Abstract
Accumulating evidence demonstrates that the expression levels of programmed cell death protein 1 (PD-1) and programmed death ligand 1 (PD-L1) are regulated at the various levels, including transcription, post-transcriptional modification and post-translational modifications (PTMs). The PTMs of PD-1/PD-L1 contain phosphorylation, ubiquitination, methylation, glycosylation and palmitoylation. Recently, PD-L1 was reported to be acetylated at Lys263 site by p300 and was deacetylated by histone deacetylase 2 (HDAC2). Acetylation of PD-L1 prevented its translocation to the nucleus and led to a reduction of the nuclear portion of PD-L1, resulting in evading immune surveillance of tumor cells. In this review article, we briefly describe the PTMs of PD-1/PD-L1 and mainly summarize the novel findings of PD-L1 acetylation in tumor cells. Moreover, we discuss the associations of PD-L1 acetylation and ubiquitination, phosphorylation and methylation. Furthermore, we highlight that targeting acetylation of PD-L1 by HDAC inhibitors might be useful for enhancing tumor immunotherapy.
Keywords: Acetylation, PD-1, PD-L1, ubiquitination, immunotherapy
Introduction
Programmed cell death protein 1 (PD-1, also known as CD279), one of coinhibitory receptors, is expressed on the surfaces of multiple types of immune cells, including B cells, natural killer (NK) T cells, CD4+ T cells, CD8+ T cells, dendritic cells and tumor infiltrating lymphocytes [1,2]. There are two ligands of PD-1: programmed death ligand 1 (PD-L1, also named CD274 or B7-H1) and PD-L2 (also called CD273 or B7-DC) [3]. PD-L1 is expressed on hematopoietic cells, including T cells, B cells, dendritic cells, macrophages and mast cells, and non-hematopoietic healthy tissue cells such as keratinocytes, vascular endothelial cells, astrocytes, islet cells, placental syncytial trophoblasts, endothelial cells and corneal epithelium. Both PD-L1 and PD-L2 can be expressed in tumor cells and tumor stroma [4]. The interaction of PD-1 and PD-L1 transmits inhibitory signals to T cells so that the tissue can maintain self-tolerance and avoid immune-mediated tissue damage [5]. PD-1 is a 288 amino acid type 1 transmembrane protein that is composed of extracellular domain, a transmembrane domain and a cytoplastic domain containing immunoreceptor tyrosine-based switch motif (ITSM) and immunoreceptor tyrosine-based inhibitory motif (ITIM), while PD-L1, a 290 amino acid type 1 transmembrane protein, consists of a short cytoplasmic tail without typical signal motif [4,5]. The interaction between the extracellular domain of PD-1 and PD-L1 leads to the conformational change of PD-1 and tyrosine phosphorylation in the cytoplasmic domain of PD-1, resulting in an increased connection between tyrosine phosphatase of SHP-2 and ITSM. SHP-2 recruitment results in a reduction of phosphorylation of TCR molecules, decreasing stimulation of downstream signals of TCR and inhibiting T cell responses. The PD-1/PD-L1 pathway eventually reduces the production of cytokines, such as IFN-γ, and cell survival proteins, such as Bcl-xl, leading to dysfunction or apoptosis of T cells [4-6]. PD-L2 is also a negative regulator of T cell activation and is highly expressed on tumor cells and antigen presenting cells [7].
PD-1/PD-L1 post-translational modifications
Evidence has demonstrated that PD-1/PD-L1 expression is regulated at different levels, such as transcription, post-transcriptional modification and post-translational modifications (PTMs) [8,9]. Regarding PTMs, PD-1/PD-L1 has been reported to be modulated by phosphorylation, ubiquitination, glycosylation, methylation, palmitoylation and acetylation [9,10]. In the following paragraphs, we will briefly describe these PTMs of PD-1/PD-L1 in human cancers.
Phosphorylation of PD-L1
AMP-activated protein kinase (AMPK) is activated by metformin, leading to PD-L1 phosphorylation at Ser195 site [11]. Knockout of AMPKα abrogated this phenotype of PD-L1 phosphorylation. Moreover, PD-L1 Ser195 phosphorylation might occur in the ER lumen and block its ER-to-Golgi translocation, leading to ER-mediated PD-L1 degradation [11]. The Janus kinase 1 (JAK1) could phosphorylate PD-L1 at Tyr112 site, resulting in enhancement of STT3A association with PD-L1 and promotion of PD-L1 glycosylation to stabilize the PD-L1 stability [12]. In addition, glycogen synthase 3 beta (GSK3β) binds and phosphorylates PD-L1 at T180 and S184 sites [13].
