Abstract
RNA-based therapeutics has attracted substantial interest from both academics and pharmaceutical companies. In this study, we investigated the function and the underlying mechanism of Gelsolin (GSN) 3’UTR in NSCLC H1299 and A549 cells. We found that transfected Flag-GSN plasmids significantly increased the proliferation, migration and invasion of NSCLC cells, whereas GSN 3’UTR could suppress the promotional effect of GSN protein on the development of NSCLC in vitro. Interestingly, we observed that these in vitro anticancer effects of GSN 3’UTR was independent of the co-expression with GSN coding sequence. Moreover, transfected GSN 3’UTR affected the actin-cytoskeleton remodeling and epithelial-mesenchymal transition (EMT) processes in H1299 and A549 cells, and targeted the co-expressed proteins to the plasma membrane. Subsequently, RNA pull-down assays have been performed to identify Tra2β protein as a GSN 3’UTR binder. We then showed that Tra2β was important for the localized protein expression mediated by GSN 3’UTR. Taken together, our results suggested that GSN 3’UTR may exert anticancer functions in NSCLC cells through regulating the subcellular localized expression of GSN protein mediated by the interaction between GSN 3’UTR-Tra2β.
Keywords: 3’untranslated regions (3’UTR), GSN, Tra2β, non-small cell lung cancer (NSCLC)
Introduction
The utilization of synthetic RNA molecules to alter cellular functions, such as COVID-19 vaccine development based on mRNA technology, is currently out of scope for traditional drug design pipelines and has attracted substantial interest from both academics and pharmaceutical companies [1-3]. Because of the important role of mRNA in gene expression regulation and their high efficiency, safe management, the potential for low-cost production, and capacity for rapid manufacture, mRNA drugs represent promising alternative strategies for the treatment of malignant tumors. Up to now, vaccine platforms against several types of cancer, such as individualized neoepitope mRNA cancer vaccines [4], have presented encouraging results in both animal models and humans [5]. So far, several 3’UTRs in eukaryotic mRNAs have been found to have tumor suppressive activity in vitro, capable to cause malignant phenotypic reversion (reduced tumorigenicity) in cultured tumor cells or other malignant cells [6-8]. For example, transfection of NF-IL6 3’UTR induced tumor suppression in human hepatoma SMMC7721 cells. Yang et al. showed that FOXO1 3’UTR inhibited the metastases of breast cancer cells via induction of E-cadherin expression [7]. Hu et al. found that the CCR2 3’UTR inhibited breast cancer metastasis by repressing EMT in vitro and in vivo [8]. 3’UTRs exerted biological functions by regulating gene expression, mRNA localization, and competitive binding to miRNA or proteins [9-14]. However, up to date, only a small number of functional 3’UTRs have been characterized and the molecular mechanisms underlying these tumor suppressor effects remain unknown.
Lung cancer is one of the highest morbidity and mortality diseases in the world, and the metastasis is the main cause of death for patients with lung cancer [15,16]. 85% of lung cancers are NSCLC, which includes lung adenocarcinoma and lung squamous cell carcinoma [17,18]. Although significant progress has been made in the treatment for patients with advanced epidermal growth factor receptor (EGFR) mutated NSCLC, represented by first line drugs Gefitinib, Erlotinib and Osimertinib, the mortality of NSCLC still remains high, and the important reasons are ascribed to the high cost of drugs, the strong metastasis, unlimited proliferation, and high heterogeneity of lung cancer cells [19-21]. Therefore, it is of great significance to further elucidate the mechanism of pathological progression of NSCLC and identify novel potential efficient and low-cost drugs for the therapy of this cancer.
Gelsolin (GSN) is one of the most abundant actin-binding proteins that mediate cellular motility and play a pivotal role in the pathogenesis of human cancers [22,23]. However, its functions and regulatory mechanisms in NSCLC have not been fully elucidated. In our previous study, we have been interested in the function of the GSN 3’UTR in NSCLC cell in vitro [24]. In pursuing our study, we found that the 3’UTR of GSN mRNA can reverse the promotional effect of GSN protein on the proliferation, migration, invasion and EMT in H1299 and A549 NSCLC cells, and had a tumor suppressive effect which could be independent of the exogenously co-expressed GSN protein. In this report, we describe these effects and investigate the underlying molecular mechanisms.
Materials and methods
Cell lines and culture
NSCLC cell lines H1299 and A549 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). These cells were maintained in high-glucose DMEM medium with 10% fetal bovine serum. All cell lines used in this study was free of mycoplasma contamination.
Plasmids and cloning
The coding sequence (CDS) of human GSN (2196 bp), and corresponding GSN 3’UTR (240 bp) sequence were amplified from total RNA by RT-PCR. The CDS of GSN flanked by BamH I and Xho I restriction sites were inserted into pCMV-flag plasmid (Flag-GSN). The GSN 3’UTR sequence flanked by Xho I and Spe I restriction sites was inserted into pCMV-flag-GSN, pCMV-N-EGFP or pCMV-Luc plasmids to construct the Flag-GSN-3’UTR, EGFP-3’UTR and Luc-3’UTR vectors, respectively. For overexpressing Tra2β, human Tra2β cDNA without the 3’UTR was cloned into pCMV-flag plasmid (Flag-Tra2β) at BamH I and Xho I sites. All plasmids were sequenced and confirmed for accuracy.
Small interfering RNA transfection
Sequences of siRNA were designed as follows: (siTRA2β-1: 5’-AGCUAAAGAACGUGCCAAUTT-3’; siTRA2β-2: 5’-CCGAUGUGUCUAUUGUAUATT-3’; siTRA2β-3: 5’-ACGCCAACACCAGGAAUUUTT-3’; siNC: 5’-UUCUCCGAACGUGUCACGUTT-3’). LipofectamineTM 2000 (Invitrogen, CA, USA) was used to transfect cells according to manufacturer’s instructions.
mRNA quantification by RT-qPCR
Total RNA of cells was purified and qRT-PCR was performed as previously described. The relative expression level of mRNA was calculated using the 2-ΔΔCt method [25,26]. The primers were synthesized as follows (Table 1).
Table 1.
The primer sequences
| Primer | Forward sequence | Reverse sequence |
|---|---|---|
| Vimentin | CCACCAGGTCCGTGTCCTCGT | CGCTGCCCAGGCTGTAGGTG |
| E-cadherin | TTGCACCGGTCGACAAAGGAC | TGGAGTCCCAGGCGTAGACCAA |
| N-cadherin | CCCTGCTTCAGGCGTCTGTA | TGCTTGCATAATGCGATTTCACC |
| Gelsolin | GATCTGGCGTGTGGAGAAGTTCG | CTCTCATCCTGGCTGCACTCATTG |
| GAPDH | GACAGTCAGCCGCATCTTCT | GCGCCCAATACGACCAAATC |
Immunofluorescence (IF) assays
IF assay was performed to confirm the subcellular localization of the indicated proteins as previously described [25]. Appropriate primary antibodies [Gelsolin (11644-2-AP, 1:200), TRA2β (900958001, 1:100)] were used to blot overnight and Alexa Flour 488 or Alexa Fluor 594-conjugated secondary antibody (ZSGB-BIO Co., Ltd., China) was used. F-actin was stained with Rhodamine-phalloidin (Invitrogen™ R415) and the nuclei were stained with DAPI (Beyotime Biotechnology) both for 30 min at room temperature. Cells were captured using Confocal Zeiss LSM 700 equipped with a Zeiss Plan-Neofluar 40 ×/0.75 Oil DIC.
Cell migration and invasion assay
Cell culture inserts (24 well, 8 μm pore size) were used for migration assays and Matrigel-coated chambers (8 µm pore size; BD Biosciences) were used for invasion assays. H1299 and A549 cells (6-8 × 104/well) in serum-free DMEM were plated into the upper chamber. Complete medium containing 10% FBS was added to the lower chamber as chemoattractants. After 8-10 h incubation, the migrated or invaded cells were fixed, stained with crystal violet, photographed and quantified in five random fields.
