Abstract
Nonhistone chromosomal proteins HMG-14 and HMG-17 are closely related nucleosomal binding proteins that unfold the higher-order chromatin structure, thereby enhancing the transcription and replication potential of chromatin. Here we report that PCAF, a transcription coactivator with intrinsic histone acetyltransferase activity, specifically acetylates HMG-17 but not HMG-14. Using mass spectrum sequence analysis, we identified the lysine at position 2 as the predominant site acetylated by PCAF. Lysine 2 is a prominent acetylation site in vivo, suggesting that this PCAF-mediated acetylation is physiologically relevant. Experiments with HMG-17 deletion mutants and competition studies with various protein fragments indicate that the specific acetylation of HMG-17 is not determined solely by the primary sequence near the acetylation site. By equilibrium dialysis we demonstrated that acetylation reduces the affinity of HMG-17 to nucleosome cores. In addition, we found that the binding of HMG-14 and HMG-17 to nucleosome cores inhibits the PCAF-mediated acetylation of histone H3. Thus, the presence of HMG-14 and HMG-17 affects the ability of PCAF to acetylate chromatin, while the acetylation of HMG-17 reduces its binding affinity to chromatin. Conceivably, in HMG-17-containing chromatin, acetylation of HMG-17 precedes the acetylation of histones.
Reversible acetylation of the N-terminal tails of histones plays a key role in the regulation of various nuclear activities such as chromatin assembly, replication, and transcription (2, 19, 29, 39, 49, 51, 52). The acetylation of lysine residues within nucleosomes weakens the interaction of the histone tails with the DNA and leads to chromatin decompaction (16, 17). These structural transitions enhance the accessibility of the underlying DNA sequence to various factors, thereby reducing the repressive effect of the nucleosome on transcription and replication. The relationship between transcriptional regulation and histone acetylation has been strengthened considerably by the discovery that certain factors associated with transcriptional activation have intrinsic histone acetylase activity (7, 20, 30, 31, 44, 53), while factors associated with transcriptional repression contain histone deacetylase activity (26, 44). It is significant that in some cases this reversible acetylation is targeted and specific. For example, Tetrahymena GCN5 preferentially acetylates residues K8 and K16 of histone H4 and K14 of histone H3 (13, 24). In contrast, in Saccharomyces cerevisiae, transcriptional repression by UME6 involves the specific deacetylation of K5 in histone H4 by the deacetylase RPD3 (40). Furthermore, the pattern of H4 acetylation in heterochromatin is unique, suggesting that specific acetylation marks discrete functional states of chromatin structure (5, 32). Taken together with other findings, these results suggest that the reversible acetylation of histones is not merely a mechanism for indiscriminately unfolding chromatin but is a key step in the selective regulation of the expression of specific genes.
Most of the studies on the effect of acetylation on the structure and function of chromatin have focused on the reversible acetylation of the core histones. However, other chromatin-associated proteins, such as the nonhistone HMG proteins (9, 10), are also reversibly modified (23). In duck erythrocytes, two acetylation sites in HMG-1 and HMG-14 and three sites in HMG-17 were identified (42, 43). In the HMG-14/-17 protein family the major acetylation site detected even in cells not treated with deacetylase inhibitors is the lysine at position 2 (43). The enzymes responsible for the reversible acetylation of HMG proteins have not been identified, and nothing is known about the functional consequences of HMG acetylation.
Chromosomal proteins HMG-14 and HMG-17 are the only nuclear proteins known to specifically bind to the 146-bp nucleosome core particle and therefore could be considered as an integral part of the chromatin fiber (9, 10). These HMG proteins specifically interact with the N termini of the core histones (11) and produce distinct footprints on the nucleosome core (1, 27, 41). Removal of the N termini of the core histones greatly reduces the binding of the proteins to the nucleosome core (11). By site-directed cross-linking we demonstrated that HMG-14 contacts the nucleosome at multiple sites (47). The N terminus of HMG-14 specifically interacts with histone H2B, while the C terminus of the protein specifically interacts with the N terminus of histone H3. In chromatin, HMG-containing nucleosomes are clustered into distinct domains, which on the average consist of six contiguous nucleosome-HMG complexes (35). The binding of HMG-14/-17 proteins to nucleosomes unfolds the higher-order chromatin structure and enhances various DNA-dependent activities, such as transcription (12, 14, 15, 34, 45, 46, 48) and replication (50).
A mechanistic view of these findings suggests that HMG-14/-17 proteins unfold the higher-order chromatin structure, thereby promoting access to nucleosomes by various regulatory factors, some of which may have histone acetyltransferase (HAT) activity. In view of the close proximity between the acetylation sites in histones (histone tails) and HMG-14/-17 proteins (47) and the recent observation that some HATs can modify transcription factors (22), it is plausible that these enzymes could also modify HMGs.
Here we report that PCAF specifically acetylates HMG-17 but not HMG-14, and we demonstrate that the specificity of acetylation may require a distinct protein conformation. We examine the ability of PCAF to acetylate HMG-nucleosome complexes and demonstrate that the presence of HMG-14/-17 proteins affects the rate of acetylation of the N termini of core histones. We studied the effect of acetylation on the interaction of HMG-17 with nucleosomes and show that acetylation of HMG-17 affects its interaction with nucleosomes. These findings represent the first identification of an acetylase capable of specifically acetylating a nonhistone structural chromosomal protein and provide insight into a mechanism whereby HATs affect transcription in the context of chromatin.
MATERIALS AND METHODS
Substrates and enzymes.
Recombinant human PCAF containing the Flag tag (Kodak) was prepared as described previously (53). Histone H1 was extracted from calf thymus with 5% trichloroacetic acid and purified by chromatography on Amberlite IRC-50 (8). Histone H3 was extracted from calf thymus with 0.1 M HCl, oxidized, and purified on Sephadex G-100 (28). Chromosomal protein HMG-1 was isolated from calf thymus by extraction with 0.35 M NaCl and purified by ion-exchange chromatography (38). Recombinant wild-type HMG-14 and HMG-17 and their N- and C-terminal truncation mutants were expressed and purified as previously described (48). Nucleosome core particles were purified from chicken erythrocytes as previously described (1, 4). Cytochrome c and acetyl-coenzyme A (CoA) were obtained from Sigma. [1-14C]acetyl-CoA (55 mCi/mmol) was obtained from Amersham. [3H]acetyl-CoA (26 Ci/mmol) was obtained from Moravek, Inc.