Glycosylation of PD-1/PD-L1
PD-L1 N-glycosylation at N192, N200 and N219 maintains stability of PD-L1 via preventing GSK3β-involved degradation of PD-L1 and inhibits T cell activity [13]. Tunicamycin, an N-linked glycosylation inhibitor, can remove the glycosylation of PD-L1 in cells. Moreover, EGF signaling pathway promoted PD-L1 glycosylation [13]. Targeting glycosylated PD-L1 prevents the interaction between PD-1 and PD-L1, and enhances PD-L1 degradation [14]. PD-1 is also N-glycosylated and maintains stability and localization of PD-1 in T cells [15]. TCR activation induces PD-1 glycosylation, especially at the N58 site, leading to enhanced PD-1 stability and membrane expression, and mediating the interaction between PD-1 and PD-L1 [15].
Ubiquitination of PD-L1
Several E3 ligases have been reported to target PD-L1 for ubiquitination and degradation, including beta-transducin repeats-containing protein (β-TrCP) [13,16], SPOP (speckle-type POZ protein) [17-19], STIP1 homology and U-box containing protein 1 (STUB1) [20], and HMG-CoA reductase degradation protein 1 (HRD1) [11,21]. F-box protein 38 (FBXO38) targets the PD-1 for ubiquitination and degradation, leading to regulating cancer immunotherapy [22]. Kelch like family member 22 (KLHL22) also participates into PD-1 degradation and regulates antitumor function of T cells and tumor progression [23]. In addition, ubiquitin specific peptidase 22 (USP22) and ubiquitin specific peptidase 9 X-linked (USP9x) induce deubiquitination of PD-L1 and maintain its stabilization [24-26].
Palmitoylation of PD-L1
Multiple investigations demonstrate that Zinc finger DHHC-type palmitoyltransferase 3 (ZDHHC3) and ZDHHC9 induce PD-L1 palmitoylation and stabilize its protein activity, leading to tumor growth promotion [27,28]. 2-bromopalmitate, a palmitoylation inhibitor, decreases the PD-L1 protein level, indicating that PD-L1 could have a palmitoylation modification. Moreover, Cys272 site is validated as a key palmitoylation site of PD-L1, contributing to PD-L1 stability and blockade of the immune surveillance of T cells [28]. In addition, PD-L1 palmitotylation is observed in cisplatin-resistance bladder cancer cells [29]. Inhibition of fatty acid synthase (FASN) repressed PD-L1 palmitoylation and its expression [29]. Targeting PD-L1 palmitoylation increases the sensitivity of tumor cells to T-cell killing and retards tumor growth [27,28].
Acetylation of PD-L1 at Lys263
Acetylation is an important modification of PTMs in which protein residues are added the acetyl group by acetyltransferases from acetyl coenzymes A [30,31]. The acetyltransferases include histone acetyltransferases (HATs), lysine acetyltransferases (KATs) and Nα-acetyltransferases (NATs) [32]. The deacetylases that can catalyze the removal of acetyl group from the acetylated proteins have histone deacetylases (HDACs) and Sirtuins (SIRTs) [33]. In the following sections, we will describe the role of PD-1/PD-L1 acetylation in regulation of its stabilization and tumor immunotherapy (Figure 1).
Figure 1.
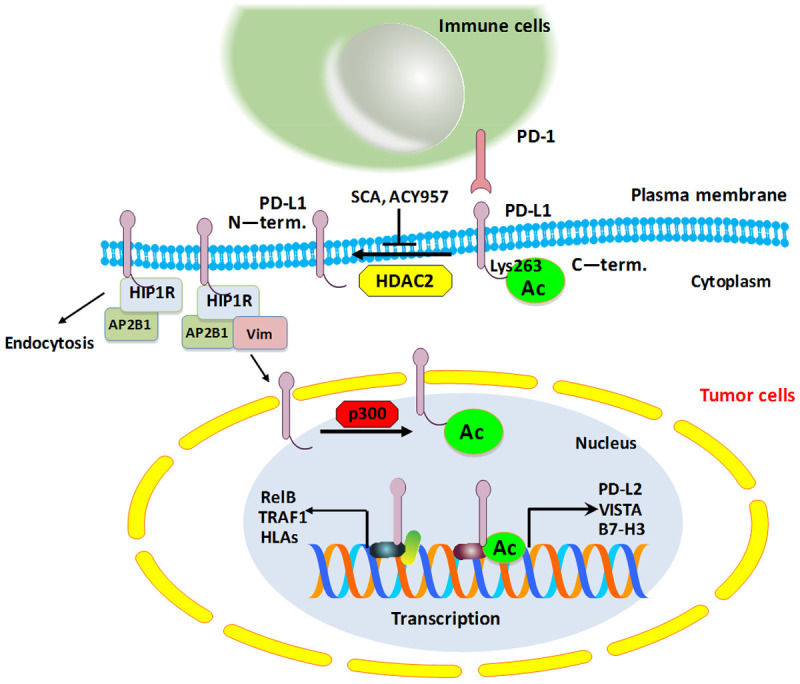
A schematic diagram showing how PD-L1 is acetylated and deacetylated in tumor cells. PD-L1 is acetylated at Lys263 site by p300, leading to preventing the translocation of PD-L1 into the nucleus from the plasma membrane. HIP1R interacts with PD-L1 and AP2B1, resulting in clathrin-dependent endocytosis. Acetylation of PD-L1 blocks the binding between PD-L1 and HIP1R. HDAC2 can reduce p300-mediated acetylation of PD-L1 and increase the nuclear portion of PD-L1, leading to regulation of immune surveillance of tumor cells.