MTT assay
Cells were seeded into 96-well plates at a density of 2 × 103 cells/well and cultured overnight. 200 µl working fluid with a final concentration of 5 mg/mL MTT was added to detect cell proliferation at each time-point by measuring the absorbance at 570 nm.
Colony formation assay
Cells were seeded into 6-well plates at a density of 1000/well and then cultured for 2 weeks and stained with crystal violet dye. Clones containing more than 50 cells were counted as one colony.
Western blotting
This assay was performed as described previously [25,26]. Primary anti-bodies against the following proteins were used: Gelsolin (11644-2-AP; ProteinTech Group), GAPDH (10494-1-AP; ProteinTech Group), E-cadherin (20874-1-AP; ProteinTech Group), Vimentin (10366-1-AP; ProteinTech Group), DDDDK-Tag (66008-3-Ig; ProteinTech Group), β-tubulin (10068-1-AP; ProteinTech Group) and TRA2β (900958001; ABclonal Group).
Biotin pull-down assay
Flag-GSN or Flag-GSN-3’UTR PCR fragments containing the T7 RNA polymerase promoter sequence were used as templates to produce the Biotinylated transcripts in vitro using T7 RNA polymerase (Invitrogen) and biotin-CTP (Perkin Elmer), and then purified with RNeasy Mini Kit (Qiagen). Biotin pull-down assays were performed by incubating 40 μg of cell lysates with 1 μg of biotinylated transcripts for 30 min at room temperature. The RNA-protein complexes were isolated with streptavidin-coated magnetic beads (Thermo), and the bound proteins were analyzed by Western blotting.
Cell motility video recordings
5 × 104 cells/well were seeded into 6-well plates which were then placed on the stage of an inverted microscope equipped with phase-contrast optics, and cultured in a carbon dioxide cell incubator (Sanyo, Japan). A computer-controlled movable stage was mounted on the microscope, allowing simultaneous recordings from several microscopic fields via Cyto-MINI software (Guangzhou Sipu Photoelectric Technology Co. LTD, China). For the observation of cell motility, images of cells were taken sequentially with an interval of 5 min, for a total duration of 20 h.
Statistical analysis
The significance of the data between the two groups were determined by paired Student’s t-tests using GraphPad Prism 6 software (GraphPad Software, USA) [27]. Significant differences between multiple groups were analyzed by one-way or two-way analysis of variance (ANOVA). P < 0.05 was considered as a significant result.
Results
GSN 3’UTR could suppress the promotional effect of GSN protein on the growth, migration and invasion of NSCLC cells in vitro
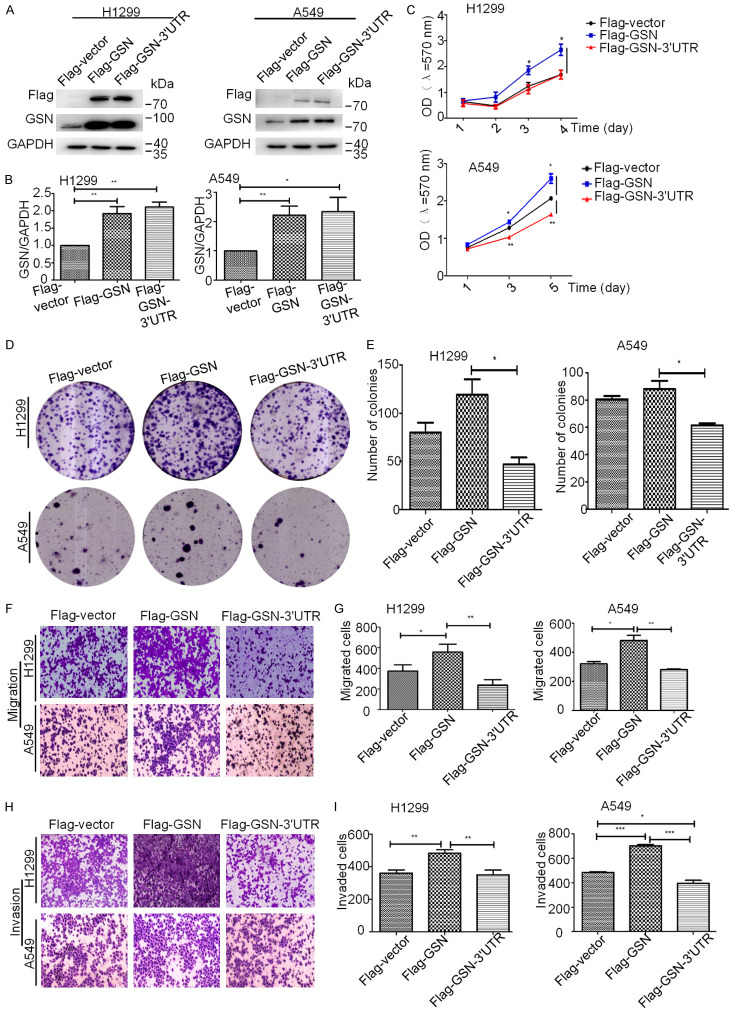
To explore the biological function in NSCLC progression of the coding and non-coding 3’UTR sequence in the GSN mRNA, we constructed Flag-GSN and Flag-GSN-3’UTR expression vectors, respectively containing the coding sequence of GSN alone or with the 3’UTR at the 3’-end right after the stop codon. H1299 and A549 cells were then either transfected with Flag-GSN or Flag-GSN-3’UTR plasmid for 24 h, collected and subjected to Western blotting analysis for the verification of GSN overexpression. We found that both H1299 and A549 cells transfected with Flag-GSN or Flag-GSN-3’UTR vectors displayed higher GSN protein levels than those transfected with control plasmids (Flag-vector) (Figure 1A and 1B). Then, MTT assay was used to evaluate the cell viability. Our results showed that Flag-GSN significantly increased the growth of both H1299 and A549 cells as compared with cells transfected with Flag-empty plasmids (Figure 1C). Surprisingly, Flag-GSN-3’UTR transfected cells displayed similar (for H1299 cells, upper diagram), or less viability (for A549 cells, lower diagram) than control cells, and even lesser than those transfected with Flag-GSN plasmids. Colony formation assays were performed to further verify these results. Our results showed that Flag-GSN overexpression did not significantly affect the number of colonies of H1299 and A549 cells in vitro, although the colony size was bigger with Flag-GSN transfected cells than those transfected with empty plasmids. In contrast, overexpression of Flag-GSN-3’UTR not only reduced the size of colonies, but also significantly suppressed the ability of H1299 and A549 cells to form colonies in vitro as compared with Flag-GSN overexpressing cells (Figure 1D and 1E).
Figure 1.
GSN 3’UTR could suppress the promotional effect of GSN protein on the growth, migration and invasion of NSCLC cells in vitro. H1299 (A) and A549 (B) cells were transfected with Flag-vector, Flag-GSN and Flag-GSN-3’UTR plasmids, and analyzed by western blotting. Signal intensities were quantified with GAPDH as internal control. (C) The effects of Flag-vector, Flag-GSN and Flag-GSN-3’UTR overexpression on the proliferation of H1299 (upper panel) and A549 cells (lower panel), analyzed by MTT assay. (D, E) H1299 and A549 cells were transfected with the indicated plasmids, and the colony formation ability was quantified by a colony formation assay. (F, G) Transwell migration assays of H1299 and A549 cells transfected with the indicated plasmids. (H, I) Transwell invasion assays of H1299 and A549 cells transfected with the indicated plasmids. Data are presented as the mean ± s.d. and are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Subsequently, we assessed the possible role of the 3’UTR of GSN on the migration and invasion abilities of NSCLC cells. Transwell migration and Matrigel invasion experiments were performed with Flag-GSN or Flag-GSN-3’UTR transfected H1299 and A549 cells as hereinabove. As shown in Figure 1F-I, we found that Flag-GSN significantly increased the cell migration and invasion in both H1299 and A549 cells. However, overexpression of Flag-GSN-3’UTR in H1299 and A549 cells led to a significant decrease in the cell migration (Figure 1F and 1G) and invasion (Figure 1H and 1I) as compared with Flag-GSN overexpressing cells, and presenting a level close to that of the control cells.