HAT assay.
All assays were performed in buffer A (50 mM Tris-HCl, pH 8.0; 10% glycerol [vol/vol]; 1 mM dithiothreitol; 0.1 mM EDTA; 10 mM butyric acid) (6). Substrate concentrations were 0.1 to 0.25 mg/ml, and the [3H]- or [1-14C]acetyl-CoA concentrations were 9 μM (unless otherwise indicated). The assay was performed at 37°C and was initiated by the addition of the protein substrate to a mixture containing the acetyltransferase and acetyl-CoA in buffer A (21). The radioactivity incorporated into the protein substrate was detected by a polyacrylamide gel assay. In this assay, the reactions were stopped by the addition of an equal volume of a sodium dodecyl sulfate (SDS) gel sample buffer (100 mM Tris-HCl, pH 6.8; 200 mM dithiothreitol; 2% SDS; 0.1% bromophenol blue; 20% glycerol) and then boiled for 5 min; the proteins were then resolved on a 15% polyacrylamide-SDS gel. The electrophoresis was performed at 15 V/cm and stopped when the bromophenol blue reached the bottom of the gel. The gels were stained with Coomassie blue to estimate the protein quantities and then soaked in Enlightening Enhancer solution (Dupont) for 30 min and vacuum dried; the radioactivity incorporated into the protein bands was then visualized on a PhosphorImager (Molecular Dynamics) and quantified with ImageQuant software. Acetylation of the nucleosome–HMG-17 complexes was performed as just described, except that the chicken nucleosomes were reconstituted with various amounts of HMG-17 prior to the acetylation reaction. In another set of experiments, the proteins were labeled with [3H]acetyl-CoA (26 Ci/mmol; Moravek, Inc.) as described above. After electrophoresis and Coomassie blue staining, the protein bands were excised and digested in 30% hydrogen peroxide (65°C, overnight), and their radioactivity was determined by liquid scintillation counting. The competition assays were performed as just described except that various amounts of competitor (a 2× to 5× molar excess above the level of HMG-17) were added. Acetylation of peptides was examined either by autoradiography of [14C]acetate-labeled peptides, by excising [3H]acetate-labeled peptides from the polyacrylamide gels, or by mass spectral analysis.
Mass spectral analysis.
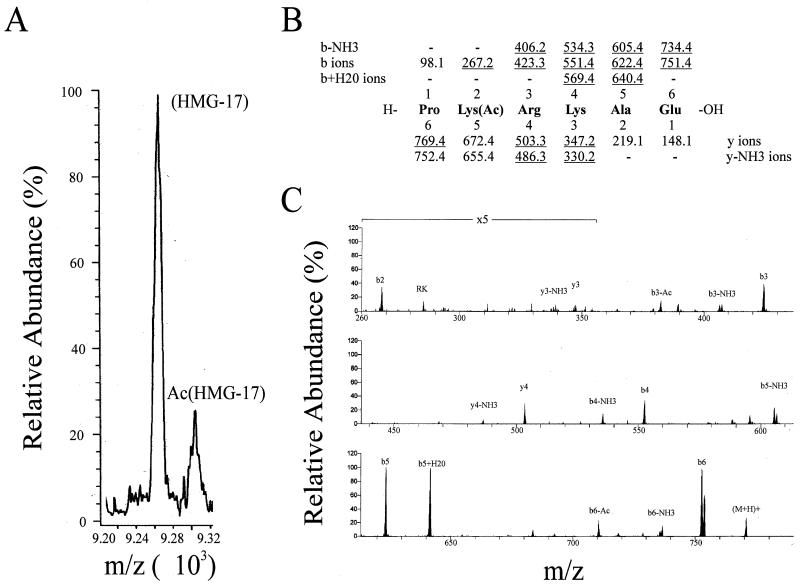
HMG-17 was acetylated by PCAF with nonradioactive acetyl-CoA as described above. To increase the yield of acetylated protein, the reaction time was extended to 4 h, with addition of fresh enzyme every hour and addition of 10 μM acetyl-CoA along with the final addition of enzyme. After acetylation, HMG-17 was purified by high-pressure liquid chromatography (HPLC) with an Aquapore butyl column (Applied Biosystems) and with a water (0.1% trifluoroacetic acid [TFA])-acetonitrile (0.1% TFA) gradient of 0 to 30% acetonitrile. HMG-17 was eluted from the column at approximately 20% acetonitrile. The HMG-17 peak was collected and subjected to mass spectral analysis to determine the level of acetylated residue. The mass of the modified protein mixture was analyzed by using a single quadrupole mass spectrometer (Finnigan SSQ-7000) equipped with an electrospray ion source. To identify the acetylation sites, we subjected the acetylated HMG to mass analysis and found that the protein incorporated a single acetyl group (see Fig. 3A). We purified the monoacetylated protein by HPLC and digested it with Glu-C (Promega) (in 50 mM ammonium bicarbonate, pH 8.0), and the resulting fragments were analyzed by nanospray ion-trap mass spectrometry (Finnigan model LCQ, equipped with a nanospray ion source). We searched for fragments with masses corresponding to monoacetylated Glu-C peptides and observed a single species corresponding to Pro(1)-Glu(6) with an M + H of 771; the unacetylated fragment would thus have a mass reduced by 42 (mass of acetyl group), i.e., an M + H of 729. The collision-induced ion products of this fragment were analyzed. Only the products consistent with acetylation of lysine 2 were observed.
FIG. 3.
Mass spectrum identification of the acetylated lysine in HMG-17. (A) Mass analysis of the full-length acetylated HMG-17. The peak designated HMG-17 is the full-length unmodified protein (m/z = 9,261), and the peak designated AcHMG-17 corresponds to HMG-17 with a single modification (one acetyl group, total m/z = 9,308). (B) Mass analysis of the Glu-C peptide (generated from the full-length acetylated species) containing the modification and identification of the modified lysine (position 2). (C) Region of the mass spectra containing the modified lysine. The mass analysis from panel B was performed from the spectra depicted in panel C.
Two-dimensional polyacrylamide gels.
