Abstract
We previously reported that the activation of the M promoter of the human choline acetyltransferase (ChAT) gene by butyrate and trapoxin in transfected CHP126 cells is blocked by PD98059, a specific mitogen-activated protein kinase kinase (MEK) inhibitor (E. Espinos and M. J. Weber, Mol. Brain Res. 56:118–124, 1998). We now report that the transcriptional effects of histone deacetylase inhibitors are mediated by an H7-sensitive serine/threonine protein kinase. Activation of the ChAT promoter by butyrate and trapoxin was blocked by 50 μM H7 in both transient- and stable-transfection assays. Overexpression of p300, a coactivator protein endowed with histone acetyltransferase activity, stimulated the ChAT promoter and had a synergistic effect on butyrate treatment. These effects were blocked by H7 and by overexpressed adenovirus E1A 12S protein. Moreover, both H7 and PD98059 suppressed the activation of the Rous sarcoma virus (RSV) and simian virus 40 promoters by butyrate in transfection experiments. Similarly, the induction of the cellular histone H10 gene by butyrate in CHP126 cells was blocked by H7 and by PD98059. Previous data (L. Cuisset, L. Tichonicky, P. Jaffray, and M. Delpech, J. Biol. Chem. 272:24148–24153, 1997) showed that the induction of the H10 gene by butyrate is blocked by okadaic acid, an inhibitor of protein phosphatases. We now show that the activation of the ChAT and RSV promoters by butyrate in transfected CHP126 cells is also blocked by 200 nM okadaic acid. Western blotting and in vivo metabolic labeling experiments showed that butyrate has a biphasic effect on histone H3 phosphorylation, i.e., depression for up to 16 h followed by stimulation. The data thus strongly suggest that the transcriptional effects of histone deacetylase inhibitors are mediated through the activation of MEK1 and of an H7-sensitive protein kinase in addition to protein phosphatases.
It has long been recognized that transcribed eukaryotic genes are associated with hyperacetylated histones (4). Our understanding of the relationship between transcription and histone acetylation has improved considerably with the purification and cloning of proteins endowed with histone acetyltransferase (HAT) or histone deacetylase (HDAC) activity (30, 43, 66). Most of these enzymes were found to be members of families of already-known transcriptional regulators. The first HAT was cloned from Tetrahymena thermophila and found to be highly homologous to the yeast transcriptional activator Gcn5p (14). A surprising variety of HAT proteins have now been cloned, including additional Gcn5 family members as well as structurally unrelated proteins such as the CREB-binding protein (CBP)/p300 coactivators and the general transcription factor TAFII250 (for reviews, see references 60 and 74). In contrast, all but one HDAC characterized so far are products of a single gene family exemplified by yeast rpd3 and human HDAC genes (44, 67).
Most importantly, both HAT and HDAC are part of multiprotein complexes containing coactivator or corepressor proteins that are tethered on promoters by specific interactions with DNA-bound transcription factors, thus providing a mechanism for local modifications at the level of histone acetylation. On the yeast INO1 gene promoter, the recruitment of Rpd3p HDAC by interaction with the Ume6p repressor has been shown to lead to the specific hypoacetylation of lysine 5 of histone H4 on two nucleosomes at most (65). Likewise, the recruitment of Gcn5p on the HIS3 promoter leads to a hyperacetylation of lysines 9 and 14 of histone H3 on the gene promoter but not on adjacent coding sequences (48). Such highly localized histone hyperacetylation is thought to subtly alter the structure of positioned nucleosomes and allow the binding of trans-acting factors. However, the study of point mutations introduced into HDAC and HAT proteins has revealed that their functional role is not entirely accounted for by their enzymatic activity. For example, the HAT enzymatic activity of CBP, but not that of the p300/CBP-associated factor (p/CAF), is required for CREB-mediated transcription, whereas the activity of p/CAF, but not that of CBP, is necessary for retinoic acid-stimulated transcription (47). Likewise, transcriptional repression caused by the recruitment of HDAC on transfected templates is either reversed by trichostatin A, a specific inhibitor of HDAC activity, or unaffected by this compound, depending on the promoter studied (54). Therefore, both HAT and HDAC proteins can regulate transcription by mechanisms that do not involve histone acetylation.
Despite this extensive knowledge, the relationship between histone hyperacetylation and transcriptional activation remains relatively unclear. It has been estimated that trichostatin A modulates the expression of only 2% of cellular genes, suggesting that only a small subset of gene promoters are sensitive to alterations in the levels of histone acetylation (72). Moreover, hyperacetylation can also occur on poised genes and thus appears to correlate with transcriptional competence rather than transcription (19, 32, 33). Although a correlation between transcription and the acetylation of specific lysine residues on core histones may exist, the available data nevertheless suggest that histone hyperacetylation is only one component of transcriptional control.
In previous reports (17, 25), we showed that the M promoter of the human choline acetyltransferase (ChAT) gene is activated by butyrate, trichostatin A, and trapoxin A, three inhibitors of HDACs, in transiently and stably transfected human neuroepithelioma CHP126 cells. This activation does not require ongoing protein synthesis, suggesting that HDAC inhibitors lead to the activation of transcription factors, e.g., by phosphorylation. We also established that butyrate and trapoxin stimulate the phosphorylation of mitogen-activated protein (MAP) kinases ERK1 and -2 in a rapid and transient manner. The blocking of the MAP kinase cascade with the MAP kinase kinase (MEK) inhibitor PD98059 or the overexpression of dominant-negative mutants of Ras and ERK2 suppressed the activation of the ChAT promoter by butyrate. Conversely, constitutive mutants of Ras and MEK1 potentiated the transcriptional effect of butyrate. We now show that the transcriptional activation of cellular and transfected genes by HDAC inhibitors is blocked by H7, an inhibitor of serine/threonine protein kinases. We have also explored the effect of overexpressed p300 and adenovirus E1A protein on ChAT promoter activity and have found that the synergistic activation of the ChAT promoter by butyrate and p300 is blocked by H7.
Interestingly, it was recently reported that the transcriptional effects of butyrate on the cellular H10 and c-myc genes are blocked by calyculin A and okadaic acid, two inhibitors of serine/thronine protein phosphatases PP1 and PP2A (20). These authors also established that butyrate increases protein phosphatase activity, most probably PP1, by 45% in HTC hepatoma cells. To further delineate the role of protein kinases and phosphatases in the mode of action of butyrate, we studied the effects of okadaic acid on the activation of the ChAT promoter and the Rous sarcoma virus RSV long terminal repeat (LTR) by butyrate, as well as the effects of H7 on the transcription of the cellular H10 gene, in CHP126 cells. The data establish that the transcriptional effects of HDAC inhibitors are mediated through the activation of both serine/threonine protein kinases and phosphatases. Accordingly, we report here that the phosphorylation of histone H3 in butyrate-treated cells is first decreased but is increased upon prolonged treatment, supporting the hypothesis of a sequential activation of protein phosphatases and kinases.
MATERIALS AND METHODS
Materials.
Trapoxin A was a generous gift from M. Yoshida (Tokyo, Japan). GF109203X (bisindolylmaleimide) was from Boehringer Mannheim. H7 [1-(5-isoquinolinylsulfonyl)-2-methylpiperazine], aphidicolin, lovastatin, sodium butyrate, and okadaic acid were purchased from Sigma. PD98059 [2-(2′-amino-3′-methoxyphenyl)-oxanaphthalene-4-one] was purchased from New England Biolabs, and KN-62 [1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl)-4-phenylpiperazine] was from Research Biochemical International.
Cell culture and transfection.
CHP126 cells (a gift from G. M. Lauro, Rome, Italy) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. For transient-transfection assays, 2 × 106 cells in 400 μl of serum-free medium were electroporated at 960 μF and 260 V, using a Gene Pulser and Capacitance Extender apparatus (Bio-Rad). To avoid potential effects on transfection efficacy, the treatment with butyrate or trapoxin was always initiated 14 to 16 h after transfection. Protein kinase inhibitors were added to cells 1 h before addition of butyrate or trapoxin. Unless otherwise indicated, enzymatic assays and RNA extraction were performed 24 h later. Cells were lysed in Reporter Lysis Buffer (Promega). Luciferase activity was measured with the Luciferase Assay System (Promega) and was expressed in relative light units (RLU). β-Galactosidase was measured with a Galacto-Light kit (Tropix) after the cell extract was heated at 48°C for 1 h to denature the endogenous enzyme. Protein concentration was measured with the Bio-Rad protein assay. Data are given as means and ranges for duplicate cultures and, for each experiment, are representative of three to four independent electroporations. Transfection efficacies in duplicate electroporations varied by less than 15%.