Taken together, our results indicated that GSN 3’UTR exerted opposite effects of exogenic GSN protein on NSCLC cells in vitro, reversing the effect of the latter.
Opposite effects of GSN and GSN 3’UTR on EMT process in H1299 and A549 cells
GSN has been reported to function as a switch that controls E- and N-cadherin conversion via Snail in human mammary epithelial cells, and to promote the EMT in hepatocellular carcinoma [28,29]. To investigate whether the 3’UTR of GSN can affect the regulation of EMT by GSN in lung cancer cells, we examined changes in expression levels of key EMT-related markers in H1299 and A549 cells transfected either by Flag-GSN or by Flag-GSN-3’UTR. The western blot analysis of H1299 cells in Figure 2A showed that the downregulation of epithelial marker E-cadherin was observed, concomitant with the upregulation of mesenchymal intermediate filament Vimentin in Flag-GSN overexpressing cells, whereas GSN-3’UTR weakened the function of GSN protein in promoting the EMT transition. In Figure 2B, western blot analysis of A549 cells ascertained the notable increase in the expression of Vimentin in Flag-GSN overexpressing cells, and as expected, Flag-GSN-3’UTR still counteracted this function. However, for the effects of Flag-GSN and Flag-GSN-3’UTR on the expression of E-cadherin, quantitative analysis did not lead to a conclusive result, probably due to the technical limitations of Western blotting as a quantitative analysis method (Figure 2A and 2B). Indeed, the mRNA levels of E-cadherin, N-cadherin, Vimentin determined by qRT-PCR confirmed the regulation of EMT process by Flag-GSN and Flag-GSN-3’UTR (Figure 2C and 2D). The mRNA levels of N-cadherin and Vimentin were both enhanced in H1299 and A549 cells transfected with Flag-GSN plasmids, whereas such a promotion of mesenchymal conversion did not occur in H1299 and A549 cells transfected with Flag-GSN-3’UTR plasmids. Taken together, these data suggest that the 3’UTR of GSN could abrogate the ability of the co-overexpressed GSN protein to promote the EMT process and blocked the cell invasion of NSCLC.
Figure 2.
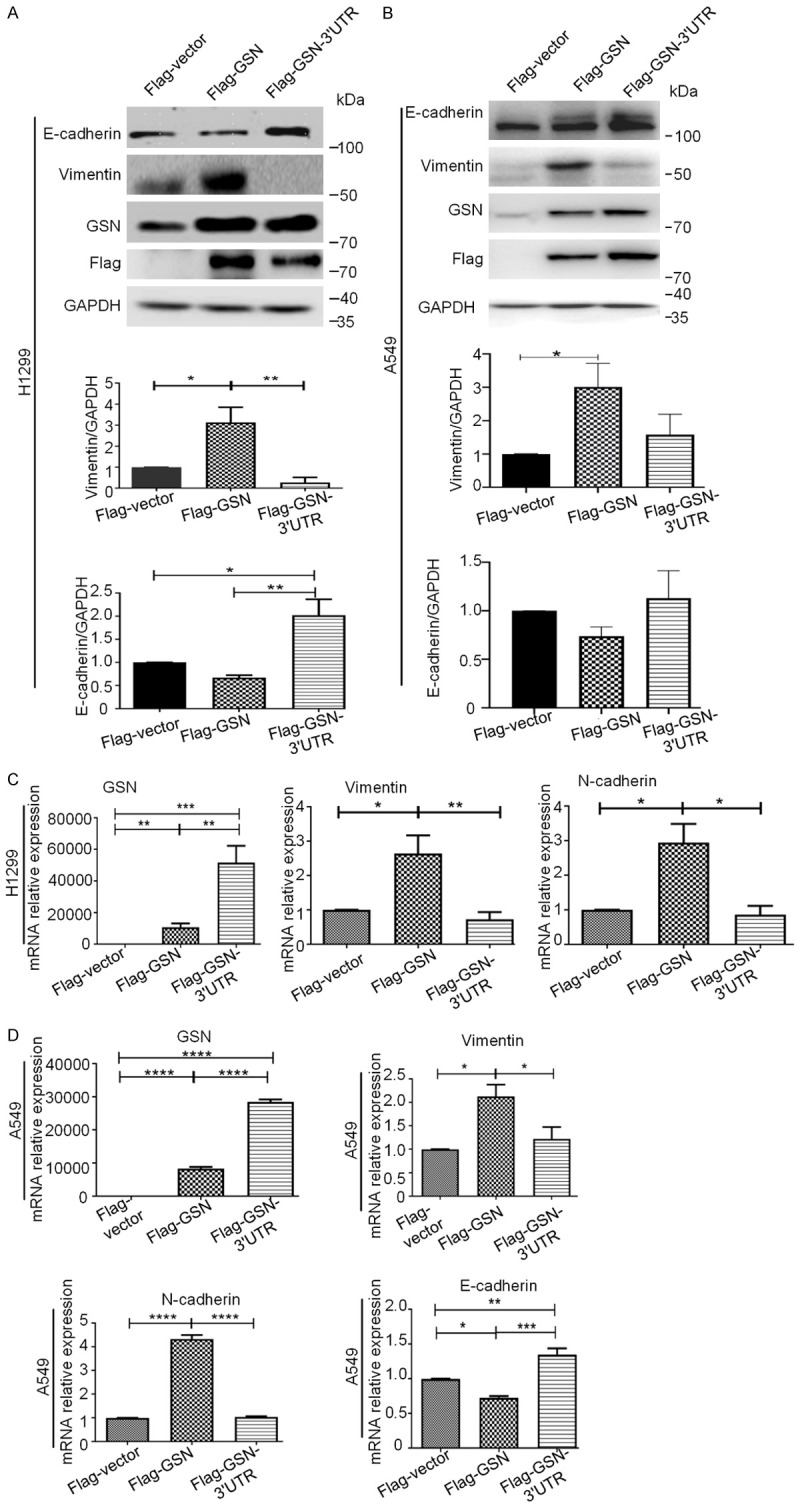
GSN or GSN 3’UTR exerted opposite effects on EMT process in H1299 and A549 cells. Protein expression of E-cadherin, Vimentin and transfected Flag-GSN expression by Western blotting in H1299 (A) and A549 (B) cells after transfected with the indicated plasmids. Signal intensities were quantified with GAPDH as internal control. (C, D) The mRNA levels of EMT marker and GSN were shown by qRT-PCR after transfected with the indicated plasmids in H1299 (C) and A549 (D) cells. Data are presented as the mean ± s.d. and are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
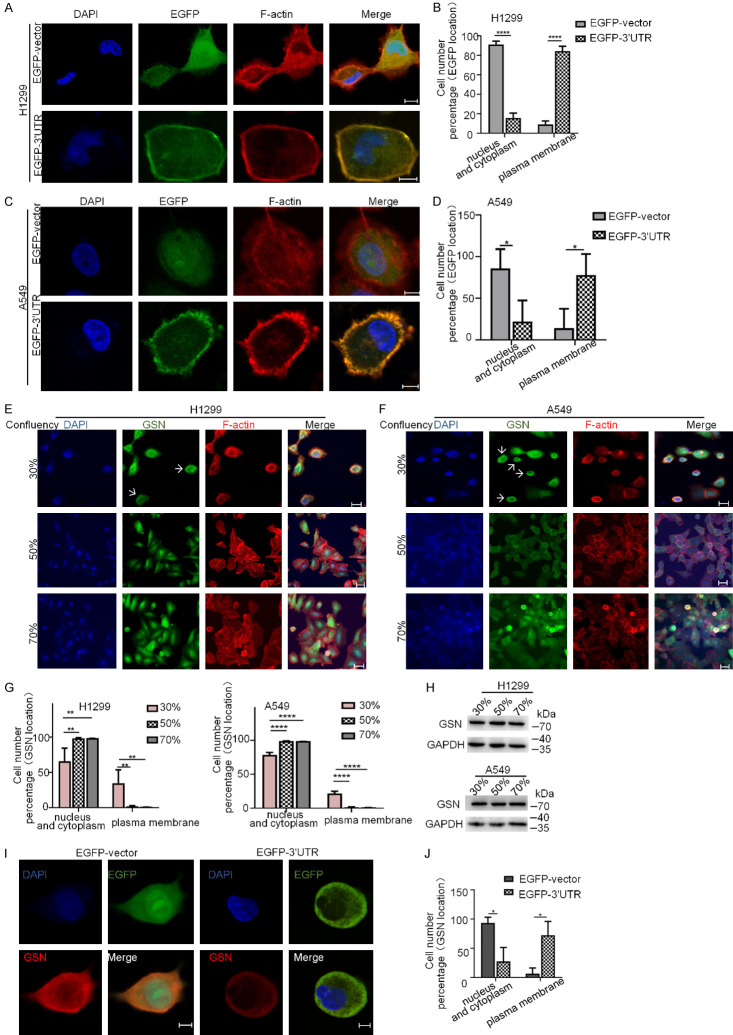
GSN 3’UTR localized the exogenously co-expressed GSN to the membrane
Our results in Figure 1A showed that the attachment of the 3’UTR to the CDS of GSN did not alter significantly its protein expression level. We then assessed if GSN 3’UTR might regulate the localization of its co-expressed GSN protein. By cell immunofluorescence analysis, we noticed that, in contrast to control (empty plasmid-transfected) cells, Flag-GSN-overexpressing H1299 cells developed stress fibers across cell body, with Flag-GSN proteins dispersed all over the whole cell body. However, Flag-GSN-3’UTR overexpressing H1299 showed more actin-rich short protrusions, with exogenous GSN protein predominately enriched at the plasma membrane, forming a ring-like structure (Figure 3A and 3C). Similarly, Flag-GSN and Flag-GSN-3’UTR overexpression in A549 cells also resulted in significantly different cytoskeletal actin remodeling, concomitant with different localization of exogenous GSN (Figure 3B and 3D). Taken together, these results suggested that the alterations of GSN subcellular distribution induced by its co-expressed 3’UTR may be directly responsible for determining the regulation of GSN on the proliferation, migration and invasion of H1299 and A549 cells.
