Abstract
Many cellular stimuli result in the induction of both the tumor suppressor p53 and NF-κB. In contrast to activation of p53, which is associated with the induction of apoptosis, stimulation of NF-κB has been shown to promote resistance to programmed cell death. These observations suggest that a regulatory mechanism must exist to integrate these opposing outcomes and coordinate this critical cellular decision-making event. Here we show that both p53 and NF-κB inhibit each other’s ability to stimulate gene expression and that this process is controlled by the relative levels of each transcription factor. Expression of either wild-type p53 or the RelA(p65) NF-κB subunit suppresses stimulation of transcription by the other factor from a reporter plasmid in vivo. Moreover, endogenous, tumor necrosis factor alpha-activated NF-κB will inhibit endogenous wild-type p53 transactivation. Following exposure to UV light, however, the converse is observed, with p53 downregulating NF-κB-mediated transcriptional activation. Both p53 and RelA(p65) interact with the transcriptional coactivator proteins p300 and CREB-binding protein (CBP), and we demonstrate that these results are consistent with competition for a limiting pool of p300/CBP complexes in vivo. These observations have many implications for regulation of the transcriptional decision-making mechanisms that govern cellular processes such as apoptosis. Furthermore, they suggest a previously unrealized mechanism through which dysregulated NF-κB can contribute to tumorigenesis and disease.
To respond and adapt to changes in its external environment, such as exposure to growth factors and cytokines or stress-inducing stimuli such as ionizing radiation, the cell must initiate a program of regulated gene expression. This response can vary and will depend on many factors such as the cell type and the number and strength of the new stimuli. To a large extent these changes are mediated by nuclear DNA-binding proteins which function in a combinatorial manner to activate or repress the transcription of target genes. An example of such a transcription factor is NF-κB, which functions as one of the principal regulators of the cellular response to stress and infection (3, 56, 64).
In most unstimulated cells, NF-κB is found in an inactive cytoplasmic form bound to its inhibitory protein, IκB (3, 64). Upon cellular stimulation with, for example, cytokines, mitogens, growth factors, and ionizing radiation, IκB is rapidly phosphorylated, which in most cases targets it for ubiquitination and subsequent degradation by the proteasome (32, 45, 68, 74). This releases NF-κB, allowing it to translocate to the nucleus, where it can induce the expression of a large number of cytokines, adhesion molecules, immunoreceptors, and other transcription factors (3, 64). In mammalian cells, there are five distinct NF-κB subunits, NF-κB1(p105/p50), NF-κB2(p100/p52), RelA(p65), RelB, and c-Rel, all of which contain a highly conserved amino-terminal DNA-binding and dimerization domain of approximately 300 amino acids, the Rel homology domain (3, 64). Of these subunits, p105 and p100 are larger precursor proteins that, as a result of IκB-like ankyrin repeat sequences in their carboxy termini, are retained in the cytoplasm and require proteolytic processing to generate their active nuclear forms, p50 and p52, respectively. RelA, RelB, and c-Rel require no additional processing and, unlike p50 and p52, contain transactivation domains in their carboxy termini. Thus, NF-κB can, with some exceptions, be composed of homo- and heterodimers formed from a variety of combinations of these proteins. Similarly, there exist at least three IκB proteins, α, β, and ɛ, although full-length p105 and p100 can also function to retain NF-κB subunits in the cytoplasm (3, 56, 64).
This complexity is utilized by the cell to control the specificity and selectivity of the genes regulated by NF-κB (56). Different NF-κB complexes have subtly different DNA-binding specificities which can result in binding to variant κB elements within different promoters and enhancers. In addition, the distinct properties of different IκB proteins or IκB–NF-κB complexes can result in the selective nuclear localization of NF-κB subunits. These differences cannot account for the overall complexity and specificity of the NF-κB response, however. Many genes are activated by only a subset of NF-κB inducers, while others will be expressed in only certain cell types. To a large extent, this can be accounted for by the interaction of NF-κB subunits with heterologous DNA-binding proteins: NF-κB has been reported to interact with a wide variety of basic region-leucine zipper-containing transcription factors such as C/EBPβ and AP-1 (Fos/Jun), the zinc finger-containing protein Sp1, and others such as serum response factor (56). These interactions, which typically occur as a result of the specific juxtaposition of their respective DNA-binding sites, bring added specificity to the genes which can be induced by NF-κB.
In addition to these interactions with DNA-binding proteins, the association of NF-κB subunits with non-DNA-binding coactivator proteins and components of the basal transcription complex confers another level of regulation on NF-κB-mediated induction of transcription. Among these reported interactions, NF-κB, similar to a wide variety of other inducible regulatory transcription factors, complexes with the coactivator proteins p300 and CREB-binding protein (CBP) (21, 58). This interaction occurs through multiple domains in both proteins and can be regulated, in part, through phosphorylation of RelA by IκB-associated protein kinase A (87). In addition, RelA-mediated transactivation can also be regulated by the cyclin-dependent kinase inhibitor p21WAF1/CIP1, a result that correlates with the direct association of p300 and CBP with specific cyclin–cyclin-dependent kinase complexes (58). This regulatory mechanism is most likely of particular relevance to NF-κB function under a subset of conditions where NF-κB activation occurs simultaneously with the induction of p21. These would include the phorbol ester-induced differentiation of HL-60 cells into macrophages, a process dependent on both the activation of NF-κB and the induction of p21 (18, 33), or cellular stimulation with ionizing radiation (15, 19, 20, 76). p21 is one of the principal target genes for the p53 tumor suppressor transcription factor, and its activation is associated strongly with the induction of cell cycle arrest following DNA damage (14, 19, 20). p53 has also been demonstrated to interact with p300 and CBP (2, 24, 43, 66), an observation which suggests that p53 might regulate itself indirectly through a positive feedback loop in which induction of p21 enhances p300/CBP activity, thus further stimulating p53 transactivation.
An implication of this putative regulatory mechanism would be that p53 could, through induction of p21, indirectly stimulate the activity of NF-κB and other transcription factors utilizing p300 and CBP. The functional consequences of induction of p53 and NF-κB are generally divergent, however. Activation of p53 is associated with the induction of apoptosis or cell cycle arrest (26, 42). Conversely, induction of NF-κB is generally associated with the promotion of resistance to programmed cell death (6, 9, 36, 44, 46, 73, 76, 81). The RelA-knockout mouse dies before birth as a result of extensive liver apoptosis (7). Fibroblasts derived from these mice show enhanced death as a result of tumor necrosis factor alpha (TNF) stimulation, a result which can be reversed by expression of RelA (6). Furthermore, inhibition of NF-κB activation enhances cell death in response to a number of stimuli, including ionizing radiation and treatment with daunorubicin, a cancer chemotherapeutic compound (76). Additionally, inhibition of constitutively active NF-κB in B cells results in cell death (81). While NF-κB has been shown to induce genes that promote resistance to apoptosis, such as A20, IEX-1L, TRAF1, TRAF2, IAP1, and IAP2 (38, 77, 82), the precise mechanisms surrounding this regulatory process have again yet to be clearly defined. Underlining this divergence of the cellular roles of NF-κB and p53, it is interesting that while many viruses encode proteins that specifically induce transcriptionally active NF-κB, most if not all oncogenic viruses also encode proteins that inactivate p53 function (3, 26, 42, 64). Consistent with the concept that p53 and NF-κB may functionally regulate each other, however, are the observations that both factors are coordinately induced by a number of similar cellular stimuli. These include DNA damage as a result of treatment with ionizing radiation (15, 26, 37, 42, 64, 76) and also stimulation by TNF (17, 25, 50, 55).