One study revealed that EGF stimulation increased tyrosine phosphorylation and acetylation of PD-L1 in A431 cells [34]. Another study reported that overexpression of p300 enhanced PD-L1 acetylation, whereas depletion of p300 or p300 inhibitor A485 reduced the acetylation of PD-L1 [35]. Moreover, Lys263 site was identified as the major acetylation site on PD-L1 by p300. Lys263 acetylation mainly blocked the translocation of PD-L1 into the nucleus from the plasma membrane, but did not reduce half-lives or dimerization of PD-L1. Nuclear PD-L1 upregulation could promote cancer cells to evade immune surveillance [35]. Huntingtin interacting protein-1 related (HIP1R) protein targeted PD-L1 for lysosomal degradation to regulate T cell-involved cytotoxicity [36]. HIP1R interacted with PD-L1 via its C-tail, whereas Lys263 acetylation of PD-L1 blocked this interaction. HIP1R was connected to PD-L1 and was also linked to Adaptin-β2 (AP2B1), leading to clathrin-dependent endocytosis [35]. Deacetylation of PD-L1 on the plasma membrane could bind with HIP1R and AP2B1 for endocytosis and bind with vimentin to traffic via the cytoskeleton, and enter into the nucleus by importin-α1. PD-L1 might bind to DNA and alter gene transcription, including interferon (IFN) signaling, NF-κB pathway, MHC class-I genes [35]. Moreover, this study identified that only HDAC2 deacetylase interacted with PD-L1 and reduced p300-mediated acetylation of PD-L1. HDAC2 inhibitor in combination with anti-PD-1 antibodies enhanced tumor growth repression and improved the survival in MC38 syngeneic mouse model [35]. Therefore, combining HDAC2 inhibitor with PD-1/PD-L1 blockade is a novel strategy for cancer immunotherapy. One HDAC2 inhibitor, santacruzamate A (SCA), and the HDAC1/2 inhibitor ACY957 upregulated PD-L1 acetylation and decreased the nuclear portion of PD-L1, alleviated the transcription of immune checkpoints, including VISTA and B7-H3, resulting in increased infiltration of CD8+ T cells in tumor microenvironment [35].
HDACs regulate histone acetylation of PD-L1 promoter region
HDACs are critically involved in regulating acetylation via removing acetyl groups from the N-acetyl lysine amino acid of histones [37]. HDACs have been classified as I, IIa, IIb, III and IV. HDAC inhibitors, including LBH589 (panobinostat), MS275 (etinostat) and MGCD0103 (mocetinostat), elevated the expression of PD-L1 in melanoma cell lines [38]. Cells obtained from patient melanomas were treated with HDAC inhibitors and demonstrated that HDAC inhibitors, especially class I HDACs inhibitors, promoted PD-L1 and PD-L2 expression [38]. LBH589 treatment increased the expression of PD-L1 and PD-L2 in C57BL/6 mice [38]. Moreover, LBH589 treatment led to higher histone acetylation at the promoter region of PD-L1 and PD-L2 in WM983A melanoma cells [38]. Furthermore, LBH589 mediated histone 3 acetylation and subsequently upregulated PD-L1 expression at mRNA, protein and gene acetylation levels. Combining LBH589 and PD-1 blocking antibody retarded tumor progression and increased survival in mice [38].