The ChAT-luciferase construct F contained a ChAT genomic fragment extending from nucleotide (nt) −2092 to nt +2483 (relative to the cap site of the alternative first exon M) linked to the luciferase gene in the pGL2basic vector (Promega), as previously described (17). The RSV-driven adenovirus type 5 E1A 12S expression vector (8) was a gift from T. Kouzarides (Cambridge, United Kingdom). The pCMVb p300-CHA expression vector (24) was a gift from D. M. Livingston and S. Bhattacharya (Boston, Mass.). Plasmid pCH110 (simian virus 40 [SV40]-lacZ) was purchased from Pharmacia. pRSV-LacZ was a gift from H. Prats (Toulouse, France).
Northern blot analysis.
A fragment of human histone H10 mRNA (nt 936 to 1519 in GenBank file X03473) was amplified by reverse transcription-PCR from total RNA of CHP126 cells, using primers 5′-GGGCCGGCAAGAAGAAG-3′ and 5′-TGGGCCCCCACTGCTCA-3′ and Taq DNA polymerase. The amplification product was cloned into the pCR-Topo vector by the use of a TA cloning kit (Invitrogen). The insert was sequenced from both ends. To make a riboprobe, the plasmid was linearized with BamHI and transcribed with T7 RNA polymerase in the presence of [α-32P]dCTP. Total RNA from subconfluent CHP126 cells was extracted by the guanidinium isothiocyanate method (18), separated on a 0.8% agarose denaturing gel, and transferred to a nylon membrane. Prehybridization was performed at 42°C for 18 h in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) 50% formamide, 5× Denhardt’s solution, 0.5% sodium dodecyl sulfate (SDS), and 20 μg of yeast tRNA/ml. Hybridization was performed in the same solution at 60°C for at least 12 h. The last washes were performed in 0.1× SSPE–0.1% SDS for 16 h at 50°C.
Gel analysis of histone acetylation and phosphorylation.
After a 30-min pretreatment with 50 μM H7 or vehicle (H2O), confluent CHP126 cell cultures (125 cm2) in RPMI culture medium were provided with 50 μCi of [32P]phosphate (New England Nuclear)/ml and 5 mM butyrate or buffer. After 8 h of culture, the cells were washed with phosphate-buffered saline (PBS) supplemented with 5 mM butyrate, 10 mM NaF, 1 mM sodium orthovanadate, 0.1 mM phenylmethylsulfonyl fluoride, and 0.1 mM benzamidine and scraped with a rubber policeman into a centrifuge tube. After centrifugation, the cells were further washed with PBS plus inhibitors and incubated for 10 min on ice in 10 ml of lysis buffer (50 mM KCl, 15 mM NaCl, 5 mM MgCl2, 250 mM sucrose, 10 mM morpholineethanesulfonic acid [MES] buffer [pH 6.5], 0.5% Triton X-100, and inhibitors as above). Nuclei were pelleted by centrifugation at 1,500 × g for 10 min, resuspended in 0.5 ml of wash buffer (60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 250 mM sucrose, 10 mM MES buffer [pH 6.5], and inhibitors) and layered on a 5-ml cushion of 30% sucrose in wash buffer. After a 10-min centrifugation at 2,600 × g, the pellets, containing the nuclei, were resuspended in 1 ml of 0.2 M H2SO4 and incubated overnight at 4°C. After centrifugation, the pellets were extracted again with 0.2 M H2SO4 for 1 h. Protein from the combined supernatants was precipitated with 20% trichloracetic acid; washed once with cold acetone–0.2 M HCl, twice with acetone-ethanol-Tris (0.2 M; pH 8) (7:2:1), and once with acetone; dried; and resuspended in a solution containing 50 μl of 8 M urea, 1 M acetic acid, 0.5 mg of pyronin Y/ml, and 0.5 mg of methyl green/ml. Acid-insoluble material was lysed in Laemmli loading buffer containing 5% β-mercaptoethanol, denatured, and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. Histone analysis on 0.75-mm-thick gels (16.5 by 26 cm) acid-urea-Triton X-100 (AUT) gels was performed as described elsewhere (63). After being stained with Coomassie blue, the gels were dried and subjected to autoradiography.
Western blot analysis.
Confluent CHP12 cells (2 cm2) were pretreated for 30 min with 50 μM H7 or vehicle (H2O) and then with 5 mM butyrate or buffer. At different time points, cells were washed with PBS plus inhibitors as described above, scraped into a centrifuge tube, centrifuged, and lysed in 50 μl of Laemmli loading buffer containing 5% β-mercaptoethanol. Proteins were separated on a 15% acrylamide gel, transferred to a nylon membrane, and subjected to Western analysis with anti-acetyl H4, anti-acetyl H3, or anti-phospho-H3 antibodies (Upstate) according to the manufacturer’s instructions.
RESULTS
Cell cycle arrest does not activate the ChAT promoter.
To study the mechanisms of transcriptional regulation by HDAC inhibitors, human CHP126 neuroepithelioma cells were transfected with ChAT-luciferase construct F, which includes nt −2092 to +2483 of the human ChAT gene relative to the cap site of the alternative first exon M. Our previous experiments have shown that luciferase expression driven by the ChAT promoter is increased by butyrate, trapoxin A, and trichostatin A in both transient- and stable-transfection assays (17). To determine whether activation of the ChAT promoter by HDAC inhibitors was a consequence of cell cycle arrest, we studied the effects of other drugs, known to block the cell cycle, on a clone of CHP126 cells stably transfected with construct F. Luciferase activity in this clone was stimulated about 20-fold by 5 mM butyrate (Fig. 1C). As shown in Table 1, both lovastatin, an inhibitor of 3-hydroxy-3-methylglutaryl coenzyme A reductase, and aphidicolin, an inhibitor of DNA polymerases α, β, and ɛ, arrested proliferating cells at G1/S but did not affect luciferase expression. Therefore, activation of the ChAT promoter by HDAC inhibitors was not triggered by cell cycle arrest. Similarly, it was established that in synchronized cells, gene induction by butyrate was independent of the progression through the cell cycle (27, 28).
FIG. 1.
Activation of the ChAT promoter by HDAC inhibitors is blocked by H7. (A) Dose-response curve for the effects of H7. CHP126 cells (8 × 106) were transfected with construct F and divided into 36 culture wells. After 16 h, the cultures were treated with the indicated concentrations of H7. After 1 h of incubation at 37°C, half of the cultures were given 40 nM trapoxin A (closed symbols) and half were given vehicle (open symbols), both in the continued presence of H7. Luciferase activities were measured after an additional 48 h. (B) Activation of the ChAT promoter by butyrate or trapoxin is blocked by H7. CHP126 cells (4 × 106) were transfected with construct F and divided into 18 culture wells. After overnight incubation at 37°C, the cells were treated with 5 mM butyrate (stippled bars) or 40 nM trapoxin A (closed bars) or left untreated (open bars) in the presence or absence of 40 μM H7, as indicated. Cells were pretreated for 1 h with H7 before the addition of butyrate, trapoxin, or vehicle. Luciferase activity and total protein concentration were measured after 48 h of incubation. (C) H7 blocks the induction of luciferase activity by butyrate in stably transfected cells. CHP126 cells were stably transfected with ChAT-luciferase construct F. Cells from clone F11 (17) were seeded into 16-mm-diameter culture wells at a density of 105/well. After overnight culture, half of the cells were treated with 40 μM H7, as indicated. After 1 h of incubation at 37°C, the culture medium was supplemented with 5 mM butyrate (stippled bars) or vehicle (open bars). Luciferase activity was measured after 24 h of treatment. For all experiments, the data correspond to the means of values for three cultures ± the SEMs and are representative of three to four experiments.
TABLE 1.
Cell cycle arrest does not activate the ChAT promotera
| Treatment | % of cells in cell cycle phase:
|
Mean luciferase activity ± SEM (RLU/μg of protein, 103) | ||
|---|---|---|---|---|
| G1 | S | G2+M | ||
| None (vehicle only) | 53.8 | 30.3 | 15.9 | 1.43 ± 0.04 |
| Lovastatin, 60 μM | 80.4 | 11.1 | 8.5 | 2.12 ± 0.01 |
| Aphidicolin, 3 μM | 67.4 | 26.6 | 6.0 | 1.10 ± 0.01 |
CHP126 cells (0.6 × 105/cm2) stably transfected with construct F were treated for 24 h with lovastatin, aphidicolin, or vehicle, as indicated. Sister cultures were used for the determination of luciferase activity and the analysis of the cell cycle by fluorescence-activated cell sorting. A total of 105 cells were analyzed.
H7 blocks activation of the ChAT promoter by HDAC inhibitors.
To study the role of serine/threonine protein kinases in the activation of the ChAT promoter by HDAC inhibitors, CHP126 cells were transiently transfected with construct F. Sixteen hours after transfection, the cells were treated for 1 h with H7, an inhibitor of serine/threonine kinases (36), and then with butyrate or trapoxin in the continued presence of H7. Under these conditions, H7 suppressed the activating effect of trapoxin, a highly specific HDAC inhibitor (81), in a dose-dependent manner, whereas the basal activity was maximally depressed by 30% (Fig. 1A). The effect of trapoxin was totally suppressed by 50 μM H7, with the half-maximal effect being observed at a concentration of 20 to 30 μM. Figure 1B further shows that H7 suppressed the activation of the ChAT promoter by butyrate. No systematic differences in the potencies of butyrate and trapoxin were observed (compare Fig. 1B and 3B). When dose-response curves were analyzed, the maximal effects of both compounds were similar (17). Although the experiments of Fig. 1A and B were performed with a 2-day expression period, subsequent experiments revealed that similar phenomena were observed after a shorter period of expression (see below).