These observations all suggest that cellular regulatory mechanisms must exist to integrate the distinct functional roles inherent in NF-κB and p53 activation. We have therefore investigated whether NF-κB and p53 transcriptionally cross-regulate each other’s activity. Interestingly, we find that the transcriptional activities of both factors are governed by their relative levels of expression: NF-κB inhibits p53-dependent transactivation, while p53 expression can also suppress NF-κB transcriptional activity, a result that contradicts the suggestion that p53 might stimulate NF-κB activity indirectly through induction of p21 but is in agreement with the divergent cellular outcomes of their induction. We demonstrate that endogenous, TNF-activated NF-κB can also inhibit p53 transactivation and that this mutual repression mechanism is consistent with a model in which both factors are competing for a limiting pool of p300/CBP coactivator protein complexes. We suggest that this regulatory mechanism contributes to cellular decision-making processes governing programmed cell death and has implications for the role of NF-κB in tumorigenesis.
MATERIALS AND METHODS
Antibodies.
Rabbit polyclonal RelA (sc-109) and p300 (sc-584) antibodies were obtained from Santa Cruz Biotechnology. Anti-p300 antibody 14991A was obtained from Pharmingen. Anti-p53 monoclonal antibody DO1 was kindly provided by Alison Sparks and David Lane (University of Dundee).
Cell culture and stimulations.
All cells were cultured as monolayers in Dulbecco modified Eagle medium supplemented with 10% filter (0.2-μm-pore-size)-sterilized fetal calf serum, 5 mM l-glutamine, 100 U of penicillin per ml, and 100 mg of streptomycin per ml. For UV stimulation, medium was removed from culture dishes and cells were exposed to 254-nm UV light (35 J/m2) in a Stratalinker (Stratagene). Cells were then cultured for various periods of time, and both adherent and detached cells were harvested for subsequent analysis. For TNF-mediated induction of NF-κB, cells were incubated with recombinant human TNF (10 ng/ml; Sigma or ICN) for 24 h.
Plasmids.
The multimerized p53 binding site reporter plasmid PG13 CAT, cytomegalovirus (CMV)-based plasmids expressing human p53 (wild type and Arg 175-to-His and Arg 248-to-Trp mutants) and mouse p53 (wild type and an amino-terminal deletion encoding amino acids 44 to 393) (referred to as CMV p53 plasmids), and pT7 p53 (for in vitro transcription-translation) were all supplied by Carol Midgley and David Lane (University of Dundee). CMV Mdm2, originally from the Vogelstein laboratory (Johns Hopkins), and a mutant with an amino-terminal 63-amino-acid deletion of Mdm2 that no longer interacts with p53, CMV Mdm2ΔXM, were also supplied by Carol Midgley and David Lane. The human Bax promoter-chloramphenicol acetyltransferase (CAT) reporter plasmid (Bax CAT) was constructed by Louise Copeland (University of Dundee) from pGL3 Bax luciferase, which originated in John Reed’s laboratory (Burnham Institute, La Jolla, Calif.) and was supplied by Tim Crook (Institute of Cancer Research, London, England). The luciferase cDNA was excised from pGL3 Bax luciferase by digestion with HpaI and SacI and replaced by an HpaI/SacI fragment containing the CAT cDNA from the BCAT reporter plasmid. The Rous sarcoma virus (RSV) RelA(p65), p50, and c-Rel expression plasmids have all been reported previously (57, 59), as have the 4× HIV κB CAT and 2× κB CAT reporter plasmids (41, 57) and transdominant negative IκBα plasmid (80), supplied by Bei-Yue Wu and Gary Nabel (University of Michigan). Plasmids Gal4 CAT and Gal4 VP16 were provided by Tom Glaser and Greg Dressler (University of Michigan). RSV CBP was provided by Richard Goodman (Vollum Institute, Portland, Oreg.).
Calcium phosphate transient transfections and reporter gene assays.
Cells growing in log phase were plated into 90-mm-diameter petri dishes at 50% confluency, unless otherwise indicated, 1 to 2 h prior to transfection to allow the cells to adhere. All transfection solutions were equilibrated to room temperature. A total of 10 to 15 μg of cesium chloride-purified, supercoiled plasmid DNA was diluted into 438 μl of water containing 61 μl of 2 M CaCl2 and added in drops with gentle agitation to 500 μl of 2× HEPES-buffered saline (0.274 M NaCl, 1.5 mM Na2HPO4, 54.6 mM HEPES [pH 7.1]). The DNA, CaCl2, and HEPES-buffered saline were mixed by pipetting and then immediately sprinkled onto the cells, ensuring that the whole dish was covered. Following a 24-h incubation, the precipitate was removed and fresh complete medium was added to the cells. For UV stimulation of transfected human foreskin fibroblasts (HFF cells), cells were exposed to precipitate for 7 to 10 h and allowed to recover overnight before stimulation the following morning. SAOS2 cells were split the day before transfection and incubated with CaPO4-DNA precipitate for only 8 h. Transfected cells were harvested at 48 h unless otherwise stated, and CAT activity was assayed on 10 to 100 μg of protein from whole-cell extracts.
Immunoprecipitation and in vitro association assays.
Nuclear extracts were prepared as described elsewhere (16). The p300-RelA interaction assay in Fig. 6B was performed essentially as described previously (58), using RelA protein synthesized in NF-κB-depleted TNT reticulocyte (Promega) extract and p300 immunoprecipitated from nuclear extracts. To determine competition between p53 and RelA for p300 binding, 200 ng of purified baculovirus-expressed recombinant mouse p53 (a gift from Ted Hupp, University of Dundee) was added simultaneously with 35S-labeled RelA.
FIG. 6.
RelA and p53 bind competitively to p300 and CBP. (A) p53 binds p300 and CBP in vitro. [35S]methionine-labeled, in vitro-translated p53 was incubated with either p300, CBP, or an IgG control immunoprecipitated from 293 cell extracts as described previously (58). After incubation and further washing, the complexes were resolved by SDS-PAGE and subjected to autoradiography. (B) Purified recombinant p53 blocks the interaction of RelA with p300 in vitro. [35S]methionine-labeled, in vitro-translated RelA was incubated with antibody-bound complexes of p300 immunoprecipitated from 293 cell extracts as described previously (58), together with 200 ng of purified p53 as indicated. After incubation and further washing, the complexes were resolved by SDS-PAGE and subjected to autoradiography. I, sample of input [35S]methionine-labeled RelA. (C) p53 blocks the interaction of RelA with p300 in nuclear extracts. 293 cells were transfected with 3 μg of RSV RelA and the indicated quantities of CMV p53. Nuclear extracts were prepared, and p300 was immunoprecipitated. The immunoprecipitates were then resolved by SDS-PAGE and immunoblotted for RelA (lanes 1 to 3). Samples of each nuclear extract were similarly immunoblotted to demonstrate equivalent starting levels of RelA (lanes 4 to 6). (D) RelA does not interact directly with p53. RelA was immunoprecipitated from nuclear extracts prepared from 293 cells transfected with RelA expression plasmid. The RelA complex (lanes 2 and 4) or control IgG-bound complex (lanes 1 and 3) were then incubated with wheat germ lysate-translated, [35S]methionine-labeled p53 or IκBα as indicated. After washing, bound proteins were resolved by SDS-PAGE, and the gel was dried and subjected to autoradiography.
To determine whether RelA and p53 bind p300 competitively in nuclear extracts (Fig. 6C), p300 was immunoprecipitated from 293 cells transfected with RelA alone or p53 expression plasmids.
To determine whether RelA interacts with p53 (Fig. 6D), IκBα and p53 proteins were 35S labeled and synthesized by in vitro translation reactions using a T7 TNT wheat germ kit (Promega). One microgram of RelA or rabbit immunoglobulin G (IgG) control antibody and 200 μg of nuclear extract from RelA-transfected 293 cells were mixed at 4°C for 2 h prior to the addition of 10 μl (packed volume) of protein G-agarose beads for a further 1 h. The beads were washed three times in low-stringency immunoprecipitation (IP) buffer containing 20 mM HEPES (pH 7.9), 75 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 0.1% Nonidet P-40, and the protease inhibitors phenylmethylsulfonyl fluoride, leupeptin, aprotinin, and pepstatin A. The beads were incubated for a further hour with 5 μl of in vitro-translated protein in 100 μl of IP buffer. Complexes were washed a further three times in IP buffer, and antibody-bound complexes were eluted by boiling in 2× sodium dodecyl sulfate (SDS) sample buffer. Supernatants were resolved on an SDS–10% polyacrylamide gel, and proteins were visualized by autoradiography.