Nucleosome–HMG-17 reconstituted complexes were acetylated by PCAF in buffer A as described above and, without further treatment, were immediately loaded, in the first dimension, onto a native 5% polyacrylamide (30:1, acrylamide–N,N′-methylene-bis-acrylamide) gel (0.75 mm by 6 cm) in 0.2× Tris-borate-EDTA (TBE). The gels were run at 20 V/cm at 4°C until the xylene cyanol dye migrated to approximately 70% of the gel length. The nucleosome complexes were visualized with ethidium bromide, and the lanes of interest were excised from the native gel and placed perpendicularly on top of a 15% polyacrylamide gel (1.5 mm by 15 cm) composed of resolving gel (1.5 mm by 8 cm) and 3% stacking gel (1.5 mm by 3.5 cm) containing SDS. The gel strip was embedded into a layer of stacking gel that was identical to the first layer of the stacking gel. Then, 0.5× SDS–gel sample buffer was layered over the embedded gel slices. The second dimension was run at 15 V/cm at room temperature until the bromophenol blue reached the bottom of the gel. The gels were stained with Coomassie blue and quantified by using a Molecular Dynamics scanning densitometer and the ImageQuant software. After the scanning, the gels were soaked in Enlightening Enhancer solution for 30 min and dried under a vacuum, and the radioactivity incorporated into each band was quantified by using a Molecular Dynamics PhosphorImager and the ImageQuant software. The specific activity of each band was determined from the densitometric and PhosphorImager analysis.
Equilibrium dialysis.
HMG-17 was acetylated in buffer A containing radioactively labeled acetyl-CoA for 3 h at 37°C (as described above). Fresh PCAF was added after each hour. After acetylation, PCAF was heat inactivated by incubating the reaction mixture at 95°C for 5 min. The heat inactivation fully abolished the acetylation activity of PCAF without affecting the ability of HMG-17 to bind to nucleosomes, as measured by gel shift assays. Gel shift assays were performed by mixing either heat-treated or untreated HMG-17 with nucleosome cores in the acetylation buffer. The complexes in the sample were then resolved by using a 5% polyacrylamide gel in 0.2× TBE; the gels were then run at 4°C for 2 h, and the complexes were visualized by using ethidium bromide. The dialysis was performed by using Teflon equilibrium chambers (Sialomed, Inc.) and 100-kDa cutoff membranes; this exclusion limit allows for the free movement of HMG-17 across the membrane while preventing the movement of the nucleosome. One chamber contained chicken core particles (50 μl [60 ng/μl] in 2× TBE); the other chamber contained various concentrations of PCAF-treated HMG-17 (heat treated) in 2× TBE. The chambers were then placed on a rocker platform at 4°C for 5 days, a time sufficient to achieve equilibrium. The protein compositions (histones and HMG-17) of the samples removed from each chamber were then analyzed on a 15% polyacrylamide gel containing SDS. The amount of HMG-17 in the samples was quantified by densitometry of the Coomassie blue-stained bands. No change in the nucleosome concentration was detected. The bands were then excised from the gel and digested in 30% hydrogen peroxide (65°C, overnight), and the radioactivity in each band was determined by scintillation counting. The specific activity was determined from the quantitation values from the Coomassie blue-stained gels and the total amounts of tritium in the gel slices.
RESULTS
PCAF specifically acetylates HMG-17 and not HMG-14.
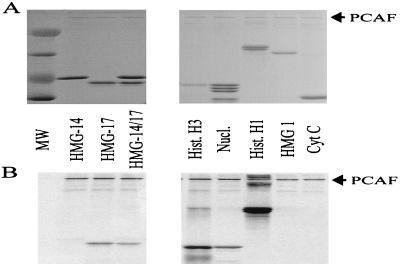
To examine whether the two closely related nonhistone proteins HMG-14 and HMG-17 are specifically acetylated by PCAF, we tested the ability of the enzyme to incorporate radioactive acetate into these proteins (Fig. 1). As we previously demonstrated, purified recombinant PCAF self-acetylates and specifically acetylates free H3, nucleosome-bound histone H3, and free linker histone H1 but not all proteins containing multiple lysines, such as cytochrome c, or the nonhistone protein HMG-1 (21). PCAF also exhibits a remarkable selectivity for the HMG proteins; it specifically acetylates HMG-17 but not HMG-14. To exclude the possibility that the HMG-14 stock solution inadvertently contained a PCAF inhibitor, we tested whether the enzyme could selectively acetylate HMG-17 in a mixture containing both HMG-14 and HMG-17. The results shown in Fig. 1 show that PCAF selectively acetylates HMG-17 even under conditions where HMG-14 is in excess and that the presence of HMG-14 has no significant effect on the level of acetylation of HMG-17. The specific activity of HMG-17 was 50-fold higher than that of HMG-14, 3-fold lower than that of nucleosome-bound H3, and about 80-fold lower than that of either free histone H3 or histone H1. No signal was obtained with either HMG-1 or cytochrome c.
FIG. 1.
Specific acetylation of HMG-17 by PCAF. The specificity of PCAF acetylation was tested by using several substrates. Panel A shows the protein gel, and panel B is the corresponding phosphorimage showing the incorporation of [14C]acetate. Substrates are as indicated (Hist.H3, histone H3; Nucl., chicken nucleosome core particles; Hist.H1, calf thymus histone H1; and Cyt. C, cytochrome c). The arrow to the right of each panel designates the position of PCAF.
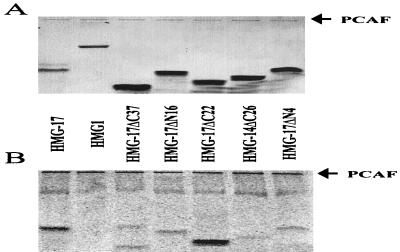
The known in vivo acetylation sites of both HMG-14 and HMG-17 are located in the N-terminal region of the proteins (42, 43) and involve lysine residues which are evolutionarily conserved among all the members of the HMG-14/-17 protein family (9, 10). Our finding that PCAF specifically acetylated HMG-17 but not HMG-14 raised the possibility that the site acetylated by PCAF is a novel, previously unidentified site. To identify the protein region containing the site acetylated by PCAF, we tested a series of HMG-17 deletion mutants for their ability to serve as substrates for this enzyme (Fig. 2). Mutants of HMG-17 lacking either 4 or 16 residues from the N terminus (designated HMG-17ΔN4 and HMG-17ΔN16) were very poor substrates for PCAF acetylation; the specific activity of these deletion mutants was 10-fold lower than that of full HMG-17. Although it is possible that PCAF acetylates more than one site, these data clearly indicate that the major PCAF-mediated acetylation site in HMG-17 is located in the N-terminal region of the protein, most probably in the first four amino acids. Consistent with this interpretation, removal of 22 residues from the C terminus of HMG-17 (producing mutant HMG-17ΔC22, containing the first 67 amino acids of HMG-17) had no discernible effect on acetylation. However, removal of 37 residues from the C terminus (i.e., mutant HMG-17ΔC37, containing the first 52 amino acids of HMG-17) reduced the acetylation to the same level as that observed with the N-terminal deletion mutants, suggesting that the acetylation site also could reside between residues 52 and 67. Nonetheless, the results suggest that only one of these regions contains an acetylation site but that both protein regions appear to be required for efficient acetylation.