FIG. 3.
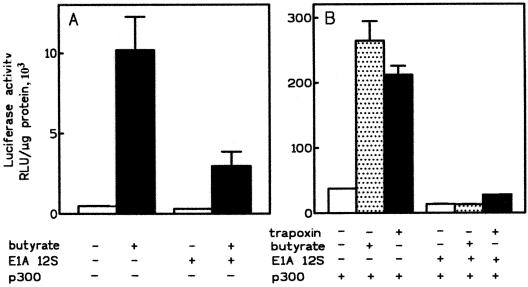
Effects of adenovirus E1A protein on ChAT promoter activity in transfection assays. (A) Activation of the ChAT promoter by butyrate is blocked by E1A. CHP126 cells were cotransfected with ChAT-luciferase construct F (5 μg) and 5 μg of an E1A 12S expression vector or the empty expression vector. Butyrate (5 mM) or vehicle (H2O) was given to the cultures for 24 h, starting 16 h after transfection. The data are means and ranges for duplicate cultures and are representative of three experiments. (B) The synergistic effects of p300 and HDAC inhibitors are blocked by E1A. CHP126 cells were cotransfected with ChAT-luciferase construct F (5 μg), 7.5 μg of p300 expression vector, and 5 μg of an E1A 12S protein expression vector or an empty vector. The cells were treated with butyrate (5 mM) or trapoxin (80 nM) as described for panel A. The data are representative of three experiments. The data in panels A and B are from separate experiments, so that a stimulation factor by p300 alone cannot be derived from a comparison of these data (see text).
To determine the effect of H7 on the ChAT promoter in a chromatin context, we used CHP126 cells stably transfected with construct F. In these cells, the increase in luciferase activity caused by butyrate was again totally suppressed by 50 μM H7 (Fig. 1C). Kinetic studies showed that the blocking effect of 50 μM H7 was complete as soon as the activation of the ChAT promoter by butyrate was significant, i.e., after 4 to 6 h of treatment with the HDAC inhibitor (data not shown).
H7 is a potent inhibitor of protein kinase C (PKC) and cyclic-nucleotide-dependent kinase activities in vitro (36). The inhibition of cellular processes by low doses of H7 has been interpreted as evidence for the involvement of PKC in the transduction of the stimulus (13, 26, 78). On the other hand, c-fos induction by phorbolmyristate acetate, a PKC activator, is insensitive to 50 μM H7 (52), suggesting that H7 does not inhibit PKC in intact cells. To study the role of PKC in the activation of the ChAT promoter by butyrate, we used bisindoylmaleimide (GF109203X), a compound that inhibits PKC isoforms at micromolar concentrations (57, 69). This compound also inhibits RSK2 and p70 S6 kinase with potencies similar to that demonstrated for the PKC isoforms (1). When CHP126 cells were transfected with construct F, bisindoylmaleimide (5 μM) increased basal luciferase activity about twofold, but the stimulatory effect of butyrate was reduced by only 33% (Table 2). KN62, an inhibitor of calcium/calmodulin-dependent kinases (CaMK) (37), maximally depressed butyrate-stimulated expression by 30% (Table 2). This suggests that the action of butyrate is at most partially dependent on conventional and novel PKC isoforms or CaMK activity. Moreover, dibutyryl cyclic AMP (cAMP) was previously shown to be a much weaker activator of ChAT promoter than butyrate (17). It is therefore unlikely that the effect of H7 results from the inhibition of protein kinase A.
TABLE 2.
PKC and CaM kinase have no effect on activation of the ChAT promoter by sodium butyratea
| Expt | Treatment | Luciferase activity (RLU/μg of protein, 103)
|
Fold stimulation | |
|---|---|---|---|---|
| Control cells | Butyrate-treated cells | |||
| A | None | 8.3 ± 2.3 | 662 ± 63 | 80 |
| GF, 5 μM | 19.8 ± 2.1 | 1,074 ± 116 | 54 | |
| B | None | 19.0 ± 0.6 | 483 ± 9 | 25 |
| KN62, 10 μM | 21.4 ± 0.8 | 311 ± 8 | 15 | |
CHP126 cells were transfected with construct F and treated for 24 h with 5 mM butyrate. Bisindoylmaleimide (GF 109203X) or KN-62 was added to the cells 1 h before addition of butyrate. Increasing the concentration of KN62 to 50 μM resulted in the same inhibition as with the 10 μM concentration (data not shown). The data represent the means of values for three cultures ± SEMs.
Synergistic effects of p300 and HDAC inhibitors on ChAT promoter activity in a transient-transfection assay.
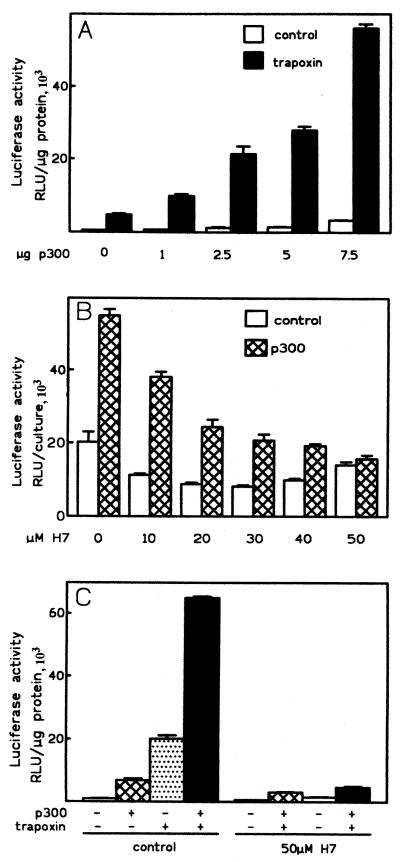
Because HDAC inhibitors stimulates ChAT promoter activity, we asked whether the overexpression of a protein endowed with HAT activity would lead to further activation. To address this question, we cotransfected CHP126 cells with construct F and increasing amounts of an expression vector for full-length p300 protein (5, 6, 24). Figure 2A shows that overexpression of p300 maximally increased luciferase expression 5- to 10-fold by itself. The mean stimulation ± the standard error of the mean (SEM) with 7.5 μg of p300 was 5.0- ± 0.7-fold (n = 6) for cells cultured in 10% serum. When the cells were serum deprived (0.5% serum) for 2 days before transfection as well as during the expression period, the mean stimulation ± the SEM was 6.6- ± 1.4-fold (n = 7), showing that serum mitogenic factors were not required. Luciferase expression in cells treated with trapoxin was also increased by p300 in a dose-dependent manner. Remarkably, trapoxin and p300 had synergistic effects on the ChAT promoter. In the experiment shown in Fig. 2A, trapoxin alone stimulated the ChAT promoter 8-fold and p300 alone stimulated it 6-fold, whereas a 94-fold activation was observed in the presence of both trapoxin and p300.
FIG. 2.
Synergistic effects of p300 and HDAC inhibitors on ChAT promoter activity. (A) p300 potentiates activation of the ChAT promoter by trapoxin. CHP126 cells were cotransfected with construct F (5 μg), increasing amounts of a pCMV-p300 expression vector, and compensating amounts of the empty expression vector. After overnight incubation, the cells were treated with 80 nM trapoxin A or vehicle (0.25% DMSO). Luciferase activities of duplicate cultures were measured 24 h later. The data are representative of four experiments. (B) Activation of the ChAT promoter by p300 is blocked by H7. CHP126 cells (4 × 106) were cotransfected with construct F (5 μg), SV40-lacZ (5 μg), and 7.5 μg of pCMV-p300 expression vector or empty expression vector and distributed into 10-mm-diameter culture wells. After overnight culture, the cells were treated with H7 as indicated. Luciferase activities were measured 24 h later. The data are means and ranges for duplicate cultures and are representative of three experiments. (C) The synergistic effects of p300 and HDAC inhibitors are suppressed by H7. CHP126 cells were transfected as described for panel B. The cultures were pretreated with 50 μM H7 for 1 h or left untreated before the addition of butyrate (5 mM) or trapoxin (80 nM). The data are representative of three experiments.