Protein immunoblotting for p53 and RelA.
Immunoblot analysis of transfected or TNF-treated cells was performed on either nuclear or whole-cell extracts. Whole-cell extracts were prepared by direct lysis of cells into SDS-gel loading buffer. Equivalent amounts of soluble protein were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) (10% gel) and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 10% milk in Tris-buffered saline (TBS) containing 0.5% Tween 20 (TBS-Tween) for 10 min at room temperature and then incubated with primary antibody in TBS– Tween–5% dried milk for 2 h at room temperature or overnight at 4°C. Following two 15-min washes in TBS-Tween containing 500 mM NaCl (high-stringency TBS), membranes were incubated for 1 h with appropriate horseradish peroxidase-conjugated secondary antibody in TBS–Tween–5% dried milk. After two more 15-min washes in high-stringency TBS-Tween, the immunoreactive proteins were visualized by enhanced chemiluminescence (Amersham).
RESULTS
Inhibition of p53-mediated transcriptional activation by NF-κB.
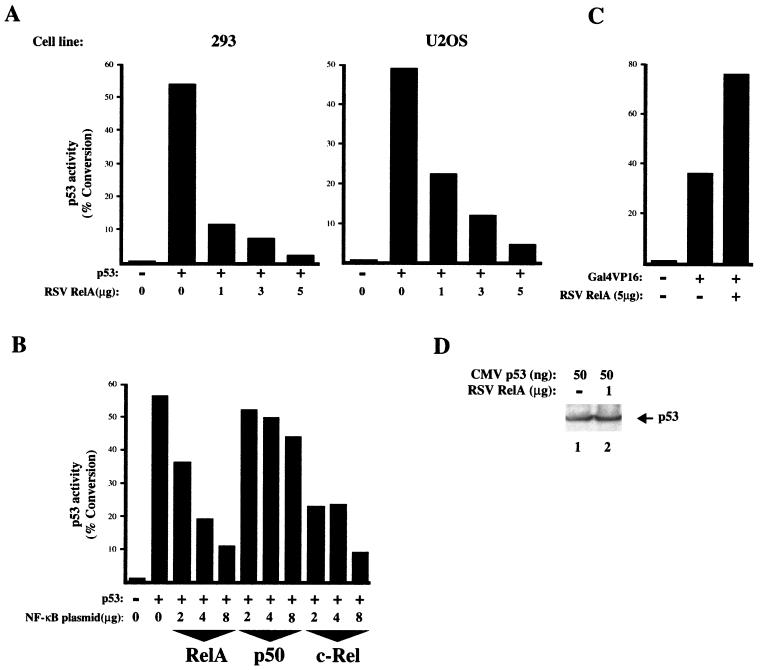
To determine whether NF-κB might affect p53 function, a p53 reporter plasmid containing the Bax promoter (Bax CAT), a known p53 target gene associated with the induction of apoptosis (49), was cotransfected with a fixed level of p53 expression plasmid and various amounts of RelA expression plasmid. As expected, p53 strongly transactivated this reporter in both 293 cells and the human osteosarcoma U2OS cell line (Fig. 1A). Interestingly, cotransfection of RelA strongly suppressed p53 transactivation of this reporter plasmid in both cell lines in a dose-dependent manner (Fig. 1A). RelA-mediated repression of p53 was also seen when a reporter plasmid containing only multiple copies of the p53 DNA-binding site, PG13 CAT, was used (data not shown). The roles of different NF-κB subunits were also examined. Cotransfection of a p50 NF-κB expression plasmid into U2OS cells had little effect on p53 transactivation, while c-Rel strongly inhibited p53, demonstrating that this effect is a general feature of transactivation domain containing NF-κB subunits and is not limited to RelA (Fig. 1B). Similar to transfection of RelA alone, cotransfection of p50 and RelA expression plasmids also suppressed p53 (data not shown). To control for the specificity of these observations, the effect of RelA on a Gal4-VP16 transactivator was analyzed. In contrast to p53, no repression of Gal4-VP16 by RelA was observed; indeed, a moderate increase in its transactivation was seen (Fig. 1C). Western blot analysis confirmed that levels of p53 remained unchanged in RelA-transfected 293 cells (Fig. 1D).
FIG. 1.
NF-κB represses p53 transactivation. (A) RelA represses p53 activation of the Bax promoter. 293 or U2OS cells were transfected with 5 μg of Bax CAT reporter plasmid, RSV RelA expression plasmid, and either 50 ng (293 cells) or 25 ng (U2OS cells) of CMV p53 expression plasmid as indicated. CMV and RSV controls were included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were harvested 48 h after transfection, and a CAT assay was performed. The results shown are representative of at least five separate experiments. (B) Repression of p53 by RelA and c-Rel but not p50. U2OS cells were transfected with 5 μg of Bax CAT reporter plasmid, RSV RelA, c-Rel, or p50 expression plasmid, and 50 ng of CMV p53 expression plasmid as indicated. CMV and RSV controls were included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were harvested 48 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (C) Gal4-VP16 is not suppressed by RelA. U2OS cells were transfected with 5 μg of Gal4 CAT reporter plasmid, 5 μg of RSV RelA expression plasmids, and 5 ng of Gal4 VP16 p53 reporter plasmid as indicated. Gal4 and RSV controls were included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were harvested 48 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (D) Western blot analysis showing that p53 levels are unaffected by cotransfected RelA. 293 cells were cotransfected with the indicated levels of CMV p53 and RSV RelA expression plasmids. After 48 h, whole-cell lysates were prepared, resolved on an SDS–10% polyacrylamide gel, and transferred to PVDF. p53 levels were determined by using the anti-p53 monoclonal antibody DO1.
Inhibition of RelA transcriptional activity by p53.
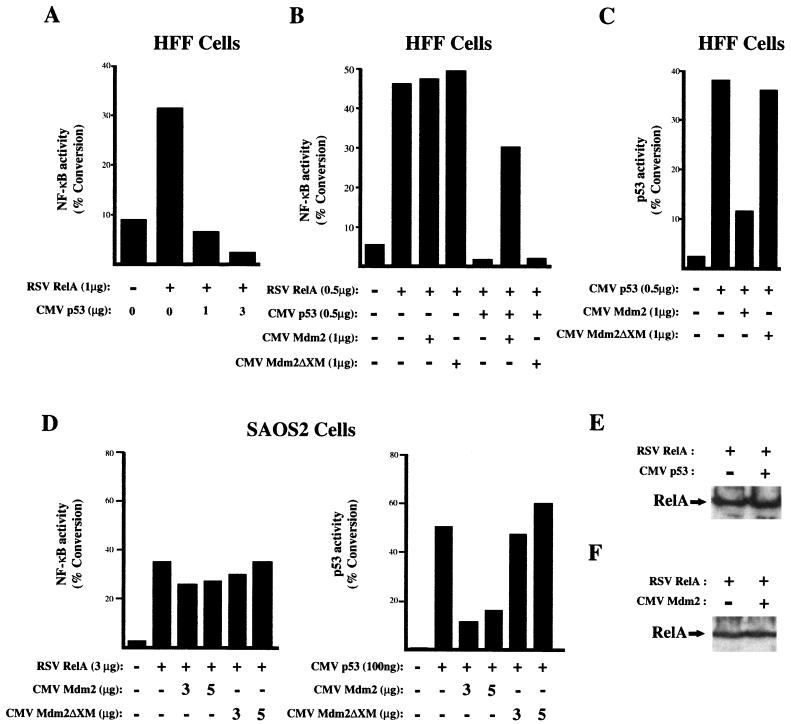
To determine whether p53 could also affect NF-κB-mediated transactivation, experiments were performed with a reporter plasmid containing multiple κB elements (4× κB CAT), a fixed level of RelA expression plasmid, and various quantities of the p53 expression plasmid cotransfected into HFF cells. Interestingly, repression of RelA-mediated transactivation was now observed (Fig. 2A). This effect was reversed by the additional coexpression of Mdm2, an inhibitor of p53 transcriptional activation (39) that binds to its amino-terminal transactivation domain (Fig. 2B and C). Since Mdm2 can interact with cellular proteins other than p53, a mutant with an amino-terminal deletion of Mdm2, Mdm2ΔXM, that abolishes this interaction was also used. This mutant was unable to reverse inhibition of RelA by p53 (Fig. 2B) and did not inhibit p53 transactivation (Fig. 2C). Furthermore, confirming that the effect of Mdm2 on RelA resulted from inhibition of p53, Mdm2 had no effect on RelA-mediated transactivation in the p53-null SAOS2 cell line (Fig. 2D). Recently, Mdm2 has also been shown to be able to interact with p300/CBP (23). The ΔXM mutant of Mdm2 still contains the p300/CBP-binding region of this protein, however, and thus it can be concluded that Mdm2 binding to p300/CBP does not affect RelA transactivation. Western blot analysis confirmed that p53 and Mdm2 did not affect the expression of RelA (Fig. 2E and F). In addition, cell viability assays indicated that transfected p53 in these experiments was not toxic, and thus the inhibition of RelA transactivation seen does not result from cell death (data not shown).
