hydrazones as therapeutics for prion disease in transgenic mice. J. Pharmacol. Exp. Ther. 2013;347:325–338. doi: 10.1124/jpet.113.205799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giles K., Berry D.B., Condello C., Hawley R.C., Gallardo-Godoy A., Bryant C., Oehler A., Elepano M., Bhardwaj S., Patel S., Silber B.M., Guan S., DeArmond S.J., Renslo A.R., Prusiner S.B. Different 2-aminothiazole therapeutics produce distinct patterns of scrapie prion neuropathology in mouse brains. J. Pharmacol. Exp. Ther. 2015;355:2–12. doi: 10.1124/jpet.115.224659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giles K., Berry D.B., Condello C., Dugger B.N., Li Z., Oehler A., Bhardwaj S., Elepano M., Guan S., Silber B.M., Olson S.H., Prusiner S.B. Optimization of aryl amides that extend survival in prion-infected mice. J. Pharmacol. Exp. Ther. 2016;358:537–547. doi: 10.1124/jpet.116.235556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tark D., Kim H., Neale M.H., Kim M., Sohn H., Lee Y., Cho I., Joo Y., Windl O. Generation of a persistently infected MDBK cell line with natural bovine spongiform encephalopathy (BSE) PLoS One. 2015;10 doi: 10.1371/journal.pone.0115939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raymond G.J., Olsen E.A., Lee K.S., Raymond L.D., Bryant P.K., 3rd, Baron G.S., Caughey W.S., Kocisko D.A., McHolland L.E., Favara C., Langeveld J.P., van Zijderveld F.G., Mayer R.T., Miller M.W., Williams E.S. Inhibition of protease-resistant prion protein formation in a transformed deer cell line infected with chronic wasting disease. J. Virol. 2006;80:596–604. doi: 10.1128/JVI.80.2.596-604.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taraboulos A., Serban D., Prusiner S.B. Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J. Cell Biol. 1990;110:2117–2132. doi: 10.1083/jcb.110.6.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bian J., Napier D., Khaychuck V., Angers R., Graham C., Telling G. Cell-based quantification of chronic wasting disease prions. J. Virol. 2010;84:8322–8326. doi: 10.1128/JVI.00633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilette D., Andreoletti O., Archer F., Madelaine M.F., Vilotte J.L., Lehmann S., Laude H. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4055–4059. doi: 10.1073/pnas.061337998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Courageot M.P., Daude N., Nonno R., Paquet S., Di Bari M.A., Le Dur A., Chapuis J., Hill A.F., Agrimi U., Laude H., Vilette D. A cell line infectible by prion strains from different species. J. Gen. Virol. 2008;89:341–347. doi: 10.1099/vir.0.83344-0. [DOI] [PubMed] [Google Scholar]
- 32.Maas E., Geissen M., Groschup M.H., Rost R., Onodera T., Schatzl H., Vorberg I.M. Scrapie infection of prion protein-deficient cell line upon ectopic expression of mutant prion proteins. J. Biol. Chem. 2007;282:18702–18710. doi: 10.1074/jbc.M701309200. [DOI] [PubMed] [Google Scholar]
- 33.Marshall K.E., Hughson A., Vascellari S., Priola S.A., Sakudo A., Onodera T., Baron G.S. PrP knockout cells expressing transmembrane PrP resist prion infection. J. Virol. 2017;91 doi: 10.1128/JVI.01686-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourkas M.E.C., Arshad H., Al-Azzawi Z.A.M., Halgas O., Shikiya R.A., Mehrabian M., Schmitt-Ulms G., Bartz J.C., Watts J.C. Engineering a murine cell line for the stable propagation of hamster prions. J. Biol. Chem. 2019;294:4911–4923. doi: 10.1074/jbc.RA118.007135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walia R., Ho C.C., Lee C., Gilch S., Schatzl H.M. Gene-edited murine cell lines for propagation of chronic wasting disease prions. Sci. Rep. 2019;9:11151. doi: 10.1038/s41598-019-47629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avar M., Heinzer D., Steinke N., Dogancay B., Moos R., Lugan S., Cosenza C., Hornemann S., Andreoletti O., Aguzzi A. Prion infection, transmission, and cytopathology modeled in a low-biohazard human cell line. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Southern P.J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 38.Prokhorova I., Altman R.B., Djumagulov M., Shrestha J.P., Urzhumtsev A., Ferguson A., Chang C.T., Yusupov M., Blanchard S.C., Yusupova G. Aminoglycoside interactions and impacts on the eukaryotic ribosome. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E10899–E10908. doi: 10.1073/pnas.1715501114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jimenez A., Davies J. Expression of a transposable antibiotic resistance element in Saccharomyces. Nature. 1980;287:869–871. doi: 10.1038/287869a0. [DOI] [PubMed] [Google Scholar]