HDAC3 suppression increased PD-L1 expression and enhanced the efficacy of anti-PD-L1 treatment in B-cell lymphomas [39]. HDAC3 inhibitor (RGFP966) and SAHA elevated histone acetylation and recruitment of bromodomain-containing protein 4 (BRD4) by B-cell lymphoma 6 protein (BCL6) at the PD-L1 gene promoter, resulting in the activation of PD-L1 transcription [39]. HDAC3 inhibitors decreased DNA methyltransferase 1 (DNMT1) expression and caused activation of PD-L1 transcription [39]. HDAC3 inhibitor in combination with anti-PD-L1 treatment promoted tumor regression in murine lymphoma model [39]. Moreover, HDAC3 inhibitor reduced the mRNA and protein levels of PD-L1 via regulation of signal transducer and activator of transcription 3 (STAT3) in pancreatic cancer cells, indicating that HDAC3 inhibitors could enhance immunotherapy [40]. Moreover, HDAC3 overexpression inhibited PD-L1 expression in non-small cell lung cancer (NSCLC) cells. The lower expression of constitutive photomorphogenic 1 (COP1) elevated the accumulation of c-Jun and consequently repressed HDAC3 expression and led to promoting histone H3 acetylation of the PD-L1 promoter, resulting in high expression of PD-L1 in drug-resistant NSCLC cells [41]. A HDAC3 inhibitor, romidepsin, upregulated PD-L1 expression via enhancing the acetylation of histones H3 and H4 and elevating BRD4 expression, leading to suppression of cellular immune function in colon cancer cells [42]. Taken together, HDACs regulate histone acetylation of PD-L1 promoter region.
PD-L1 acetylation and other PTMs
The associations of PD-1/PD-L1 PTMs are also needed to be clarified. Epidermal growth factor (EGF) stimulation induced upregulation of phosphorylation, acetylation and ubiquitination of PD-L1, but not SUMOylation, in A431 cells [34]. PD-L1 palmitoylation enhanced PD-L1 stability via preventing its ubiquitination and repressing its subsequent degradation by lysosomes [27]. PD-L1 activity is regulated by ubiquitination and N-glycosylation. GSK3β binds with PD-L1 and activates β-TrCP-mediated degradation of PD-L1, whereas glycosylation (at N192, N200 and N219) of PD-L1 antagonizes GSK3β interaction. EGF led to enhanced PD-L1 stability via inactivation of GSK3β in breast cancer, resulting in reduction of antitumor T cell immunity and efficacy of anti-PD-1 therapy in syngeneic mouse models [13]. N-glycosylation (at N35, N192, N200 and N219) and ubiquitinylation (at K178) of PD-L1 cannot affect its interaction with the biphenyl drug BMS-202 and did not change PD-L1 dimer stability [43]. PD-L1 phosphorylation is linked to its glycosylation and stabilization. Metformin treatment led to PD-L1 phosphorylation at Ser195 site that was phosphorylates by AMPK (AMP-activated protein kinase), resulting in abnormal PD-L1 glycosylation and subsequent accumulation of PD-L1 in the ER (endoplasmic reticulum) and ERAD (ER-associated protein degradation)-mediated destruction [11]. Moreover, IL-6 stimulated JAK1 to cause PD-L1 phosphorylation at Tyr112, leading to PD-L1 glycosylation by N-glycoltransferase STT3A and promotion of PD-L1 stability [12]. Interestingly, PD-L1 glycosylation did not affect its acetylation and nuclear translocation [13]. PD-L1 glycosylation by B3GNT3 blocked cell surface internalization and GSK3β-involved degradation of PD-L1 [14].
Lysine specific demethylase (LSD) is recruited to the PD-L1 gene locus by B lymphocyte induced maturation protein 1 (Blmp-1) and inhibits the expression of PD-L1 via removing H3K4 methylation of PD-L1 following ex vivo stimulation, such as acute viral infection [44]. LSD1, an H3K4 and H3K9 demethylase, can govern eomesoderin (EOMES) nuclear dynamics through switching demethylation and acetylation of EOMES residues in PD-1+CD8+ T cells [45]. This observation suggests that demethylation and acetylation can be switched in special conditions. HDAC3 inhibitors alleviated DNA methyltransferase DNMT1 expression, leading to activation of PD-L1 transcription [39]. Therefore, studies are underway to explore whether PD-L1 acetylation is correlated with its methylation.
