Abstract
Background
Post-translational modifications (PTMs) on proteins can be targeted by antibodies associated with autoimmunity. Despite a growing appreciation for their intrinsic role in disease, there is a lack of highly multiplexed serological assays to characterize the fine specificities of PTM-directed autoantibodies.
Methods
In this study, we used the programmable phage display technology, Phage ImmunoPrecipitation Sequencing (PhIP-Seq), to profile rheumatoid arthritis (RA) associated anti-citrullinated protein antibody (ACPA) reactivities.
Findings
Using both unmodified and peptidylarginine deiminase (PAD)-modified phage display libraries consisting of ~250,000 overlapping 90 amino acid peptide tiles spanning the human proteome, PTM PhIP-Seq robustly identified antibodies to citrulline-dependent epitopes.
Interpretation
PTM PhIP-Seq was used to quantify key differences among RA patients, including PAD isoform specific ACPA profiles, and thus represents a powerful tool for proteome-scale antibody-binding analyses.
Funding
This research is based upon work supported in part by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA). The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of ODNI, IARPA, or the US Government. The US Government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation therein. This study was made possible by a National Institute of General Medical Sciences (NIGMS) grant R01 GM136724 (HBL). MFK was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grant T32AR048522. ED was supported by the Rheumatology Research Foundation.
Keywords: Phage ImmunoPrecipitation Sequencing, Citrullination, Peptidylarginine deiminase, Rheumatoid arthritis
Research in context.
Evidence before this study
Despite the established importance of autoantibody reactivities that are directed at protein post-translational modifications, efficient serological assays capable of characterizing disease-specific reactivities at proteome scale are lacking. Anti-citrullinated protein antibodies (ACPAs) are well-established biomarkers for rheumatoid arthritis (RA), which are believed to arise in the context of aberrant peptidyl arginine deiminase (PAD) citrullination. Of the five human isoforms, PAD2 and PAD4 are considered most likely to be involved in disease pathogenesis. With PAD-specific inhibitors in pre-clinical development, it is unknown whether some RA patients exhibit a discernable preference for PAD2- or PAD4-specific epitopes.
Added value of this study
To address the unmet need for proteome-wide technologies to profile anti-PTM autoantibodies, we adapted the programmable phage display assay Phage ImmunoPrecipitation Sequencing (PhIP-Seq) with a 90 amino acid human peptidome library to the characterization of ACPAs in a cohort of RA patients. PAD PhIP-Seq enabled the quantitative comparison of each patient's antibody reactivity against every unmodified human peptide versus the corresponding PAD2- or PAD4-modified peptides. We identified a subset of human peptides with PAD isoform mono-specific reactivity, and examined the preference of each individual's antibody repertoire toward PAD2-modified versus PAD4-modified peptides. A subset of individuals appeared to have significant preference for citrullinated peptides generated by one enzyme over the other.
Implication of all the available evidence
Examination of mono-reactive peptides revealed underlying PAD isoform-specific preference in some individuals. This finding may have implications for the appropriate matching of emerging PAD inhibitors with patients most likely to derive benefit. The methods presented here are broadly useful for studies of PTM-dependent antibodies.
Alt-text: Unlabelled box
1. Introduction
Phage ImmunoPrecipitation Sequencing (PhIP-Seq) is a programmable phage display technology that enables unbiased analysis of antibody binding specificities [1], [2], [3]. An oligonucleotide library encoding the complete human proteome as ~250,000 overlapping 90 amino acid peptides was cloned into the mid-copy T7 phage display vector (~5–15 copies of displayed peptide per phage particle) and is immunoprecipitated by serum antibodies for analysis by high throughput DNA sequencing. This enables serum antibody profiling against hundreds of thousands of peptide epitopes at low cost [2,4]. Since the phage library is produced in E. coli, the displayed peptides lack post-translational modifications (PTMs) relevant to human disease [5,6]. Here, we enzymatically modify the phage-displayed human peptidome, such that PTM-dependent autoantibody reactivities can be precisely assessed by quantitative comparison to the unmodified library.
Citrullination is the post-translational conversion of arginine in proteins to citrulline, which in humans is catalyzed by the calcium-dependent peptidylarginine deiminase (PAD) family of enzymes [7,8]. This modification can impart antigenicity to self-proteins, and in the case of rheumatoid arthritis (RA), results in the production of anti-citrullinated protein antibodies (ACPAs) [9]. These antibodies are believed to participate in disease pathogenesis by binding citrullinated proteins in the synovial joint [10,11]. The discovery of autoantibodies to peptidylcitrulline epitopes in RA serum led to the development of a diagnostic Enzyme Linked Immunosorbent Assay (ELISA) using synthetic cyclic citrullinated peptides (CCP) [12]. Since this anti-CCP antibody test can achieve diagnostic sensitivity of ~80% and a specificity up to 98%, anti-CCP antibodies have become a routinely utilized biomarker for RA [13,14].
Of the five known PAD isoforms, PAD2 and PAD4 make up the majority of expressed PAD in inflamed synovial tissue [15]. Studies using recombinantly expressed enzymes suggest that PAD2 and PAD4 can generate distinct, yet overlapping sets of epitopes that can be targeted by ACPAs [16], [17], [18]. ACPAs that recognize these substrates are known to cross-react, making it difficult to distinguish the relative contributions of the different PAD isoforms to the expression of ACPAs [19], [20], [21]. While aberrant citrullination is best known for its role in RA pathology, it is also implicated in other disease processes, including multiple sclerosis, Alzheimer's disease, and cancer [22,23]. Therefore, unbiased identification of antibody reactivities to citrulline epitopes is of broad interest.
2. Methods
2.1. Ethics committee approval and human subjects
Sera from 30 individuals with RA from a cross-sectional cohort and 10 demographically matched healthy controls were studied. Patient serum was collected under a study protocol approved by the Office of Human Subjects Research Institutional Review Boards of the Johns Hopkins University School of Medicine (IRB00036200); written consent was obtained prior to participation. Healthy control sera were obtained from volunteers who self-identified, were not pregnant, and did not have a history of cancer, autoimmune disease, or active tuberculosis/HIV/hepatitis infection. RA patients fulfilled 2010 ACR classification criteria for RA, as described [24]. Anti-CCP antibody levels were determined by the QUANTA Lite® CCP3 IgG ELISA (Inova Diagnostics, #704535). For one RA patient, serum was available at several time points pre- and post-treatment with the chimeric anti-CD20 monoclonal antibody rituximab. At baseline (day 0), the patient was treated with methotrexate and glucocorticoids with insufficient control of arthritis symptoms. Two doses of rituximab were administered on days 22 and 35, respectively. On day 0, anti-CCP and RF normalized units were 252 and 139, respectively.
2.2. Expression and purification of peptidylarginine deiminases
Recombinant human PAD4 and PAD2 containing N-terminal 6 x histidine tags were expressed in BL21(DE3) pLysS competent cells and purified by metal affinity chromatography with a nickel resin, as previously described [25,26].