FIG. 2.
p53 represses RelA-mediated transactivation. (A) p53 represses RelA activation of a κB reporter plasmid. HFF cells were transfected with 3 μg of 4× κB CAT reporter plasmid, 1 μg of RSV RelA expression plasmid, and CMV human p53 expression plasmid as indicated. CMV and RSV controls were included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were harvested 36 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (B) Mdm2 blocks p53-mediated repression of RelA. HFF cells were transfected as described above except that CMV Mdm2 or CMV Mdm2ΔXM expression plasmid was included as indicated. (C) Mdm2 inhibits p53-mediated transactivation. HFF cells were transfected as for panel B except that RelA plasmid was excluded and the PG13 CAT p53 reporter plasmid was used in place of 4× κB CAT. (D) Mdm2 does not affect RelA-mediated transactivation in p53-null SAOS2 cells. SAOS2 cells were transfected with CMV Mdm2 or CMV Mdm2ΔXM expression plasmid as indicated, together with either RelA and 2× κB CAT reporter plasmid or p53 and Bax CAT reporter plasmid. (E) Western blot analysis showing that RelA levels are unaffected by cotransfected p53. HFF cells were cotransfected with 3 μg of CMV p53 and 1 μg of RSV RelA expression plasmids. After 36 h, whole-cell lysates were prepared, resolved on an SDS–10% polyacrylamide gel, and transferred to PVDF before incubation with anti-RelA antibody. (F) Western blot analysis showing that RelA levels are unaffected by cotransfected Mdm2. HFF cells were cotransfected with 3 μg of CMV Mdm2 and 1 μg of RSV RelA expression plasmids. After 36 h, whole-cell lysates were prepared, resolved on an SDS–10% polyacrylamide gel, and transferred to PVDF before incubation with anti-RelA antibody.
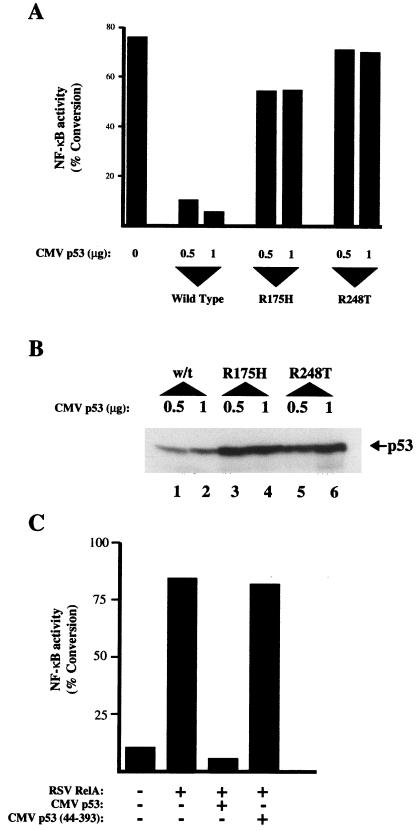
Point mutants of p53 that abolish its transcriptional activity through altering the conformation of its core DNA-binding domain (31, 75) were also analyzed for the ability to repress RelA-mediated transactivation. Strikingly, both the Arg 175-to-His and Arg 248-to-Trp mutants were severely compromised in their ability to repress RelA relative to wild type (Fig. 3A), although some inhibition was observed at higher levels (data not shown). Furthermore, the mutant forms of p53 were expressed at higher levels than the wild type, as might be expected from their increased stability (39) (Fig. 3B).
FIG. 3.
p53 mutants do not inhibit RelA. (A) Mutant p53 proteins do not inhibit RelA transactivation. HFF cells were transfected as for Fig. 2B with 3 μg of 4× κB CAT reporter plasmid except that the R175H and R248T mutant p53 expression plasmids were included as indicated. All lanes shown included 1 μg of RSV RelA expression plasmid. (B) Mutant p53 proteins are expressed at higher levels than wild-type p53. The indicated levels of the wild-type (w/t) and the R175H and R248T mutant p53 expression plasmids were cotransfected into U2OS cells. After 48 h, whole-cell lysates were prepared, resolved on an SDS–10% polyacrylamide gel, and transferred to PVDF. p53 levels were determined by using the anti-p53 monoclonal antibody DO1. (C) p53 lacking its amino-terminal transactivation domain does not inhibit RelA. HFF cells were transfected with 3 μg of 4× κB CAT reporter plasmid and 0.5 μg of RSV RelA expression plasmid as indicated; 3 μg of CMV murine p53 and CMV murine p53 (amino acids 44 to 393) was included as indicated.
To further analyze p53’s ability to repress RelA, experiments were performed to determine the involvement of the p53 amino-terminal transactivation domain, required for its interaction with basal transcription factors and the coactivator proteins p300 and CBP (references 2, 24, 43 and 66 and data not shown). These experiments were performed with murine p53, which like human p53 strongly inhibited RelA-mediated transactivation (Fig. 3C). Deletion of the amino-terminal 43 amino acids abolished inhibition of RelA by p53, indicating a requirement for the transactivation domain in addition to a wild-type protein conformation.
p53 transactivation is inhibited by endogenous, TNF-activated NF-κB.
While the experiments performed above demonstrated the potential for NF-κB-mediated suppression of p53, they relied on the expression of exogenous RelA or c-Rel. Thus, the results obtained could be interpreted as being dependent on overexpression of p53 or NF-κB and might not reflect the situation with endogenous levels of these transcription factors. To address this important question, we analyzed whether endogenous NF-κB could also inhibit endogenous p53-mediated transcriptional activation. HFF cells were plated at high cell density and transfected with PG13 CAT, a reporter plasmid containing multimerized p53 binding sites. In these cells, growth at high cell density elicits a wild-type p53 transcriptional response resulting from cellular contact inhibition that can be inhibited by Mdm2 (Fig. 4A and data not shown). Under these growth conditions, stimulation with TNF resulted in a strong activation of NF-κB (Fig. 4B) and significant repression of p53 transcriptional activity (Fig. 4C). Exposure to TNF initiates a number of signalling cascades and activates transcription factors other than NF-κB, however. Cotransfection of a transdominant negative IκBα expression plasmid reversed both the TNF-mediated activation of NF-κB (Fig. 4B) and inhibition of p53 (Fig. 4C). Thus, suppression of p53 transcriptional activity by TNF is dependent on the induction of NF-κB. Furthermore, Western blot analysis confirmed that TNF stimulation did not affect the overall levels of p53 protein (Fig. 4D). Similar results have been observed in U2OS cells, where TNF can also repress the activity of transfected p53 (data not shown).
FIG. 4.