FIG. 2.
Specificity of the acetylation reaction with HMG-17 truncation mutants. Panel A shows the protein gel; panel B is the corresponding phosphorimage showing the incorporation of [14C]acetate. Substrates are as indicated (HMG-17ΔC37, an HMG-17 truncation mutant lacking 37 residues from the C terminus; HMG-17ΔN16, an HMG-17 truncation mutant lacking 16 residues from the N terminus; HMG-17ΔC22, an HMG-17 truncation mutant lacking 22 residues from the C terminus; HMG-14ΔC26, an HMG-14 truncation mutant lacking 26 residues from the C terminus; and HMG-17ΔN4, an HMG-17 truncation mutant lacking the first 4 residues from the N terminus).
Identification of the acetylated lysine.
Analysis of the truncation mutants indicated two regions as possible targets for the acetylation of HMG-17 by PCAF in vitro. One region resides in the first four N-terminal amino acids and represents a previously identified site of modification in vivo (42, 43). Importantly, the first four residues, PKRK, are conserved among all the members of this protein family (9, 10), including HMG-14, which is not acetylated by PCAF. The second region, located between residues 52 and 67, has not been previously shown to contain an acetylation site in vivo. Nevertheless, this region is a sequence with significant divergence between HMG-14 and HMG-17 (9, 10). To unequivocally identify the acetylation site, the full-length acetylated protein was subjected to mass spectrum sequence analysis. Mass analysis of the modified protein detected only two molecular species: unacetylated and monoacetylated forms of HMG-17 (Fig. 3A). Mass spectrum analysis of a Glu-C digest of the protein revealed a single acetylation site, the lysine at position 2 (Fig. 3B and C; see Materials and Methods for the rationale of this analysis). No acetylation sites were detected beyond residue 47. These data, in combination with the truncation mutant analysis, indicate that the region between residues 52 and 67 is required for acetylation in the N-terminal region (lysine 2), although this region is not acetylated.
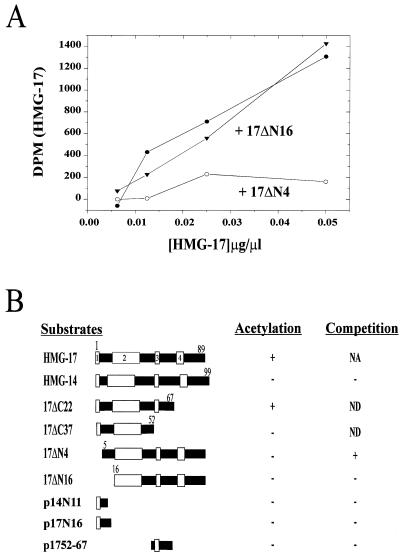
Specific acetylation of the evolutionarily conserved residue within a tetrapeptide present in both HMG-14 and HMG-17 suggests that the presence of the acetylation site is not sufficient to confer this specificity. We therefore used competition assays to identify which regions of the protein are required for the specific acetylation of HMG-17 by PCAF. As summarized in Fig. 4B, neither HMG-14, which contains the conserved sequence for the acetylation, nor mutants containing this site efficiently competed for the specific acetylation of HMG-17 by PCAF. Likewise, PCAF failed to acetylate any of the peptides containing this acetylation site. However, the HMG-17ΔN4 mutant, which lacked the acetylation site, could efficiently compete for acetylation of HMG-17, whereas the HMG-17ΔN16 mutant (lacking the first 16 amino acids) could not (Fig. 4A). Therefore, we conclude that the region between residues 5 and 16 may play a role in the specific acetylation of HMG-17 by PCAF. However, additional regions may be required because mutant HMG-17ΔC37, which lacks the 37 C-terminal residues and contains an intact N terminus that includes the acetylation site, was not acetylated (see also Fig. 2). In addition, a peptide containing residues 52 to 67 (present in HMG-17ΔC22 and absent HMG-17ΔC37) could not effectively compete for acetylation. We therefore conclude that the primary sequences near the acetylation site are not the only factors responsible for the specific acetylation of HMG-17 by PCAF.
FIG. 4.
Determinants for the specific acetylation of HMG-17. (A) The acetylation of HMG-17 is inhibited by a deletion mutant lacking the first 4 amino acids but is not inhibited by a deletion mutant lacking the first 16 amino acids. In these experiments the acetylation of HMG-17 was performed in the presence of 0.2 μg of competitor per μl. (B) Schematic diagram summarizing the studies indicating that residues 5 to 16 and a peptide longer than 52 residues of HMG-17 are required for the specific acetylation of HMG-17. Acetylation and competition studies were done as described in Materials and Methods. The designations for the truncation mutants are as indicated in Fig. 2. Three peptides were used: p14N11, a peptide containing the first 11 residues of HMG-14 (contains the conserved Lys-2); p17N16, a peptide containing the first 16 residues of HMG-17 (including the acetylated Lys-2); and p1752-67, a peptide containing residues 52 to 67 of HMG-17 (residues included in the acetylated HMG-17ΔC22 mutant but absent in the HMG-17ΔC37 mutant that is not a substrate). The HMG-17ΔC22 mutant was not tested (ND) as a competitor since it can function as a substrate. The HMG-17ΔC37 mutant was not tested as a competitor since it contains a fully intact N terminus, including the conserved acetylated Lys-2, yet it cannot function as a substrate. It is also indicated whether the substrate could (+) or could not (−) function as a substrate for acetylation or as a competitor for the acetylation of HMG-17. The boxes in the diagram indicate the evolutionarily conserved domains in HMG-14/-17. NA, not applicable.
Acetylation of the nucleosome–HMG-17 complex.