Figure 3.
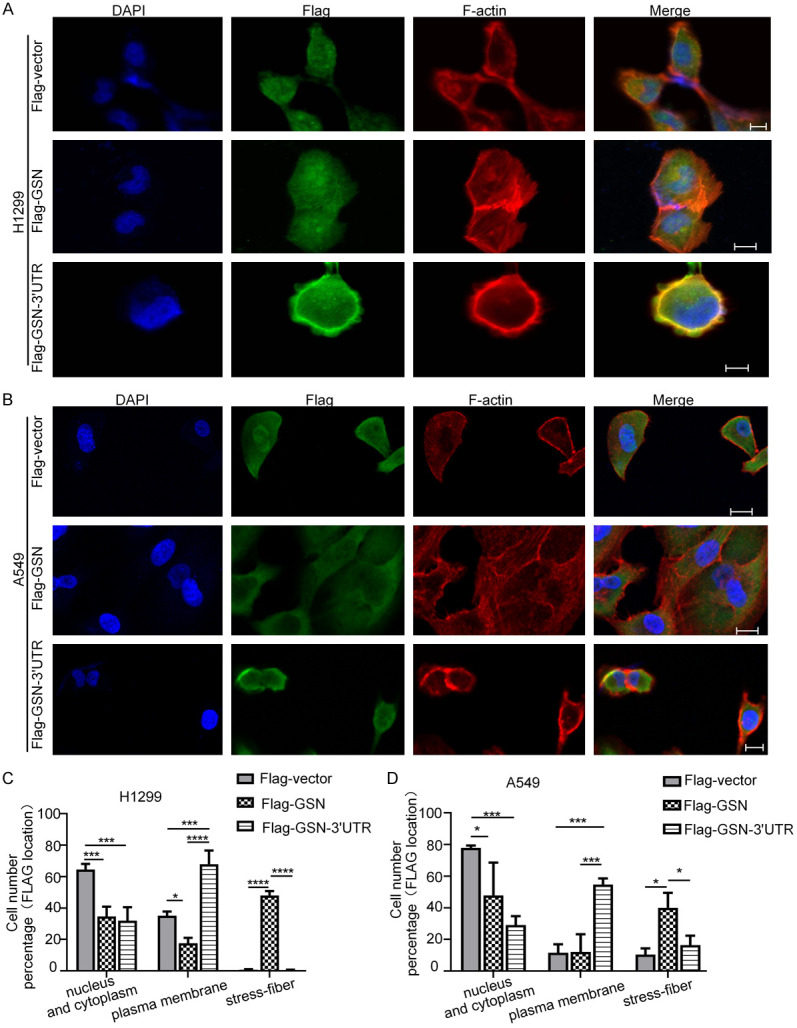
GSN 3’UTR localized the exogenously co-expressed GSN to the membrane. Subcellular location of Flag-GSN protein by immunofluorescence confocal microscopy. H1299 (A) and A549 (B) cells transfected with the indicated plasmids were stained with appropriate primary and secondary antibodies. DAPI was used for nuclear staining. F-actin was stained using rhodamine-conjugated phalloidin. Scale bars: 10 μm. (C, D) The counting of cells displaying various Flag-GSN location shown in (A and B). Data shown were from three independent experiments (> 15 cells each, n=3).
The functions of GSN 3’UTR could be independent of the exogenously co-expressed GSN protein
We then addressed whether the functions of GSN 3’UTR were dependent of the exogenously co-expressed GSN protein. To this end, we have replaced the co-expressed GSN CDS sequence by the firefly luciferase CDS sequence. The 3’UTR of GSN was inserted right after the stop codon of luciferase CDS in a pCMV-Luciferase vector. H1299 and A549 cells transfected with the yielded vector (Luc-3’UTR) or the control vector (Luc-vector, expressing Luc CDS without GSN 3’UTR) were then analyzed by transwell migration, matrigel invasion, and MTT assays. Both H1299 and A549 cells transfected with the GSN 3’UTR had a significantly decreased level of cell migration, metastasis, and proliferation in vitro (Figure 4A-E). These phenotype changes were again associated with the upregulation of E-cadherin expression and the downregulation of Vimentin and N-cadherin as determined by western blot analysis (Figure 4F). Quantitative real-time PCR showed that GSN 3’UTR consistently reduced the expression of mesenchymal markers, N-cadherin, and Vimentin (Figure 4G). Together, our data indicated that GSN 3’UTR could exert its anticancer functions in vitro in a manner independent of the co-expressed GSN protein.
Figure 4.