- 40.Suri C., Fung B.P., Tischler A.S., Chikaraishi D.M. Catecholaminergic cell lines from the brain and adrenal glands of tyrosine hydroxylase-SV40 T antigen transgenic mice. J. Neurosci. 1993;13:1280–1291. doi: 10.1523/JNEUROSCI.13-03-01280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi Y., Wang J.K., McMillian M., Chikaraishi D.M. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oelschlegel A.M., Fallahi M., Ortiz-Umpierre S., Weissmann C. The extended cell panel assay characterizes the relationship of prion strains RML, 79A, and 139A and reveals conversion of 139A to 79A-like prions in cell culture. J. Virol. 2012;86:5297–5303. doi: 10.1128/JVI.00181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishida N., Harris D.A., Vilette D., Laude H., Frobert Y., Grassi J., Casanova D., Milhavet O., Lehmann S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 2000;74:320–325. doi: 10.1128/jvi.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Korth C., May B.C.H., Cohen F.E., Prusiner S.B. Acridine and phenothiazine derivatives as pharmacotherapeutics for prion disease. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9836–9841. doi: 10.1073/pnas.161274798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bosque P.J., Prusiner S.B. Cultured cell sublines highly susceptible to prion infection. J. Virol. 2000;74:4377–4386. doi: 10.1128/jvi.74.9.4377-4386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kung M., Stadelmann B., Brodbeck U., Butikofer P. Addition of G418 and other aminoglycoside antibiotics to mammalian cells results in the release of GPI-anchored proteins. FEBS Lett. 1997;409:333–338. doi: 10.1016/s0014-5793(97)00452-3. [DOI] [PubMed] [Google Scholar]
- 47.Westergard L., Turnbaugh J.A., Harris D.A. A naturally occurring C-terminal fragment of the prion protein (PrP) delays disease and acts as a dominant-negative inhibitor of PrPSc formation. J. Biol. Chem. 2011;286:44234–44242. doi: 10.1074/jbc.M111.286195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altmeppen H.C., Puig B., Dohler F., Thurm D.K., Falker C., Krasemann S., Glatzel M. Proteolytic processing of the prion protein in health and disease. Am. J. Neurodegener. Dis. 2012;1:15–31. [PMC free article] [PubMed] [Google Scholar]
- 49.Liang J., Kong Q. alpha-Cleavage of cellular prion protein. Prion. 2012;6:453–460. doi: 10.4161/pri.22511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poncet-Montange G., St.-Martin S.J., Bogatova O.V., Prusiner S.B., Shoichet B.K., Ghaemmaghami S. A survey of antiprion compounds reveals the prevalence of non-PrP molecular targets. J. Biol. Chem. 2011;286:27718–27728. doi: 10.1074/jbc.M111.234393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reidenbach A.G., Mesleh M.F., Casalena D., Vallabh S.M., Dahlin J.L., Leed A.J., Chan A.I., Usanov D.L., Yehl J.B., Lemke C.T., Campbell A.J., Shah R.N., Shrestha O.K., Sacher J.R., Rangel V.L. Multimodal small-molecule screening for human prion protein binders. J. Biol. Chem. 2020;295:13516–13531. doi: 10.1074/jbc.RA120.014905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niesen F.H., Berglund H., Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 53.Wille H., Bian W., McDonald M., Kendall A., Colby D.W., Bloch L., Ollesch J., Boronvinskiy A.L., Cohen F.E., Prusiner S.B., Stubbs G. Natural and synthetic prion structure from X-ray fiber diffraction. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilham J.M., Orru C.D., Bessen R.A., Atarashi R., Sano K., Race B., Meade-White K.D., Taubner L.M., Timmes A., Caughey B. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duesberg P., Stindl R., Hehlmann R. Explaining the high mutation rates of cancer cells to drug and multidrug resistance by chromosome reassortments that are catalyzed by aneuploidy. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14295–14300. doi: 10.1073/pnas.97.26.14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mange A., Nishida N., Milhavet O., McMahon H.E., Casanova D., Lehmann S. Amphotericin B inhibits the generation of the scrapie isoform of the prion protein in infected cultures. J. Virol. 