In chromatin, HMG-17 and the closely related protein HMG-14 form specific complexes with nucleosomes (9, 10, 35–37). In these complexes, the C-terminal region of HMG-14 makes specific contacts with the N-terminal region of histone H3 (11, 47), which is preferentially acetylated by PCAF (53). We therefore tested the activity of PCAF on nucleosomes containing HMG-17. Specifically, we examined whether PCAF can acetylate the nucleosome-bound HMG-17 and, in addition, whether HMG-17 affects the ability of PCAF to acetylate the N-terminal region of histone H3. In these experiments (Fig. 5) the concentrations of nucleosome-bound HMG-17 were in the same range as those used for the acetylation of free HMG-17 (Fig. 1).
FIG. 5.
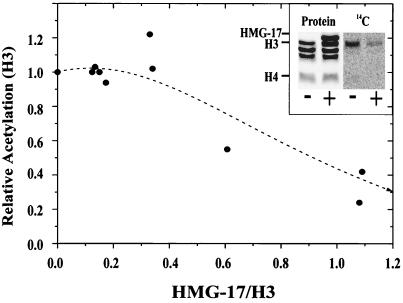
HMG-17 inhibits the acetylation of nucleosomal H3. Nucleosomes reconstituted with various amounts of HMG-17 were acetylated by PCAF, and the proteins were resolved on SDS-polyacrylamide gels. The incorporation of [14C]acetate into each protein was determined by phosphorimaging. The figure depicts the decrease in specific activity of nucleosomal H3 as a function of added HMG-17. The inset shows the protein gel (left panel) and the corresponding phosphorimage (right panel) showing 14C incorporation either into the nucleosomes alone (−) or into nucleosomes containing stoichiometric amounts of HMG-17 (+).
The results indicate that the nucleosome-bound HMG-17 is less efficiently acetylated by PCAF than is the free HMG-17 (inset in Fig. 5). This conclusion is based on our finding that the specific activity of free HMG-17 is only threefold lower than that of nucleosome-bound H3 (Fig. 1) and on the inability to detect radioactivity in the nucleosome-bound HMG-17 (compare the “−” and “+” lanes in the inset in Fig. 5). In addition, we found that the presence of HMG-17 in nucleosomes decreased the efficiency of H3 acetylation by PCAF (Fig. 5). An incremental increase in the ratio of HMG-17 protein to the nucleosome core results in an incremental decrease in the acetylation of histone H3. In addition, increasing the time of acetylation of the complexes enhanced the acetylation of H3 proportionately with time but did not alter the percentage of inhibition (not shown). These results suggest that the binding of HMG to nucleosomes interferes with the PCAF-mediated acetylation of nucleosomal H3. Similar results were obtained by analyzing the complexes by using a two-dimensional gel system (not shown). In the two-dimensional gel assay, the acetylated complexes are first resolved on a native polyacrylamide gel; the components of the resolved complexes (consisting of free nucleosome, nucleosome plus one bound HMG, and the final complex of nucleosome plus two bound HMGs) are then analyzed by using a denaturing gel containing SDS. The components of each complex are determined, and the specific activity of each species in the complex is determined. The specific activity of the nucleosomal H3 (specific activity, 214 dpm/optical density [OD] unit) was threefold lower than that of H3 in the complex containing two bound HMGs/nucleosome (specific activity, 67 dpm/OD unit). These results are consistent with the one-dimensional analysis (Fig. 4) and suggest that the inhibition of H3 acetylation is mediated by the binding of HMG-17 to nucleosome.
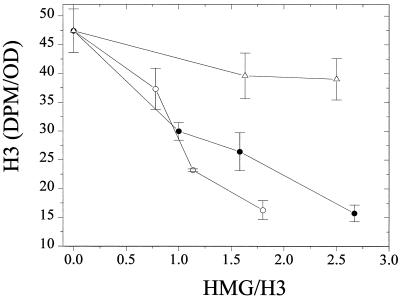
The titration data (Fig. 5) and the two-dimensional gel analyses (not shown) indicated that HMG-17 impedes but does not prevent the acetylation of histone H3. These results suggest that the binding of HMG-17 to nucleosome cores affects the kinetics of H3 acetylation, most probably by sterically hindering the accessibility of PCAF to the N-terminal region of H3. Since HMG-14 is not acetylated but its interaction with nucleosomes is indistinguishable from that of HMG-17, we tested whether HMG-14 inhibited the acetylation of the nucleosomal H3 to the same degree as HMG-17. The results indicate that both proteins inhibited the PCAF-mediated acetylation of nucleosomal H3 (Fig. 6). In contrast, the HMG-17ΔC37 truncation mutant, which lacks the C-terminal region but binds with equal affinity compared to the full-length protein (36), exhibits a reduced ability to inhibit H3 acetylation (Fig. 6). These results are in agreement with previous observations that the C terminus of HMG-14 contacts the N-terminal tail of histone H3 (47) in the region where it exits from the nucleosome (25), i.e., very close to the acetylation sites. We conclude therefore that interactions between the C-terminal domain of HMG-14/-17 proteins and the N-terminal tail of H3 reduce the rate of acetylation of the H3 by PCAF.
FIG. 6.
The C-terminal region of HMG-17 hinders the acetylation of nucleosomal H3. A gel assay was used to monitor the acetylation of nucleosomal H3 as a function of added HMG-17 (●), HMG-14 (○), or HMG17ΔC37 (▵, a truncation mutant of HMG-17 lacking 37 residues from the C terminus). HMG-14 and HMG-17 inhibited H3 acetylation; in contrast, the C-terminal truncation mutant of HMG-17 was a poor inhibitor of nucleosomal H3 acetylation.
Binding of acetylated HMG-17 to nucleosome core particles.
Chromosomal HMG-14/-17 proteins bind to nucleosome core particles in a highly specific manner, forming unique complexes that yield distinct DNA footprints (1, 27, 41). The interaction of these proteins with nucleosome cores can be disrupted by single point mutations, especially by mutations in the nucleosomal binding domain of the proteins (48). Our finding that PCAF fails to acetylate the nucleosome-bound HMG-17 raises the possibility that the lysine at position 2 is also in close contact with a nucleosomal component and that acetylation of this residue may affect the binding of HMG-17 nucleosomes. We therefore examined whether acetylation of HMG-17 by PCAF altered its interaction with core particles.