As shown on Fig. 2B, activation of the ChAT promoter by p300 protein alone was blocked by H7 in a dose-dependent manner. The effect of p300 was totally suppressed by 50 μM H7, with 50% inhibition being observed at a concentration of around 15 μM. Therefore, activation of the ChAT promoter by overexpressed p300 requires an H7-sensitive serine/threonine kinase activity, as observed for activation by HDAC inhibitors. H7 blocked the effects of p300 and of HDAC inhibitors in the same range of concentration (compare Fig. 1A and 2B). Accordingly, the synergistic activation of the ChAT promoter triggered by trapoxin and p300 was suppressed by 50 μM H7 (Fig. 2C). Results similar to those shown in Fig. 2A to C were also observed by overexpressing full-length CBP (data not shown), but experiments with other proteins endowed with HAT activity, like p/CAF and hGCN5, are currently under way.
Adenovirus E1A protein depresses ChAT promoter activation by HDAC inhibitors.
Since HDAC inhibitors and overexpressed p300 protein have synergistic effects on the ChAT promoter, we attempted to delineate the role of endogenous CBP/p300 protein in the activation by butyrate. As a first approach, we overexpressed E1A 12S protein, an alternative splicing product of the adenovirus E1A gene that lacks the CR3 trans-activating domain and blocks CBP/p300-dependent transcription at many promoters (5, 8, 62). E1A binds CBP/p300 at three sites, including the cysteine-histidine-rich region (C/H3) that also binds pp90rsk and numerous transcription factors (for a review, see reference 39) and a C-terminal site that also binds the coactivators p300/CBP interacting protein (p/CIP) and nuclear receptor coactivator (N-CoA) (50). When CHP126 cells were cotransfected with plasmid F and an E1A 12S expression vector, basal luciferase expression was not significantly affected (−21% ± 29%; n = 3), but butyrate-stimulated expression was depressed by 56% ± 7% (70% in the experiment shown in Fig. 3A). When the same type of experiment was repeated in the additional presence of p300, the synergistic effects between p300 and butyrate or trapoxin were also suppressed by E1A (Fig. 3B). Therefore, the activation of the ChAT promoter by HDAC inhibitors is blocked by the E1A protein, suggesting that CBP/p300 is required for activation. Moreover, these data establish striking similarities in the effects of the E1A protein and the protein kinase inhibitor H7 on ChAT promoter activation by HDAC inhibitors and/or p300 protein.
Role of protein kinases in the activation of viral promoters by butyrate.
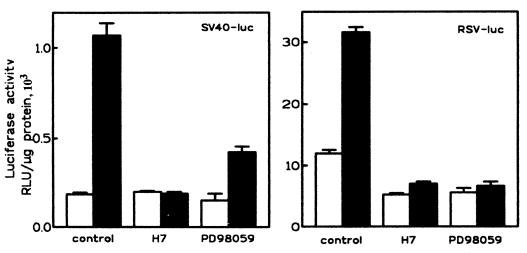
The minimal promoter of SV40 and the LTR of RSV are known to be activated by HDAC inhibitors (10, 29). We thus asked whether activation of these viral promoters by butyrate would also require protein kinase activity. When CHP126 cells were transfected with the plasmid pGL2promoter (Promega), in which the luciferase gene is fused to an SV40 minimal promoter, or with a pRSV-luciferase construct (11), a three- to sixfold increase in luciferase activity was observed after a 6-h treatment with 5 mM butyrate. In both cases, the activation of promoter activity by butyrate was totally suppressed by 50 μM H7 (Fig. 4). A similar observation was made with the SV40-lacZ plasmid pCH110 (data not shown). Moreover, activation of the RSV promoter by trapoxin was also blocked by H7 (data not shown). We previously reported that activation of the ChAT promoter by HDAC inhibitors is blocked by PD98059, a compound that selectively blocks the activation of MEK1 by Raf kinases (25). As shown in Fig. 4, activation of the RSV LTR by butyrate was totally blocked by 80 μM PD59098, whereas that of the SV40 promoter was depressed about threefold. As was already shown for the ChAT promoter, these data further suggest that the activation of both the SV40 and RSV promoters by HDAC inhibitors requires the activation of both MEK1 and an H7-sensitive protein kinase.
FIG. 4.
H7 and PD98059 block activation of viral promoters by butyrate. CHP126 cells (4 × 106) were transfected with 10 μg of the pGL2promoter (left panel) or of an RSV-luciferase (luc) plasmid (right panel) and distributed into 16 culture wells. After overnight incubation, the cells were treated for 1 h with 50 μM H7, 80 μM PD98059, or vehicle (0.24% DMSO), as indicated. Half of the cultures were then treated for 6 h with 5 mM butyrate (filled bars) or left untreated (open bars) in the continued presence of kinase inhibitor.
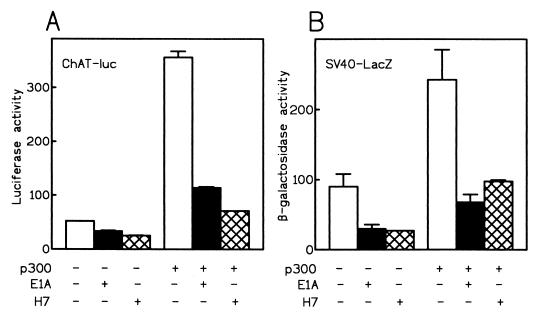
We next studied the effects of p300 overexpression on the SV40 promoter. For a direct comparison, CHP126 cells were cotransfected with construct F and the SV40-lacZ plasmid in the presence or absence of E1A and p300 expression vectors. As expected from our previous results, ChAT-driven luciferase activity was stimulated by overexpressed p300 protein (Fig. 5A). The activation of the ChAT promoter by p300 was suppressed by E1A protein as well as by H7. It is well established that the E1A 12S protein represses transcription from the SV40 early promoter (73). Accordingly, β-galactosidase expression driven by the SV40 promoter was depressed three- to fourfold by E1A protein (Fig. 5B). An identical level of transcriptional repression was obtained with 50 μM H7. Moreover, the SV40 promoter was activated about threefold by p300. As observed for the ChAT promoter, the activating effect of p300 was largely suppressed by E1A protein or by H7. Taken together, the data obtained from studies with two unrelated promoters establish that E1A-sensitive transcriptional activation by the p300 protein requires an H7-sensitive protein kinase activity.
FIG. 5.
Comparison of the effects of H7 and E1A on the ChAT and SV40 promoters. CHP126 cells were cotransfected with construct F (5 μg), SV40-lacZ (5 μg), with (+) or without (−) p300 expression vector (7.5 μg); E1A 12S expression vector (5 μg); or equivalent molar amounts of empty expression vectors. The total amount of plasmid DNA was adjusted to 31 μg with pBLCAT3 plasmid. Enzymatic activities were measured 40 h after transfection. (A) Luciferase (luc) activities; (B) β-galactosidase activities.
Induction of the histone H10 gene by butyrate is blocked by PD98059 and H7.
We next asked whether protein kinase inhibitors would similarly affect the induction of cellular genes by butyrate. We selected the histone variant H10 gene because it is induced by butyrate in many cell types (20, 28, 84). Run-on experiments have shown that butyrate stimulates H10 gene transcription (64), and transient-transfection assays have delineated in the H10 gene promoter two regions essential for butyrate activation that correspond to elements also required for basal expression (45). CHP126 cells were treated for 6 h with 5 mM butyrate in the absence or presence of 50 μM H7 or PD98059. Total RNA was then extracted and analyzed by Northern blotting with a specific H10 cRNA probe. Figure 6 shows that butyrate increased the level of H10 mRNA about fivefold. H7 or PD98059 alone had a modest effect on the mRNA level in the absence of butyrate but totally suppressed the induction by butyrate. Therefore, the induction of a cellular gene by butyrate also requires the activities of MEK1 and an H7-sensitive protein kinase.
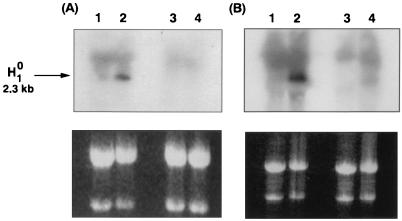
FIG. 6.
H7 suppresses induction of the H10 gene by butyrate. (A) CHP126 cells (10 × 106, in 100-mm-diameter dishes) were treated for 1 h with 50 μM H7 (lanes 3 and 4) or left untreated (lanes 1 and 2) and then were treated for 6 h with 5 mM sodium butyrate (lanes 2 and 4) or vehicle (lanes 1 and 3) in the continued presence of H7. Total RNA was extracted and analyzed by Northern blotting with a human H10 cRNA probe. Twenty micrograms of RNA was deposited in each lane. The exposure time was 4 h at −70°C. (B) The experiment was repeated with 50 μM PD98059 instead of H7. In both experiments, the upper panels show quantification of H10 mRNA by autoradiography while the lower panels are stained with ethidium bromide to verify equal loading of the lanes. The signal seen above the specific H10 band possibly results from cross-hybridization with a related gene product.
Activation of the ChAT promoter and of the RSV LTR by butyrate is blocked by okadaic acid.