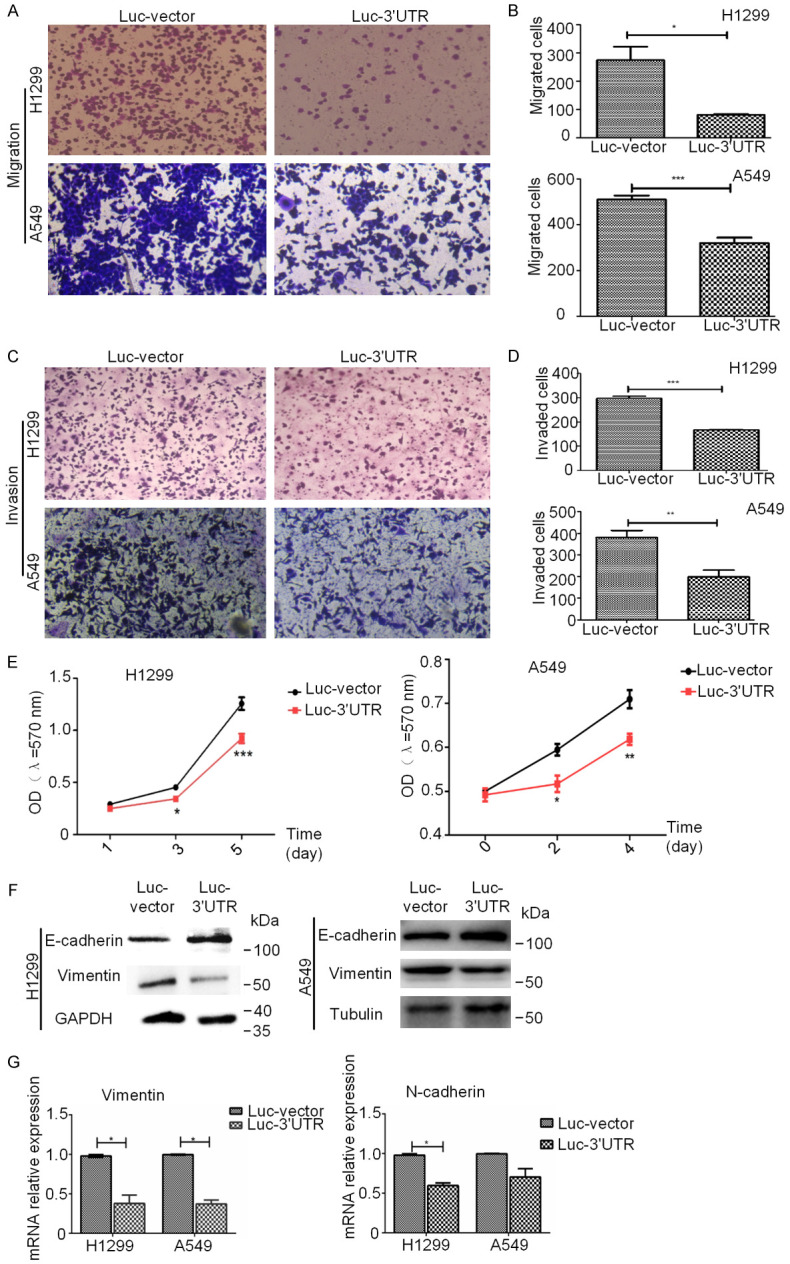
The functions of GSN 3’UTR could be independent of the exogenously co-expressed GSN protein. A, B. Cell migration ability of H1299 and A549 cells transfected with Luc-vector or Luc-3’UTR was analyzed by transwell assay. C, D. Invasion ability of H1299 and A549 cells transfected with Luc-vector or Luc-3’UTR was measured by use of transwell assay. E. Transfected of GSN 3’UTR in H1299 and A549 cells decreased the proliferation rate by MTT assay. F. The level of E-cadherin, Vimentin and GAPDH was examined by western blotting analysis in H1299 and A549 cells transfected with Luc-vector or Luc-3’UTR. G. The level of Vimentin/GAPDH and N-cadherin/GAPDH was examined by RT-qPCR. Data are presented as the mean ± s.d. and are representative of at least three independent EXPERIMENTS. *P < 0.05, **P < 0.01, ***P < 0.001.
We then asked if GSN 3’UTR was also capable to target another protein than GSN to the peripheral region of the cell. To this end, GSN 3’UTR were attached to the open reading frame of enhanced green fluorescence protein (EGFP) and then expressed in H1299 and A549 cells by transfection. Cell immunofluorescence experiments revealed that GSN 3’UTR could target the co-expressed EGFP to the proximity of the plasma membrane in co-localization with the F-actin forming a ring-like structure. As commonly observed in previous studies, EGFP protein encoded by EGFP CDS in the control cells displayed a diffused pattern of expression (Figure 5A-D). This result clearly indicated that GSN 3’UTR could target the expression of the upstream CDSs (including that of GSN and other genes such as EGFP) to the proximity of the plasma membrane.
Figure 5.
The 3’UTR of GSN mRNA is sufficient for the mRNA localization in the proximity of the plasma membrane. (A-D) Immunofluorescence confocal microscopy and corresponding quantification analyses of EGFP subcellular localization in H1299 (A, B) and A549 (C, D) cells transfected with EGFP-vector and EGFP-3’UTR plasmids. (E-H) Immunofluorescence confocal microscopy and corresponding quantification analyses of endogenous GSN subcellular localization in H1299 (E, F) and A549 (G, H) cells at various confluences. Cells were seeded at 1 × 104 cells/cm2 and cultured for a total of 24, 48 or 72 h in standard culture medium. At the indicated confluence, cells were fixed and immunolabelled with antibodies for detection of GSN. DAPI was used for nuclear staining. F-actin was stained using rhodamine-conjugated phalloidin. Arrows: endogenous GSN proteins at the proximity of the plasma membrane. (I, J) Immunofluorescence analyses of the localization of transfected EGFP and endogenous GSN proteins in H1299 cells transfected with the indicated plasmids. Quantification analysis was performed based on counting of cells with diverse protein distribution patterns from three independent experiments (> 15 cells each, n=3). Scale bars: 10 μm.
Transfected GSN 3’UTR disturbed the subcellular localization of endogenous GSN protein in NSCLC cells
During our study, we have observed that endogenous GSN proteins were differently distributed at different cell density in culture. As shown in Figure 5E-H, while a considerable portion of cells expressed GSN proteins at the proximity of the plasma membrane when cells were sparse (at 30% of confluence), in the cells at higher densities (50% and 70% of confluence), GSN protein were expressed in the nucleus and cytoplasm. Moreover, in reminiscence of previous reported findings [30], we have observed that cells in vitro culture displayed different mobility at different confluence. In order to demonstrate this observation, we have recorded the motility of cells at different density of culture in the same and also in separated microscopic vision fields during 20 h by taking a photograph per 5 min. In the accelerated (4000 folds) slideshow of these photos, we could observe that cells at low confluence (about 30% of confluence) were less prone to migration than other cells at higher confluence in the surrounding area. However, in the second half of the recording, when cell density became higher, these cells also were subjected to cell division and migration (Supplementary Video 1, cells denoted by curved arrows). This suggests that the plasma membrane localization of endogenous GSN, mainly occurring when the cell density was low, might be related to the decrease in cell motility. Consistently, in EGFP-vector transfected H1299 and A549 cells, similarly as observed for EGFP protein, endogenous GSN were evenly distributed throughout the cell body. However, when transfected with EGFP-3’UTR plasmid, a large proportion of cells expressed GSN mainly at the cell membrane, in co-localization with EGFP protein (Figure 5I and 5J). These results suggested that the exogenously expressed 3’UTR of GSN mRNA could exert its functions through disturbing the subcellular distribution of the endogenous GSN protein.