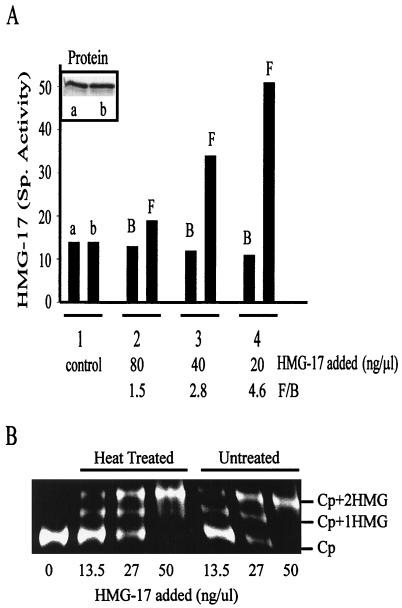
Mobility shift assays indicated that the acetylated HMG-17 protein could bind to nucleosomes (not shown). We examined, by using equilibrium dialysis, whether the acetylated form bound to nucleosomes with the same affinity as did the unmodified form. In these studies we first acetylated HMG-17 with PCAF and then heat inactivated the PCAF. Heat treatment of HMG-17 had no effect on its ability to bind to nucleosome, as measured by gel shift experiments (Fig. 7B). Since only a fraction of the HMG-17 was being acetylated, the reaction mixture contained both modified and unmodified HMG-17. Control experiments, without nucleosomes, demonstrated that the acetylation does not affect the partitioning of the proteins across the membrane, since both the amount of and the specific activity of the protein was identical in both chambers (Fig. 7A). In contrast, when one of the dialysis chambers contained nucleosome cores, the specific activity of the free HMG-17 in the chamber devoid of nucleosome cores was greater than that of the HMG-17 in the chamber containing nucleosome cores (Fig. 7A). Furthermore, at nucleosome concentrations that approach the HMG-nucleosome dissociation constant (about 10−7 M; see reference 36) the difference in the specific activity between the free and bound HMG-17 fractions varied as a function of HMG-17 concentration and was most pronounced at low concentrations of HMG proteins. The data indicate that the acetyl-HMG-17 binds to nucleosomes with a reduced affinity compared to the unmodified form of the protein. We conclude therefore that the specific acetylation of HMG-17 by PCAF affects its interaction with nucleosome cores.
FIG. 7.
Acetylation of HMG-17 reduces its binding affinity to nucleosome cores. Equilibrium dialysis (described in Materials and Methods) was used to determine the relative binding affinities of acetylated versus unacetylated HMG-17 to nucleosomes. (A) A mixture of acetylated and unacetylated HMG-17 (set 1, control, 80 ng/ml; set 2, 80 ng/μl; set 3, 40 ng/μl; and set 4, 20 ng/μl) were placed in one chamber, and the adjoining chambers were filled with either buffer (2× TBE [set 1, control]) or chicken nucleosome cores (60 ng/μl [sets 2 to 4]). The inset shows the quantity of HMG-17 in the control chambers of set 1 (with no nucleosome added). The specific activity of the HMG-17 in these control chambers is shown as set 1, columns a and b. Note that the specific activity in both chambers was identical. The specific activity of HMG-17 in each of the two chambers in the remaining sets is given as either bound (B) or free (F). Note that the specific activity of the free form was always greater than that of the bound form, indicating that the unmodified HMG bound with greater affinity. The ratio, F/B, is an indication of the increase in specific activity of the unbound HMG-17. (B) Nucleoprotein gel assay showing that heat-treated (95°C, 5 min) HMG-17 bound to core particles (Cp) in a manner that is indistinguishable from the untreated HMG-17. The samples were mixed on ice in the acetylation buffer and contained 0.43 μg of the nucleosome core particles per ml and the indicated amounts of HMG-17. Labels: Cp, core particle with no HMG-17 added; Cp+1HMG, core particle with one HMG-17 bound; and Cp+2HMG, saturated complex of core particle with two bound HMG-17s.
DISCUSSION
We report here that PCAF specifically acetylates HMG-17 but not the closely related protein HMG-14, that this acetylation reduces the affinity of HMG-17 for nucleosome cores, and that the presence of HMG-14/-17 proteins on nucleosome cores affects the ability of PCAF to acetylate histone H3. These findings broaden the scope of the molecular interactions affected by PCAF and raise the possibility that PCAF affects transcription in the context of chromatin, not only by acetylating the conserved lysine residues in the N termini of the core histones (2, 19, 29, 39, 49, 51, 52) or general transcription factors (22) but also by modifying specifically and selectively nonhistone structural proteins. In addition, the data suggest a possible mechanism involved in selectively regulating the interaction of HMG-14/-17 with chromatin.
Specific acetylation of HMG-17.
Previous studies with cells and tissue slices incubated with radioactively labeled acetyl-CoA suggested that all of the HMG proteins, including HMG-14 and HMG-17, could be acetylated (23). Analysis of radioactively labeled HMG proteins extracted from unbutyrated duck erythrocytes revealed that the lysine residue at position 2 is the only residue that is constitutively acetylated in both HMG-14 and HMG-17 proteins (42, 43). Thus, the site modified in vitro by PCAF, i.e., lysine 2 in HMG-17, is the same as the in vivo modification site and is therefore physiologically relevant.
Our finding that PCAF specifically acetylates the lysine at position 2 only in HMG-17 is surprising since the first four amino acid residues are identical in both HMG-14 and HMG-17. We used deletion mutant analysis and competition studies to gain insights into the structural features that render HMG-17 a suitable substrate for PCAF. The HMG-14/-17 protein family contains several highly conserved protein domains (see Fig. 4) (9, 10). The region between the first and the second conserved domains is highly divergent between HMG-14 and HMG-17. This region contains 8 residues in HMG-14 (VSSAEGAA) and 12 residues in HMG-17 (AEGDAKGDKAKV). Analysis of various deletion mutants indicated that this region is required for the specific acetylation of HMG-17 (Fig. 4). Interestingly, this sequence is reminiscent of the sequence motif [--K--G(G/A)K-(not G)-K--, where hyphens represent any residue] that is acetylated in H3 by the closely related yeast GCN5 (24). We have found that human GCN5 and PCAF exhibit identical substrate specificities (unpublished data). Although in HMG-17 this sequence is not acetylated in vitro by PCAF or in unbutyrated duck erythrocytes it is acetylated in butyrate-treated duck erythrocytes (42, 43). We therefore conclude that this region, where the sequence of the two HMG proteins is divergent, is involved in the selective acetylation of HMG-17. However, our results also indicate that additional regions in the protein are involved in conferring selectivity to the acetylation process. A peptide containing both the acetylation site and this HMG-17-specific region was not acetylated and did not inhibit the acetylation of the entire protein (Fig. 4). Most strikingly, a deletion mutant lacking only the 22 C-terminal amino acids was efficiently acetylated, while a deletion mutant lacking the 37 C-terminal amino acids was not acetylated (Fig. 2 and 4). These findings suggest that the protein region spanning residues 52 to 67 contributes to the acetylation in the N-terminal region of the protein. However, a peptide spanning this region did not inhibit the acetylation and does not contain an acetylation site.