A recent report has shown that the induction of H10 and the repression of c-myc transcription by butyrate in HTC hepatoma cells are blocked by okadaic acid and calyculin A, two inhibitors of protein phosphatases PP1 and PP2A (20). These results thus suggest that the transcriptional effects of butyrate on cellular genes are mediated by the activation of protein phosphatases, in an apparent contradiction of our present results. To address this discrepancy, we compared the effects of H7 and okadaic acid on transcriptional activation by butyrate in transfection assays. CHP126 cells were transiently transfected with both construct F and pRSV-LacZ. After overnight culture, cells were treated with 200 nM okadaic acid or vehicle (0.2% dimethyl sulfoxide [DMSO]) for 30 min and then with 5 mM butyrate in the continued presence of okadaic acid. Since prolonged treatment (12 h) with okadaic acid induces apoptosis in a variety of cells, we reduced the length of treatment with butyrate and/or okadaic acid to 8 h. Under these conditions, the level of activation of the ChAT promoter was low but significant (Table 3). Galactosidase activity driven by the RSV LTR was increased about fivefold by this short treatment with butyrate. Interestingly, the stimulating effect of butyrate on both the ChAT promoter and the RSV LTR was totally suppressed by okadaic acid. Therefore, okadaic acid, an inhibitor of serine/threonine protein phosphatases, and H7, an inhibitor of serine/threonine protein kinases, were both able to suppress the transcriptional effects of butyrate in transfection experiments and to block induction of the cellular H10 gene.
TABLE 3.
Okadaic acid blocks the activation of the ChAT promoter and of the RSV LTR by butyratea
| Concn of okadaic acid (nM) | Luciferase activity with ChAT promoter (RLU/μg of protein, 103)
|
β-Galactosidase activity with RSV LTR (RLU/min/μg of protein, 103)
|
||
|---|---|---|---|---|
| − Butyrate | + Butyrate | − Butyrate | + Butyrate | |
| 0 | 4.02 ± 0.15 | 6.75 ± 0.10 | 1.07 ± 0.17 | 11.1 ± 1.6 |
| 200 | 5.03 ± 0.36 | 5.24 ± 0.17 | 0.48 ± 0.08 | 0.93 ± 0.38 |
CHP126 cells were cotransfected with construct F and RSV-lacZ (5 μg of each). Sixteen hours after transfection, sister cultures were treated with 200 nM okadaic acid or vehicle (0.25% DMSO). After a 30-min incubation, cultures were provided with 5 mM butyrate, as indicated, in the continued presence of okadaic acid. The cells were lysed 8 h later and tested for luciferase and β-galactosidase activities.
Effects of butyrate and H7 on histone acetylation and phosphorylation.
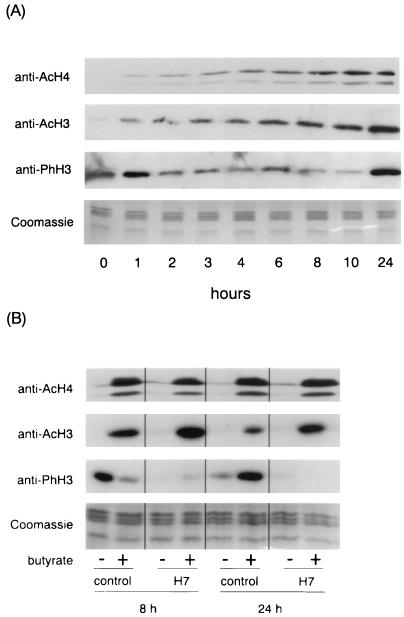
Histone H3 was initially reported to be phosphorylated in butyrate-treated nuclei (75, 76), but the phosphorylated histone was later identified as the variant H2AX (15). We studied the effects of butyrate on histone H3 phosphorylation by Western blot analysis with an antibody that specifically recognizes histone H3 phosphorylated on the conserved serine 10 residue. For comparison, the Western analysis was also performed with an antibody that recognizes histone H3 acetylated on lysine 9 and/or 14 and with a third antibody that recognizes tetraacetylated histone H4 and cross-reacts with acetylated histone H2B. When CHP126 cells were treated with 5 mM butyrate, acetylation of H3, H4, and H2B was significantly increased after 1 h of treatment and was maximal after 8 to 24 h (Fig. 7A). By contrast, phosphorylation of histone H3 progressively decreased up to 10 h but increased between 10 and 24 h. Further kinetics experiments showed that the minimal level of phosphorylation of histone H3 was observed after 16 h of treatment (data not shown). Figure 7B shows that after 8 and 24 h of treatment with butyrate, H7 had no effect on histone acetylation. This experiment confirmed the biphasic effect of butyrate on histone H3 phosphorylation and further showed that H7 blocks the phosphorylation of H3 in both butyrate-treated and untreated cells. Results similar to those shown in Fig. 7 were obtained with a new, highly potent inhibitor of HDACs (data not shown).
FIG. 7.
Western blot analysis of histone acetylation and phosphorylation in butyrate-treated CHP126 cells. (A) Time course. The culture medium of confluent CHP126 cells was supplemented with 5 mM butyrate. After the indicated length of culture, nuclei were purified and then extensively digested with micrococcal nuclease. Histones were acid extracted, separated on a 15% acrylamide gel, and subjected to Western analysis with anti-acetyl-H4 (anti-AcH4), anti-acetyl-H3 (anti-AcH3), and anti-phospho-H3 (anti-PhH3) antibodies, as indicated. Coomassie blue staining of an identical gel verified equal loading of the lanes. (B) Comparison of the effects of butyrate and H7. CHP126 cells were pretreated for 30 min with 50 μM H7 or vehicle (control) and then treated (+) or not treated (−) with 5 mM for 8 or 24 h, as indicated. Western blot analysis was performed as described for panel A.
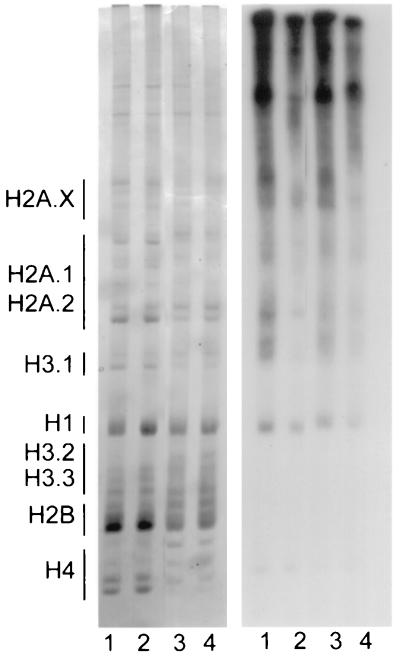
To further analyze the effects of butyrate on histone phosphorylation, CHP126 cells were labeled for 8 h with [32P]phosphate in the presence or absence of 5 mM butyrate and/or 50 μM H7. Nuclei were then purified, extensively digested with micrococcal nuclease, and acid extracted. After the acid-insoluble protein was submitted to SDS-polyacrylamide gel electrophoresis, autoradiography revealed in this fraction a weak labeling of histones that was unaffected by treatment with butyrate and/or H7 (data not shown). Acid-extracted histones were analyzed on AUT gels and subjected to autoradiography after being stained with Coomassie blue. The staining of the AUT gels with Coomassie blue clearly showed that H7 did not affect histone hyperacetylation induced by butyrate (Fig. 8). This suggests that H7 blocks the transcriptional effect of butyrate downstream of histone hyperacetylation. Moreover, these results establish that bulk histone hyperacetylation in butyrate-treated cells is not sufficient to activate the ChAT promoter. They do not, however, exclude the possibility that H7 selectively affects the acetylation status of histones in butyrate-sensitive promoter regions.
FIG. 8.
Effects of butyrate and H7 on histone phosphorylation. CHP126 cells were cultured for 8 h with [32P]phosphate in the presence of 50 μM H7 (lane 2), 5 mM butyrate (lane 3), H7 and butyrate (lane 4), or vehicle (lane 1). Histones were acid extracted from purified nuclei and analyzed on a 12% AUT gel. Left panel, Coomassie blue staining; right panel, autoradiograph of the gel.
Autoradiography of the AUT gels showed that phosphorylation of histones H4, H2B, H3.2, and H3.3 was very weak or absent. Phosphorylation of histone H1 was affected little by treatment with butyrate and was slightly depressed by H7. The phosphorylation of histones H3.1, H2A.1, and H2A.2 was clearly depressed in butyrate-treated cells and almost suppressed by H7. Interestingly, the phosphorylation of these histones mostly resided in the hyperacetylated pools. It is interesting that the sequence surrounding Ser-10 differs in H3.1 (KSTGGK) and H3.3 (KS-GGK). This suggests that different kinases are implicated in their phosphorylation and may explain the phosphorylation of H3.1 but not of H3.2 or H3.3 in the experiment shown in Fig. 8.
DISCUSSION
Transcriptional activation by HDAC inhibitors requires an H7-sensitive protein kinase activity.