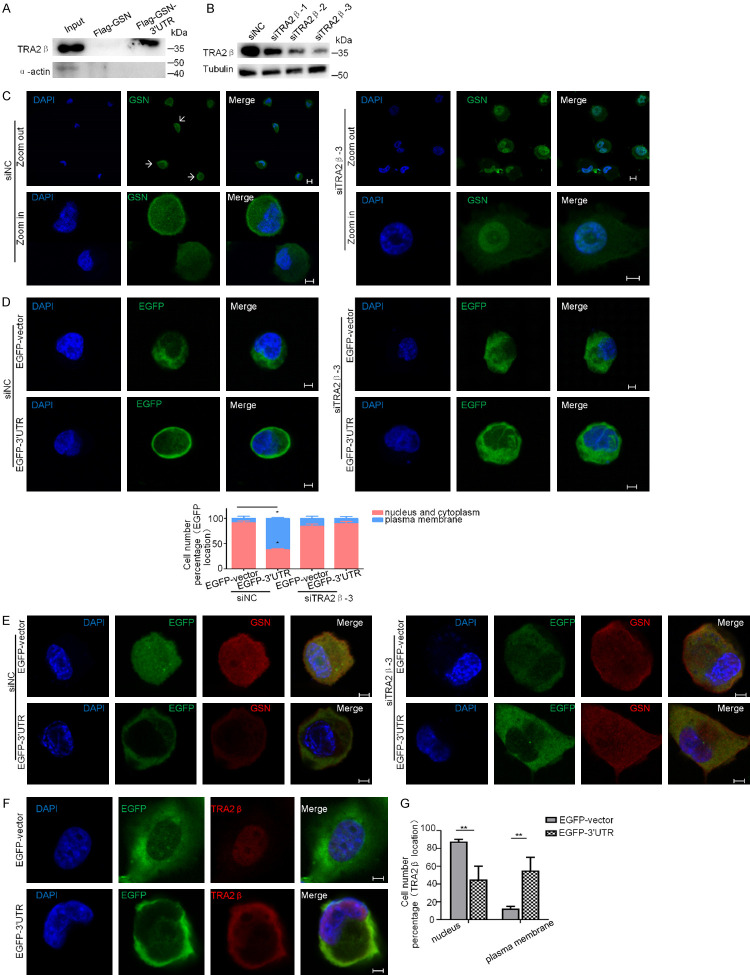
Tra2β bound to GSN 3’UTR and was important for the localized expression of GSN
It is widely acknowledged that the 3’UTRs are sites for interactions with trans-acting effector factors. In order to identify proteins interacting with GSN 3’UTR, we have performed an RNA pull-down assay using in vitro synthesized Flag-GSN-3’UTR transcript with H1299 cell lysates. A total of 66 interacting proteins were then identified using liquid chromatography-mass spectrometry (LC-MS) (Supplementary Figure 1A). 54 of them also bound to Flag-GSN (CDS) transcript, thus excluded from further analysis. The remaining 12 3’UTR-specific binders are listed in Supplementary Figure 1B, most of which are reported in the literature as RNA binding proteins. Among these proteins, we were particularly interested by Tra2β (Transformer-2 protein homolog beta).
Tra2β is recognized as a modulator of gene function and associated with cancer. It interacts with RNA targets through its RNA recognition motifs (RRMs) [31,32]. To directly confirm the association of Tra2β with the 3’UTR of GSN, biotinylated transcripts of Flag-GSN or Flag-GSN-3’UTR was incubated with H1299 cell lysates, the RNA-protein complexes were precipitated with streptavidin-coated beads, and the precipitated Tra2β or α-actin (as a negative control) was examined by western blotting. As shown in Figure 6A, biotinylated transcript of Flag-GSN-3’UTR specifically interacted with Tra2β, but not with α-actin.
Figure 6.
Tra2β bound to GSN 3’UTR and was important for the localized expression of GSN. (A) RNA pull-down assay. H1299 cell lysate was incubated with in vitro synthesized Flag-GSN transcript or Flag-GSN-3’UTR transcript as described in Materials and methods. Western blot analysis was performed to detect Tra2β protein bound specifically to GSN 3’UTR. α-actin was used as a negative control. (B) Verification of Tra2β knockdown in H1299 cells using three siRNAs by Western blotting. β-tubulin was used for internal control. (C) H1299 cells were transfected with siTra2β-3 or siNC, and then immunostained with antibodies against GSN (green). Nuclear DNA was stained with DAPI (blue). Arrows: endogenous GSN proteins at the proximity of the plasma membrane. (D, E) H1299 cells were subsequently transfected with siNC or siTra2β-3 for 24 h, and with EGFP-3’UTR or EGFP-vector for another 24 h, then subjected to immunofluorescence confocal microscopy for the subcellular localization of EGFP (D) or endogenous GSN protein (E). (F, G) Immunofluorescence analyses the subcellular localization of EGFP and endogenous Tra2β protein in H1299 cells transfected with EGFP-vector and EGFP-3’UTR plasmids. The cell number with each indicated expression pattern was quantified from three independent experiments (> 15 cells each, n=3). Scale bars: 10 μm.
Next, we examined whether Tra2β knockdown affected the localization of GSN. Three specific siRNA against Tra2β (siTra2β-1, -2 and -3) were designed and tested as described for their efficiency in H1299 cells (Figure 6B). We performed cell immunofluorescence experiments to examine the subcellular distribution of GSN in H1299 cells transfected with siTra2β or siNC. Interestingly, we found that in siNC-treated control cells, endogenous GSN was either located near the plasma membrane or evenly distributed throughout the cell body. However, in Tra2β knockdown cells, endogenous GSN was mainly located in the nucleus (Figure 6C). Subsequently, the role of Tra2β in GSN 3’UTR-mediated mRNA localization was investigated. H1299 cells were transfected either with specific Tra2β-targeting siRNA or with siNC for 24 h, and then transfected with EGFP-3’UTR or control plasmids for another 24 h. The subcellular localization of EGFP protein was again examined by confocal microscopy. When transfected with EGFP-3’UTR, EGFP and endogenous GSN displayed an expected ring-like localization pattern in siNC treated cells, whereas this localization pattern was much less frequently in Tra2β knockdown cells, regardless of whether cells were transfected with EGFP-3’UTR or not (Figure 6D, 6E and Supplementary Figure 1C). This phenotype could be reversed through the reconstitution of Tra2β (Supplementary Figure 1D). Re-expression of Tra2β increased the metastatic capacity of Tra2β knockdown cells, in the case of co-transfection with EGFP-3’UTR (Supplementary Figure 1E).
Furthermore, as shown in Figure 6F and 6G, Tra2β was mainly localized in the nucleus of EGFP-vector transfected H1299 cells, but in the EGFP-3’UTR transfected cells, a large number of Tra2β has been translocated to cytoplasm, precisely in the cell periphery, forming a ring-like structure at the proximity of the plasma membrane. These results suggested that the GSN 3’UTR-induced translocation of EGFP protein or endogenous GSN protein was mediated by the interaction between GSN 3’UTR and its protein partner Tra2β.
Discussion
GSN expression is downregulated in 60-90% of tumors during carcinogenesis of the breast [33], colon [34], stomach [35], bladder [36], prostate [37], and lung [38]. Literature reported that GSN may be a tumor suppressor that exerts a crucial role in the carcinogenic process [23,35,39]. Hiroki et al. showed that GSN functions as a switch that controls conversion of cadherin from the E- and N-type, and demonstrated that its knockdown induces EMT in human mammary epithelial MCF10A cells [40]. Conversely, Zhang et al. showed that GSN promotes HCC progression by influencing the EMT process [28]. These conflicting observations hinted on the versatile functions and complex regulation mechanisms of GSN. Conceivably, the activities of GSN may be regulated not only by its expression level but also by its subcellular localizations, where different locations elicit different functions. For instance, besides its role in the cytoplasm by severing, capping, nucleating or bundling actin filaments, GSN could also be endowed of unexpected nuclear functions such as transcription regulation [23,41,42]. Particularly, the role of the 3’UTR of GSN mRNA has not yet been investigated. Meanwhile, during the last decades, there have been studies concerning the association of 3’UTR functions with diverse diseases, including cancer [11,12,43,44]. In our study, when H1299 and A549 cells were transfected with plasmids containing Flag-GSN coding sequence, Flag-GSN was uniformly distributed in the cytoplasm, accompanied by the appearance of stress fiber and increased cell migration and invasion. However, the insertion of non-coding 3’UTR sequence in the above expression vector led to the Flag-GSN accumulation at the proximity of the plasma membrane accompanied by distinct changes in actin cytoskeleton and reduced cell growth, migration and invasion (Figures 1, 2 and 3). These results suggested that GSN 3’UTR might counteract the function of the co-expressed GSN protein to promote cancer development in vitro. Interestingly, our further investigation indicated that the effect of GSN 3’UTR could be independent of the exogenously co-expressed GSN (Figure 4). We found that transfected GSN 3’UTR could perturb the localization of endogenous GSN protein. Consistent with this finding, differential localization of GSN was a characteristic associated with different cell confluence of in vitro culture, and could be correlated with different cell migration potential (Figure 5 and Supplementary Video 1).