2.3. Phage modification quality control
A negative control clone that lacks a displayed peptide (due to a premature stop codon, introduced as a frameshift mutation of the DNA insert) and a positive control clone that displays a 90-mer arginine-containing peptide from POU3F1 were isolated by randomly picking plaques from the human peptidome phage library. Peptide sequences were determined by Sanger sequencing. PAD modification was performed as described below. Dialysis, plaque assay, and western blotting were performed according to standard molecular biology protocols. Histone citrullination was detected using anti-citrullinated histone antibody (citrulline R2 + R8 + R17, Abcam #ab5103, RRID: AB_304752) 1:1,000 in TBST and visualized by chemiluminescence. Quantitative PCR assays were performed using SYBR green on a 1:1,000 dilution of product obtained from 10 cycles of PCR1 [1].
2.4. Human peptidome T7 phage library citrullination
Citrullination was carried out in 42 mL containing buffer alone or 150 μg of either recombinant PAD4 or recombinant PAD2, 1012 pfu human peptidome T7 phage library (pre-dialyzed into 100 mM Tris-HCl, pH 7.5 using a Spectra/Por 7 RC 50 kDa molecular weight cut off dialysis membrane, Spectrum Labs, #132130), 5 mM CaCl2 and 2 mM DTT. The reactions were incubated at 37 °C for 3 h with end-over-end rotation and quenched by dialysis against 100 mM Tris-HCl, pH 7.5 in order to remove free Ca2+ ions using a 10 kDa molecular weight cutoff dialysis membrane (Thermo Fisher, #88245). A second round of dialysis against TBS, pH 7.4 was performed in order to prepare the libraries for antibody binding and immunoprecipitation. Phage viability after modification was assessed by plaque assay.
2.5. Phage immunoPrecipitation Sequencing (PhIP-Seq)
Screening, PCR, and peptide read count data generation was performed as previously described [1], Briefly, 0.2 μL of sample (~2 μg IgG) was added to the phage library (1010 pfu) in TBS, pH 7.4. The phage/serum mixtures were rotated overnight at 4 °C after which 40 μL of a 1:1 Protein A/G coated magnetic bead mixture (Invitrogen, #10002D and #10004D) was added to each well and rotated for an additional 4 h at 4 °C to capture all IgG. The beads were then washed three times with TBS, pH 7.4 containing 0.1% NP-40 and resuspended in 20 μL of a Herculase II Fusion Polymerase (Agilent, #600679) PCR1 master mix to amplify library inserts. Sample-specific barcoding and the Illumina P5/P7 adapters were then incorporated during a subsequent PCR2 reaction. PCR2 amplicons were pooled and sequenced using an Illumina NextSeq 500 SE 1 × 50 protocol. Reads were demultiplexed and aligned to the human peptide library using exact matching.
2.6. Pairwise analysis of PTM PhIP-Seq data
We used pairwise enrichment analysis to identify peptides that were differentially reactive between treatment conditions. Robust regression of the top 1000, by abundance, unmodified read counts (Unmod) was used to calculate the ‘expected’ modified read counts (PAD2 & PAD4). The observed modified read counts minus the expected modified read counts for each peptide was calculated to determine peptide read count residuals. Peptides were grouped in bins according to unmodified read counts and a standard deviation was calculated for all peptides in each bin. From these binned standard deviations, a linear regression was developed and used to assign each peptide an expected standard deviation. Each peptide's residual was normalized to its expected standard deviation, in order to calculate a 'pairwise z-score'. Peptide reactivities were considered positive for pairwise z-scores ≥7.
2.7. Statistical analyses
Differences among the total number of peptide enrichments between sample groups was calculated using a Wilcoxon test (Fig. 4). Differences among peptide read counts were calculated using a Kruskal-Wallis test with Dunn's correction (Fig. S4). To characterize the interconnectedness of each individual's PAD specific peptide reactivities, we constructed undirected network graphs using the igraph software package in R. We constructed a database of all peptide-peptide alignments in the human peptidome library with an E value <1; each node represents a reactive peptide and each edge represents a peptide-peptide alignment. We used these network graphs to calculate the number of independent peptide reactivities detected for each individual, defined as the maximal number of reactive peptides with no sequence homology using the igraph independence.number function. Differences between the total number of anti-CCP+ patient specific PAD2- and PAD4-mono-reactivities were determined using an exact binomial test assuming a null probability of 0.5 for independent PAD2/4-specific peptides (Fig. 5e).
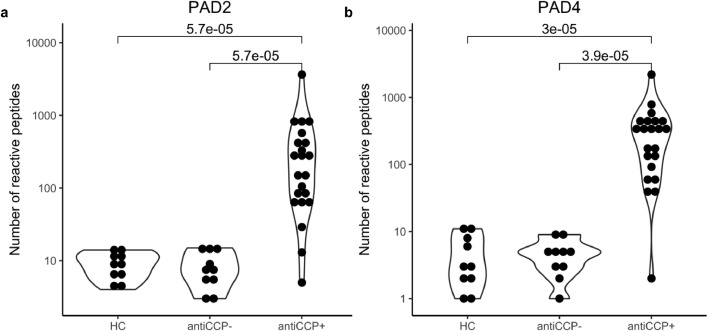
Fig. 4.
Cross sectional concordance of PAD PhIP-Seq with the clinical CCP assay. a The number of differentially reactive PAD2-dependent and b PAD4-dependent epitopes, per individual across the three groups: healthy controls (HC, n = 10), anti-CCP- RA (n = 10), and anti-CCP+ RA (n = 21).
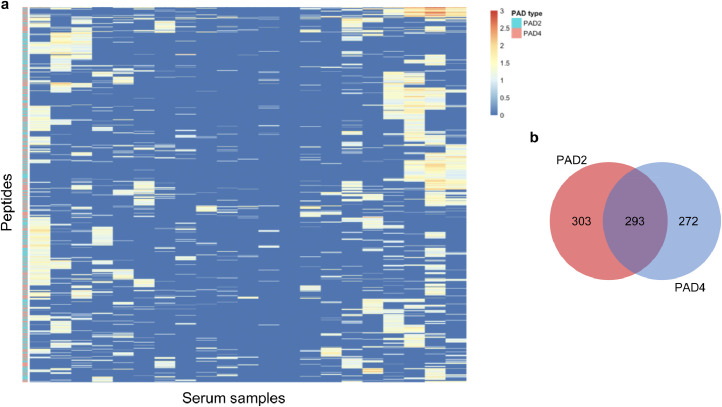
Fig. 5.
Reactivity profile of anti-CCP+ individuals. a Clustering of ACPA reactivity to citrulline-containing epitopes. Rows are peptides recognized by at least three anti-CCP+ sera (columns). b Distribution of the citrulline-dependent epitopes identified in a.
2.8. Role of funders
The funders were not involved in the study design, data collection and analysis, interpretation, writing of the manuscript, or in the decision to submit the paper for publication.