Endogenous p53 transcriptional activity is repressed by TNF activated NF-κB. (A) HFF cells grown at high cell density contain endogenous p53 transcriptional activity that is repressed by expression of Mdm2. HFF cells grown at high density were transfected with 3 μg of the PG13 CAT p53 reporter plasmid and the indicated quantities of the CMV Mdm2 expression plasmid. CMV control plasmid was included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were harvested 48 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (B) TNF stimulates NF-κB activity in HFF cells which is inhibited by a transdominant negative IκB. HFF cells grown at high density were transfected with 5 μg of the 4× κB CAT reporter plasmid and the indicated quantities of the transdominant negative RSV IκB expression plasmid. RSV control plasmid was included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were stimulated with TNF (10 ng/ml) after 24 h as indicated and harvested 40 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (C) TNF represses endogenous p53 transcriptional activity in an NF-κB-dependent manner. HFF cells grown at high density were transfected with 3 μg of the PG13 CAT p53 reporter plasmid and the indicated quantities of the transdominant negative RSV IκB expression plasmid. RSV control plasmid was included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were stimulated with TNF (10 ng/ml) after 24 h as indicated and harvested 40 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (D) Endogenous p53 levels are not affected by TNF stimulation. Whole-cell lysates were prepared from untransfected high-density HFF cells treated with TNF as indicated and as described for panel C. Protein samples were resolved on an SDS–10% polyacrylamide gel and transferred to PVDF. p53 levels were determined by using the anti-p53 monoclonal antibody DO1. The results from two separate experiments (Exp.) are shown.
Following UV light stimulation, p53 can suppress NF-κB transactivation.
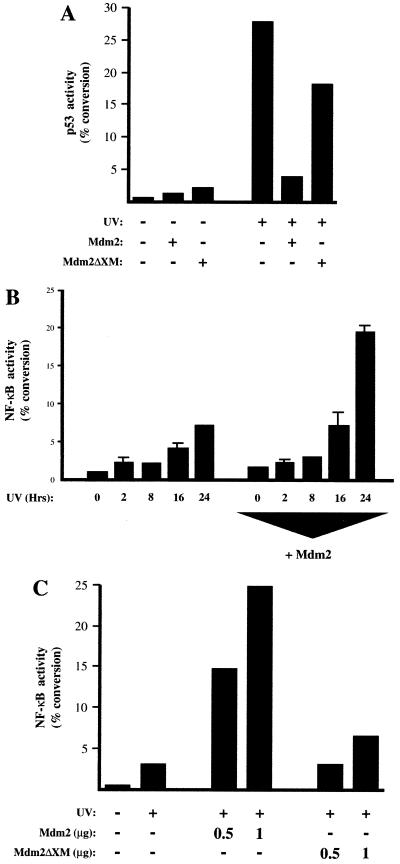
Both p53 and NF-κB are induced following exposure to ionizing radiation. The results obtained above suggested that under these circumstances transcriptional cross talk between NF-κB and p53 may occur. To determine whether this was the case, subconfluent HFF cells were exposed to UV radiation. Under the growth conditions used in this experiment, we observed a strong induction of p53 transcriptional activation following UV stimulation (Fig. 5A), accompanied by a relatively weak stimulation of NF-κB transactivation (Fig. 5B). Cotransfection of an Mdm2 expression plasmid, while strongly inhibiting the p53 transcriptional response (Fig. 5A), significantly enhanced expression from the NF-κB reporter plasmid at later time points following UV stimulation (Fig. 5B). In contrast, the Mdm2ΔXM mutant, which failed to inhibit UV-activated p53 (Fig. 5A), also failed to stimulate NF-κB activity (Fig. 5C). Although the involvement of other Mdm2-binding proteins in this effect cannot be ruled out, these results are consistent with the transcriptional activity of endogenous NF-κB being suppressed by endogenous levels of p53.
FIG. 5.
Following UV stimulation, NF-κB activity is suppressed by p53. (A) UV treatment induces endogenous p53 transcriptional activity in HFF cells. HFF cells were transfected with 5 μg of the PG13 CAT p53 reporter plasmid and 500 ng of CMV Mdm2 or Mdm2ΔXM reporter plasmid as indicated. After 24 h, the cells were exposed to UV light (35 J/m2). Cells and harvested after a further 24 h. Control CMV plasmid was included such that all samples contained the same quantity of expression plasmid. The results shown are representative of three separate experiments. (B) UV-induced NF-κB transcriptional activity is stimulated by cotransfected Mdm2. Cells were transfected as for panel A except that 5 μg of the 4× κB CAT reporter plasmid was included in place of PG13 CAT. The results shown are the averages of three separate experiments. The error bars represent calculated standard deviations. (C) UV-induced NF-κB transcriptional activity is not stimulated by an Mdm2 mutant unable to interact with p53. Cells were transfected with 5 μg of 4× κB CAT reporter plasmid and the indicated quantities of CMV Mdm2 or CMV Mdm2ΔXM. After 24 h, the cells were exposed to UV light (35 J/m2). Cells were harvested after a further 24 h. Control CMV plasmid was included such that all samples contained the same quantity of expression plasmid.
p53 and NF-κB compete for limiting quantities of p300 and CBP.
The experiments described above demonstrate that the principal factor determining whether p53 or NF-κB is the dominant inducer of gene expression is the relative level of each factor. Since deletion of the p53 transactivation abolished repression of RelA (Fig. 3C), this observation would be explained by both transcription factors competing for a limiting pool of common transcriptional coactivators, such as p300 and CBP (2, 21, 24, 43, 58, 66). A feature of p300 and CBP is that both are present in limiting quantities within the nucleus, and quantitation of the number of p300 molecules in the nucleus has shown it to be at significantly lower levels than RelA (30). To determine, therefore, whether the bindings of both RelA and p53 to p300 or CBP are mutually exclusive, these interactions were analyzed in vitro. As expected, in vitro-translated p53 interacted with antibody-bound complexes of both immunoprecipitated p300 and CBP (Fig. 6A). Similarly, and as has been shown previously (59), p300 efficiently bound in vitro-translated RelA. Addition of purified recombinant p53 into this assay completely blocked RelA binding to p300 (Fig. 6B, lanes 3 and 4). To further address this question, p300 was immunoprecipitated from nuclear extracts prepared from 293 cells transfected with RelA expression plasmid and increasing concentrations of p53 followed by subsequent Western blotting for RelA. Again, and in the context of other nuclear proteins, p53 inhibited binding of RelA to p300, while the levels of RelA itself remained unaffected (Fig. 6C). In addition, no direct interaction between p53 and RelA themselves could be detected (Fig. 6D), and p53 did not affect RelA DNA binding as assessed by electrophoretic mobility shift assay (data not shown).
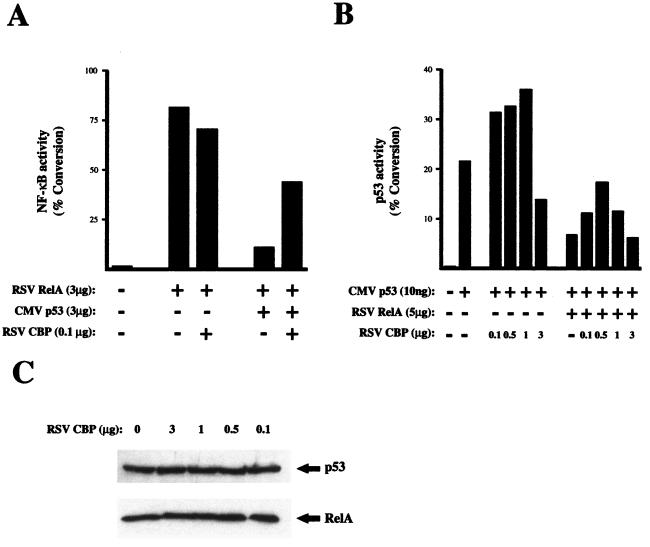
These observations suggested that suppression of p53 transactivation by RelA, or vice versa, might occur through sequestration of p300 and CBP and thus should be abolished by the expression of additional amounts of these coactivators. To address this question, a CBP expression plasmid was cotransfected with an NF-κB reporter plasmid together with p53 and RelA expression plasmids. As observed previously (Fig. 2), p53 inhibited RelA transcriptional activity in HFF cells (Fig. 7A). Significantly, coexpression of CBP reversed the inhibition of RelA by p53 (Fig. 7A). Similar results were observed in U2OS cells with Bax reporter plasmid and RelA-mediated inhibition of p53 transactivation (Fig. 7B). Interestingly recovery of p53 or NF-κB activity was observed only with lower levels of CBP expression plasmid; higher levels failed to reverse this effect and could even be slightly inhibitory (Fig. 7B and data not shown). Western blot analysis demonstrated that CBP had no effect on the level of either RelA or p53 (Fig. 7C). Taken together, these results indicate that corepression of transcriptional activity by p53 and NF-κB is consistent with sequestration of a limiting pool of p300/CBP complexes.