By using RNA-pull down assay followed by LC-MS, we identified Tra2β as a GSN 3’UTR-binder. Knockdown of Tra2β prevented GSN 3’UTR-mediated membrane localization of both EGFP and endogenous GSN proteins (Figure 6 and Supplementary Figure 1). Furthermore, we observed that transfection of GSN 3’UTR resulted in the translocation of endogenous Tra2β protein from nucleus to cytoplasm (Figure 6). Taken together, all these data converge towards a hypothesis where the transfected GSN 3’UTR perturb the subcellular localization of GSN and thereby influence the migration and invasion of NSCLC cell in vitro (Figure 7). Our findings also point out the possible role of Tra2β in the mRNA trafficking leading to the localized expression of protein which could influence cancer development. Future investigations would be meaningful to further delineate the precise molecular mechanisms by which the interaction between GSN 3’UTR-Tra2β regulates the subcellular localization of GSN protein.
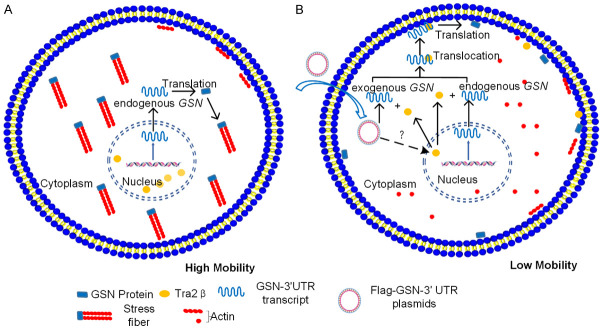
Figure 7.
Exogenous GSN 3’UTR inhibits the EMT process and migration/invasion abilities of NSCLC cell through interaction with Tra2β and perturbation of endogenous GSN protein localization. A. Without GSN 3’UTR transfection, endogenous GSN proteins are distributed in the cytoplasm, Tra2β protein is exclusively located in the nucleus, with actin-rich stress fibers across the cell body, and cells display mesenchymal characteristics with high mobility. B. Upon GSN 3’UTR transfection, through an unknown mechanism, Tra2β protein is translocated from the nucleus to the cytoplasm, where it can interact with the endogenous or exogenous GSN 3’UTR, bringing the corresponding mRNA to the proximity of the plasma membrane and eliciting the localized translation of the latter. This in turn perturbs the remodeling of actin-rich cytoskeletons, causing disappearance of stress fibers, and inhibits the EMT process, thereby decreases the mobility of the cell.
Acknowledgements
This work was supported by the Natural Science Foundation of Guangdong Province (Nos. 2019A1515010182, 2018A030313544), the National Natural Science Foundation of China (No. 81800675), the Guangzhou Science and Technology Project (No. 201804010066).
Disclosure of conflict of interest
None.
Supplementary Figure 1
Supplementary Video 1
References
- 1.Wang Y, Hoinka J, Liang Y, Adamus T, Swiderski P, Przytycka TM. AptaBlocks: designing RNA complexes and accelerating RNA-based drug delivery systems. Nucleic Acids Res. 2018;46:8133–8142. doi: 10.1093/nar/gky577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Zuroske T, Watts JK. RNA therapeutics on the rise. Nat Rev Drug Discov. 2020;19:441–442. doi: 10.1038/d41573-020-00078-0. [DOI] [PubMed] [Google Scholar]
- 3.L Garner A. RNA-targeted drug discovery: moving beyond promiscuous small-molecule scaffolds. Future Med Chem. 2019;11:2487–2490. doi: 10.4155/fmc-2019-0200. [DOI] [PubMed] [Google Scholar]
- 4.Türeci Ö, Vormehr M, Diken M, Kreiter S, Huber C, Sahin U. Targeting the heterogeneity of cancer with individualized neoepitope vaccines. Clin Cancer Res. 2016;22:1885–1896. doi: 10.1158/1078-0432.CCR-15-1509. [DOI] [PubMed] [Google Scholar]
- 5.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines-a new era in vaccinology. Nature Reviews Drug Discovery. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu DG, Jiang QH, Wei YY, Sun L, Fu BB, Zhao FK, Zhou Q. Gene expression profile favoring phenotypic reversion: a clue for mechanism of tumor suppression by NF-IL6 3’UTR. Cell Res. 2003;13:509–514. doi: 10.1038/sj.cr.7290195. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Li T, Gao C, Lv X, Liu K, Song H, Xing Y, Xi T. FOXO1 3’UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett. 2014;588:3218–3224. doi: 10.1016/j.febslet.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Hu J, Li X, Guo X, Guo Q, Xiang C, Zhang Z, Xing Y, Xi T, Zheng L. The CCR2 3’UTR functions as a competing endogenous RNA to inhibit breast cancer metastasis. J Cell Sci. 2017;130:3399–3413. doi: 10.1242/jcs.202127. [DOI] [PubMed] [Google Scholar]
- 9.Park HJ, Ji P, Kim S, Xia Z, Rodriguez B, Li L, Su J, Chen K, Masamha CP, Baillat D, Fontes-Garfias CR, Shyu AB, Neilson JR, Wagner EJ, Li W. 3’UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet. 2018;50:783–789. doi: 10.1038/s41588-018-0118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berkovits BD, Mayr C. Alternative 3’UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, Maeda T, Nagata Y, Kitanaka A, Mizuno S, Tanaka H, Chiba K, Ito S, Watatani Y, Kakiuchi N, Suzuki H, Yoshizato T, Yoshida K, Sanada M, Itonaga H, Imaizumi Y, Totoki Y, Munakata W, Nakamura H, Hama N, Shide K, Kubuki Y, Hidaka T, Kameda T, Masuda K, Minato N, Kashiwase K, Izutsu K, Takaori-Kondo A, Miyazaki Y, Takahashi S, Shibata T, Kawamoto H, Akatsuka Y, Shimoda K, Takeuchi K, Seya T, Miyano S, Ogawa S. Aberrant PD-L1 expression through 3’-UTR disruption in multiple cancers. Nature. 2016;534:402–406. doi: 10.1038/nature18294. [DOI] [PubMed] [Google Scholar]
- 12.Chaudhury A, Hussey GS, Howe PH. 3’-UTR-mediated post-transcriptional regulation of cancer metastasis: beginning at the end. RNA Biol. 2011;8:595–599. doi: 10.4161/rna.8.4.16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr C. Regulation by 3’-Untranslated Regions. Annu Rev Genet. 2017;51:171–194. doi: 10.1146/annurev-genet-120116-024704. [DOI] [PubMed] [Google Scholar]
- 14.Mayr C. What are 3’UTRs doing? Cold Spring Harb Perspect Biol. 2019;11:a034728. doi: 10.1101/cshperspect.a034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islami F, Goding Sauer A, Miller KD, Siegel RL, Fedewa SA, Jacobs EJ, McCullough ML, Patel AV, Ma J, Soerjomataram I, Flanders WD, Brawley OW, Gapstur SM, Jemal A. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin. 2018;68:31–54. doi: 10.3322/caac.21440. [DOI] [PubMed] [Google Scholar]
- 16.Tong L, Shen S, Huang Q, Fu J, Wang T, Pan L, Zhang P, Chen G, Huang T, Li K, Liu Q, Xie S, Yang X, Moses RE, Li X, Li L. Proteasome-dependent degradation of Smad7 is critical for lung cancer metastasis. Cell Death Differ. 2020;27:1795–1806. doi: 10.1038/s41418-019-0459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Brien TD, Jia P, Caporaso NE, Landi MT, Zhao Z. Weak sharing of genetic association signals in three lung cancer subtypes: evidence at the SNP, gene, regulation, and pathway levels. Genome Med. 2018;10:16. doi: 10.1186/s13073-018-0522-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Relli V, Trerotola M, Guerra E, Alberti S. Abandoning the notion of non-small cell lung cancer. Trends Mol Med. 2019;25:585–594. doi: 10.1016/j.molmed.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Leighl NB, Karaseva N, Nakagawa K, Cho BC, Gray JE, Hovey T, Walding A, Rydén A, Novello S. Patient-reported outcomes from FLAURA: Osimertinib versus erlotinib or gefitinib in patients with EGFR-mutated advanced non-small-cell lung cancer. Eur J Cancer. 2020;125:49–57. doi: 10.1016/j.ejca.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Liu J, Cai X, Pan Z, Liu J, Yin W, Chen H, Xie Z, Liang H, Wang W, Guo Z, Zhao S, Liang W, He J. Efficacy and safety of first line treatments for patients with advanced epidermal growth factor receptor mutated, non-small cell lung cancer: systematic review and network meta-analysis. BMJ. 2019;367:l5460. doi: 10.1136/bmj.l5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguiar PN Jr, Haaland B, Park W, San Tan P, Del Giglio A, de Lima Lopes G Jr. Cost-effectiveness of osimertinib in the first-line treatment of patients with EGFR-mutated advanced non-small cell lung cancer. JAMA Oncol. 2018;4:1080–1084. doi: 10.1001/jamaoncol.2018.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldt J, Schicht M, Garreis F, Welss J, Schneider UW, Paulsen F. Structure, regulation and related diseases of the actin-binding protein gelsolin. Expert Rev Mol Med. 2019;20:e7. doi: 10.1017/erm.2018.7. [DOI] [PubMed] [Google Scholar]