We showed in this study that the transcriptional effects of butyrate and trapoxin, two HDAC inhibitors, are suppressed by the serine/threonine protein kinase inhibitor H7 and by PD98059, a highly specific inhibitor of the activation of MEK1 by Raf and MEKK1 (2). We presented evidence for this sensitivity to kinase inhibitors under three sets of experimental conditions in which the chromatin structure of the DNA template differed: transient transfection for the M promoter of the human ChAT gene, the SV40 minimal promoter, and the RSV LTR; stable transfection for the ChAT promoter; and a natural chromatin environment for an endogenous gene, H10. Although transcriptional activation by butyrate was in some cases found to be strictly dependent on chromosomal integration (71), several promoters are activated by butyrate in transient-transfection assays. Moreover, HDAC targeted to a transfected template by fusion with heterologous DNA-binding domains represses transcription in a trichostatin-sensitive manner (79). Since transfected plasmids do not show a canonical nucleosomal structure (7, 41), data obtained in transient-transfection assays have been sometimes interpreted as indicating that relevant targets of HDACs are not histones but rather are nonhistone nuclear proteins. Alternatively, selective and dynamic changes in the acetylation of histones may regulate the expression not only of genes in their chromatin environment but also of transfected templates. In particular, HDAC targeted to transfected templates through interactions with the retinoblastoma protein Rb or with the Mad protein has been shown to depress the acetylation of histone H3 associated with episomal DNA, a phenomenon correlated with transcriptional repression (54).
We previously showed that PD98059 blocks the activation of the ChAT promoter by trapoxin or butyrate (25). Two other previous reports showed that cellular effects of butyrate are sensitive to a kinase inhibitor. First, the apoptosis of thymocytes induced by butyrate is decreased by H7 (49). Second, the induction of core 2 acetylglucosaminyltransferase activity in CHO cells by butyrate is blocked by 50 μM H7 (21). In that case, butyrate mostly acts by increasing the maximal velocity of the enzymatic reaction, probably through cAMP-dependent phosphorylation, but potential effects of butyrate and H7 on acetylglucosaminyltransferase gene expression were not explored. Our results, to our knowledge, constitute the first report of suppression of the transcriptional effects of HDAC inhibitors by protein kinase inhibitors.
H7 and E1A have similar effects on transcriptional activation by butyrate and p300.
An intriguing aspect of the present work is the similarity in the effects of adenovirus E1A protein and H7. Both have a limited effect on ChAT and SV40 promoters in untreated cells but suppress their activation by butyrate. In agreement with these data, E1A suppresses the activation of the hsp70 promoter by trichostatin A (53). In addition, overexpression of the p300 protein had a relatively small effect on ChAT promoter activity by itself but strongly synergized with HDAC inhibitors. As expected, the stimulating effect of p300 was suppressed by E1A, but, unexpectedly, it was also blocked by H7. These data suggest, first, that activation of the ChAT promoter by HDAC inhibitors requires p300/CBP activity that is blocked by E1A and, second, that p300/CBP function requires the activity of an H7-sensitive kinase. This strongly suggests that derepression by blockage of HDAC activity and activation by the recruitment of HATs are not separable steps in transcriptional regulation. Rather, the effect of HDAC inhibitors is to unmask the activity of HATs already present on the promoter. It has been established that transcription factors YY1 (51, 79) and E2F (12, 54, 55, 70) as well as retinoic acid and thyroid hormone receptors (3, 34, 58) can interact with both HAT and HDAC complexes. Hormone binding disrupts interactions between nuclear receptors and the corepressor NCoR, allowing the release of the HDAC complex and the recruitment of an activating complex containing CBP/p300, P/CAF, and coactivators (47, 58). In these cases, HDAC inhibitors by themselves have little effect on transcription but strongly potentiate hormonal induction (27, 40, 58), a situation different from that of butyrate-inducible genes. What regulates the recruitment of HAT and HDAC complexes by YY1 and E2F is less clear. Whereas E2F directly interacts with p300 (70), its interaction with HDAC1 is mediated by the Rb (12, 54, 55). Interestingly, HDAC1 preferentially interacts with hypophosphorylated, rather than hyperphosphorylated, Rb protein (54). Therefore, the cell cycle-regulated phosphorylation of Rb may lead to a displacement of HDAC1 complexes and the recruitment of HAT-activating complexes. This suggests that phosphorylation events are involved in regulating the balance between deacetylase and acetyltransferase activities that controls transcription.
Both H7 and okadaic acid block transcriptional activation by butyrate.
Our data seem at odds with those of Cuisset et al. (20), who showed that induction of the H10 gene and repression of the c-myc gene by butyrate in Morris hepatoma cells are blocked by okadaic acid and calyculin A, two inhibitors of PP1 and PP2A serine/threonine protein phosphatases. Using the same protocol with CHP126 cells, we established that the induction of the H10 gene by butyrate is totally blocked by H7 or PD98059. Furthermore, our transient-transfection assays showed that activation of both the ChAT promoter and the RSV LTR by butyrate is blocked by okadaic acid, as it is by the two kinase inhibitors. For cellular genes and transfected templates, the transcriptional effect of butyrate thus appears to require both serine/threonine kinase and phosphatase activities. This paradoxical result could be explained by assuming that the effector of HDAC inhibitors is a phosphatase, activated by the H7-sensitive kinase. This kinase could be located either downstream of ERK kinases or on an alternative signaling pathway.
Effects of butyrate and H7 on histone H3 phosphorylation.
The phosphorylation of histone H3 on the Ser-10 residue has been implicated in chromosome condensation at mitosis (35, 42, 68). Moreover, growth factors, phorbol esters, and okadaic acid trigger H3 phosphorylation (56). Interestingly, mitogen-stimulated H3 phosphorylation occurs in a hyperacetylated pool (9), suggesting a possible link with transcription. Indeed, the phosphorylation of a linker histone in the Tetrahymena micronucleus was correlated with transcriptional activation (35). As a first approach in elucidating the role of H3 phosphorylation in transcription, we studied the effects of butyrate and H7 on that process. Western blot experiments with anti-phospho-H3 antibodies revealed that butyrate first caused a decrease in H3 phosphorylation but that after 16 h of treatment an actual increase was observed. In contrast, acetylation of histones H3, H2A, and H4 slowly increased up to 24 h. Similar observations were made with a new, highly selective inhibitor of HDACs (data not shown). Therefore, the effects of butyrate on H3 phosphorylation do not result from nonspecific effects unrelated to HDAC inhibition. These data are thus in agreement with the hypothesis of a sequential activation of protein phosphatases and kinases by inhibitors of HDACs. It was, however, conceivable that the initial decrease in H3 phosphorylation was in fact due to the masking of the epitope by the acetylation of neighbor lysines, in particular Lys-9. This appears unlikely, however, since H3 phosphorylation, as revealed by Western blotting, was increased after 24 h of treatment with butyrate, when H3 acetylation was maximal. Moreover, in vivo labeling of histones followed by an autoradiographic analysis of AUT gels revealed that butyrate decreased the phosphorylation of H2A and H3.1. In addition, the phosphorylation of H3.1 resided mostly in the hyperacetylated pool. Therefore, the data strongly suggest that the anti-phospho-H3 antibody can recognize the acetylated forms of the histone.
The Western and AUT analyses both revealed that H7 blocks histone H3 phosphorylation. Taken together, our observations did not unravel a clear relationship between transcriptional activation and H3 phosphorylation. First, during the initial hours of treatment, butyrate and H7 both decreased H3 phosphorylation but had opposite effects on transcription. Second, H3 phosphorylation was first decreased and then increased by butyrate, whereas transcription was increased after both short (8 h) and long (24 h) treatments with butyrate. Third, short treatments with H7 and okadaic acid had opposite effects on H3 phosphorylation (56), whereas both compounds blocked butyrate-activated transcription.
Potential role of CBP/p300-associated kinases in transcriptional activation by HDAC inhibitors.
Although our data establish that transcriptional regulation by butyrate and p300 is sensitive to H7, the nature of the serine/threonine kinase(s) involved remains unclear. In a previous study (25), we showed that butyrate and trapoxin transiently stimulate the phosphorylation of ERK1 and -2 in CHP126 cells and that activation of the MEK/ERK MAP kinase cascade is required for activation of the ChAT promoter by HDAC inhibitors. In particular, ChAT promoter activation is blocked by PD98059, a highly specific inhibitor of MEK1, and by an ERK2 dominant-negative mutant. We have now established that activation of two viral promoters and induction of the H10 gene by butyrate are also blocked by PD98059.