FIG. 7.
Coexpression of CBP recovers p53 and RelA transcriptional repression. (A) Repression of RelA-mediated transactivation by p53 is recovered by CBP. HFF cells were transfected with 5 μg of 4× κB CAT reporter plasmid, 3 μg of RSV RelA expression plasmid, 3 μg of CMV p53 expression plasmid, and 0.1 μg or RSV CBP expression plasmid as indicated. CMV and RSV controls were included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were harvested 36 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (B) Repression of p53-mediated transactivation by RelA is recovered by CBP. U2OS cells were transfected with 5 μg of Bax CAT reporter plasmid, 10 ng of CMV p53 expression plasmid, 3 μg of RSV RelA expression plasmid, and the indicated quantities of RSV CBP expression plasmid. CMV and RSV controls were included as appropriate such that all transfections had equivalent levels of expression plasmid. Cells were harvested 36 h after transfection, and a CAT assay was performed. The results shown are representative of at least three separate experiments. (C) Western blot analysis showing that RelA and p53 levels are unaffected by cotransfected CBP. HFF cells were cotransfected with the indicated quantities of RSV CBP expression plasmid and either 1 μg of CMV p53 or 1 μg of RSV RelA expression plasmid. After 36 h, whole-cell lysates were prepared, resolved on an SDS–10% polyacrylamide gel, and transferred to PVDF before incubation with anti-RelA or anti-p53 antibody.
DISCUSSION
In this report, we have demonstrated previously unrealized transcriptional cross talk between the tumor suppressor protein p53 and NF-κB. This regulatory mechanism provides a molecular explanation for how the divergent functional consequences of p53 and NF-κB activation can be integrated by the cell. In these experiments, we show that the ability of p53 or RelA to stimulate transcription is strongly influenced by the relative level of the other protein. Importantly, we have been able to demonstrate that this effect can be observed both with cotransfected expression plasmids (Fig. 1 and 2) and with endogenous levels of these transcription factors (Fig. 4 and 5), indicating its biological relevance and significance. Furthermore, we demonstrate that these results are consistent with competition between p53 and NF-κB for limiting quantities of complexes containing the p300 and CBP coactivator proteins (Fig. 6 and 7). While this report was under review, Ravi et al. similarly reported repression of RelA by p53 through a p300-dependent mechanism (61).
The implications of these observations are numerous. Both p53 and NF-κB can be induced by similar stimuli, such as ionizing radiation or TNF (15, 17, 25, 26, 37, 42, 50, 55, 64, 76), but while p53 is generally a proapoptotic transcription factor, strong evidence exists to indicate that under most circumstances, NF-κB promotes resistance to programmed cell death (6, 9, 26, 36, 42, 44, 46, 73, 76, 81). Our results suggest that one factor influencing cell fate following such stimulation is the ability of NF-κB to suppress p53 transactivation and thus inhibit the transcriptionally dependent induction of apoptosis by p53. In this model, NF-κB could be viewed as having a direct transcriptional effect, inducing genes that promote resistance to apoptotis (e.g., A20, IEX-1L, TRAF1, TRAF2, IAP1, and IAP2 [38, 77, 82]) while also indirectly suppressing factors, such as p53, capable of stimulating the production of proteins, such as Bax, that could counter this effect (49). Evidence exists that p53 can also regulate cell death in a non-transcription-dependent manner (1, 11, 83) and we predict that such a mechanism would not be directly affected by NF-κB and would thus serve as a means with which NF-κB’s antiapoptotic functions could be overridden. We have also shown that p53 can suppress NF-κB activity and that this effect can be observed with the endogenous proteins following DNA damage induced by UV radiation in HFF cells (Fig. 5). Thus, it appears likely that the outcome of cross talk between NF-κB and p53 will depend on the nature of the stimuli, the growth conditions, and the cell type. Further investigation will be required to assess how this effect may vary with other forms of DNA damage-inducing agents and in other cell types.
Sequestration of p300 and CBP is likely to be understood as an increasingly common mechanism regulating inducible gene expression. Repression of AP-1-induced gene expression by nuclear hormone receptors, STAT proteins, or p53 has been shown to result from sequestration of p300 and CBP (2, 29, 35), while other researchers have shown that cross talk between E2F and p53 (40) or STAT2 and NF-κB (29) occurs through a similar mechanism. Although we have shown that p53 and RelA can bind p300 and CBP competitively (Fig. 6), this mutually exclusive binding might only partially contribute to the transcriptional cross talk observed. Interestingly, a previous study using a temperature-sensitive mutant of p53 found that while a wild-type p53 conformation was required for the p300/CBP-mediated suppression of AP-1 activity, p53 in the mutant conformation was still capable of binding these coactivators (2). Similarly, we have found that mutant forms of p53 which still have intact transactivation domains bind competitively with RelA to p300/CBP and, although greatly impaired in the ability to inhibit RelA, will do so when expressed at concentrations higher than those shown in Fig. 3 (data not shown). These observations suggest that coactivator sequestration has a requirement for DNA binding. Such a mechanism would be similar to the report that Gal4-VP16-induced toxicity in yeast required the integrity of both the transactivation and DNA-binding domains of the protein (8). Since the interaction of NF-κB with p300/CBP can be regulated by phosphorylation of RelA (88), we cannot rule out the involvement of a more indirect mechanism, similar to the report that activation of the glucocorticoid receptor leads to inhibition of the JNK signalling pathway, resulting in the absence of c-Jun phosphorylation and consequently an inability of AP-1 to bind p300/CBP (10).
Interestingly, we observed recovery of p53 or NF-κB transcriptional activity only with relatively low levels of CBP plasmid and found that higher levels tended to be inhibitory in their own right (Fig. 7B and data not shown). Generally, many experiments with p300 and CBP have relied on using levels of plasmid many orders of magnitude higher than those used here. Although it is hard to make a direct comparison between different experimental systems and cell lines, the observations presented here suggest that at least in the case of p53 and NF-κB, limiting quantities of p300 and CBP per se are probably not the whole explanation for the cross talk that occurs between these transcription factors. Instead, they are consistent with there being, in addition, a limiting pool of a p300/CBP-associated cofactor that is also required for efficient transcriptional activation by p53 or NF-κB. An example of such a complex is found with the nuclear hormone receptors which interact with both p300/CBP and other, non-DNA-binding coactivator proteins such as SRC1 and ACTR (12, 13, 27, 35, 65, 67). SRC1 and ACTR also interact with p300 and CBP, thus potentially forming a stable complex (13, 35, 85). Interestingly, a recent report has shown that the p50 NF-κB subunit, but not RelA, can also interact with SRC1 (51). Many non-DNA-binding transcriptional regulatory proteins, such as P/CAF and P/CIP, as well as kinases such as pp90rsk and cyclin E-Cdk2, have also been shown to complex with p300 and CBP, and it is likely that others remain to be identified (52, 58, 72, 84). Whether any of these will also participate in transcriptional regulation by p53 will be the subject of future investigations.