2000;74:3135–3140. doi: 10.1128/jvi.74.7.3135-3140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKenzie D., Kaczkowski J., Marsh R., Aiken J. Amphotericin B delays both scrapie agent replication and PrP-res accumulation early in infection. J. Virol. 1994;68:7534–7536. doi: 10.1128/jvi.68.11.7534-7536.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Demaimay R., Adjou K.T., Beringue V., Demart S., Lasmezas C.I., Deslys J.P., Seman M., Dormont D. Late treatment with polyene antibiotics can prolong the survival time of scrapie-infected animals. J. Virol. 1997;71:9685–9689. doi: 10.1128/jvi.71.12.9685-9689.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forloni G., Iussich S., Awan T., Colombo L., Angeretti N., Girola L., Bertani I., Poli G., Caramelli M., Grazia Bruzzone M., Farina L., Limido L., Rossi G., Giaccone G., Ironside J.W. Tetracyclines affect prion infectivity. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10849–10854. doi: 10.1073/pnas.162195499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Luigi A., Colombo L., Diomede L., Capobianco R., Mangieri M., Miccolo C., Limido L., Forloni G., Tagliavini F., Salmona M. The efficacy of tetracyclines in peripheral and intracerebral prion infection. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masullo C., Macchi G., Xi Y.G., Pocchiari M. Failure to ameliorate Creutzfeldt-Jakob disease with amphotericin B therapy. J. Infect. Dis. 1992;165:784–785. doi: 10.1093/infdis/165.4.784. [DOI] [PubMed] [Google Scholar]
- 62.Haik S., Marcon G., Mallet A., Tettamanti M., Welaratne A., Giaccone G., Azimi S., Pietrini V., Fabreguettes J.R., Imperiale D., Cesaro P., Buffa C., Aucan C., Lucca U., Peckeu L. Doxycycline in Creutzfeldt-Jakob disease: A phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2014;13:150–158. doi: 10.1016/S1474-4422(13)70307-7. [DOI] [PubMed] [Google Scholar]
- 63.Varges D., Manthey H., Heinemann U., Ponto C., Schmitz M., Schulz-Schaeffer W.J., Krasnianski A., Breithaupt M., Fincke F., Kramer K., Friede T., Zerr I. Doxycycline in early CJD: A double-blinded randomised phase II and observational study. J. Neurol. Neurosurg. Psychiatry. 2017;88:119–125. doi: 10.1136/jnnp-2016-313541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bidou L., Allamand V., Rousset J.P., Namy O. Sense from nonsense: Therapies for premature stop codon diseases. Trends Mol. Med. 2012;18:679–688. doi: 10.1016/j.molmed.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 65.Wangen J.R., Green R. Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. Elife. 2020;9 doi: 10.7554/eLife.52611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hatin I., Bidou L., Cullin C., Rousset J.P. Translational errors as an early event in prion conversion. Cell. Mol. Biol. (Noisy-le-grand) 2001;47:OL23–OL28. [PubMed] [Google Scholar]
- 67.Ghaemmaghami S., Ahn M., Lessard P., Giles K., Legname G., DeArmond S.J., Prusiner S.B. Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J., Browning S., Mahal S.P., Oelschlegel A.M., Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010;327:869–872. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oelschlegel A.M., Weissmann C. Acquisition of drug resistance and dependence by prions. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Safar J.G., Geschwind M.D., Deering C., Didorenko S., Sattavat M., Sanchez H., Serban A., Vey M., Baron H., Giles K., Miller B.L., DeArmond S.J., Prusiner S.B. Diagnosis of human prion disease. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3501–3506. doi: 10.1073/pnas.0409651102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sajnani G., Silva C.J., Ramos A., Pastrana M.A., Onisko B.C., Erickson M.L., Antaki E.M., Dynin I., Vazquez-Fernandez E., Sigurdson C.J., Carter J.M., Requena J.R. PK-sensitive PrP is infectious and shares basic structural features with PK-resistant PrP. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trevitt C.R., Collinge J. A systematic review of prion therapeutics in experimental models. Brain. 2006;129:2241–2265. doi: 10.1093/brain/awl150. [DOI] [PubMed] [Google Scholar]
- 73.Silveira J.R., Raymond G.J., Hughson A.G., Race R.E., Sim V.L., Hayes S.F., Caughey B. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kanu N., Imokawa Y., Drechsel D.N., Williamson R.A., Birkett C.R., Bostock C.J., Brockes J.P. Transfer of scrapie prion infectivity by cell contact in culture. Curr. Biol. 2002;12:523–530. doi: 10.1016/s0960-9822(02)00722-4. [DOI] [PubMed] [Google Scholar]