Taken together, these results suggest that the specific acetylation of HMG-17 by PCAF is not solely dependent on the primary amino acid sequence in the vicinity of the acetylated lysine. These results therefore raise the possibility that the HMG-17 protein but not the HMG-14 protein adopts a conformation recognizable by PCAF. Although previous studies failed to detect ordered structures in free HMG-14/-17 proteins, we note that there are 7 proline residues in the 14-amino-acid region spanning residues 30 to 44 of HMG-17. In the homologous region of HMG-14 there are only three proline residues. The high proline content in HMG-17 could reduce the conformational freedom of the polypeptide chain and perhaps promote a conformation recognizable by PCAF.
Acetylation of the HMG-17–nucleosome complex.
PCAF and GCN5 acetylate free histones more efficiently than the nucleosome-bound histones (24, 53). Similarly, PCAF acetylates free HMG-17 more efficiently than nucleosome-bound HMG-17. For GCN5, efficient acetylation of histones in nucleosomes requires additional cofactors such as the Ada complexes found in S. cerevisiae (18). A similar situation may be applicable to the PCAF mediated acetylation of HMG-17–nucleosome core complexes, where coactivators may enhance the efficiency of acetylation of HMG-17 in the nucleosome complex. In addition, the redundancy in histone acetylases suggests that a similar situation may exist for the acetylation of HMG-17, where PCAF is probably not the only HAT to function on HMG-17.
HMG-14 and HMG-17 form specific complexes with and stabilize the structure of the nucleosome cores (11; for a review, see reference 9). In these complexes, the C-terminal region of HMG-14/-17 proteins is in close proximity to the N-terminal region of histone H3, which is the main nucleosomal target of PCAF (53). We found that the presence of both HMG-14 and HMG-17 inhibits histone H3 acetylation. The C-terminally deleted form of HMG-17, which binds to nucleosomes with the same affinity as did the intact protein (36), inhibits the acetylation of H3 to a significantly lower degree than did intact HMG-17 (Fig. 6). Therefore, we suggest that in the HMG-nucleosome complex the C terminus of HMG-17 (and HMG-14) sterically hinders the interaction between PCAF and H3. The reversible acetylation of the N termini of the histones is a key step in the reorganization of the chromatin structure leading to transcriptional activation (3, 19, 29, 52). Our finding that the presence of HMG-14/-17 affects the acetylation of H3 points to an additional mechanism whereby these proteins may affect DNA-dependent processes occurring in chromatin.
Role of HMG acetylation in chromatin function.
Results from several laboratories indicate that HMG-14/-17 proteins enhance transcription (12, 14, 15, 34, 45, 46, 48) and replication (50), but only from chromatin and not from DNA templates. The enhancement of these DNA-dependent activities is associated with the unfolding of the higher-order chromatin structure and is due to the interaction of the proteins with the N termini of the core histones (11, 47) and with histone H1 (14). By unfolding the higher-order chromatin structure, these HMG proteins facilitate the access of various chromatin-modifying factors to the underlying oligonucleosomal chain, i.e., to the primary level of DNA packing in chromatin.
On the other hand, it is well documented that the binding of HMG-14/-17 stabilizes the structure of the nucleosomes (11; for a review see reference 9), a situation that seems inconsistent with transcriptional activation. It is generally accepted that nucleosomes repress transcription and that destabilizing this structure is a prerequisite for transcriptional activation (33). Our finding that PCAF acetylates a known in vivo acetylation site and that this modification reduces the affinity of HMG-17 to nucleosomes suggests a possible mechanism to resolve this conflict of HMG. Conceivably, the binding of HMG-17 to nucleosomes unfolds the higher-order chromatin fiber and enhances the accessibility of various factors, including HATs, to the primary level of chromatin organization, i.e., the nucleosomes in the 10-nm chromatin fiber. The temporary stabilization of the nucleosome in this first stage of HMG-17 action may promote transcription from chromatin, especially if certain components of the transcriptional machinery preferentially recognize the nucleosome core particle. In a later stage a HAT-containing complex acetylates the protein, thereby reducing its affinity to nucleosomes and alleviating its stabilizing affects on the nucleosome structure. This model provides a unifying concept for the effect of HATs on the interaction of nuclear protein with their targets. Specific acetylation of the structural nonhistone protein HMG-17 reduces its binding affinity to nucleosome cores much as the specific acetylation of the N termini of histones reduces their affinity for the nucleosomal DNA.
ACKNOWLEDGMENTS
We thank Y. Postinikov, J. Wagner, K. Marsh, C. Laufer, and F. Friedman for helpful discussions and S. Smith for discussion and review of the manuscript.