We have demonstrated mutual suppression of transcriptional activation between two p300/CBP-binding DNA-binding proteins. Many other inducible transcription factors also interact with p300 and CBP, however (22, 63), and it is certain that many of these will also be active at the same time as p53 and NF-κB. This leaves the question of how p300 and CBP can simultaneously integrate the diverse functions of all of these proteins. Part of the answer to this problem may lie in the utilization of different p300/CBP complexes, discussed above, being used by distinct subsets of DNA-binding proteins, thus functionally separating them and limiting cross talk to certain regulatory families of transcription factors. It is probable, however, that the promoter context in which a transcription factor finds itself will also determine its ability to access the limiting pool of p300/CBP complexes. For example, in the case of the beta interferon promoter, part of the function of the assembly of NF-κB with other transcription factors into the enhanceosome complex appears to be the cooperative recruitment of p300 and CBP (47, 78). Other DNA-binding proteins such as C/EBP and AP-1(Fos/Jun) also interact with p300 and CBP (4, 28, 35, 48) and have been shown to interact with and bind DNA cooperatively with NF-κB (69, 70). It might be anticipated that at promoters where binding sites for NF-κB and these other transcription factors are correctly juxtaposed, they will also cooperatively recruit p300 and CBP. At promoters where these factors are found in isolation or in combination with transcription factors unable to interact with p300 and CBP, it could be expected that their ability to stimulate transcription would be more susceptible to inhibition through coactivator sequestration. Thus, we favor a model in which NF-κB does not completely suppress p53 function but rather modulates it, limiting p53-mediated transactivation to a subset of promoters where it can function cooperatively with other proteins to recruit p300 and CBP. That mechanisms exist to limit p53-mediated induction or repression of transcription to specific promoters is suggested by a recent study demonstrating that only a relatively limited number of genes were actively regulated by p53 during apoptosis (60). It has been recently questioned whether general transcription factors are limiting with respect to the regulation of endogenous genes as opposed to transiently transfected reporter plasmids (54). It is therefore important to note that single-allele knockouts of both p300 and CBP, which result in 50% of the level of expression of wild-type mice, result in profound developmental defects in mice, confirming that these coactivator proteins are at limiting concentrations in vivo (71, 86).
We have also shown that mutant p53 proteins do not inhibit NF-κB function (Fig. 3), and it is worth noting that many previous experiments on NF-κB transcriptional regulation have been performed in cell lines with mutated p53 or in which it has been functionally inactivated due to the expression of viral proteins. We have also demonstrated that the concentration of a transcriptional activator in the cell can influence whether it is susceptible to cross talk from other regulatory proteins. It is likely, therefore, that experiments relying on the overexpression of p53 or NF-κB, while important, must be viewed with some caution as to whether they reflect the true function of the protein when it is present at its normal cellular levels. Additionally, results obtained from experiments performed on NF-κB in virally transformed or p53-null cells might differ if performed in cell lines capable of mounting a wild-type p53 response.
In most cell types, activation of NF-κB, and of complexes containing the RelA subunit in particular, is transient and self-limiting (56). This would be consistent with NF-κB having a modulatory role on p53, and over the time course of activation, as the relative levels of the two factors fluctuated, it is possible that the genes being induced by each would alter. In some diseases, however, such as ataxia telangiectasia, Hodgkin’s lymphoma, and breast cancer (5, 34, 53, 79), NF-κB has been reported to be aberrantly constitutively active, and its inhibition can reverse the phenotype of cells derived from patients. Furthermore, activation of NF-κB by Bcr-Abl is required for cellular transformation (62). We speculate that suppression of p53 function might contribute to NF-κB’s role in tumorigenesis and other diseases. Furthermore, it is possible that novel cancer therapies based on reactivation of wild-type p53 function might benefit from coordinate suppression of NF-κB in order to promote a beneficial outcome.
ACKNOWLEDGMENTS
We thank Lisa Anderson, Neil Chapman, Louise Copeland, and Andrew Snowden for assistance with this project; Carol Midgley, Alison Sparks, Tim Crook, Bei-Yue Wu, Gary Nabel, Alan Prescott, Richard Goodman, Ted Hupp, and David Lane for providing invaluable reagents; and members of the Division of Gene Regulation and Expression at the University of Dundee, Stefan Roberts, Tom Owen-Hughes, and Julian Blow, for helpful comments, support, and critical reading of the manuscript.
N.D.P. is funded by a Royal Society University fellowship, and G.A.W. is supported by a grant from the Medical Research Council. This work was also supported in part by a grant from TENOVUS.
REFERENCES
- 1.Allday M J, Inman G J, Crawford D H, Farrell P J. DNA-damage in human B-cells can induce apoptosis, proceeding from G(1)/S when P53 is transactivation competent and G(2)/M when it is transactivation defective. EMBO J. 1995;14:4994–5005. doi: 10.1002/j.1460-2075.1995.tb00182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin A S. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–681. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP, c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 5.Bargou R C, Emmerich F, Krappmann D, Bommert K, Mapara M Y, Arnold W, Royer H D, Grinstein E, Greiner A, Scheidereit C, Dorken B. Constitutive nuclear factor-kappa B-RelA activation is required for proliferation and survival of Hodgkin’s disease tumor cells. J Clin Investig. 1997;100:2961–2969. doi: 10.1172/JCI119849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beg A A, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 7.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 8.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of Ada2—a potential transcriptional adapter required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand F, Atfi A, Cadoret A, Lallemain G, Robin H, Lascols O, Capeau J, Cherqui G. A role for nuclear factor kappa B in the antiapoptotic function of insulin. J Biol Chem. 1998;273:2931–2938. doi: 10.1074/jbc.273.5.2931. [DOI] [PubMed] [Google Scholar]
- 10.Caelles C, Gonzalez-Sancho J M, Munoz A. Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes Dev. 1997;11:3351–3364. doi: 10.1101/gad.11.24.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caelles C, Helmberg A, Karin M. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–223. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 12.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon T, Montminy M, Evans R M. Role of CBP/p300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complexed with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 15.Devary Y, Rosette C, DiDonato J A, Karin M. NF-κB activation by ultraviolet light is not dependent on a nuclear signal. Science. 1993;261:1442–1445. doi: 10.1126/science.8367725. [DOI] [PubMed] [Google Scholar]
- 16.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donato N J, Perez M. Tumor necrosis factor-induced apoptosis stimulates p53 accumulation and p21WAF1 proteolysis in ME-180 cells. J Biol Chem. 1998;273:5067–5072. doi: 10.1074/jbc.273.9.5067. [DOI] [PubMed] [Google Scholar]
- 18.Eck S L, Perkins N D, Carr D P, Nabel G J. Inhibition of phorbol ester-induced cellular adhesion by competitive binding of NF-κB in vivo. Mol Cell Biol. 1993;13:6530–6536. doi: 10.1128/mcb.13.10.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.el-Deiry W S, Harper J W, O’Connor P M, Velculescu V E, Canman C E, Jackman J, Pietenpol J A, Burrell M, Hill D E, Wang Y, Wiman K G, Mercer W E, Kastan M B, Kohn K W, Elledge S J, Kinzler K W, Vogelstein B. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 20.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 21.Gerritsen M E, Williams A J, Neish A S, Moore S, Shi Y, Collins T. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc Natl Acad Sci USA. 1997;94:2927–2932. doi: 10.1073/pnas.94.7.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 23.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 24.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 25.Gualberto A, Hixon M L, Finco T S, Perkins N D, Nabel G J, Baldwin A S., Jr A proliferative p53-responsive element mediates tumor necrosis factor alpha induction of the human immunodeficiency virus type 1 long terminal repeat. Mol Cell Biol. 1995;15:3450–3459. doi: 10.1128/mcb.15.6.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall P A, Meek D, Lane D P. p53—integrating the complexity. J Pathol. 1996;180:1–5. doi: 10.1002/(SICI)1096-9896(199609)180:1<1::AID-PATH712>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptor. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 28.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hottiger M O, Felzien L K, Nabel G J. Modulation of cytokine-induced HIV gene expression by the competitive binding of transcription factors to the coactivator p300. EMBO J. 1998;17:3124–3134. doi: 10.1093/emboj/17.11.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hupp T R, Meek D W, Midgley C A, Lane D P. Activation of the cryptic DNA-binding function of mutant forms of p53. Nucleic Acids Res. 1993;21:3167–3174. doi: 10.1093/nar/21.14.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Israel A. Signal transduction—IκB kinase all zipped up. Nature. 1997;388:519–521. doi: 10.1038/41433. [DOI] [PubMed] [Google Scholar]
- 33.Jiang H, Lin J, Su Z, Collart F R, Huberman E, Fisher P B. Induction of differentiation in human promyelocytic HL-60 leukemia cells activates p21, WAF1/CIP1, expression in the absence of p53. Oncogene. 1994;9:3397–3406. [PubMed] [Google Scholar]
- 34.Jung M, Zhang Y, Lee S, Dritschilo A. Correction of radiation sensitivity in ataxia-telangiectasia cells by a truncated IκB-α. Science. 1995;268:1619–1621. doi: 10.1126/science.7777860. [DOI] [PubMed] [Google Scholar]
- 35.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 36.Klefstrom J, Arighi E, Littlewood T, Jaattela M, Saksela E, Evan G I, Alitalo K. Induction of TNF-sensitive cellular phenotype by c-myc involves p53 and impaired NF-κB activation. EMBO J. 1997;16:7382–7392. doi: 10.1093/emboj/16.24.7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 38.Krikos A, Laherty C D, Dixit V M. Transcriptional activation of the tumor-necrosis-factor α inducible zinc finger protein, A20, is mediated by κB elements. J Biol Chem. 1992;267:17971–17976. [PubMed] [Google Scholar]
- 39.Lane D P, Hall P A. MDM2—arbiter of p53’s destruction. Trends Biochem Sci. 1997;22:372–374. doi: 10.1016/s0968-0004(97)01119-5. [DOI] [PubMed] [Google Scholar]
- 40.Lee C W, Sorensen T S, Shikama N, LaThangue N B. Functional interplay between p53 and E2F through co-activator p300. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- 41.Leung K, Nabel G J. HTLV-I transactivator induces interleukin-2 receptor expression through an NF-κB-like factor. Nature. 1988;333:776–778. doi: 10.1038/333776a0. [DOI] [PubMed] [Google Scholar]
- 42.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 43.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 44.Liu Z, Hsu H, Goeddel D V, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 45.Maniatis T. Signal transduction—catalysis by a multiprotein IκB kinase complex. Science. 1997;278:818–819. doi: 10.1126/science.278.5339.818. [DOI] [PubMed] [Google Scholar]
- 46.Mayo M W, Wang C Y, Cogswell P C, Rogers-Graham K S, Lowe S W, Der C J, Baldwin A S., Jr Requirement of NF-κB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 47.Merika M, Williams A J, Chen G Y, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 48.Mink S, Haenig B, Klempnauer K H. Interaction and functional collaboration of p300 and C/EBPβ. Mol Cell Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyashita T, Reed J C. Tumor-suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 50.Mukhopadhyay T, Roth J A, Maxwell S A. Induction of p53 DNA-binding activity by tumor-necrosis-factor-alpha. Int J Oncol. 1996;9:715–720. [PubMed] [Google Scholar]
- 51.Na S Y, Lee S K, Han S J, Choi H S, Im S Y, Lee J W. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor kappa B-mediated transactivations. J Biol Chem. 1998;273:10831–10834. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. The signal-dependent coactivator CBP is a nuclear target for pp90rsk. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 53.Nakshatri H, Bhat-Nakshatri P, Martin D A, Goulet R J, Jr, Sledge G W., Jr Constitutive activation of NF-κB during progression of breast cancer to hormone-independent growth. Mol Cell Biol. 1997;17:3629–3639. doi: 10.1128/mcb.17.7.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Natesan S, Rivera V M, Molinari E, Gilman M. Transcriptional squelching re-examined. Nature. 1997;390:349–350. doi: 10.1038/37019. [DOI] [PubMed] [Google Scholar]
- 55.Osborn L, Kunkel S, Nabel G J. Tumor necrosis factor a and interleukin-1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor κB. Proc Natl Acad Sci USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perkins N D. Achieving transcriptional specificity with NF-kappaB. Int J Biochem Cell Biol. 1997;29:1433–1448. doi: 10.1016/s1357-2725(97)00088-5. [DOI] [PubMed] [Google Scholar]
- 57.Perkins N D, Agranoff A B, Pascal E, Nabel G J. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 59.Perkins N D, Schmid R M, Duckett C S, Leung K, Rice N R, Nabel G J. Distinct combinations of NF-κB subunits determine the specificity of transcriptional activation. Proc Natl Acad Sci USA. 1992;89:1529–1533. doi: 10.1073/pnas.89.5.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 61.Ravi R, Mookerjee B, van Hensbergen Y, Bedi G C, Giordano A, El-Deiry W S, Fuchs E J, Bedi A. p53-mediated repression of nuclear factor-kappa B RelA via the transcriptional integrator p300. Cancer Res. 1998;58:4531–4536. [PubMed] [Google Scholar]
- 62.Reuther J Y, Reuther G W, Cortez D, Pendergast A M, Baldwin A S. A requirement for NF-κB activation in Bcr-Abl-mediated transformation. Genes Dev. 1998;12:968–981. doi: 10.1101/gad.12.7.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Curr Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 64.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 65.Smith C L, Onate S A, Tsai M J, O’Malley B W. CREB binding-protein acts synergistically with steroid-receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93:8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 67.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J X, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 68.Stancovski I, Baltimore D. NF-κB activation: the IκB kinase revealed? Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 69.Stein B, Baldwin A S, Jr, Ballard D W, Greene W C, Angel P, Herrlich P. Cross-coupling of the NF-κB p65 and Fos/Jun transcription factors produces potentiated biological function. EMBO J. 1993;12:3879–3891. doi: 10.1002/j.1460-2075.1993.tb06066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stein B, Cogswell P C, Baldwin A S., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain-bZIP interaction. Mol Cell Biol. 1993;13:3964–3974. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc Natl Acad Sci USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional coactivator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 73.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 74.Verma I M, Stevenson J. IκB kinase: beginning, not the end. Proc Natl Acad Sci USA. 1997;94:11758–11760. doi: 10.1073/pnas.94.22.11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vojtesek B, Dolezalova H, Lauerova L, Svitakova M, Havlis P, Kovarik J, Midgley C A, Lane D P. Conformational-changes in p53 analyzed using new antibodies to the core DNA-binding domain of the protein. Oncogene. 1995;10:389–393. [PubMed] [Google Scholar]
- 76.Wang C, Mayo M W, Baldwin A S. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 77.Wang C Y, Mayo M W, Korneluk R G, Goeddel D V, Baldwin A S. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 78.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 79.Wood K M, Roff M, Hay R T. Defective IκBα in Hodgkin cell lines with constitutively active NF-κB. Oncogene. 1998;16:2131–2139. doi: 10.1038/sj.onc.1201735. [DOI] [PubMed] [Google Scholar]
- 80.Wu B Y, Woffendin C, MacLachlan I, Nabel G J. Distinct domains of IκB-α inhibit human immunodeficiency virus type 1 replication through NF-κB and Rev. J Virol. 1997;71:3161–3167. doi: 10.1128/jvi.71.4.3161-3167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu M, Lee H, Bellas R E, Schauer S L, Arsura M, Katz D, FitzGerald M J, Rothstein T L, Sherr D H, Sonenshein G E. Inhibition of NF-κB/Rel induces apoptosis of murine B cells. EMBO J. 1996;15:4682–4690. [PMC free article] [PubMed] [Google Scholar]
- 82.Wu M X, Ao Z H, Prasad K V S, Wu R L, Schlossman S F. IEX-1L, an apoptosis inhibitor involved in NF-κB-mediated cell survival. Science. 1998;281:998–1001. doi: 10.1126/science.281.5379.998. [DOI] [PubMed] [Google Scholar]
- 83.Yan Y, Shay J W, Wright W E, Mumby M C. Inhibition of protein phosphatase activity induces p53-dependent apoptosis in the absence of p53 transactivation. J Biol Chem. 1997;272:15220–15226. doi: 10.1074/jbc.272.24.15220. [DOI] [PubMed] [Google Scholar]
- 84.Yang X, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 85.Yao T, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch’ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 87.Zhong H H, Voll R E, Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]