Protein acetylation has been reported to antagonize its ubiquitination to regulate stability and subcellular localization of proteins [46,47]. For example, S-phase kinase associated protein 2 (Skp2) was acetylated by p300 and acetylation of Skp2 blocked its proteolysis by CDH1, leading to enhanced oncogenic activity [46]. USP7 regulated the degradation of Tip60, one of HATs, and forkhead box P3 (Foxp3) expression [48]. Tip60 promoted Foxp3 acetylation and dimerization [49,50]. In the absence of acetylation, several lysines on Foxp3 could be ubiquitinated and caused its destruction [51]. Hyperacetylation of Foxp3 blocked its ubiquitination and degradation and increased Foxp3 protein levels, providing a rapid temporal regulation of Foxp3 expression levels [51]. It is necessary to determine whether PD-L1 acetylation prevents its ubiquitination in cancer cells. Entinostat, class I HDAC inhibitor, increased STAT3 acetylation and subsequently suppressed STAT3 phosphorylation and activity, and resultantly inhibited Foxp3 expression in Foxp3+ Treg cells [52]. Similarly, entinostat enhanced STAT3 acetylation in myeloid-derived suppressor cells (MDSCs)-like cells [53]. It is necessary to define the relationship between PD-L1 acetylation and its phosphorylation.
Targeting acetylation pathway for enhancing PD-1/PD-L1 therapy
Antibodies targeting PD-1/PD-L1 pathway have been used and exhibited impressive outcomes in several types of cancers [54]. In the past decade, Food and Drug Administration (FDA) has approved several antibodies targeting cytotoxic T-lymphocyte antigen-4 (CTLA-4), PD-1 or PD-L1 for the treatment of a wide spectrum of cancers, including melanoma, hepatocellular carcinoma, renal cell carcinoma (RCC), NSCLC, head and neck cancer, and bladder cancer [55-60]. Anti-PD-1 antibodies have pembrolizumab and nivolumab, while anti-PD-L1 antibodies have atezolizumab and durvalumab, and Ipilimumab is a monoclonal antibody against CTLA-4 [61,62]. Unfortunately, the response rate is low in some tumors, such as the prostate and colon, and nearly 1/3 of respondents will relapse [6,63]. Some patients develop resistance to immune checkpoint blockade, leading to poor prognosis [64]. Therefore, targeting PD-1/PD-L1 PTMs might be useful to improve the efficacy of anti-PD-1/PD-L1 therapy.
Selenium nanoparticles inhibited the expression of PD-1 and upregulated the expression of cytotoxicity factors such as CD16, interferon-γ (IFN-γ) and natural killer group 2, member D (NKG2D), and increased tubulin-α acetylation in γδ cells, leading to potentiating antitumor cytotoxicity [65]. HDAC6 upregulated PD-L1 expression via regulation of STAT3 pathway, and suppression of HDAC6 inhibited tumor progression in mice [66]. MPT0G612, an inhibitor of HDAC6, blocked IFN-γ-mediated upregulation of PD-L1 and stimulated apoptosis via inhibition of autophagy [67]. Several studies have shown that HDAC inhibitors, including VPA, nexturastat A, increased the efficacy of anti-PD-L1 antibody via activation of immune surveillance [68,69]. HDAC3 inhibitor romidepsin in combination with anti-PD-1 therapy enhanced antitumor effects on colon cancer cells [42]. SCA and ACY957 elevated PD-L1 acetylation and the expression of immune checkpoints, leading to enhancement of immunotherapy [35]. It is important to mention that not all HDAC inhibitors can target acetylation of PD-L1. Moreover, HDAC inhibitors cannot specifically target acetylation of PD-L1. Targeting HDAC might be an effective approach to improve immune checkpoint blockade in cancer cells.