REFERENCES
- 1.Alfonso P J, Crippa M P, Hayes J J, Bustin M. The footprint of chromosomal proteins HMG-14 and HMG-17 on chromatin subunits. J Mol Biol. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- 2.Allfrey V G, Faulkner R, Mirsky A E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. doi: 10.1016/s0959-437x(98)80137-8. [DOI] [PubMed] [Google Scholar]
- 4.Ausio J, Dong F, van Holde K E. Use of selectively trypsinized nucleosome core particles to analyze the role of histone “tails” in the stabilization of nucleosomes. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 5.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell J E, Allis C D. An activity gel assay detects a single, catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownell J E, Allis C D. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 8.Bustin M. Arrangement of histones in chromatin. Nat New Biol. 1973;245:207–209. doi: 10.1038/newbio245207a0. [DOI] [PubMed] [Google Scholar]
- 9.Bustin M, Lehn D A, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049:231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 10.Bustin M, Reeves R. High mobility group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 11.Crippa M P, Alfonso P J, Bustin M. Nucleosome core binding region of chromosomal protein HMG-17 acts as an independent functional domain. J Mol Biol. 1992;228:442–449. doi: 10.1016/0022-2836(92)90833-6. [DOI] [PubMed] [Google Scholar]
- 12.Crippa M P, Trieschmann L, Alfonso P J, Wolffe A P, Bustin M. Deposition of chromosomal protein HMG-17 during replication affects the nucleosomal ladder and transcriptional potential of nascent chromatin. EMBO J. 1993;12:3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davie J R. Covalent modification of histones: expression from chromatin templates. Curr Opin Genet Dev. 1998;8:173–178. doi: 10.1016/s0959-437x(98)80138-x. [DOI] [PubMed] [Google Scholar]
- 14.Ding H-F, Bustin M, Hansen U. Alleviation of histone H1-mediated transcriptional repression and chromatin compaction by the acidic activation region of chromosomal protein HMG-14. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding H F, Rimsky S, Batson S C, Bustin M, Hansen U. Stimulation of RNA polymerase II elongation by chromosomal protein HMG-14. Science. 1994;265:796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher T M, Hansen J C. The nucleosomal array: structure/function relationships. Crit Rev Eukaryot Gene Expr. 1996;6:149–188. doi: 10.1615/critreveukargeneexpr.v6.i2-3.40. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Ramirez M, Rocchini C, Ausio J. Modulation of chromatin folding by histone acetylation. J Biol Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 18.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. yGCN5 function within multisubunit ADA and SPT/ADA adapter complexes to acetylate nucleosomal histones. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 19.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 20.Hansen J C, Ausio J. Chromatin dynamics and the modulation of genetic activity. Trends Biochem Sci. 1992;17:187–191. doi: 10.1016/0968-0004(92)90264-a. [DOI] [PubMed] [Google Scholar]
- 21.Herrera J E, Bergel M, Yang X J, Nakatani Y, Bustin M. The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J Biol Chem. 1997;272:27253–27258. doi: 10.1074/jbc.272.43.27253. [DOI] [PubMed] [Google Scholar]
- 22.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 23.Johns E W. The HMG chromosomal proteins. London, United Kingdom: Academic Press, Inc.; 1982. [Google Scholar]
- 24.Kuo M H, Brownell J E, Sobel R E, Ranalli T A, Cook R G, Edmondson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 25.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 26.Magnaghi-Jaulin L, Grosiman R, Naguibneva I, Robin P, Lorains S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 27.Mardian J K, Paton A E, Bunick G J, Olins D E. Nucleosome cores have two specific binding sites for nonhistone chromosomal proteins HMG 14 and HMG 17. Science. 1980;209:1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- 28.Marzluff W F, Jr, Sanders L A, Miller D M, McCarty K S. Two chemically and metabolically distinct forms of calf thymus histone F3. J Biol Chem. 1972;247:2026–2033. [PubMed] [Google Scholar]
- 29.Mizzen C A, Allis C D. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizzen C A, Yang X-J, Kokuba T, Brownell J E, Banister A J, Owen-Hughes T, Workman J C, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. The TAF250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko V V, Sciltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 32.O’Neill L P, Turner B M. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent but transcription-independent manner. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 34.Paranjape S M, Krumm A, Kadonaga J T. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 35.Postnikov Y V, Herrera J E, Hock R, Scheer U, Bustin M. Clusters of nucleosomes containing chromosomal protein HMG-17 in chromatin. J Mol Biol. 1997;274:454–465. doi: 10.1006/jmbi.1997.1391. [DOI] [PubMed] [Google Scholar]
- 36.Postnikov Y V, Lehn D A, Robinson R C, Friedman F K, Shiloach J, Bustin M. The cooperative binding of chromosomal protein HMG-14 to nucleosome cores is reduced by single point mutations in the nucleosomal binding domain. Nucleic Acids Res. 1994;22:4520–4526. doi: 10.1093/nar/22.21.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postnikov Y V, Trieschmann L, Rickers A, Bustin M. Homodimers of chromosomal proteins HMG-14 and HMG-17 in nucleosome cores. J Mol Biol. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 38.Romani M, Rodman T C, Vidali G, Bustin M. Serological analysis of the specificity of HMG chromosomal proteins. J Biol Chem. 1979;254:2918–2922. [PubMed] [Google Scholar]
- 39.Roth S Y, Allis C D. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 40.Rundlet S E, Carmen A A, Noriyuki S, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 41.Sandeen G, Wood W I, Felsenfeld G. The interaction of high mobility proteins HMG14 and 17 with nucleosomes. Nucleic Acids Res. 1980;8:3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sterner R, Vidali G, Allfrey V G. Studies on the acetylation and deacetylation of HMG proteins. J Biol Chem. 1979;254:11577–11583. [PubMed] [Google Scholar]
- 43.Sterner R, Vidali G, Allfrey V G. Studies on the acetylation and deacetylation of HMG proteins: identification of the sites of acetylation of HMG-14 and HMG-17. J Biol Chem. 1981;256:8892–8895. [PubMed] [Google Scholar]
- 44.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 45.Tremethick D J, Hyman L. High mobility group proteins 14 and 17 can prevent the close packing of nucleosomes by increasing the strength of protein contacts in the linker DNA. J Biol Chem. 1996;271:12009–12016. doi: 10.1074/jbc.271.20.12009. [DOI] [PubMed] [Google Scholar]
- 46.Trieschmann L, Alfonso P J, Crippa M P, Wolffe A P, Bustin M. Incorporation of chromosomal proteins HMG-14/-17 into nascent nucleosomes induces an extended chromatin conformation and enhances the utilization of active transcription complexes. EMBO J. 1995;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trieschmann L, Martin B, Bustin M. The chromatin unfolding domain of chromosomal protein HMG-14 targets the N-terminal tail of histone H3 in nucleosomes. Proc Natl Acad Sci USA. 1998;95:5468–5473. doi: 10.1073/pnas.95.10.5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trieschmann L, Postnikov Y V, Rickers A, Bustin M. Modular structure of chromosomal proteins HMG-14 and HMG-17: definition of a transcriptional activation domain distinct from the nucleosomal binding domain. Mol Cell Biol. 1995;15:6663–6669. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner B M, O’Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 50.Vestner B, Bustin M, Gruss C. Stimulation of replication efficiency of a chromatin template by chromosomal protein HMG-17. J Biol Chem. 1998;273:9409–9414. doi: 10.1074/jbc.273.16.9409. [DOI] [PubMed] [Google Scholar]
- 51.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 52.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 53.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]