- 75.Ghaemmaghami S., Phuan P.W., Perkins B., Ullman J., May B.C., Cohen F.E., Prusiner S.B. Cell division modulates prion accumulation in cultured cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17971–17976. doi: 10.1073/pnas.0708372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gousset K., Schiff E., Langevin C., Marijanovic Z., Caputo A., Browman D.T., Chenouard N., de Chaumont F., Martino A., Enninga J., Olivo-Marin J.C., Mannel D., Zurzolo C. Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol. 2009;11:328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- 77.Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., Raposo G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandberg M.K., Al-Doujaily H., Sharps B., Clarke A.R., Collinge J. Prion propagation and toxicity in vivo occur in two distinct mechanistic phases. Nature. 2011;470:540–542. doi: 10.1038/nature09768. [DOI] [PubMed] [Google Scholar]
- 79.Sandberg M.K., Al-Doujaily H., Sharps B., De Oliveira M.W., Schmidt C., Richard-Londt A., Lyall S., Linehan J.M., Brandner S., Wadsworth J.D., Clarke A.R., Collinge J. Prion neuropathology follows the accumulation of alternate prion protein isoforms after infective titre has peaked. Nat. Commun. 2014;5:4347. doi: 10.1038/ncomms5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fang C., Imberdis T., Garza M.C., Wille H., Harris D.A. A neuronal culture system to detect prion synaptotoxicity. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Massignan T., Stewart R.S., Biasini E., Solomon I.H., Bonetto V., Chiesa R., Harris D.A. A novel, drug-based, cellular assay for the activity of neurotoxic mutants of the prion protein. J. Biol. Chem. 2010;285:7752–7765. doi: 10.1074/jbc.M109.064949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Imberdis T., Heeres J.T., Yueh H., Fang C., Zhen J., Rich C.B., Glicksman M., Beeler A.B., Harris D.A. Identification of anti-prion compounds using a novel cellular assay. J. Biol. Chem. 2016;291:26164–26176. doi: 10.1074/jbc.M116.745612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Solomon I.H., Huettner J.E., Harris D.A. Neurotoxic mutants of the prion protein induce spontaneous ionic currents in cultured cells. J. Biol. Chem. 2010;285:26719–26726. doi: 10.1074/jbc.M110.134619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Biasini E., Unterberger U., Solomon I.H., Massignan T., Senatore A., Bian H., Voigtlaender T., Bowman F.P., Bonetto V., Chiesa R., Luebke J., Toselli P., Harris D.A. A mutant prion protein sensitizes neurons to glutamate-induced excitotoxicity. J. Neurosci. 2013;33:2408–2418. doi: 10.1523/JNEUROSCI.3406-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mehrabian M., Brethour D., MacIsaac S., Kim J.K., Gunawardana C.G., Wang H., Schmitt-Ulms G. CRISPR-Cas9-based knockout of the prion protein and its effect on the proteome. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Williamson R.A., Peretz D., Pinilla C., Ball H., Bastidas R.B., Rozenshteyn R., Houghten R.A., Prusiner S.B., Burton D.R. Mapping the prion protein using recombinant antibodies. J. Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Safar J.G., Scott M., Monaghan J., Deering C., Didorenko S., Vergara J., Ball H., Legname G., Leclerc E., Solforosi L., Serban H., Groth D., Burton D.R., Prusiner S.B., Williamson R.A. Measuring prions causing bovine spongiform encephalopathy or chronic wasting disease by immunoassays and transgenic mice. Nat. Biotechnol. 2002;20:1147–1150. doi: 10.1038/nbt748. [DOI] [PubMed] [Google Scholar]
- 88.Polymenidou M., Moos R., Scott M., Sigurdson C., Shi Y.Z., Yajima B., Hafner-Bratkovic I., Jerala R., Hornemann S., Wuthrich K., Bellon A., Vey M., Garen G., James M.N., Kav N. The POM monoclonals: A comprehensive set of antibodies to non-overlapping prion protein epitopes. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Orru C.D., Hughson A.G., Groveman B.R., Campbell K.J., Anson K.J., Manca M., Kraus A., Caughey B. Factors that improve RT-QuIC detection of prion seeding activity. Viruses. 2016;8:140. doi: 10.3390/v8050140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arosio P., Knowles T.P., Linse S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.