Several lines of data suggested that members of the pp90rsk family of ribosomal S6 protein kinases (RSK) might be involved in transcriptional activation by butyrate. First, among the different kinases of the MEK/ERK cascade, only RSK is sensitive to H7 (80). Second, RSK phosphorylated by ERK binds the C/H3 domain of CBP in mitogen-stimulated cells (59). E1A and RSK might mutually exclude one another since their binding domains on CBP/p300 are largely overlapping. If CBP/p300-associated RSK activity were required for transcriptional activation by HDAC inhibitors, it should be blocked by H7, which inhibits kinase activity, and by E1A, which antagonizes the binding of RSK on CBP/p300. It has been shown that the binding of RSK on CBP blocks cAMP-stimulated transcription in a manner similar to E1A (59). Since a kinase-negative RSK mutant is as effective as the wild type, RSK may simply act by displacing a coactivator like P/CAF, which is required for CREB-dependent transcription (47). It was suggested, however, that RSK may play a positive role in Ras-dependent transcription (59). Third, RSK2, and to a lesser extent RSK3, phosphorylates histone H3 (16, 83). Fourth, RSK2 phosphorylates and activates protein phosphatase PP-1 (23), in agreement with the hypothesis of a sequential activation of kinases and phosphatases by butyrate.
However, GF109203X, which inhibits RSK2 as well as PKC isoforms (1), did not block the activation of the ChAT promoter by butyrate (Table 2). Moreover, overexpression of dominant-negative mutants of RSK1 or RSK2 in CHP126 cells did not affect the activation of the ChAT promoter by butyrate (data not shown). This contrasts with our previous experiments in which the overexpression of dominant-negative mutants of Ras or ERK1 suppressed ChAT promoter activation (25). Therefore, these experiments failed to implicate RSK kinases in the mode of action of butyrate.
Other possible candidates are the mitogen- and stress-activated protein kinases MSK1 and MSK2 (22). However, stressful stimuli (e.g., arsenite and d-sorbitol) did not reproduce the transcriptional effect of butyrate; moreover, SB203580, a specific inhibitor of SAPK2a/p38 and SAPK2b/p38b that blocks the activation of MSK by stressful stimuli, has no effect on butyrate action (25). Nevertheless, a potential role for MSK in transcriptional regulation by butyrate remains to be studied.
Another kinase associated with p300 is the cyclin-dependent kinase cdk2, which is implicated in the phosphorylation of p300 during the cell cycle (77) and during the differentiation of F9 cells that is triggered by retinoic acid or E1A (46). The effects of p300 phosphorylation on transcription remain unclear, however (39), and relevant targets of p300-associated cdk probably include several transcription factors. In particular, the p65 (RelA) subunit of NF-κB binds the N-terminal region of p300, where it is phosphorylated by the cyclin E-CDK complex. The overexpression of p21 (WAF-1, CIP-1), an inhibitor of all CDKs, stimulates NF-κB-dependent transcription, most probably by preventing RelA phosphorylation (61). We found that overexpression of p21 in CHP126 cells had no effect on ChAT promoter activity in the absence or presence of butyrate (data not shown). It is therefore unlikely that p300-associated CDKs participate in ChAT promoter activation by HDAC inhibitors.
Potential targets for phosphorylation in transcriptional control by butyrate.
The present and previous data clearly suggest a necessary role for protein kinases and phosphatases in the regulation of transcription by inhibitors of HDACs (20, 25). Potential targets could be histones, although we found no clear relationship between H3 phosphorylation and transcriptional activation. This nevertheless evokes the possibility that efficient transcription requires modifications in both the acetylation status and the phosphorylation status of core histones. On the other hand, CBP/p300 is also known to acetylate a growing list of specific transcription factors, including p53, NF-Y, GATA-1, and EKLF (31, 53, 82) as well as the general transcription factors TFIIEβ and TFIIF (38). It is therefore conceivable that the activation of the MAP kinase cascade by butyrate or trapoxin modifies the phosphorylation status of transcription factors or other, nonhistone chromatin-associated proteins. This would alter the DNA-binding properties of transcription factors and/or the recruitment of multiprotein complexes endowed with HAT or HDAC activity. This raises the question of whether protein kinases and phosphatases are members of such complexes and can be coimmunoprecipitated with CBP/p300 and HDAC.
ACKNOWLEDGMENTS
We thank M. Yoshida (Tokyo) for the gift of trapoxin A and G. M. Lauro (Rome, Italy) for CHP126 cells. We also thank D. M. Livingston and S. Bhattacharya (Boston, Mass.) for the CMV-p300 plasmids and T. Kouzarides (Cambridge, United Kingdom) and D. Trouche (Toulouse, France) for E1A and CBP expression vectors. We thank E. Käs (Toulouse) for careful reading of the manuscript and A. Amiche (Toulouse) for help with histone analysis.
This work was supported by funds from the Association Française contre les Myopathies and by the Région Midi-Pyrénées.
REFERENCES
- 1.Alessi D R. The protein kinase C inhibitors Ro 318220 and GF 109203X are equally potent inhibitors of MAPKAP kinase-1β (Rsk-2) and p70 S6 kinase. FEBS Lett. 1997;402:121–123. doi: 10.1016/s0014-5793(96)01510-4. [DOI] [PubMed] [Google Scholar]
- 2.Alessi D R, Cuenda A, Cohen P, Dudley D T, Saltiel A R. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J Biol Chem. 1995;270:27489–27494. doi: 10.1074/jbc.270.46.27489. [DOI] [PubMed] [Google Scholar]
- 3.Alland L, Muhle R, Hou H J, Potes J, Chin L, Schreiber A N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 4.Allfrey V G, Faulkner R, Mirsky A E. Acetylation and methylation of histones and their possible role in regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 6.Arany Z, Sellers W R, Livingston D M, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. . (Letter.). [DOI] [PubMed] [Google Scholar]
- 7.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. . (Erratum, 256:161.) [DOI] [PubMed] [Google Scholar]
- 8.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barratt M J, Hazzalin C A, Cano E, Mahadevan L C. Mitogen-stimulated phosphorylation of histone H3 is targeted to a small hyperacetylation-sensitive fraction. Proc Natl Acad Sci USA. 1994;91:4781–4785. doi: 10.1073/pnas.91.11.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartl S, Taplick J, Lagger G, Khier H, Kuchler K, Seiser C. Identification of mouse histone deacetylase 1 as a growth factor-inducible gene. Mol Cell Biol. 1997;17:5033–5043. doi: 10.1128/mcb.17.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boronat S, Richard-Foy H, Pina B. Specific deactivation of the mouse mammary tumor virus long terminal repeat promoter upon continuous hormone treatment. J Biol Chem. 1997;272:21803–21810. doi: 10.1074/jbc.272.35.21803. [DOI] [PubMed] [Google Scholar]
- 12.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 13.Brenner D A, O’Hara M, Angel P, Chojkier M, Karin M. Prolonged activation of jun and collagenase genes by tumour necrosis factor-alpha. Nature. 1989;337:661–663. doi: 10.1038/337661a0. [DOI] [PubMed] [Google Scholar]
- 14.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 15.Cano E, Barratt M J, Mahadevan L C. Which histone kinase? Nature. 1992;360:116. doi: 10.1038/360116a0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 16.Chen R-H, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chireux M, Espinos E, Bloch S, Yoshida M, Weber M J. Histone hyperacetylating agents stimulate promoter activity of human choline acetyltransferase gene in transfection experiment. Mol Brain Res. 1996;39:68–78. doi: 10.1016/0169-328x(96)00006-x. [DOI] [PubMed] [Google Scholar]
- 18.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 19.Clayton A L, Hebbes T R, Thorne A W, Crane R C. Histone acetylation and gene induction in human cells. FEBS Lett. 1993;336:23–26. doi: 10.1016/0014-5793(93)81601-u. [DOI] [PubMed] [Google Scholar]
- 20.Cuisset L, Tichonicky L, Jaffray P, Delpech M. The effects of sodium butyrate on transcription are mediated through activation of a protein phosphatase. J Biol Chem. 1997;272:24148–24153. doi: 10.1074/jbc.272.39.24148. [DOI] [PubMed] [Google Scholar]
- 21.Datti A, Dennis J W. Regulation of UDP-GlcNAc:Gal β1-3GalNAc-R β 1-6-N-acetylglucosaminyltransferase (GlcNAc to GalNAc) in Chinese hamster ovary cells. J Biol Chem. 1993;268:5409–5416. [PubMed] [Google Scholar]
- 22.Deak M, Clifton A D, Lucocq L M, Alessi D R. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dent P, Lavoinne A, Nakielny S, Caudwell F B, Watt P, Cohen P. The molecular mechanism by which insulin stimulates glycogen synthesis in mammalian skeletal muscle. Nature. 1990;348:302–308. doi: 10.1038/348302a0. [DOI] [PubMed] [Google Scholar]
- 24.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCaprio J A, Lawrence J B, Livingston D M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 25.Espinos E, Weber M J. Activation of the MAP kinase cascade by histone deacetylase inhibitors is required for the stimulation of choline acetyltransferase gene promoter. Mol Brain Res. 1998;56:118–124. doi: 10.1016/s0169-328x(98)00036-9. [DOI] [PubMed] [Google Scholar]