3. Results
3.1. Citrullination of the T7 phage-displayed human peptidome library
In a typical PhIP-Seq experiment, the phage display library is mixed with 0.2 μl of serum or plasma and immunoprecipitated (IPed) using magnetic beads coated with protein A and protein G, which together capture all IgG subclasses. The DNA sequences from the immunoprecipitated phage clones are then amplified by PCR and the abundance of each clone is quantified by high throughput DNA sequencing. To detect citrullinated peptide autoreactivities, we first developed a method to enzymatically citrullinate the human 90-mer phage library [27] in vitro using recombinant human PAD2 and PAD4 enzymes (Methods). Sequencing read count data from PhIP-Seq with a citrullinated library can be directly compared against count data from PhIP-Seq with an unmodified library. Fig. 1 illustrates how this type of analysis is able to identify citrulline-dependent autoantibody binding specificities.
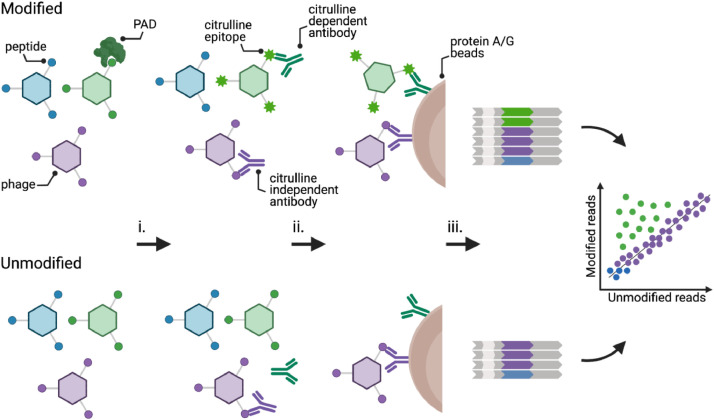
Fig. 1.
Experimental design of PAD PhIP-Seq. (i) PAD-modified (top) and unmodified (bottom) phage libraries are mixed with patient serum. (ii) The resulting phage-antibody immunocomplexes are then precipitated on Protein A/G beads and unbound phage washed away. (iii) Peptide inserts of the immunoprecipitated clones are PCR amplified and analyzed by high throughput DNA sequencing. Abundant clones along the diagonal (purple) represent reactive peptides that do not depend upon modification. Off-diagonal peptides (green) are immunoprecipitated only after being post-translationally modified. Low abundant clones and non-reactive library members cluster at the origin (blue).
3.2. Phage display library modification and quality assessment
We reasoned it would be important to establish a rigorous and generalizable quality control pipeline for assessing the integrity of PTM phage libraries. We first confirmed that buffer exchange using standard dialysis into two common buffers (PBS and TBS) did not impact phage viability (Fig. S1a). Second, we assessed phage viability after enzymatic modification. The 90-mer library, a control clone that does not display a peptide, and a second control clone that displays a peptide lacking any arginine residues, were all found to retain their infectivity after treatment with PAD enzyme (Fig. S1b). We also confirmed by western blot that the buffer and conditions used to citrullinate the phage display library were compatible with robust PAD activity (Fig. S1c).
We next sought to confirm that PAD does not create reactive citrulline epitopes on the phage coat proteins. If it did, the entire library might be non-specifically immunoprecipitated by ACPAs. To this end, we assessed the binding of anti-CCP+ versus anti-CCP- sera to a phage clone without a displayed peptide, which was either subjected to PAD4 modification or not. RA51, the serum sample with the highest level of anti-CCP+ antibodies, did not precipitate this modified phage clone (Fig. S1d). Lastly, we confirmed that treatment with PAD enzyme does not interfere with PCR amplification of the library inserts (Fig. S1e). In summary, the phage library can be successfully citrullinated in vitro using recombinant PAD enzyme without loss of viability and without the formation of phage capsid citrulline epitopes, while remaining compatible with downstream amplification for sequencing. A flow chart illustrating this quality control pipeline, which is readily generalizable to other PTM PhIP-Seq studies, is shown in Fig. S2.
We next analyzed negative control (“mock”) IPs that omitted antibody input in order to characterize the background binding of each modified and unmodified library peptide to the protein A/G-coated magnetic beads. The mock IPed libraries were sequenced to a depth of ~20-fold. The mock IPs of the PAD2- and PAD4-modified libraries exhibited an essentially indistinguishable background binding profile compared to the unmodified library (Fig. S3). Conveniently, pairwise comparison of modified and unmodified libraries is therefore not confounded by bias in background binding to the beads, simplifying downstream analyses.
3.3. PAD PhIP-Seq robustly identifies antibodies to citrulline modified epitopes
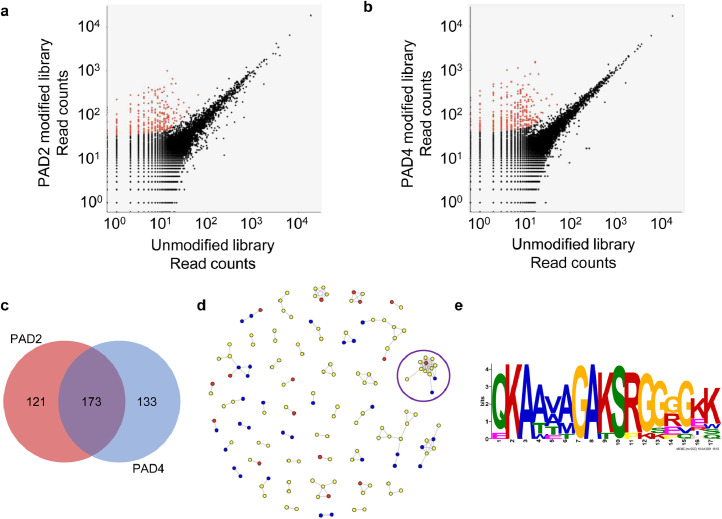
We next sought to detect binding of PAD-dependent peptide epitopes by ACPAs from anti-CCP+ RA sera. Unmodified, PAD2-modified, and PAD4-modified libraries were separately immunoprecipitated with RA51. The PhIP-Seq read count data of RA51 was then subjected to a pairwise analysis pipeline, as described in Methods and presented in pre-print [28], to identify PAD-dependent antibody reactivities. Comparison of the unmodified versus PAD-modified libraries identified 294 PAD2- and 306 PAD4-dependent reactivities (Fig. 2a,b, Table S1). In total, 427 unique peptides were found to exhibit PAD-dependent reactivity; 121 modified peptide reactivities were specific to PAD2, whereas 133 were specific to PAD4. The remaining 173 modified peptide reactivities were shared among both enzymes (Fig. 2c).
Fig. 2.