- 23.Li GH, Arora PD, Chen Y, McCulloch CA, Liu P. Multifunctional roles of gelsolin in health and diseases. Med Res Rev. 2012;32:999–1025. doi: 10.1002/med.20231. [DOI] [PubMed] [Google Scholar]
- 24.Liu XH, Ma J, Feng JX, Feng Y, Zhang YF, Liu LX. Regulation and related mechanism of GSN mRNA level by hnRNPK in lung adenocarcinoma cells. Biol Chem. 2019;400:951–963. doi: 10.1515/hsz-2018-0417. [DOI] [PubMed] [Google Scholar]
- 25.Li Z, Liu X, Ma J, Zhang T, Gao X, Liu L. hnRNPK modulates selective quality-control autophagy by downregulating the expression of HDAC6 in 293 cells. Int J Oncol. 2018;53:2200–2212. doi: 10.3892/ijo.2018.4517. [DOI] [PubMed] [Google Scholar]
- 26.Liu XH, Ma J, Feng JX, Feng Y, Zhang YF, Liu LX. Regulation and related mechanism of GSN mRNA level by hnRNPK in lung adenocarcinoma cells. Biol Chem. 2019;400:951–963. doi: 10.1515/hsz-2018-0417. [DOI] [PubMed] [Google Scholar]
- 27.Ni J, Hou X, Wang X, Shi Y, Xu L, Zheng X, Liu N, Qiu A, Zhuang S. 3-deazaneplanocin A protects against cisplatin-induced renal tubular cell apoptosis and acute kidney injury by restoration of E-cadherin expression. Cell Death Dis. 2019;10:355. doi: 10.1038/s41419-019-1589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Luo X, Lin J, Fu S, Feng P, Su H, He X, Liang X, Liu K, Deng W. Gelsolin promotes cancer progression by regulating epithelial-mesenchymal transition in hepatocellular carcinoma and correlates with a poor prognosis. J Oncol. 2020;2020:1980368. doi: 10.1155/2020/1980368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka H, Shirkoohi R, Nakagawa K, Qiao H, Fujita H, Okada F, Hamada J, Kuzumaki S, Takimoto M, Kuzumaki N. siRNA gelsolin knockdown induces epithelial-mesenchymal transition with a cadherin switch in human mammary epithelial cells. International Journal of Cancer. 2006;118:1680–1691. doi: 10.1002/ijc.21559. [DOI] [PubMed] [Google Scholar]
- 30.Hartmann-Petersen R, Walmod PS, Berezin A, Berezin V, Bock E. Individual cell motility studied by time-lapse video recording: influence of experimental conditions. Cytometry. 2000;40:260–270. doi: 10.1002/1097-0320(20000801)40:4<260::aid-cyto2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 31.Grellscheid SN, Dalgliesh C, Rozanska A, Grellscheid D, Bourgeois CF, Stévenin J, Elliott DJ. Molecular design of a splicing switch responsive to the RNA binding protein Tra2β. Nucleic Acids Res. 2011;39:8092–8104. doi: 10.1093/nar/gkr495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Best A, Dagliesh C, Ehrmann I, Kheirollahi-Kouhestani M, Tyson-Capper A, Elliott DJ. Expression of Tra2 β in cancer cells as a potential contributory factor to neoplasia and metastasis. Int J Cell Biol. 2013;2013:843781. doi: 10.1155/2013/843781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baig RM, Mahjabeen I, Sabir M, Masood N, Ali K, Malik FA, Kayani MA. Mutational spectrum of gelsolin and its down regulation is associated with breast cancer. Dis Markers. 2013;34:71–80. doi: 10.3233/DMA-120952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Li K, Yin X, Li H, Li Y, Zhang Q, Wang H, Qiu Y. Lower expression of gelsolin in colon cancer and its diagnostic value in colon cancer patients. J Cancer. 2019;10:1288–1296. doi: 10.7150/jca.28529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang HC, Chen CW, Yang CL, Tsai IM, Hou YC, Chen CJ, Shan YS. Tumor-associated macrophages promote epigenetic silencing of gelsolin through DNA methyltransferase 1 in gastric cancer cells. Cancer Immunol Res. 2017;5:885–897. doi: 10.1158/2326-6066.CIR-16-0295. [DOI] [PubMed] [Google Scholar]
- 36.Kuzumaki N. Progress of research on tumor suppressor genes. Hokkaido Igaku Zasshi. 1996;71:133–137. [PubMed] [Google Scholar]
- 37.Lee HK, Driscoll D, Asch H, Asch B, Zhang PJ. Downregulated gelsolin expression in hyperplastic and neoplastic lesions of the prostate. Prostate. 1999;40:14–19. doi: 10.1002/(sici)1097-0045(19990615)40:1<14::aid-pros2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 38.Kuzumaki N, Tanaka M, Sakai N, Fujita H. Tumor suppressive function of gelsolin. Gan To Kagaku Ryoho. 1997;24:1436–1441. [PubMed] [Google Scholar]
- 39.Xiao Y, Liu G, Sun Y, Gao Y, Ouyang X, Chang C, Gong L, Yeh S. Targeting the estrogen receptor alpha (ERα)-mediated circ-SMG1.72/miR-141-3p/Gelsolin signaling to better suppress the HCC cell invasion. Oncogene. 2020;39:2493–2508. doi: 10.1038/s41388-019-1150-6. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka H, Shirkoohi R, Nakagawa K, Qiao H, Fujita H, Okada F, Hamada J, Kuzumaki S, Takimoto M, Kuzumaki N. siRNA gelsolin knockdown induces epithelial-mesenchymal transition with a cadherin switch in human mammary epithelial cells. Int J Cancer. 2006;118:1680–1691. doi: 10.1002/ijc.21559. [DOI] [PubMed] [Google Scholar]
- 41.Kwiatkowski DJ. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr Opin Cell Biol. 1999;11:103–108. doi: 10.1016/s0955-0674(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 42.Archer SK, Claudianos C, Campbell HD. Evolution of the gelsolin family of actin-binding proteins as novel transcriptional coactivators. Bioessays. 2005;27:388–396. doi: 10.1002/bies.20200. [DOI] [PubMed] [Google Scholar]
- 43.Weng J, Han X, Liu K, Yang J, Wei S, Zhang Y, Zeng F, Li Y, Shen L, Gao Y. CD44 3’-untranslated region functions as a competing endogenous RNA to enhance NK sensitivity of liver cancer stem cell by regulating ULBP2 expression. Int J Biol Sci. 2019;15:1664–1675. doi: 10.7150/ijbs.35216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y, Ye Y, Lou Y, Yang Y, Cai C, Zhang Z, Mills T, Chen NY, Kim Y, Muge Ozguc F, Diao L, Karmouty-Quintana H, Xia Y, Kellems RE, Chen Z, Blackburn MR, Yoo SH, Shyu AB, Mills GB, Han L. Comprehensive characterization of alternative polyadenylation in human cancer. J Natl Cancer Inst. 2018;110:379–389. doi: 10.1093/jnci/djx223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.