- 26.Fan X D, Goldberg M, Bloom B R. Interferon-gamma-induced transcriptional activation is mediated by protein kinase C. Proc Natl Acad Sci USA. 1988;85:5122–5125. doi: 10.1073/pnas.85.14.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia V P, Jimenez L A, Castillo A I, Aranda A. Histone acetylation influences thyroid hormone and retinoic acid-mediated gene expression. DNA Cell Biol. 1997;16:421–431. doi: 10.1089/dna.1997.16.421. [DOI] [PubMed] [Google Scholar]
- 28.Girardot V, Rabilloud T, Yoshida M, Beppu T, Lawrence J J, Khochbin S. Relationship between core histone acetylation and histone H10 gene activity. Eur J Biochem. 1994;224:885–892. doi: 10.1111/j.1432-1033.1994.00885.x. [DOI] [PubMed] [Google Scholar]
- 29.Gorman C M, Howard B H, Reeves R. Expression of recombinant plasmids in mammalian cells is enhanced by sodium butyrate. Nucleic Acids Res. 1983;11:7631–7648. doi: 10.1093/nar/11.21.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 31.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 32.Hebbes T R, Thorne A W, Clayton A L, Crane R C. Histone acetylation and globin gene switching. Nucleic Acids Res. 1992;20:1017–1022. doi: 10.1093/nar/20.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hebbes T R, Thorne A W, Crane R C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 35.Hendzel M J, Wei Y, Mancini M A, Van Hooser A, Ranalli T, Brinkley B R, Bazett-Jones D P, Allis C D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 36.Hidaka H, Inagaki M, Kawamoto S, Sasaki Y. Isoquinolinesulfonamides, novel and potent inhibitors of cyclic nucleotide dependent protein kinase and protein kinase C. Biochemistry. 1984;23:5036–5041. doi: 10.1021/bi00316a032. [DOI] [PubMed] [Google Scholar]
- 37.Hidaka H, Yokokura H. Molecular and cellular pharmacology of a calcium/calmodulin-dependent protein kinase II (CaM kinase II) inhibitor, KN-62, and proposal of CaM kinase phosphorylation cascades. Adv Pharmacol. 1996;36:193–219. doi: 10.1016/s1054-3589(08)60583-9. [DOI] [PubMed] [Google Scholar]
- 38.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 39.Janknecht R, Hunter T. Versatile molecular glue. Transcriptional control. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 40.Jenster G, Spencer T E, Burcin M M, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor induction of gene transcription: a two-step model. Proc Natl Acad Sci USA. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong S, Stein A. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 1994;22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juan G, Traganos F, James W M, Ray J M, Roberge M, Sauve D M, Anderson H, Darzynkiewicz Z. Histone H3 phosphorylation and expression of cyclins A and B1 measured in individual cells during their progression through G2 and mitosis. Cytometry. 1998;32:71–77. doi: 10.1002/(sici)1097-0320(19980601)32:2<71::aid-cyto1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 43.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 44.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khochbin S, Wolffe A P. Developmental regulation and butyrate-inducible transcription of the Xenopus histone H10 promoter. Gene. 1993;128:173–180. doi: 10.1016/0378-1119(93)90560-p. [DOI] [PubMed] [Google Scholar]
- 46.Kitabayashi I, Eckner R, Arany Z, Chiu R, Gachelin G, Livingston D M, Yokoyama K K. Phosphorylation of the adenovirus E1A-associated 300 kDa protein in response to retinoic acid and E1A during the differentiation of F9 cells. EMBO J. 1995;14:3496–3509. doi: 10.1002/j.1460-2075.1995.tb07356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 48.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurita-Ochiai T, Fukushima K, Ochiai K. Butyric acid-induced apoptosis of murine thymocytes, splenic T cells, and human Jurkat T cells. Infect Immun. 1997;65:35–41. doi: 10.1128/iai.65.1.35-41.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 51.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. . (Erratum, 9:1948–1949.) [DOI] [PubMed] [Google Scholar]
- 52.Lew D J, Decker T, Darnell J E., Jr Alpha interferon and gamma interferon stimulate transcription of a single gene through different signal transduction pathways. Mol Cell Biol. 1989;9:5404–5411. doi: 10.1128/mcb.9.12.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 55.Magnaghi J L, Groisman R, Naguibneva I, Robin P, Lorain S, Le V J, Troalen F, Trouche D, Harel B A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 56.Mahadevan L C, Willis A C, Barratt M J. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- 57.Martiny-Baron G, Kazanietz M G, Mischak H, Blumberg P M, Kochs G, Hug H, Marme D, Schachtele C. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J Biol Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 58.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 59.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. The signal-dependent coactivator CBP is a nuclear target for pp90rsk. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 60.Neuwald A F, Landsman D. GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast SPT10 protein. Trends Biochem Sci. 1997;22:154–155. doi: 10.1016/s0968-0004(97)01034-7. [DOI] [PubMed] [Google Scholar]
- 61.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 62.Rochette E C, Fromental C, Chambon P. General repression of enhanson activity by the adenovirus-2 E1A proteins. Genes Dev. 1990;4:137–150. doi: 10.1101/gad.4.1.137. [DOI] [PubMed] [Google Scholar]
- 63.Rogakou E P, Pilch D R, Orr A H, Ivanova V S, Bonner W M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 64.Rousseau D, Khochbin S, Gorka C, Lawrence J J. Induction of H10-gene expression in B16 murine melanoma cells. Eur J Biochem. 1992;208:775–779. doi: 10.1111/j.1432-1033.1992.tb17247.x. [DOI] [PubMed] [Google Scholar]
- 65.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 67.Taunton J, Hassig C A, Schreiber S L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 68.Th’ng J P, Guo X W, Swank R A, Crissman H A, Bradbury E M. Inhibition of histone phosphorylation by staurosporine leads to chromosome decondensation. J Biol Chem. 1994;269:9568–9573. [PubMed] [Google Scholar]
- 69.Toullec D, Pianetti P, Coste H, Bellevergue P, Grand P T, Ajakane M, Baudet V, Boissin P, Boursier E, Loriolle F, et al. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J Biol Chem. 1991;266:15771–15781. [PubMed] [Google Scholar]
- 70.Trouche D, Kouzarides T. E2F1 and E1A(12S) have a homologous activation domain regulated by RB and CBP. Proc Natl Acad Sci USA. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Lint C, Emiliani S, Ott M, Verdin E. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 1996;15:1112–1120. [PMC free article] [PubMed] [Google Scholar]
- 72.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 73.Velcich A, Ziff E. Adenovirus E1a proteins repress transcription from the SV40 early promoter. Cell. 1985;40:705–716. doi: 10.1016/0092-8674(85)90219-3. [DOI] [PubMed] [Google Scholar]
- 74.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 75.Whitlock J P, Jr, Augustine R, Schulman H. Calcium-dependent phosphorylation of histone H3 in butyrate-treated HeLa cells. Nature. 1980;287:74–76. doi: 10.1038/287074a0. [DOI] [PubMed] [Google Scholar]
- 76.Whitlock J P, Jr, Galeazzi D, Schulman H. Acetylation and calcium-dependent phosphorylation of histone H3 in nuclei from butyrate-treated HeLa cells. J Biol Chem. 1983;258:1299–1304. [PubMed] [Google Scholar]
- 77.Yaciuk P, Moran E. Analysis with specific polyclonal antiserum indicates that the E1A-associated 300-kDa product is a stable nuclear phosphoprotein that undergoes cell cycle phase-specific modification. Mol Cell Biol. 1991;11:5389–5397. doi: 10.1128/mcb.11.11.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang C H, Shi W, Basu L, Murti A, Constantinescu S N, Blatt L, Croze E, Mullersman J E, Pfeffer L M. Direct association of STAT3 with the IFNAR-1 chain of the human type I interferon receptor. J Biol Chem. 1996;271:8057–8061. doi: 10.1074/jbc.271.14.8057. [DOI] [PubMed] [Google Scholar]
- 79.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yin T, Yang Y C. Mitogen-activated protein kinases and ribosomal S6 protein kinases are involved in signaling pathways shared by interleukin-11, interleukin-6, leukemia inhibitory factor, and oncostatin M in mouse 3T3-L1 cells. J Biol Chem. 1994;269:3731–3738. [PubMed] [Google Scholar]
- 81.Yoshida H, Sugita K. A novel tetracyclic peptide, trapoxin, induces phenotypic change from transformed to normal in sis-oncogene-transformed NIH3T3 cells. Jpn J Cancer Res. 1992;83:324–328. doi: 10.1111/j.1349-7006.1992.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang W, Bieker J J. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao Y, Bjørbaek C, Weremowicz S, Morton C C, Moller D E. RSK3 encodes a novel pp90rsk isoform with a unique N-terminal sequence: growth factor-stimulated kinase function and nuclear translocation. Mol Cell Biol. 1995;15:4353–4363. doi: 10.1128/mcb.15.8.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zlatanova J, Doenecke D. Histone H10: a major player in cell differentiation? FASEB J. 1994;8:1260–1268. doi: 10.1096/fasebj.8.15.8001738. [DOI] [PubMed] [Google Scholar]