Profiling PAD-dependent antibody reactivity. a Scatter plot comparison of PAD PhIP-Seq data from separate IPs of the unmodified and the PAD2-citrullinated and b PAD4-citrullinated library using serum from RA51, an anti-CCP+ RA patient. Citrulline-dependent peptide reactivities are colored red. c Pie chart demonstrating the distribution of citrulline-dependent epitopes identified from PAD PhIP-Seq of RA51. d Network graph of all reactive peptides in a and b. Nodes represent reactive peptides from the PAD2 library (red), PAD4 library (blue), or both PAD2 and PAD4 libraries (yellow). Nodes are linked if the peptides share sequence homology. The purple circle indicates the largest cluster. e A multiple sequence alignment logo generated from the largest cluster in d.
All RA51 serum reactive peptides were searched for shared sequences using the shared epitope detection algorithm epitopefindr [29]. The results are visualized as a network graph in which peptides are represented as nodes and a potentially shared epitope as edges (Fig. 2d). Peptides in the largest cluster were then analyzed using the motif discovery software MEME [30], which revealed a conserved sequence modified by both PAD2 and PAD4. The motif features -RGGGGK- and likely contains the citrullinated arginine (Fig. 2e) [30]. Crystallographic data suggests that PAD4 recognizes five successive residues with a ØXRXX consensus where Ø represents amino acids with small side-chains and X denotes any amino acid [31]. This is in agreement with an in silico -RXXXXK- PAD2 model and a -RGXXXX- PAD4 model (with a strong preference for glycine at the +1, +2, and +3 positions [32], as well as the experimentally determined PAD4 citrullination sequences -RG/RGG- of FET proteins [33] and -SGRGK- of histone H2A [34]. PTM PhIP-Seq can thus be used to characterize the substrate specificity of enzymes imparting post-translational modifications.
3.4. Longitudinal analysis of ACPA fine specificities using PAD PhIP-Seq
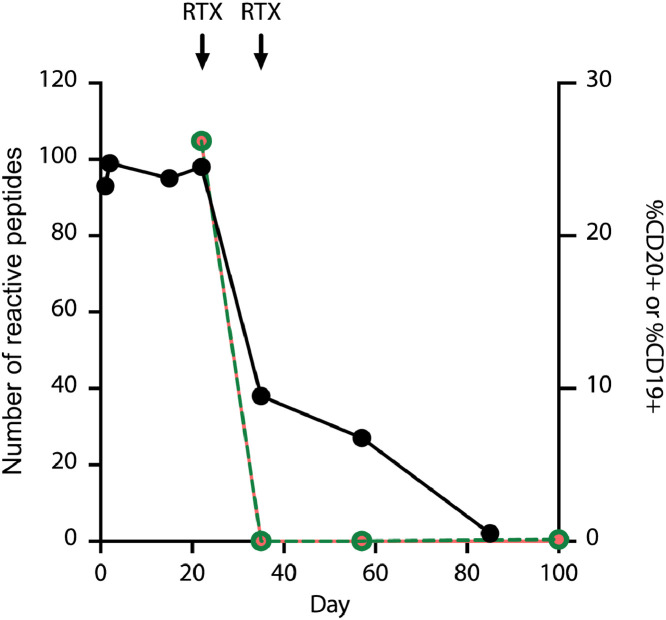
ACPA are expressed at all stages of RA and can precede the onset of clinical disease by over a decade [35]. Clinically, ACPAs are routinely used to aid in the diagnosis of RA and provide prognostic information about disease severity and phenotype (e.g., risk of erosive disease and interstitial lung disease) [36]. Moreover, changes in anti-CCP levels correlate with treatment responses [37]. By monitoring ACPA fine specificities, however, it may be possible to increase the sensitivity and specificity of this diagnostic modality. As a proof of concept, we longitudinally profiled serum from an anti-CCP+ RA patient with high disease activity. PAD4 PhIP-Seq was employed to probe the individual's ACPA repertoire at seven timepoints, both pre- and post-treatment with the B cell depletion therapy, rituximab (Figs. 3, S4). Reactivity profiles obtained prior to treatment harbored consistent levels of PAD4-dependent reactivity, directed at ~100 citrullinated peptides comprising ~80 distinct protein targets. Subsequent to B cell depletion, the number of PAD4-dependent reactivities declined at a rate of approximately 9 peptides per week for 9 weeks (R2 = 0.8 during this time period). This fall in citrulline-dependent antibody levels and diversity was followed by clinical improvement of synovitis as measured by Clinical Disease Activity Index (36 on Day 1 and 10.5 a month after the first rituximab treatment). These data indicate that PAD PhIP-Seq can be used to characterize the temporal evolution of citrulline-dependent autoantibody fine specificities in response to treatment.
Fig. 3.
Longitudinal monitoring of ACPA reactivity in a patient with RA using PAD4 PhIP-seq. The total number of immunoprecipitated citrullinated peptides (black) declines following B cell depletion with rituximab (arrows) starting on Day 22, at which point a steady decline in citrulline-dependent antibody reactivity is observed at a rate of y=-1.3 x + 106.9 (R2=0.8) over the course of 9 weeks. The percentages of CD19+ (magenta) and CD20+ (green) cells of total lymphocytes in peripheral blood are shown on the secondary axis. Immunoprecipitated peptides on days 0-22 are reflective of baseline treatment with methotrexate and glucocorticoids.
3.5. PAD PhIP-Seq positivity is concordant with clinical CCP test results
We next asked how well the PAD PhIP-Seq assay correlates with clinical anti-CCP serostatus. Unmodified, PAD2-modified and PAD4-modified libraries were separately immunoprecipitated using sera from a cohort of 31 RA patients and 10 demographically matched healthy controls. Upon sample unblinding, 20 of 21 anti-CCP+ patients were found to exhibit significant reactivity to a large set of PAD2 and/or PAD4-dependent peptide epitopes (Fig. 4a,b, Table S1). Using a cutoff of ≥15 PAD-dependent reactive peptides, PAD PhIP-Seq positivity was highly concordant with the clinical antigen-agnostic CCP assay; only one anti-CCP+ serum recognized neither PAD2 nor PAD4 PhIP-Seq peptides, and one additional anti-CCP+ serum recognized PAD4 PhIP-Seq peptides, but did not recognize PAD2 PhIP-Seq peptides above threshold (Table S1). It is important to note that the observed discordance may be due to CCP testing and PAD PhIP-Seq analysis not always being performed on the same sample timepoints. None of the sera from the 10 healthy controls, nor that of a Sjogren's Syndrome patient, reacted with PAD PhIP-Seq peptides above threshold (Table S1). In this cohort, the PAD PhIP-Seq assay performed with a sensitivity of 90–95% and a specificity of 100%, when compared to the standard-of-care clinical assay that globally captures ACPAs using artificial CCP peptides.