Conclusions and perspectives
To the end, this review article proposes a combination therapy of HDAC inhibition and PD-1/PD-L1 blockade, which makes up for the deficiency of PD-1/PD-L1 blockade resistance, and provides a theoretical basis for this new tumor immunotherapy. However, a couple of critical concerns still need to be addressed in follow-up studies. The expression of nuclear PD-L1 in metastatic tumors is higher than that in primary tumors, but it is not clear how nuclear PD-L1 increases tumor invasiveness [35]. Without a doubt, further investigations are wanted to determine whether PD-L1 nuclear translocation inhibitors can be used as a complementary therapy for malignant tumors. Lys263 site was reported as the key acetylation site on PD-L1 by p300. Whether there are other acetylation sites on PD-L1 needs to be determined. Do other acetyltransferases involve in regulation of PD-L1 acetylation? It is also necessary to define whether other deacetylases, besides HDAC2, could reduce acetylation of PD-L1. High expression of PD-L1 was associated with cisplatin resistance and ADR resistance in non-small cell lung cancer [41]. It is unclear whether PD-L1 acetylation is associated with drug resistance, which is required to be fully explored. Since PD-L1 has multiple types of PTMs, it is pivotal to determine the associations among PTMs of PD-L1. Discovery of the regulatory mechanisms how PD-L1 acetylation is involved in regulating other PTMs might be useful to find the new approaches for targeting PD-L1 acetylation other than HDACs inhibitors. One group showed that knockdown of transcription factor PU.1 decreased the expression and H3K27 acetylation of PD-L2 via interacting with p300 in dendritic cells [70]. Depletion of p300 reduced the expression and PD-L2 acetylation in dendritic cells [70]. Moreover, PU.1 could bind to IRF4 on an Ets-IRF composite element (EICE) sequence of PD-L2 to regulate p300-mediated PD-L2 acetylation in dendritic cells [70]. It is elusive whether targeting PD-L2 acetylation is helpful to improve the tumor immunotherapy. In conclusion, it is required to further investigate the role and molecular mechanism of PD-1/PD-L1 acetylation in tumor cells for improving immunotherapy.
Disclosure of conflict of interest
None.
References
- 1.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 2.Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, Wistuba II, Rimm DL, Tsao MS, Hirsch FR. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18:345–362. doi: 10.1038/s41571-021-00473-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Dang F, Ren J, Wei W. Biochemical aspects of PD-L1 regulation in cancer immunotherapy. Trends Biochem Sci. 2018;43:1014–1032. doi: 10.1016/j.tibs.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH. Coinhibitory pathways in immunotherapy for cancer. Annu Rev Immunol. 2016;34:539–573. doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 7.Bardhan K, Anagnostou T, Boussiotis VA. The PD1: PD-L1/2 pathway from discovery to clinical implementation. Front Immunol. 2016;7:550. doi: 10.3389/fimmu.2016.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu JM, Li CW, Lai YJ, Hung MC. Posttranslational modifications of PD-L1 and their applications in cancer therapy. Cancer Res. 2018;78:6349–6353. doi: 10.1158/0008-5472.CAN-18-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha JH, Chan LC, Li CW, Hsu JL, Hung MC. Mechanisms controlling PD-L1 expression in cancer. Mol Cell. 2019;76:359–370. doi: 10.1016/j.molcel.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang YN, Lee HH, Hsu JL, Yu D, Hung MC. The impact of PD-L1 N-linked glycosylation on cancer therapy and clinical diagnosis. J Biomed Sci. 2020;27:77. doi: 10.1186/s12929-020-00670-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha JH, Yang WH, Xia W, Wei Y, Chan LC, Lim SO, Li CW, Kim T, Chang SS, Lee HH, Hsu JL, Wang HL, Kuo CW, Chang WC, Hadad S, Purdie CA, McCoy AM, Cai S, Tu Y, Litton JK, Mittendorf EA, Moulder SL, Symmans WF, Thompson AM, Piwnica-Worms H, Chen CH, Khoo KH, Hung MC. Metformin promotes antitumor immunity via endoplasmic-reticulum-associated degradation of PD-L1. Mol Cell. 2018;71:606–620. e7. doi: 10.1016/j.molcel.2018.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan LC, Li CW, Xia W, Hsu JM, Lee HH, Cha JH, Wang HL, Yang WH, Yen EY, Chang WC, Zha Z, Lim SO, Lai YJ, Liu C, Liu J, Dong Q, Yang Y, Sun L, Wei Y, Nie L, Hsu JL, Li H, Ye Q, Hassan MM, Amin HM, Kaseb AO, Lin X, Wang SC, Hung MC. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J Clin Invest. 2019;129:3324–3338. doi: 10.1172/JCI126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN, Hung MC. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li CW, Lim SO, Chung EM, Kim YS, Park AH, Yao J, Cha JH, Xia W, Chan LC, Kim T, Chang SS, Lee HH, Chou CK, Liu YL, Yeh HC, Perillo EP, Dunn AK, Kuo CW, Khoo KH, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Huang TH, Sahin AA, Hortobagyi GN, Yoo SS, Hung MC. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33:187–201. e110. doi: 10.1016/j.ccell.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Li CW, Chung EM, Yang R, Kim YS, Park AH, Lai YJ, Yang Y, Wang YH, Liu J, Qiu Y, Khoo KH, Yao J, Hsu JL, Cha JH, Chan LC, Hsu JM, Lee HH, Yoo SS, Hung MC. Targeting glycosylated PD-1 induces potent antitumor immunity. Cancer Res. 2020;80:2298–2310. doi: 10.1158/0008-5472.CAN-19-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng L, Qian G, Zhang S, Zheng H, Fan S, Lesinski GB, Owonikoko TK, Ramalingam SS, Sun SY. Inhibition of mTOR complex 1/p70 S6 kinase signaling elevates PD-L1 levels in human cancer cells through enhancing protein stabilization accompanied with enhanced beta-TrCP degradation. Oncogene. 2019;38:6270–6282. doi: 10.1038/s41388-019-0877-4. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, Tan Y, Ci Y, Wu F, Dai X, Guo J, Huang YH, Fan C, Ren S, Sun Y, Freeman GJ, Sicinski P, Wei W. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553:91–95. doi: 10.1038/nature25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Y, Xu Y, Pan C, Yan L, Wang ZW, Zhu X. The emerging role of SPOP protein in tumorigenesis and cancer therapy. Mol Cancer. 2020;19:2. doi: 10.1186/s12943-019-1124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Song Y, Ye M, Dai X, Zhu X, Wei W. The diverse roles of SPOP in prostate cancer and kidney cancer. Nat Rev Urol. 2020;17:339–350. doi: 10.1038/s41585-020-0314-z. [DOI] [PubMed] [Google Scholar]
- 20.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, Broeks A, Horlings HM, Wessels LFA, Blank CU, Xiao Y, Heck AJR, Borst J, Brummelkamp TR, Schumacher TNM. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:106–110. doi: 10.1038/nature23669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, Wang J, Chu M, Liu Y, Wang ZW, Zhu X. Emerging role of ubiquitination in the regulation of PD-1/PD-L1 in cancer immunotherapy. Mol Ther. 2021;29:908–919. doi: 10.1016/j.ymthe.2020.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng X, Liu X, Guo X, Jiang S, Chen T, Hu Z, Liu H, Bai Y, Xue M, Hu R, Sun SC, Zhou P, Huang X, Wei L, Yang W, Xu C. FBXO38 mediates PD-1 ubiquitination and regulates anti-tumour immunity of T cells. Nature. 2018;564:130–135. doi: 10.1038/s41586-018-0756-0. [DOI] [PubMed] [Google Scholar]
- 23.Zhou XA, Zhou J, Zhao L, Yu G, Zhan J, Shi C, Yuan R, Wang Y, Chen C, Zhang W, Xu D, Ye Y, Wang W, Shen Z, Wang J. KLHL22 maintains PD-1 homeostasis and prevents excessive T cell suppression. Proc Natl Acad Sci U S A. 2020;117:28239–28250. doi: 10.1073/pnas.2004570117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Zhang Q, Lou Y, Wang J, Zhao X, Wang L, Zhang X, Li S, Zhao Y, Chen Q, Liang T, Bai X. USP22 deubiquitinates CD274 to suppress anticancer immunity. Cancer Immunol Res. 2019;7:1580–1590. doi: 10.1158/2326-6066.CIR-18-0910. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Sun Q, Mu N, Sun X, Fan S, Su L, Liu X. The deubiquitinase USP22 regulates PD-L1 degradation in human cancer cells. Cell Commun Signal. 2020;18:112. doi: 10.1186/s12964-020-00612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jingjing W, Wenzheng G, Donghua W, Guangyu H, Aiping Z, Wenjuan W. Deubiquitination and stabilization of programmed cell death ligand 1 by ubiquitin-specific peptidase 9, X-linked in oral squamous cell carcinoma. Cancer Med. 2018;7:4004–4011. doi: 10.1002/cam4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, Lu H, Fang C, Zhang Y, Liang L, Zhou X, Wang C, Xue Y, Cui Y, Xu J. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat Biomed Eng. 2019;3:306–317. doi: 10.1038/s41551-019-0375-6. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Hsu JM, Sun L, Chan LC, Li CW, Hsu JL, Wei Y, Xia W, Hou J, Qiu Y, Hung MC. Palmitoylation stabilizes PD-L1 to promote breast tumor growth. Cell Res. 2019;29:83–86. doi: 10.1038/s41422-018-0124-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahid M, Kim M, Jin P, Zhou B, Wang Y, Yang W, You S, Kim J. S-palmitoylation as a functional regulator of proteins associated with cisplatin resistance in bladder cancer. Int J Biol Sci. 2020;16:2490–2505. doi: 10.7150/ijbs.45640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 31.Kaypee S, Sudarshan D, Shanmugam MK, Mukherjee D, Sethi G, Kundu TK. Aberrant lysine acetylation in tumorigenesis: implications in the development of therapeutics. Pharmacol Ther. 2016;162:98–119. doi: 10.1016/j.pharmthera.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Drazic A, Myklebust LM, Ree R, Arnesen T. The world of protein acetylation. Biochim Biophys Acta. 2016;1864:1372–1401. doi: 10.1016/j.bbapap.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Wu D, Qiu Y, Jiao Y, Qiu Z, Liu D. Small molecules targeting HATs, HDACs, and BRDs in cancer therapy. Front Oncol. 2020;10:560487. doi: 10.3389/fonc.2020.560487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horita H, Law A, Hong S, Middleton K. Identifying regulatory posttranslational modifications of PD-L1: a focus on monoubiquitinaton. Neoplasia. 2017;19:346–353. doi: 10.1016/j.neo.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Nihira NT, Bu X, Chu C, Zhang J, Kolodziejczyk A, Fan Y, Chan NT, Ma L, Liu J, Wang D, Dai X, Liu H, Ono M, Nakanishi A, Inuzuka H, North BJ, Huang YH, Sharma S, Geng Y, Xu W, Liu XS, Li L, Miki Y, Sicinski P, Freeman GJ, Wei W. Acetylation-dependent regulation of PD-L1 nuclear translocation dictates the efficacy of anti-PD-1 immunotherapy. Nat Cell Biol. 2020;22:1064–1075. doi: 10.1038/s41556-020-0562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Yao H, Li C, Shi H, Lan J, Li Z, Zhang Y, Liang L, Fang JY, Xu J. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat Chem Biol. 2019;15:42–50. doi: 10.1038/s41589-018-0161-x. [DOI] [PubMed] [Google Scholar]
- 37.Hesham HM, Lasheen DS, Abouzid KAM. Chimeric HDAC inhibitors: comprehensive review on the HDAC-based strategies developed to combat cancer. Med Res Rev. 2018;38:2058–2109. doi: 10.1002/med.21505. [DOI] [PubMed] [Google Scholar]
- 38.Woods DM, Sodre AL, Villagra A, Sarnaik A, Sotomayor EM, Weber J. HDAC inhibition upregulates PD-1 ligands in melanoma and augments immunotherapy with PD-1 blockade. Cancer Immunol Res. 2015;3:1375–1385. doi: 10.1158/2326-6066.CIR-15-0077-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng S, Hu Q, Zhang H, Yang F, Peng C, Huang C. HDAC3 inhibition upregulates PD-L1 expression in B-cell lymphomas and augments the efficacy of anti-PD-L1 therapy. Mol Cancer Ther. 2019;18:900–908. doi: 10.1158/1535-7163.MCT-18-1068. [DOI] [PubMed] [Google Scholar]
- 40.Hu G, He N, Cai C, Cai F, Fan P, Zheng Z, Jin X. HDAC3 modulates cancer immunity via increasing PD-L1 expression in pancreatic cancer. Pancreatology. 2019;19:383–389. doi: 10.1016/j.pan.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Fu C, Du J, He R, Yin X, Li H, Li X, Li K, Zheng L, Liu Z, Qiu Y. Enhanced histone H3 acetylation of the PD-L1 promoter via the COP1/c-Jun/HDAC3 axis is required for PD-L1 expression in drug-resistant cancer cells. J Exp Clin Cancer Res. 2020;39:29. doi: 10.1186/s13046-020-1536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi Y, Fu Y, Zhang X, Zhao G, Yao Y, Guo Y, Ma G, Bai S, Li H. Romidepsin (FK228) regulates the expression of the immune checkpoint ligand PD-L1 and suppresses cellular immune functions in colon cancer. Cancer Immunol Immunother. 2021;70:61–73. doi: 10.1007/s00262-020-02653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bailly C, Vergoten G. N-glycosylation and ubiquitinylation of PD-L1 do not restrict interaction with BMS-202: a molecular modeling study. Comput Biol Chem. 2020;88:107362. doi: 10.1016/j.compbiolchem.2020.107362. [DOI] [PubMed] [Google Scholar]
- 44.Bally APR, Neeld DK, Lu P, Majumder P, Tang Y, Barwick BG, Wang Q, Boss JM. PD-1 expression during acute infection is repressed through an LSD1-Blimp-1 axis. J Immunol. 2020;204:449–458. doi: 10.4049/jimmunol.1900601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu WJ, McCuaig RD, Tan AHY, Hardy K, Seddiki N, Ali S, Dahlstrom JE, Bean EG, