Abstract
The transcription factor GATA-1 is a key regulator of erythroid-cell differentiation and survival. We have previously shown that the transcriptional cofactor CREB-binding protein (CBP) binds to the zinc finger domain of GATA-1, markedly stimulates the transcriptional activity of GATA-1, and is required for erythroid differentiation. Here we report that CBP, but not p/CAF, acetylates GATA-1 at two highly conserved lysine-rich motifs present at the C-terminal tails of both zinc fingers. Using [3H]acetate labelling experiments and anti-acetyl lysine immunoprecipitations, we show that GATA-1 is acetylated in vivo at the same sites acetylated by CBP in vitro. In addition, we show that CBP stimulates GATA-1 acetylation in vivo in an E1A-sensitive manner, thus establishing a correlation between acetylation and transcriptional activity of GATA-1. Acetylation in vitro did not alter the ability of GATA-1 to bind DNA, and mutations in either motif did not affect DNA binding of GATA-1 expressed in mammalian cells. Since certain functions of GATA-1 are revealed only in an erythroid environment, GATA-1 constructs were examined for their ability to trigger terminal differentiation when introduced into a GATA-1-deficient erythroid cell line. We found that mutations in either acetylation motif partially impaired the ability of GATA-1 to induce differentiation while mutations in both motifs abrogated it completely. Taken together, these data indicate that CBP is an important cofactor for GATA-1 and suggest a novel mechanism in which acetylation by CBP regulates GATA-1 activity in erythroid cells.
Erythrocyte development is among the best-studied model systems used to define the mechanisms responsible for lineage commitment, cellular differentiation, and tissue-specific gene expression. The lineage-restricted transcription factor GATA-1 orchestrates several key aspects of erythroid development. Functionally important binding sites for GATA-1 are found in the regulatory regions of essentially all erythroid-cell-specific genes (30). Mice deficient for GATA-1 succumb to fatal anemia due to an inability of erythroid precursor cells to survive and mature (14, 43, 44). In addition, GATA-1 induces terminal erythroid maturation when introduced into the GATA-1-deficient erythroid cell line G1E (45).
Tissue-specific gene expression is the result of a concerted action of tissue-restricted and ubiquitous transcription factors. Our previous studies indicated that a widely expressed transcriptional cofactor, CREB-binding protein (CBP), is a potent coactivator of GATA-1 (4). CBP associates with GATA-1 in vitro and in vivo and markedly stimulates its activity. Expression of the adenovirus oncoprotein E1A, which interferes with the action of CBP, blocks the effects of CBP on GATA-1 activity, inhibits erythroid differentiation, and reduces the expression of several GATA-1-dependent genes (4).
Multiple mechanisms have been invoked to account for the activity of CBP and its close relative p300. First, CBP interacts with TFIIB (23), TATA-binding protein (1, 12, 37, 40), and RNA polymerase II (10, 21, 27) and could thus serve as a bridging molecule between DNA-bound transcription factors and the basal transcription machinery. Second, CBP interacts with numerous transcription factors via dedicated domains and could thus provide a scaffold for the assembly of a high-molecular-weight “enhanceosome” complex (for a review, see reference 38). Such multifunctional interactions may account for the observed synergy between transcription factors that use CBP and p300 as cofactors. Third, CBP and p300 possess intrinsic and associated histone acetyltransferase activity (3, 9, 28, 39, 46). Acetylated histones are associated with an open chromatin configuration, which in turn might facilitate the accessibility of DNA to other transcription factors (for a review, see references 31 and 35). Finally, CBP and p300 were recently shown to acetylate nonhistone proteins such as the tumor suppressor protein p53 (16), the erythroid Kruppel-like factor (EKLF) (48), and the basal transcription factors, TFIIE and TFIIF (19). In the case of p53, acetylation of the regulatory domain led to a dramatic increase in DNA binding (16) in vitro, whereas the functional consequences of EKLF, TFIIE, and TFIIF acetylation remain to be explored.
In this report, we further define the mechanisms of transcriptional activation of GATA-1 by CBP. We found that CBP acetylates GATA-1 at two highly conserved lysine-rich motifs near the zinc finger domains. In contrast to a recent report which appeared while this paper was under review (7), we found that acetylation of GATA-1 did not affect its ability to bind DNA and that substitutions or deletions in these motifs did not affect DNA binding of mammalian-cell-expressed GATA-1. We also found that mutations in the C-terminal acetylation motif, but not in the N-terminal motif, reduce the binding to CBP and diminish the response to CBP in transient-transfection assays. We further demonstrate that CBP stimulates GATA-1 acetylation in vivo and that coexpression of E1A abolishes GATA-1 acetylation.
Certain functions of GATA-1 critically depend on an erythroid environment. For example, the N-terminal zinc finger of GATA-1 is required for function in erythroid cells but is dispensable for transactivation of some GATA-1-dependent reporter constructs in transiently transfected NIH 3T3 cells (45). Conversely, a potent activation domain at the N terminus of GATA-1 is required for transactivation in the NIH 3T3 cell assay while this domain is dispensable for function in erythroid and megakaryocytic cells (6, 25, 42, 45). Therefore, we examined the function of the acetylation sites in an assay that measures GATA-1 activity by its ability to trigger terminal differentiation when introduced into the GATA-1-deficient erythroid-cell line G1E. This assay revealed that mutations in either motif reduce GATA-1 function without affecting its ability to bind DNA. These results indicate that the acetylated motifs are of biological importance independent of a role in DNA binding. They further suggest that acetylation of GATA-1 might provide a novel mechanism of controlling GATA-1 activity.
MATERIALS AND METHODS
Plasmids.
Vectors expressing CBP (pCMV5-CBP) (a gift from M. Rosenfeld), GATA-1 (pXM-G1), E1A (pEF1α-E1A), and GATA-1 reporter construct M1αGH have been described previously (20, 25). To construct CBP HAT− (a histone acetyltransferase [HAT]-deficient version of CBP), Leu1690 and Cys1691 were changed to Lys1690 and Leu1691 by overlapping PCR (22). The retroviral construct expressing GATA-1, MFG–GATA-1, and various glutathione-S-transferase (GST)–GATA-1 fusion constructs have been described previously (11, 45). GST-CBP-HAT containing the HAT domain of murine CBP (amino acids [aa] 1196 to 1718) (3) was generated by cloning a PCR-amplified fragment into pGEX4T. GST-p/CAF-HAT (a gift from M. Montminy) contains the HAT domain of human p/CAF (46) (aa 352 to 832). To introduce Lys-to-Ala mutations into the acetylation motifs of GATA-1, overlapping PCR was used. Sequence analysis confirmed the absence of PCR errors. The N-terminal acetylation motif (LIRPKKRMIVSKRA; aa 241 to 254) was mutated to LIRAARMIVSKRA (N-mut). The C-terminal motif (KASGKGKKKRGSNLAG; aa 308 to 323) was mutated to KASGAGAAARGSNLAG (C-mut). In the construct termed NC-mut, both motifs were mutated. The mutant GATA-1 constructs were inserted into pXM, pGEX2T, MFG, and pBluescript.
Acetylation site mapping.
Peptides corresponding to residues 236 to 255 and 304 to 323 of murine GATA-1 were synthesized with an Applied Biosystems 430A synthesizer. The purity was estimated to be 85% based on high-pressure liquid chromatography (HPLC) analysis. Twenty micrograms of peptide was incubated with 1 μg of GST-CBP-HAT at 30°C for 2 h in 60 μl of acetyltransferase reaction buffer (50 mM Tris-HCl [pH 8], 10% glycerol, 50 mM NaCl, 2.5 mM MgCl2, 10 mM sodium butyrate, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.08 μCi of [14C]acetyl coenzyme A [CoA]). The peptide was separated from GST-CBP-HAT and free [14C]acetyl-CoA by reverse-phase HPLC. Acetylated peptide was subjected to Edman degradation; the first three cycles were used to confirm the amino acid sequence, and the remaining cycles were collected for measurement of 14C incorporation by scintillation counting.
In vivo sodium [3H]acetate labeling and immunoprecipitation experiments.
Twenty million exponentially growing MEL cells were centrifuged and resuspended in 2 ml of medium containing 1 mCi of sodium [3H]acetate per ml (16 Ci/mmol; ICN) and 50 nM Trichostatin (Sigma) and incubated for 90 min. The cells were lysed in 350 mM NaCl–50 mM Tris-HCl (pH 7.5)–0.5% Igepal–1 mM EDTA–0.5 mM DTT–10 mM sodium butyrate–1 μg of aprotinin per ml–1 μg of pepstatin per ml–1 μg of leupeptin per ml–0.2 mM PMSF. Following centrifugation at 10,000 × g for 5 min, the supernatants were diluted to 150 mM NaCl with H2O containing 1 mM EDTA, 0.5 mM DTT, 10 mM sodium butyrate, 1 μg of aprotinin per ml, 1 μg of pepstatin per ml, 1 μg of leupeptin per ml, and 0.2 mM PMSF and immunoprecipitated with monoclonal GATA-1 antibodies (N6; Santa Cruz) or irrelevant isotype-matched antibodies. The antibodies were captured by adding 30 μl of a 50% slurry of protein G-Sepharose. Immunoprecipitated samples were washed five times in 150 mM NaCl–50 mM Tris-HCl (pH 7.5)–0.5% Igepal–1 mM EDTA–0.5 mM DTT–10 mM sodium butyrate–1 μg of aprotinin per ml–1 μg of pepstatin per ml–1 μg of leupeptin per ml–0.2 mM PMSF, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, fixed, enhanced with Autofluor (National Diagnostics), and autoradiographed.
Immunoprecipitations with anti-acetyl lysine (anti-AK) antibodies (18) were performed under the same conditions as described above. Anti-AK antibodies were a gift from Colyn Crane-Robinson.
Retroviral infections.
BOSC23 cells (a gift from W. Pear) (32) were used to generate retrovirus harboring GATA-1 cRNAs. BOSC23 cells grown in T25 flasks to 80% confluence were transiently transfected with 8 μg of DNA per flask by the calcium phosphate precipitation method in the presence of 25 μM chloroquine. Fresh medium was added 10 h following transfection. At 24 h later, G1E cells (1.5 × 106/flask) were cocultivated with virus-producing BOSC23 cells for 42 h. Infected G1E cells were transferred to fresh medium and grown for an additional 48 h before being subjected to benzidine staining of hemoglobin (45).
Transient transfections, gel shift assays, and GST pull-down experiments.
Transient transfections, gel shift assays, and GST pull-down experiments were performed as described previously (5, 11).
Acetyltransferase assays.
Acetyltransferase assays were performed exactly as reported by Bannister and Kouzarides (3).
RESULTS
CBP acetylates GATA-1 at two conserved lysine-rich motifs.
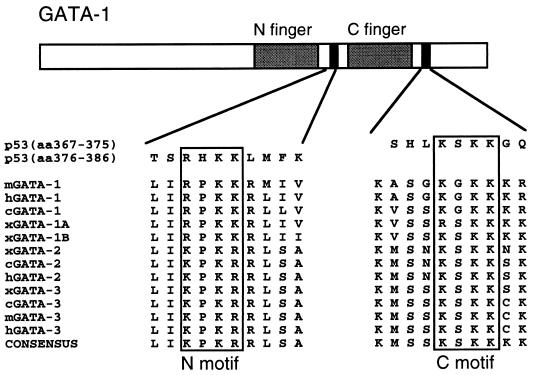
We have previously shown that CBP interacts with GATA-1 and stimulates its activity (4). GATA-1 contains two lysine-rich motifs located just C-terminal to each of its two zinc fingers, referred to below as the N-motif and C-motif. These motifs, which are highly conserved among GATA-1 proteins from different species and among other members of the GATA family (Fig. 1), are very similar to a sequence found in the C terminus of p53 which is directly acetylated by p300 (16) (Fig. 1). This raised the possibility that CBP also acetylates GATA-1. Indeed, the bacterially expressed HAT domain of CBP (CBP-HAT) acetylates full-length recombinant GST–GATA-1 but not GST alone (Fig. 2). Acetylation occurs with an efficiency comparable to that of histones (results not shown), and scintillation counting indicates an average incorporation of two [14C]acetate molecules per GATA-1 molecule (results not shown). Deletion of the zinc finger region, which removes both lysine-rich motifs (aa 198 to 316), abolishes acetylation while the isolated finger region (aa 177 to 333) is acetylated as strongly as wild-type GATA-1 (Fig. 2). Therefore, little or no acetylation occurs outside the zinc finger region. To determine whether the conserved lysine-rich motifs are the actual sites of acetylation, we replaced lysines with alanines in either motif alone or in combination. Since acetylation of p53 by CBP involved all three lysines in its KXKK motif (16), we began by replacing all the lysine residues in the lysine-rich motifs of GATA-1. Substitutions in the N-terminal motif (N-mut) or the C-terminal motif (C-mut) reduced acetylation to 50 to 60% of the wild-type level, as determined by phosphorimager analysis, while substitutions in both motifs (NC-mut) abrogated acetylation almost completely (Fig. 2); the residual acetylation is most probably attributable to lysine 252 next to the N-motif (see below).
FIG. 1.
Conservation of two lysine-rich motifs among GATA-1 family members. The lysine-rich motifs C-terminal to both GATA-1 zinc fingers are conserved among species and among different members of the GATA family. These motifs resemble those in p53 found to be acetylated by CBP.
FIG. 2.
In vitro acetylation of GATA-1 by CBP-HAT. (Top) GATA-1 constructs used as substrates for CBP-HAT. All recombinant proteins were expressed as GST fusion proteins. GATA-1, full-length GATA-1; GATA-1Δf, GATA-1 lacking the zinc finger region and the lysine-rich motifs (aa 198 to 316 deleted); f(GATA-1), the zinc finger region of GATA-1 including the lysine-rich motifs (aa 177 to 333); N-mut, K-to-A substitutions in the N-terminal lysine-rich motif in the context of full-length GATA-1; C-mut, K-to-A substitutions at the C-terminal motif; NC-mut, both motifs mutated. (Bottom) Acetylation of GATA-1 constructs by the HAT domain of CBP (CBP-HAT aa 1196 to 1718). The high-molecular-weight bands present in all lanes represent autoacetylated CBP-HAT. p/CAF-HAT, the HAT domain of p/CAF from aa 352 to 832.
The p300/CBP-associated histone acetyltransferase p/CAF (46) did not detectably acetylate GATA-1 (Fig. 2, right panel) but acetylated free histones with efficiency comparable to that of CBP (data not shown). Taken together, these results show that GATA-1 is specifically acetylated by CBP in vitro at the two lysine-rich motifs and are consistent with a high degree of substrate specificity among different acetyltransferases (36).
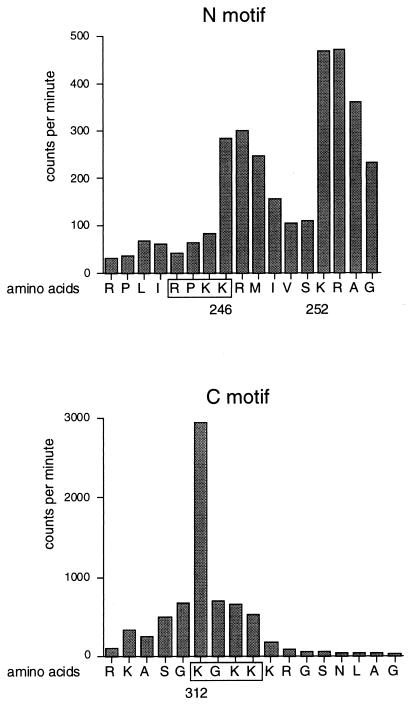
To pinpoint the amino acid residues of GATA-1 acetylated by CBP without introducing mutations which might affect binding to CBP (see below) we used synthetic peptides containing either acetylation motif as substrates for CBP-HAT. Following in vitro acetylation in the presence of [14C]acetyl-CoA and Edman degradation, individual amino acids were analyzed by scintillation counting for [14C]acetate incorporation. The results show that the most strongly acetylated residues in the N-motif were lysines 246 and 252 (Fig. 3, top). This finding was somewhat unexpected, since it indicates that acetylation can occur at residues outside the RXKK motif. The high levels of counts observed in some nonacetylated amino acids following acetylated lysine residues might represent carryovers from incomplete Edman degradation from preceding cycles. In the C-motif, the major acetylation site is lysine 312, which also represents the residue with the largest absolute 14C incorporation into the GATA-1 peptides (Fig. 3, bottom). Other residues in the C-motif were not acetylated to a significant degree. Of note, in both motifs the pattern of acetylation is different from that observed in p53 (16), indicating that the amino acid context surrounding the acetylation sites directs substrate specificity.
FIG. 3.
Mapping of acetylation sites by using synthetic GATA-1 peptides. Peptides corresponding to residues 236 to 255 containing the N-motif (top) and residues 304 to 323 containing the C-motif (bottom) of murine GATA-1 were acetylated by CBP-HAT in vitro and sequenced. Following purification and Edman degradation, [14C]acetate incorporation into individual amino acids was determined by scintillation counting. The RXKK and KXKK motifs are boxed. Note that the absolute amounts of [14C]acetate incorporation is significantly higher in the C-terminal peptide.
Acetylation of the peptide spanning the C-motif occurred with higher efficiency than that of the peptide spanning the N-motif, as judged by the absolute number of incorporated acetate residues (Fig. 3, compare the top and bottom panels). Given that both motifs are acetylated with comparable efficiency in the context of full length GATA-1 (Fig. 2), this suggests that other portions of GATA-1 contribute to substrate recognition by CBP-HAT. Residue 252, which is conserved among different members of the GATA family, remained intact in the NC-mut construct and might account for its residual acetylation (Fig. 2).
Given the importance of the amino acid context in directing acetylation toward specific lysine residues, we asked whether a substitution at lysine 312, which resides in the C-motif, leads to reduced acetylation in the context of full-length N-mut (N-mut 312A). Surprisingly, we found that N-mut 312A is acetylated to a similar degree to N-mut (data not shown), indicating that lysine residues adjacent to lysine 312 can serve as alternative substrates and that the specificity of CBP-HAT toward certain residues is not absolute. Therefore, to ensure complete loss of acetylation for our functional studies, we used GATA-1 mutants which contain multiple lysine-to-alanine substitutions in each motif, as shown in Fig. 2.
In vivo acetylation of GATA-1.
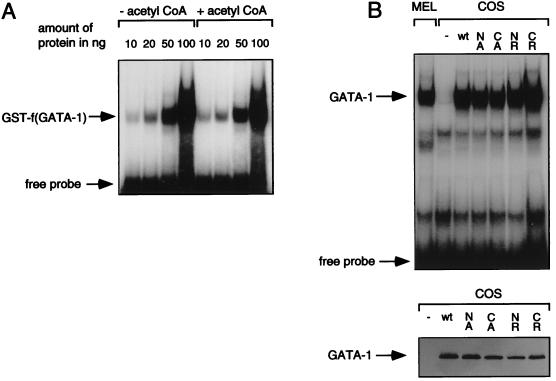
To determine whether acetylation of GATA-1 occurs in vivo, we pulse-labeled MEL cells with [3H]acetate for 90 min and then subjected them to cell lysis and immunoprecipitation with monoclonal GATA-1 antibodies. The autoradiograph shown in Fig. 4A demonstrates that GATA-1 antibodies, but not isotype-matched control antibodies, precipitate [3H]acetate-labelled GATA-1, indicating that acetylation of GATA-1 occurs in intact erythroid cells. Similar results were obtained with [3H]acetate-labelled COS cells transfected with GATA-1 (data not shown). A large number of cellular proteins are acetylated at their N terminus, which could account at least in part for the observed [3H]acetate incorporation. To ensure that acetylation of GATA-1 occurs in vivo at the same sites acetylated by CBP in vitro, we used an antibody, anti-AK, which reacts with acetyl-lysine residues but not with unacetylated lysine residues (18). Lysates from transfected COS cells were immunoprecipitated with anti-AK and then subjected to Western blotting with anti-GATA-1 antibodies. Anti-AK precipitated significant amounts of wild-type GATA-1 (Fig. 4B, top), while precipitation of N-mut and C-mut was reduced and only very little NC-mut was precipitated (Fig. 4B, top). Control Western blots demonstrate the presence of comparable amounts of transfected GATA-1 constructs in the lysates used for immunoprecipitation (Fig. 4B, bottom). These results indicate that GATA-1 is acetylated in vivo at the same sites acetylated by CBP in vitro. In addition, both lysine-rich motifs appear to contribute equally to total acetylation, again similar to what is observed with in vitro-acetylated GATA-1. As in the in vitro acetylation experiments, residual acetylation observed with NC-mut might be due to acetylation of lysine 252.
FIG. 4.
Acetylation of GATA-1 in vivo. (A) Lysates from MEL cells pulse-labeled with 1 mCi of [3H]acetate per ml in the presence of 50 nM Trichostatin A were immunoprecipitated with anti-GATA-1 antibodies or isotype-matched irrelevant antibodies (ctr). Immunoprecipitated samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and enhanced by fluorgraphy. (B) (Top) Whole-cell lysates from transfected COS cells were immunoprecipitated (I.P.) with anti-AK antibodies and analyzed for the presence of wild-type or mutant GATA-1 by Western blotting. Mutant contructs are as described in Fig. 2. ctr, untransfected COS cells; wt, wild type. (Bottom) Western blotting of unprecipitated lysates confirms the presence of similar amounts of GATA-1 proteins. (C) Whole-cell lysates from transfected NIH 3T3 cells were immunoprecipitated as in panel B.
To determine whether CBP can stimulate GATA-1 acetylation in vivo, we cotransfected NIH 3T3 cells with GATA-1 and CBP and then subjected them to anti-AK immunoprecipitation and anti-GATA-1 Western blotting. The results demonstrate that coexpression of CBP leads to a strong increase in acetylation of wild-type GATA-1 but not of NC-mut (Fig. 4C). Thus, CBP induces GATA-1 acetylation in vivo at the same site as it does in vitro. Of note, coexpression of E1A, but not of mutant E1A defective for CBP binding (E1AΔ2–36), abolished in vivo acetylation of GATA-1 (Fig. 4C). Control Western blots showed that neither CBP nor E1A expression significantly altered GATA-1 protein levels (data not shown). This establishes a strong correlation between acetylation of GATA-1 and its transcriptional activity (4).
The failure to detect acetylated GATA-1 in NIH 3T3 cells in the absence of cotransfected CBP is due to the presence of substantially lower basal levels of acetylated GATA-1 protein than in COS cells (data not shown).
Acetylation does not affect DNA binding of GATA-1.
In contrast to p53, where acetylation occurs outside the DNA-binding domain (16), the acetylation motifs in GATA-1 are directly adjacent to the two zinc fingers (Fig. 1). Since acetylation leads to a change in the charge and size of the lysine residues, it seemed possible that acetylation results in altered DNA binding of GATA-1 (25). We tested this possibility in gel mobility shift assays with bacterially expressed GST–GATA-1 finger constructs acetylated in vitro. As shown in Fig. 5A, there was no detectable difference in DNA binding upon acetylation of the GATA-1 fingers over a broad range of protein concentrations. Similar results were obtained when full-length GST–GATA-1 was used (data not shown). As noted above, in vitro acetylation of GATA-1 occurred at high efficiency with incorporation of approximately two acetate residues per GATA-1 molecule. Therefore, it is unlikely that the failure to detect a difference in DNA binding is due to substoichiometric acetylation of GATA-1.
FIG. 5.
(A) Acetylation of GATA-1 does not affect DNA binding. Gel mobility shift assays of acetylated and nonacetylated GST–f(GATA-1). Acetylation reactions were performed in the presence or absence of acetyl-CoA. The indicated amounts of protein were used in a gel mobility shift assay with a probe containing a single GATA site. (B) (Top) The lysine-rich motifs of GATA-1 do not participate in DNA binding. Gel mobility shift assays with wild-type (wt) and mutant GATA-1 proteins expressed in COS cells. NA and CA represent GATA-1 mutants bearing alanine substitutions in the N- and C-motifs, respectively; NR and CR represent analogous mutants with arginine substitutions; MEL cell nuclear extracts served as positive control. (Bottom) A control Western blot shows similar levels of GATA-1 expression.
As additional evidence that modifications at the acetylation motifs do not alter DNA binding, we tested various GATA-1 mutants expressed in mammalian cells. We transfected GATA-1 constructs bearing either alanine or arginine substitutions at the relevant sites into COS-1 cells and tested nuclear extracts in gel mobility shift assays. The arginine substitutions should maintain the positive charge originally provided by lysine residues, while the alanine substitutions are neutral in charge. Neither the arginine substitutions nor the alanine substitutions affected the ability of GATA-1 to bind DNA (Fig. 5B). Furthermore, dissociation rates (off-rates) of wild-type and mutant GATA-1 proteins from both single and palindromic GATA sites were indistinguishable (data not shown). Western blots of nuclear extracts confirmed the presence of equal amounts of GATA proteins in all samples and implied that mutations did not significantly affect protein stability. Nuclear localization of all GATA-1 mutants was confirmed by immunofluorescence microscopy (data not shown).
Together, these results indicate that CBP does not appear to regulate GATA-1 activity through acetylation-induced changes in its DNA-binding ability.
The C-terminal acetylation motif of GATA-1 is required for binding to and stimulation by CBP in transient-transfection assays.
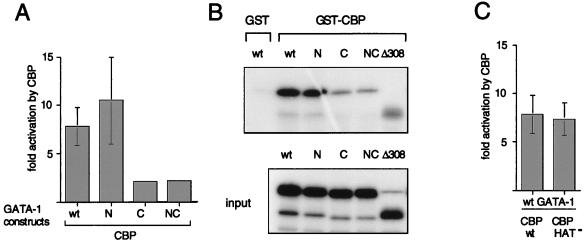
To test whether acetylation by CBP affects transcriptional activity of GATA-1, we cotransfected GATA-1 acetylation mutants together with CBP and a synthetic GATA site-containing reporter plasmid into NIH 3T3 cells. CBP expression increased wild-type GATA-1 and N-mut activity about eightfold, in agreement with our previous studies (Fig. 6A) (4). In contrast, C-mut and NC-mut were severely impaired in their response to CBP. Since CBP interacts with the zinc finger region of GATA-1 (4), we considered the possibility that mutations in the C-terminal motif affect interaction with CBP. To address this question, we performed in vitro protein-binding assays with bacterially expressed GST-CBP containing the GATA-1-binding domain (aa 1805 to 1891) and in vitro-translated GATA-1 constructs. The results show that C-mut and NC-mut bind GST-CBP less avidly than wild-type GATA-1 and N-mut do (Fig. 6B). This establishes a correlation between the ability of CBP to bind to GATA-1 and to enhance its activity, indicating that a direct interaction between these molecules is required for transcriptional stimulation.
FIG. 6.
(A) Stimulation of GATA-1 activity by CBP depends on an intact C-motif. In transient-transfection experiments with NIH 3T3 cells, a GATA-binding site-containing artificial promoter-reporter, M1αGH (1 μg), was cotransfected with 2 μg of wild-type (wt) or mutant (K-to-A substitutions) GATA-1 in the presence or absence of 7 μg of pCMV5-CBP. Fold activation represents the ratio of activity obtained in the presence and absence of CBP. (B) The C-motif of GATA-1 participates in CBP binding. GST pull-down assays were performed with wild-type and mutant in vitro-translated [35S]methionine-labeled GATA-1. GST-CBP, which represents the GATA-1-binding domain (aa 1805 to 1891), served as an affinity reagent. (C) The acetyltransferase activity of CBP is dispensable for GATA-1 activation in transient-transfection assays. Experiments were performed as in panel A. CBP-HAT− represents an acetyltransferase-defective mutant of CBP bearing a double point mutation in the HAT domain.
Since mutations in the C-motif do not distinguish between the effects of binding to and acetylation by CBP, we directly tested the requirement of the acetyltransferase domain of CBP for GATA-1 activation. We introduced a double point mutation into the acetyltransferase domain of CBP (CBP-HAT−), which completely abolishes in vitro acetylation of free histones (22) and GST-GATA-1 (results not shown). In transient-transfection assays, CBP-HAT− activated GATA-1 to the same extent as wild-type CBP did (Fig. 6C). Similar results were obtained with a CBP construct bearing a deletion in the acetyltransferase domain (Δ1458–1475) (reference 26 and data not shown). Thus, in transient-transfection assays of nonerythroid cells with an artificial reporter construct, GATA-1 activation by CBP does not require intrinsic acetyltransferase activity of CBP. This suggests that acetylation might be mediated by other acetyltransferases associated with CBP or that GATA-1 activation might involve an acetylation-independent mechanism (see Discussion).
The following additional considerations have to be made when assaying GATA-1 and CBP functions. First, the chromatin structure of stably integrated genes regulated by GATA-1 and CBP is likely to be important, especially when evaluating the role of histone and factor acetylation. Second, regulation of GATA-1 and CBP activity might require the architecture of natural GATA-1-dependent gene promoters. Third, it is established that some GATA-1 functions are critically dependent on an erythroid environment (see below). To address these issues, we used the GATA-1-deficient erythroid cell line G1E, which can be induced to terminally differentiate upon introduction of GATA-1.
Both N and C acetylation motifs are required to trigger erythroid differentiation.
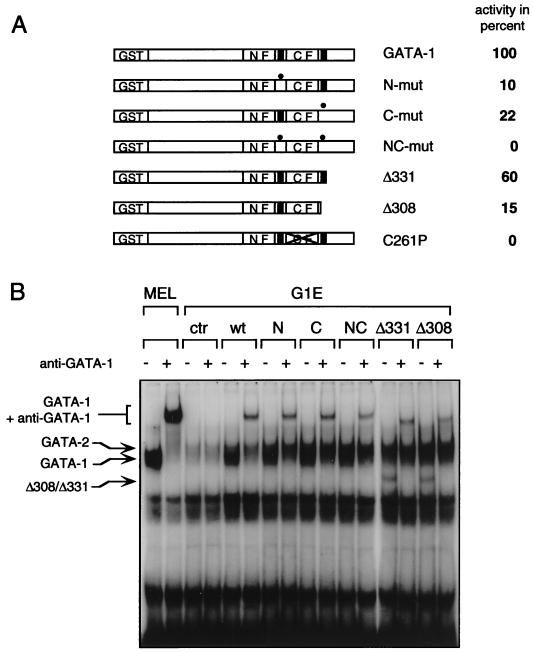
GATA-1-deficient G1E cells proliferate as immature erythroblasts and undergo terminal differentiation and hemoglobinization upon infection with GATA-1-expressing retrovirus (45). Previous structure-function studies of GATA-1 yielded important differences in domain requirements depending on whether these studies were performed with NIH 3T3 cells or in G1E cells (25, 45) (see the introduction). To assess the role of GATA-1 acetylation in an erythroid context with chromatinized GATA-1 target genes, we determined the ability of various GATA-1 mutants to induce G1E cell differentiation. Retroviruses harboring wild-type and mutant GATA-1 constructs were used to infect G1E cells, and the extent of erythroid differentiation was monitored by benzidine staining for hemoglobin. Infection with wild-type GATA-1-carrying virus induced hemoglobinization in about 7% of cells, which approximates the proportion of infected cells, similar to what has been reported previously (45). The number of benzidine-positive cells obtained with GATA-1 was used as a standard (100%) against which to compare the other GATA-1 constructs. A substantially reduced number of benzidine-positive cells was seen when either N-mut or C-mut was expressed (10% and 22% of wild-type levels, respectively) (Fig. 7A), while NC-mut was completely inactive, with no detectable benzidine-positive cells. Consistent with these findings, a GATA-1 mutant which lacks the entire C terminus (Δ308, residues 308 to 413 deleted), including the C-terminal acetylation motif, showed an activity comparable to that of C-mut (Fig. 7A). In contrast, a similar deletion construct (Δ331, residues 331 to 413 deleted) which leaves the C motif intact was almost as active as wild-type GATA-1. A non-DNA-binding mutant of GATA-1, which bears a mutation in the C-terminal zinc finger (C261P) (25), showed no activity in this assay (Fig. 7A). To confirm that expression levels and DNA binding of all GATA-1 constructs were comparable in erythroid cells, nuclear extracts were prepared from the infected G1E cells and analyzed by gel mobility shift assays with a GATA-1-binding site as probe. To distinguish GATA-1 from endogenous GATA-2, which migrates at a similar position, monoclonal anti-GATA-1 antibodies were used to generate a GATA-1-specific supershift. The amounts of GATA-1 supershifted with GATA-1 antibodies were comparable in all samples (Fig. 7B). Together, these experiments establish the requirement of both acetylation motifs for GATA-1 activity in erythroid cells. These results contrast with those obtained in NIH 3T3 cells, where mutations at the N-motif had no detectable effect, and underscore the importance of the cellular environment for the analysis of transcription factor activity.
FIG. 7.
Both N and C acetylation motifs are required for GATA-1-induced erythroid-cell differentiation. (A) G1E cells infected with a retrovirus carrying the indicated GATA-1 constructs were stained for hemoglobin with the dye benzidine. The number of hemoglobinized cells obtained with wild-type GATA-1 (∼7% of all cells in a representative field) was defined as 100%. N-mut and C-mut are as described in the legend to Fig. 2. C261P, GATA-1 mutant lacking DNA-binding activity; Δ308, GATA-1 mutant lacking the C terminus from aa 308 to 413; Δ331, GATA-1 mutant lacking the C terminus from aa 331 to 413. The critical difference between Δ331 and Δ308 is the presence or absence of the C-terminal acetylation motif. (B) Control gel shift from infected G1E cells demonstrating comparable expression and DNA binding of GATA-1 constructs. MEL cells which express high levels of GATA-1 but no GATA-2 served as controls. GATA-2 levels are high in G1E cells. To distinguish between GATA-1 and GATA-2, anti-GATA-1 antibodies were used where indicated to generate a supershift. wt, wild type.
DISCUSSION
It has long been known that acetylated histones are associated with transcriptionally active chromatin (2, 33). A strong correlation between histone acetylation and DNase I sensitivity has been demonstrated at the chicken β globin gene (17). Moreover, a number of transcription factors have recently been added to the list of acetyltransferase substrates (16, 19, 48). We have previously shown that the HAT CBP physically interacts with GATA-1 and stimulates its activity in an E1A-sensitive manner (4). Interference with CBP function by forced expression of E1A led to a block in erythroid differentiation and failure to induce GATA-1-dependent genes, including the α- and β-globin genes. Deletion of the N-terminal 23 aa of E1A, which abrogates binding to CBP/p300 but not to Rb family proteins, led to loss of the ability of E1A to block GATA-1 activity and erythroid differentiation (4). Of note, this E1A mutant retained the critical amino acids required for binding to p/CAF, which can also directly bind to E1A (34), suggesting that the effects of E1A are largely due to specific inhibition of CBP/p300 function. Since GATA-1-binding sites in the locus control regions of the globin genes contribute to maintaining DNase I hypersensitivity, this raised the possibility that GATA-1 acts by recruiting CBP to the locus control regions, thereby locally increasing histone acetylation. The results of the present study suggest that the interaction between CBP and GATA-1 entails an additional function, namely, acetylation of GATA-1 itself.
We demonstrated that CBP acetylates murine GATA-1 in vitro at two conserved lysine-rich motifs near the two zinc fingers. Using the CBP-related protein p300, Boyes et al. (7) mapped the major in vitro acetylation sites to the analogous motifs in chicken GATA-1, demonstrating the high degree of conservation between chicken and mouse GATA-1 in CBP- and p300-mediated acetylation.
Using anti-AK antibodies and in vivo labeling experiments we found that GATA-1 is acetylated in vivo, consistent with the findings of Boyes et al. (7). Using site-specific GATA-1 mutants, we further showed that acetylation in intact cells occurred at the same sites acetylated by CBP in vitro.
We also demonstrate that CBP can stimulate GATA-1 acetylation in vivo at the relevant sites and that expression of E1A abolishes acetylation completely. This demonstrates that CBP can strongly stimulate the acetylation of a transcription factor in vivo and that this activity is inhibited by E1A. These data further demonstrate that GATA-1 activity correlates well with its acetylation status (4).
While it is possible that other acetyltransferases such as SRC-1 and ACTR or unknown acetyltransferases acetylate GATA-1, several observations suggest that CBP is a major GATA-1 acetyltransferase. First, we have previously shown that CBP and GATA-1 associate in vivo; second, p/CAF does not acetylate GATA-1; third, there is remarkable specificity among different acetyltransferases regarding substrate and residue specificity (24, 36, 48).
To examine the mechanism by which CBP regulates GATA-1 activity, we showed that in vitro acetylation did not detectably alter DNA binding of either full-length GATA-1 or the zinc finger region alone. Substitutions of lysine residues with arginine or alanine residues, which maintain and neutralize the positive charge, respectively, did not affect DNA-binding and dissociation rates of GATA-1 constructs expressed in mammalian cells, further supporting the lack of involvement of the acetylated motifs in DNA binding. These findings are consistent with the observation that GATA-1 mutants bearing a deletion of the entire C terminus including the C-motif (Δ308) bind DNA normally (25, 42). Furthermore, the solution nuclear magnetic resonance spectroscopy structure of the C-terminal zinc finger of chicken GATA-1 bound to DNA demonstrated that the site corresponding to the C-terminal acetylation motif of murine GATA-1 does not make direct contact with DNA (29). Our findings contrast with those of Boyes et al., who reported a significant change in DNA binding upon in vitro acetylation (7). These discrepancies might be the result of differences between chicken and mouse GATA-1 and between CBP and p300. However, in agreement with our findings, mutations in the chicken GATA-1 acetylation sites do not alter DNA binding (7). Clearly, the strong acetylation-induced increase in DNA binding of a peptide spanning the zinc finger region of chicken GATA-1 observed by Boyes et al. in vitro is not reflected in the moderate stimulation of chicken GATA-1 activity by p300 in transient-activation assays (7). It is possible that the in vitro conditions do not reflect the actual changes in GATA-1 function triggered upon acetylation.
Sequence analysis of in vitro-acetylated peptides revealed that lysine 312 is the major acetylated residue in the C-motif while lysine 252 and lysine 246 are the predominant sites in the N-motif. Lysine 252, which is conserved among GATA-1 of different species, residues outside the canonical RXKK motif and might account for the residual acetylation observed in vitro with the NC-mut construct. We are in the process of testing the function of GATA-1 carrying a point mutation at this site. Nevertheless, even with lysine 252 intact, mutations at the N-motif lead to reduced acetylation and diminished biological function, indicating that the N-terminal acetylation motif is important.
While sequencing of acetylated synthetic peptides allows the identification of acetylation sites in the context of an intact amino acid sequence, i.e., without the potential artifacts which might result from the introduction of mutations, the results obtained by this method must be interpreted in the context of other experimental approaches. For example, we noted that acetylation of the peptide spanning the N-motif occurred with lower efficiency than did that of the peptide spanning the C-motif. However, when assayed in the context of full-length GATA-1, disruption of the C-motif reduced acetylation to only about half of that observed with wild-type GATA-1, indicating that the extents of acetylation at the N-motif and at the C-motif are comparable. Thus, sequences outside of the stretch of amino acids contained in the peptides might contribute to maximal acetylation efficiency, perhaps by increasing the binding affinity between GATA-1 and CBP.
In performing in vitro acetylation reactions, it is important to keep in mind that recombinant, purified acetyltransferases might display a substrate spectrum distinct from that observed in the presence of additional cofactors. For example, the recombinant purified yeast acetyltransferase Gcn5 acetylates only free histones but not histones packaged into nucleosomes (8). However, when assayed as a multicomponent complex, Gcn5 acetylates nucleosomal histones as well (for example, see reference 15). Therefore, cofactors bound to CBP or to GATA-1 might modulate the specificity and/or efficiency of the CBP acetyltransferase.
Transient-transfection experiments designed to test the functional role of acetylation showed that the C-motif but not the N-motif of GATA-1 is required for stimulation by CBP. Since the C-motif contributes to CBP binding, this suggests that physical association or subsequent acetylation or both are important for stimulating GATA-1 activity. Mutations in the C-motif do not interfere with acetylation at the N-motif in vitro (Fig. 2) or in vivo (Fig. 4), suggesting that the interaction between C-mut and CBP is sufficient for efficient catalysis. To address directly whether CBP-mediated acetylation is required for GATA-1 activation, we used two CBP mutants which lack HAT activity, one containing a point mutation and the other bearing a deletion in the HAT domain. Both constructs failed to acetylate GATA-1 and histones in vitro but activated GATA-1 with an efficiency comparable to that of wild-type CBP in transiently transfected NIH 3T3 cells. Together, these findings indicate that in transient-expression assays, acetylation of GATA-1 and histones by CBP is not required for GATA-1 stimulation. At least two explanations could account for this result. First, it is important to note that in transiently transfected nonerythroid cells with artificial reporter genes, CBP might stimulate GATA-1 activity by a mechanism different from that operational in erythroid cells with fully chromatinized GATA-1 target genes. The reporter gene used in the transient-transfection assays contains a single GATA site positioned closely to a TATA box. Therefore, in this setting, it is possible that CBP augments GATA-1 activity by serving as a link between GATA-1 and the basal transcription machinery. A functional domain analysis of CBP is being used to address this question. Second, it is possible that acetylation is mediated by p300/CBP-associated acetyltransferases, including ACTR (9) and SRC-1 (39, 47). Experiments are under way to test the role of the CBP-associated acetyltransferases during GATA-1 activation.
In performing similar transient-transfection experiments, Boyes et al. observed that a p300 mutant bearing a 50-aa deletion in the HAT domain failed to stimulate chicken GATA-1 activity (7). It is possible that such a deletion abrogated functions of p300 unrelated to its HAT activity.
While Boyes et al. did not test the function of the C-motif (the strongest acetylation site), mutations in the chicken GATA-1 N-motif resulted in a reduced response to p300, leading the authors to establish a correlation between chicken GATA-1 acetylation at the N-motif and transcriptional activation. Two open questions remain regarding this interpretation: first, the observed reduction in the p300 response is marginal (2.5-fold in wild-type GATA-1 compared to 1.4- and 1.9-fold in two acetylation mutants), especially in a transient-transfection assay. Second, the authors did not provide information about the acetylation status of these sites in vivo. In our experiments, mutations in the N-motif had no effect on DNA binding or transactivation by CBP in transient-transfection assays. Furthermore, binding of GATA-1 to a single GATA-1 site does not require the N-terminal zinc finger (25). Therefore, we believe that the mechanism by which the N-motif contributes to the GATA-1 function remains to be elucidated but is unlikely to involve regulation of DNA binding. In evaluating these findings, we point out that a biological function of the N-motif required the assay of GATA-1 constructs in erythroid cells with GATA-1-dependent genes present in their natural state (see below).
Since critical GATA-1 functions were uncovered only in maturing erythroid cells (6, 41, 45), we used the GATA-1-deficient cell line G1E to study the functional role of the acetylation sites. Mutations in either the N- or C-motif significantly reduced activity, while mutations in both motifs abrogated activity entirely. Thus, only in erythroid cells did we uncover a requirement for the N-motif. Mutations in the N-motif did not interfere with binding to CBP or to Fog (data not shown), the recently identified cofactor which selectively binds to the N-terminal zinc finger of GATA-1 (41), excluding the possibility that mutations simply disrupt the interaction of GATA-1 with its known cofactors. Deletion of the C terminus of GATA-1 leaving the C-motif intact (Δ331) resulted only in a small loss of activity, whereas a slightly larger deletion which removes the C-motif (Δ308) led to a much more pronounced loss of function without affecting DNA binding. Together, these results demonstrate that mutations in the acetylation motifs impair biological activity without affecting DNA binding, which demonstrates that the DNA-binding and acetylation functions can be uncoupled.
Interestingly, the ability of GATA-1 to induce megakaryocytic conversion of the early myeloid cell line 416B also depends in part on an intact C-motif (42). In these assays, Δ308 was significantly less active than Δ331, which raises the possibility that CBP also synergizes with GATA-1 in the regulation of megakaryocytic gene expression. Together, these results indicate that the C-terminal acetylation site is of great biological importance and are consistent with a requirement of CBP for GATA-1 function in vivo.
Acetylation of lysine residues neutralizes their positive charge and changes the size of the residue. In general, such changes could conceivably entail alterations in protein function similar to those incurred upon phosphorylation. For example, acetylation could lead to changes in protein conformation. Such a mechanism has been proposed for p53, where acetylation in the regulatory domain leads to increased DNA-binding activity (16). Alternatively, acetylation could directly influence protein-DNA interactions, as has been suggested for binding of histone tails to DNA, or it could affect protein-protein interactions, as described for binding of histone tails to the yeast transcriptional repressor Tup1 (13).
The mechanism by which either acetylation motif in GATA-1 exerts its function in G1E cells remains to be determined. Gel shift analysis with nuclear extracts of the virally infected G1E cells demonstrated the presence of equal amounts of DNA-bound GATA-1 proteins, and immunofluorescence microscopy confirmed the nuclear localization of all constructs. This suggests that acetylation in either motif does not regulate nuclear localization, DNA binding, or protein stability. Several additional mechanisms by which acetylation regulates GATA-1 activity could be envisioned. It is conceivable that acetylation leads to a conformational change in GATA-1, resulting in increased activity through exposure of an activation domain. Alternatively, the acetylation motifs might represent docking sites for still unidentified coactivators or repressors, and acetylation might positively or negatively regulate their binding affinity. More extensive studies, including structural analyses, will be required to determine the mechanistic role of GATA-1 acetylation.
ACKNOWLEDGMENTS
We thank Merlin Crossley, Colyn Crane-Robinson, Warren Pear, Marc Montminy, and Michael Rosenfeld for providing plasmids, antibodies, and cell lines. We are grateful to Dawn Eastmond and Toshio Asakura for assistance with HPLC separation of GATA-1 peptides. Peptide synthesis and protein-sequencing services were provided by the Protein Chemistry Laboratory of the Medical School of the University of Pennsylvania. We thank Margaret Chou, Merlin Crossley, Stuart Orkin, and Morty Poncz for helpful suggestions and for critically reading the manuscript.
This work was supported by the Baldasare Award (G.A.B.), the American Society of Hematology Scholar Award (G.A.B.) and the Cooley’s Anemia Foundation (G.A.B.). Services provided by the Protein Chemistry Laboratory of the Medical School of the University of Pennsylvania were supported by core grants of the Diabetes and Cancer Centers (DK-19525 and CA-16520).
REFERENCES
- 1.Abraham S E, Lobo S, Yaciuk P, Wang H G, Moran E. p300, and p300-associated proteins, are components of TATA-binding protein (TBP) complexes. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 2.Allfrey V, Faulkner R M, Mirsky A E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 4.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. CREB-binding protein (CBP) cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blobel G A, Sieff C A, Orkin S H. Ligand-dependent repression of the erythroid transcription factor GATA-1 by the estrogen receptor. Mol Cell Biol. 1995;15:3147–3153. doi: 10.1128/mcb.15.6.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blobel G A, Simon M C, Orkin S H. Rescue of GATA-1-deficient embryonic stem cells by heterologous GATA-binding proteins. Mol Cell Biol. 1995;15:626–633. doi: 10.1128/mcb.15.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 8.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crossley M, Merika M, Orkin S H. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallas P B, Yaciuk P, Moran E. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J Virol. 1997;71:1726–1731. doi: 10.1128/jvi.71.2.1726-1731.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edmondson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histone H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 14.Fujiwara Y, Browne C P, Cunniff K, Goff S C, Orkin S H. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones:characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 16.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 17.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hebbes T R, Turner C H, Thorne A W, Crane-Robinson C. A “minimal epitope” anti-protein antibody that recognises a single modified amino acid. Mol Immunol. 1989;26:865–873. doi: 10.1016/0161-5890(89)90143-0. [DOI] [PubMed] [Google Scholar]
- 19.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolfe A P, Ge H. Acetylation of general transcription factors by histone acetyltransferases. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 20.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S, Heyman R A, Rose D, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 21.Kee B, Arias J, Montminy M. Adaptor mediated recruitment of RNA polymerase II to a signal dependent activator. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 22.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T-M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 23.Kwok R P, Lunblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D I, Orkin S H. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf1. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Balbas M, Bannister A, Martin K, Haus-Seuffert P, Meisternst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakajima T, Uchida C, Anderson S F, Lee C-G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 28.Ogryzko V V, Schiltz L R, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 29.Omichinsky J G, Clore G M, Schaad O, Felsenfeld G, Trainor C, Appella E, Stahl S J, Gronenborn A M. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science. 1993;261:438–446. doi: 10.1126/science.8332909. [DOI] [PubMed] [Google Scholar]
- 30.Orkin S H. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 31.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 32.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogo P G T, Allfrey V G, Mirsky A E. RNA synthesis and histone acetylation during the course of gene activation in lymphocytes. Proc Natl Acad Sci USA. 1966;55:805–812. doi: 10.1073/pnas.55.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid J L, Bannister A J, Zegerman P, Martinez-Balbas M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth S Y, Allis C D. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 36.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sang N, Avantaggiati M L, Giordano A. Roles of p300, pocket proteins, and hTBP in E1A-mediated transcriptional regulation and inhibition of p53 transactivation activity. J Cell Biochem. 1997;66:277–285. doi: 10.1002/(sici)1097-4644(19970901)66:3<277::aid-jcb1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 38.Shikama N, Lyon J, LaThangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 39.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 40.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 41.Tsang A P, Visvader E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 42.Visvader J E, Crossley M, Hill J, Orkin S H. The C-terminal zinc finger of GATA-1 or GATA-2 is sufficient to induce differentiation of an early myeloid cell line. Mol Cell Biol. 1995;15:634–641. doi: 10.1128/mcb.15.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss M J, Keller G, Orkin S H. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1− embryonic stem cells. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 44.Weiss M J, Orkin S H. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc Natl Acad Sci USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss M J, Yu C, Orkin S H. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene targeted cell line. Mol Cell Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 47.Yao T-P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W, Bieker J J. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]