3.6. PAD PhIP-Seq reveals PAD2- and PAD4-dependent autoantibody reactivity signatures
Small molecule inhibitors of PAD enzymatic activity have emerged as promising candidates for the treatment of RA [18,38,39]. Pan-PAD inhibitors and those with distinct inhibitory profiles against PAD2 and PAD4 have been developed [40]. It is therefore of great interest to determine whether subgroups of RA patients harbor ACPAs that exhibit a discernable preference for PAD2- versus PAD4-modified epitopes. Isoform-preferring antibody signatures, if they exist, may have utility in distinguishing subgroups of individuals more likely to benefit from one class of PAD inhibitors over another.
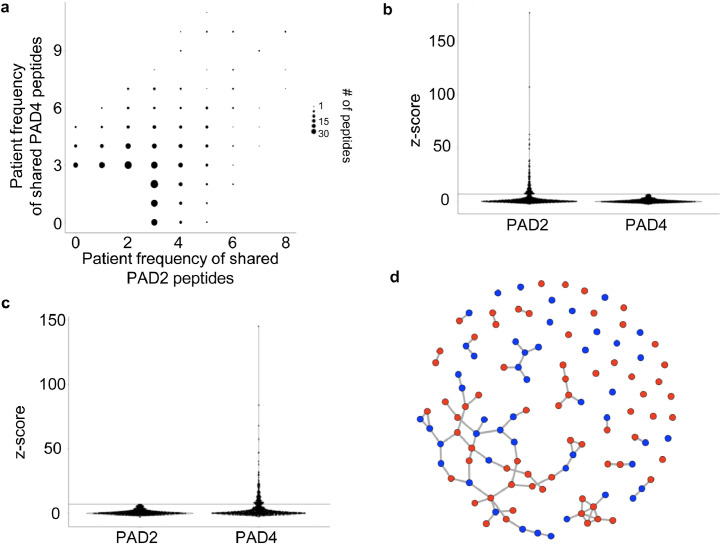
To identify potential PAD2- and PAD4-specific antibody signatures, we hierarchically clustered the anti-CCP+ patient subset based on their reactivity profiles (Fig. 5a). While we observe diverse reactivity to both PAD2- and PAD4-modified peptides, the clustering suggested the existence of distinct, patient-specific reactivity preferences among PAD2 versus PAD4 substrates. A total of 1,161 PAD-dependent reactivities were detected in at least three sera; 303 were unique to PAD2, 272 were unique to PAD4, and 293 peptides were reactive after modification by either enzyme (Fig. 5b). Among these dominant peptides are the known PAD substrates hornerin (HNNR) [41], keratin (KRT4) [42], and collagen (COL2A1) [43]. Other well-known substrates, however, including histone, vimentin, and fibrinogen, were not represented. The absence of these likely reflect a requirement for a higher level of conformational structure to serve as a PAD substrate and/or an antibody epitope, compared to the 90 amino acid peptides presented on the surface of the T7 phage.
Similar to the protein citrullination patterns observed in PAD transfected cells [16,44], many of the dominant peptide reactivities were due to modification by either PAD2 or PAD4. The number of anti-CCP+ individuals reactive to a given PAD modified peptide is shown in Fig. 6a, illustrating peptide seroprevalence as a function of PAD2 versus PAD4 modification. Approximately 2.5% of all peptides were reactive in at least one anti-CCP+ individual. Several peptides, however, were converted into citrullinated epitopes exclusively by one or the other enzyme: 65 by PAD2 and 50 by PAD4 (Table S2). We reasoned that these particular peptides could be used to probe the PAD specificity profile of ACPAs. A second potential application for these peptides would be as sensors for PAD2 or PAD4 enzymatic activity in a biological matrix.
Fig. 6.
Identifying PAD2 and PAD4 reactivity signatures. a Number of anti-CCP+ individuals reactive to a PAD modified peptide. Data is shown for peptides (dots) reactive in at least 3 people. b Distribution of PAD2-specific and c PAD4-specific reactivities for all anti-CCP+ individuals. d Network graph of all PAD2 (red) and PAD4 (blue) specific reactive peptides. Nodes are peptides and are linked if they share sequence similarity.
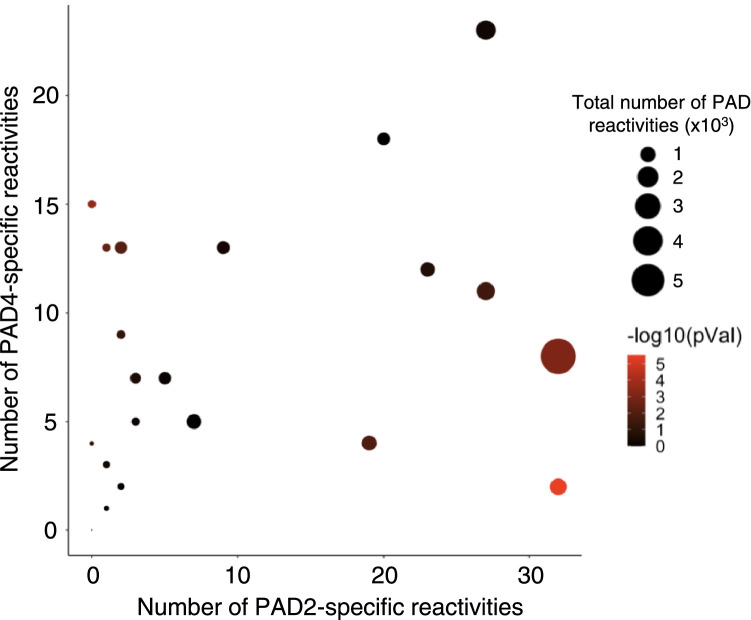
The number of PAD2- and PAD4-specific peptide reactivities across all the anti-CCP+ individuals are shown in Fig. 6b,c. Many peptides are strongly reactive after modification by one, but not the other, PAD enzyme (“mono-reactive”). A shared epitope network graph was used to characterize the degree of sequence homology among these 115 mono-reactive peptides. Approximately 50% of the mono-reactive peptides form nodal networks of >3 edges with the largest network consisting of 42 peptides (Fig. 6d, Fig. S5). A total of 32 mono-reactive peptides remains as singletons and do not align to any other mono-reactive peptide. A comparison of the number of PAD2- versus PAD4-specific reactivities detected in each anti-CCP+ individual is shown in Fig. 7. Overall preference for PAD2 versus PAD4 was quantified using an exact binomial test. A significant differential PAD2/4 “preference” was detected in some anti-CCP+ individuals (Table S3). These data support further investigation into specific PAD isoform-associated ACPA profiles and their potential relevance for prognostication and/or PAD-inhibitor companion diagnostic testing.
Fig. 7.
Number of PAD specific reactivities for all anti-CCP+ individuals. Dot size is proportional to the total number of PAD2 and PAD4 reactivities. Statistical significance was determined using an exact binomial test assuming a hypothesized null probability of 0.5 for independent peptides and is represented as a color gradient from black (not significant) to red (significant).
4. Discussion
Serum antibody profiling can inform understanding of human immune processes, as well as suggest disease-specific antibody biomarkers. Despite advances in highly multiplexed serological analyses, there exists an unmet need for methods that can characterize antibodies to post-translationally modified or haptenized epitopes. Here, we adapted the antibody profiling platform PhIP-Seq to the quantitative analysis of antibody reactivity towards an enzymatically citrullinated human peptidome library. This allowed us to characterize and track the fine specificities of ACPA responses of individual RA patients over time, without bias, in high-throughput and at relatively low cost.
There are important caveats to the use of phage-displayed 90-mer peptides as surrogates for full length proteins. First, peptides may not contain the necessary conformational requirements for binding to ACPAs, thus contributing to false negative results. However, ACPAs are thought to behave to some extent like anti-hapten antibodies such that citrulline is recognized as the primary point of contact [45], while the surrounding amino acids underlie each ACPA's substrate binding profile. This model is supported by a significant monoclonal cross-reactivity among citrullinated substrates [46], and the fact that non-physiologic anti-cyclic citrullinated peptide (CCP) assays perform with high clinical accuracy.
Second, many 90-mer peptides used in this study may not form the necessary secondary or tertiary structure required for appropriate PAD recognition. This could also contribute to false negatives. Non-physiologic availability of normally inaccessible arginine residues, however, may lead to false-positive results. While it is true that PAD enzymes would typically citrullinate fully folded protein substrates initially, in vitro assays indicate that modification of solvent-exposed arginines can lead to a stepwise unfolding that reveals buried arginines, which can be subsequently citrullinated and serve as antibody epitopes [47,48]. This process is thought to contribute to the structural changes associated with aberrant citrullination and autoantigenicity. Therefore, sequential citrulline-associated unfolding of full-length proteins may reveal cryptic arginine residues that are also exposed in the 90-mer peptide library.
Despite such caveats, because the complete RA citrullinome and associated ACPA targets remains unclear, unbiased approaches can serve as powerful hypothesis-generating tools. Novel reactivities of interest should be confirmed by orthogonal assays using full length proteins, preferably expressed in mammalian cells under relevant physiologic conditions.
The use of PTM PhIP-Seq is broadly applicable to studies of diverse PTMs, potentially adaptable to studies of glycosylation [49], carbamylation [50], phosphorylation [51], deamidation [52], and other modifications [6]. PTMs of the phage-displayed human peptidome may also be used to profile PTM-dependent protease substrate preference via the proteome-wide SEPARATE assay [53,54]. PTM PhIP-Seq is therefore a powerful addition to the programmable phage display toolkit with broad utility for genome-wide proteomic analyses.
Contributors
Conceptualization: GDRM, RSQ, MA, ED, HBL; Data curation: GDRM, DRM; Formal analysis: GDRM, DRM, MFK, ED, HBL; Funding acquisition: MA, HBL; Methodology: GDRM, HBL; Resources: JMM, RSQ, MFK, MA, ED, HBL; Software: DRM; Visualization: GDRM, DRM, MFK, ED, HBL; Writing - original draft: GDRM, HBL; Writing - review and editing: DRM, JMM, MFK, MA, ED, HBL. All authors read and approved the final version of the manuscript. GDRM, DRM, MFK, and HBL verified the underlying data.
Declaration of Competing Interest
HBL is a founder of Portal Bioscience, Alchemab and ImmuneID, and is an advisor to TScan Therapeutics. ED is an inventor on a licensed patent (US patent no. 8,975,033) and licensed provisional patent (US patent no. 62/481,158) related to the use of antibodies to PAD3 and PAD2, respectively, in identifying clinically informative disease subsets in RA, has received consulting fees from Celgene and Bristol Myers Squibb, and has received research support from Pfizer, Celgene, and Bristol Myers Squibb outside of this work. MFK has received consulting fees from Celltrion outside of this work. MA is an inventor on a patent (US patent no. 10,415,032) related to detection of antibodies associated with chemical exposure. GDRM, DRM, ED, and HBL are inventors on a provisional patent application related the work described herein.
Acknowledgments
Funding
This research is based upon work supported in part by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA). The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of ODNI, IARPA, or the US Government. The US Government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation therein. This study was made possible by a National Institute of General Medical Sciences (NIGMS) grant R01 GM136724 (HBL). MFK was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grant T32AR048522. ED was supported by the Rheumatology Research Foundation.
Data sharing
Data are available from the corresponding authors upon request.
Acknowledgments
We are grateful to Stephen J. Elledge (Harvard Medical School) for generously providing the human peptidome library used in this study.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103506.
Contributor Information
Erika Darrah, Email: edarrah1@jhmi.edu.
H. Benjamin Larman, Email: hlarman1@jhmi.edu.
Appendix. Supplementary materials
References
- 1.Mohan D., Wansley D.L., Sie B.M., Noon M.S., Baer A.N., Laserson U. PhIP-Seq characterization of serum antibodies using oligonucleotide-encoded peptidomes. Nat Protoc. 2018;13(9):1958–1978. doi: 10.1038/s41596-018-0025-6. Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larman H.B., Laserson U., Querol L., Verhaeghen K., Solimini N.L., Xu G.J. PhIP-Seq characterization of autoantibodies from patients with multiple sclerosis, type 1 diabetes and rheumatoid arthritis. J Autoimmun. 2013;43:1–9. doi: 10.1016/j.jaut.2013.01.013. Jun 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monaco D.R., Sie B.M., Nirschl T.R., Knight A.C., Sampson H.A., Nowak-Wegrzyn A. Profiling serum antibodies with a pan allergen phage library identifies key wheat allergy epitopes. Nat Commun. 2021;12(1):379. doi: 10.1038/s41467-020-20622-1. Jan 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larman H.B., Zhao Z., Laserson U., Li M.Z., Ciccia A., Gakidis M.A.M. Autoantigen discovery with a synthetic human peptidome. Nat Biotechnol. 2011;29(6):535–541. doi: 10.1038/nbt.1856. May 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster R.E. Academic Press; Burlington: 1996. Chapter 1 - Biology of the Filamentous Bacteriophage; pp. 1–20. Kay BK, Winter J, McCafferty JPhage Display of Peptides and Proteins [Internet][cited 2020 Oct 4]Available from: http://www.sciencedirect.com/science/article/pii/B9780124023802500034. [Google Scholar]
- 6.Doyle H.A., Mamula MJ. Autoantigenesis: the evolution of protein modifications in autoimmune disease. Curr Opin Immunol. 2012;24(1):112–118. doi: 10.1016/j.coi.2011.12.003. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen H., James E.A. Immune recognition of citrullinated epitopes. Immunology. 2016;149(2):131–138. doi: 10.1111/imm.12640. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mondal S., Thompson PR. Protein arginine deiminases (PADs): biochemistry and chemical biology of protein citrullination. Acc Chem Res. 2019;52(3):818–832. doi: 10.1021/acs.accounts.9b00024. Mar 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kristyanto H., Blomberg N.J., Slot L.M., Voort E.I.H., van der, Kerkman P.F., Bakker A. Persistently activated, proliferative memory autoreactive B cells promote inflammation in rheumatoid arthritis. Sci Transl Med. 2020;12(570):5327. doi: 10.1126/scitranslmed.aaz5327. [Internet]Nov 18 [cited 2020 Nov 30]Available from: https://stm.sciencemag.org/content/12/570/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinloch A., Lundberg K., Wait R., Wegner N., Lim N.H., Zendman A.J.W. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum. 2008;58(8):2287–2295. doi: 10.1002/art.23618. Aug. [DOI] [PubMed] [Google Scholar]
- 11.Darrah E., Andrade F. Rheumatoid arthritis and citrullination. Curr Opin Rheumatol. 2018;30(1):72–78. doi: 10.1097/BOR.0000000000000452. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Venrooij W.J., van Beers J., Pruijn GJM. Anti-CCP antibodies: the past, the present and the future. Nat Rev Rheumatol. 2011;7(7):391–398. doi: 10.1038/nrrheum.2011.76. Jul. [DOI] [PubMed] [Google Scholar]
- 13.Matsui T., Shimada K., Ozawa N., Hayakawa H., Hagiwara F., Nakayama H. Diagnostic utility of anti-cyclic citrullinated peptide antibodies for very early rheumatoid arthritis. J Rheumatol. 2006;33(12):2390–2397. Dec 1. [PubMed] [Google Scholar]
- 14.Braschi E., Shojania K., Allan GM. Anti-CCP: a truly helpful rheumatoid arthritis test? Can Fam Phys. 2016;62(3):234. Mar. [PMC free article] [PubMed] [Google Scholar]
- 15.Foulquier C., Sebbag M., Clavel C., Chapuy-Regaud S., Al Badine R., Méchin M.C. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56(11):3541–3553. doi: 10.1002/art.22983. Nov. [DOI] [PubMed] [Google Scholar]
- 16.Romero V., Darrah E., Andrade F. Generation of distinct patterns of rheumatoid arthritis autoantigens by peptidylarginine deiminase types 2 and 4 during perforin-induced cell damage. Arthritis Rheumatol Hoboken NJ. 2020;72(6):912–918. doi: 10.1002/art.41196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darrah E., Rosen A., Giles J.T., Andrade F. Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Ann Rheum Dis. 2012;71(1):92–98. doi: 10.1136/ard.2011.151712. Jan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willis V.C., Banda N.K., Cordova K.N., Chandra P.E., Robinson W.H., Cooper D.C. Protein arginine deiminase 4 inhibition is sufficient for the amelioration of collagen-induced arthritis. Clin Exp Immunol. 2017;188(2):263–274. doi: 10.1111/cei.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damgaard D., Bawadekar M., Senolt L., Stensballe A., Shelef M.A., Nielsen CH. Relative efficiencies of peptidylarginine deiminase 2 and 4 in generating target sites for anti-citrullinated protein antibodies in fibrinogen, alpha-enolase and histone H3. PLOS ONE. 2018;13(8) doi: 10.1371/journal.pone.0203214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge C., Xu B., Liang B., Lönnblom E., Lundström S.L., Zubarev R.A. Structural basis of cross-reactivity of anti–citrullinated protein antibodies. Arthritis Rheumatol. 2019;71(2):210–221. doi: 10.1002/art.40698. [DOI] [PubMed] [Google Scholar]
- 21.Steen J., Forsström B., Sahlström P., Odowd V., Israelsson L., Krishnamurthy A. Recognition of amino acid motifs, rather than specific proteins, by human plasma cell–derived monoclonal antibodies to posttranslational modified proteins in rheumatoid arthritis. Arthritis Rheumatol Hoboken NJ. 2019;71(2):196. doi: 10.1002/art.40699. Feb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witalison E.E., Thompson P.R., Hofseth LJ. Protein Arginine deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr Drug Targets. 2015;16(7):700–710. doi: 10.2174/1389450116666150202160954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuzhalin AE. Citrullination in cancer. Cancer Res. 2019;79(7):1274–1284. doi: 10.1158/0008-5472.CAN-18-2797. Apr 1. [DOI] [PubMed] [Google Scholar]
- 24.Kay J., Upchurch KS. ACR/EULAR 2010 rheumatoid arthritis classification criteria. Rheumatology. 2012;51(suppl_6):vi5–vi9. doi: 10.1093/rheumatology/kes279. Dec 1. [DOI] [PubMed] [Google Scholar]
- 25.Andrade F., Darrah E., Gucek M., Cole R.N., Rosen A., Zhu X. Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010;62(6):1630–1640. doi: 10.1002/art.27439. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darrah E., Giles J.T., Davis R.L., Naik P., Wang H., Konig M.F. Autoantibodies to peptidylarginine deiminase 2 are associated with less severe disease in rheumatoid arthritis. Front Immunol. 2018;9:2696. doi: 10.3389/fimmu.2018.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu G.J., Shah A.A., Li M.Z., Xu Q., Rosen A., Casciola-Rosen L. Systematic autoantigen analysis identifies a distinct subtype of scleroderma with coincident cancer. Proc Natl Acad Sci U S A. 2016;113(47):E7526–E7534. doi: 10.1073/pnas.1615990113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monaco D.R., Kottapalli S.V., Yuan T., Breitwieser F.P., Anderson D.E., Wijaya L. Deconvoluting virome-wide antiviral antibody profiling data. bioRxiv. 2018;30 [Google Scholar]
- 29.Minimal Overlaps from BLAST Alignments [Internet]. [cited 2020 Mar 24]. Available from: https://brandonsie.github.io/epitopefindr/
- 30.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L. MEME suite: tools for motif discovery and searching. Nucl Acids Res. 2009;37(suppl_2):W202–W208. doi: 10.1093/nar/gkp335. Jul 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arita K., Shimizu T., Hashimoto H., Hidaka Y., Yamada M., Sato M. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci. 2006;103(14):5291–5296. doi: 10.1073/pnas.0509639103. Apr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson J.S., Lubner J.M., Meyer D.J., Grant JE. An in silico analysis of primary and secondary structure specificity determinants for human peptidylarginine deiminase types 2 and 4. Comput Biol Chem. 2017;70:107–115. doi: 10.1016/j.compbiolchem.2017.08.001. Oct. [DOI] [PubMed] [Google Scholar]
- 33.Tanikawa C., Ueda K., Suzuki A., Iida A., Nakamura R., Atsuta N. Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 2018;22(6):1473–1483. doi: 10.1016/j.celrep.2018.01.031. Feb. [DOI] [PubMed] [Google Scholar]
- 34.Hagiwara T., Hidaka Y., Yamada M. Deimination of histone H2A and H4 at arginine 3 in HL-60 granulocytes. Biochemistry. 2005;44(15):5827–5834. doi: 10.1021/bi047505c. Apr 1. [DOI] [PubMed] [Google Scholar]
- 35.Curran A.M., Naik P., Giles J.T., Darrah E. PAD enzymes in rheumatoid arthritis: pathogenic effectors and autoimmune targets. Nat Rev Rheumatol. 2020;16(6):301–315. doi: 10.1038/s41584-020-0409-1. Jun. [DOI] [PubMed] [Google Scholar]
- 36.Derksen VF a M., Huizinga T.W.J., Woude D.V.D. The role of autoantibodies in the pathophysiology of rheumatoid arthritis. Semin Immunopathol. 2017;39(4):437. doi: 10.1007/s00281-017-0627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iannone F., Tampoia M., Giannini M., Lopalco G., Cantarini L., Villalta C.D. Changes in anti-cyclic citrullinated peptide antibodies and rheumatoid factor isotypes serum levels in patients with rheumatoid arthritis following treatment with different biological drugs. Clin Exp Rheumatol. 2016;34(3):424–429. Jun. [PubMed] [Google Scholar]
- 38.Willis V.C., Gizinski A.M., Banda N.K., Causey C.P., Knuckley B., Cordova K.N. N-α-benzoyl-N5-(2-chloro-1-iminoethyl)-L-ornithine amide, a protein arginine deiminase inhibitor, reduces the severity of murine collagen-induced arthritis. J Immunol. 2011;186(7):4396–4404. doi: 10.4049/jimmunol.1001620. Apr 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawalkowska J., Quirke A.M., Ghari F., Davis S., Subramanian V., Thompson P.R. Abrogation of collagen-induced arthritis by a peptidyl arginine deiminase inhibitor is associated with modulation of T cell-mediated immune responses. Sci Rep. 2016;6:26430. doi: 10.1038/srep26430. May 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muth A., Subramanian V., Beaumont E., Nagar M., Kerry P., McEwan P. Development of a selective inhibitor of protein arginine deiminase 2. J Med Chem. 2017;60(7):3198–3211. doi: 10.1021/acs.jmedchem.7b00274. Apr 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu C.Y., Gasc G., Raymond A.-.A., Burlet-Schiltz O., Takahara H., Serre G. Deimination of human hornerin enhances its processing by calpain-1 and its cross-linking by transglutaminases. J Invest Dermatol. 2017;137(2):422–429. doi: 10.1016/j.jid.2016.09.030. Feb 1. [DOI] [PubMed] [Google Scholar]
- 42.Chang X., Jian X., Yan X. Expression and citrullination of keratin in synovial tissue of rheumatoid arthritis. Rheumatol Int. 2009;29(11):1337–1342. doi: 10.1007/s00296-009-0863-1. Sep 1. [DOI] [PubMed] [Google Scholar]
- 43.Yoshida M., Tsuji M., Kurosaka D., Kurosaka D., Yasuda J., Ito Y. Autoimmunity to citrullinated type II collagen in rheumatoid arthritis. Mod Rheumatol. 2006;16(5):276–281. doi: 10.1007/s10165-006-0498-y. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Assohou-Luty C., Raijmakers R., Benckhuijsen W.E., Stammen-Vogelzangs J., de Ru A., van Veelen P.A. The human peptidylarginine deiminases types 2 and 4 have distinct substrate specificities. Biochim Biophys Acta. 2014;1844(4):829–836. doi: 10.1016/j.bbapap.2014.02.019. Apr. [DOI] [PubMed] [Google Scholar]
- 45.Trejo-Zambrano M.I., Gómez-Bañuelos E., Andrade F. Redox-mediated carbamylation as a hapten model applied to the origin of antibodies to modified proteins in rheumatoid arthritis. Antioxid Redox Signal. 2021 doi: 10.1089/ars.2021.0064. https://www.liebertpub.com/doi/10.1089/ars.2021.0064 Ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kongpachith S., Lingampalli N., Ju C.H., Blum L.K., Lu D.R., Elliott S.E. Affinity maturation of the anti-citrullinated protein antibody paratope drives epitope spreading and polyreactivity in rheumatoid arthritis. Arthritis Rheumatol Hoboken NJ. 2019;71(4):507–517. doi: 10.1002/art.40760. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarcsa E., Marekov L.N., Mei G., Melino G., Lee S.-.C., Steinert PM. Protein unfolding by peptidylarginine deiminase: substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin*. J Biol Chem. 1996;271(48):30709–30716. doi: 10.1074/jbc.271.48.30709. Nov 29. [DOI] [PubMed] [Google Scholar]
- 48.Travers T.S., Harlow L., Rosas I.O., Gochuico B.R., Mikuls T.R., Bhattacharya S.K. Extensive citrullination promotes immunogenicity of HSP90 through protein unfolding and exposure of cryptic epitopes. J Immunol Baltim Md 1950. 2016;197(5):1926–1936. doi: 10.4049/jimmunol.1600162. Sep 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bäcklund J, Treschow A, Bockermann R, Holm B, Holm L, Issazadeh-Navikas S, Kihlberg J. and Holmdahl R. (2002), Glycosylation of type II collagen is of major importance for T cell tolerance and pathology in collagen-induced arthritis. Eur. J. Immunol, 32: 3776-3784. 10.1002/1521-4141(200212)32 [DOI] [PubMed]
- 50.Gan R.W., Trouw L.A., Shi J., Toes R.E.M., Huizinga T.W.J., Demoruelle M.K. Anti-carbamylated protein antibodies are present prior to rheumatoid arthritis and are associated with its future diagnosis. The Journal of Rheumatology. 2015 doi: 10.3899/jrheum.140767. https://www.jrheum.org/content/early/2015/01/07/jrheum.140767 [Internet]Jan 15 [cited 2020 Nov 15]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neugebauer K.M., Merrill J.T., Wener M.H., Lahita R.G., Roth MB. SR proteins are autoantigens in patients with systemic lupus erythematosus. Importance of phosphoepitopes. Arthritis Rheum. 2000;43(8):1768–1778. doi: 10.1002/1529-0131(200008)43:8<1768::AID-ANR13>3.0.CO;2-9. Aug. [DOI] [PubMed] [Google Scholar]
- 52.Caja S., Mäki M., Kaukinen K., Lindfors K. Antibodies in celiac disease: implications beyond diagnostics. Cell Mol Immunol. 2011;8(2):103–109. doi: 10.1038/cmi.2010.65. Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Román-Meléndez G.D., Venkataraman T., Monaco D.R., Larman H.B. Protease activity profiling via programmable phage display of comprehensive proteome-scale peptide libraries. Cell Syst. 2020;11(4):375–381.e4. doi: 10.1016/j.cels.2020.08.013. Oct 21. [DOI] [PubMed] [Google Scholar]
- 54.Zhou J., Li S., Leung K.K., O'Donovan B., Zou J.Y., DeRisi J.L. Deep profiling of protease substrate specificity enabled by dual random and scanned human proteome substrate phage libraries. Proc Natl Acad Sci. 2020;117(41):25464–25475. doi: 10.1073/pnas.2009279117. Oct 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.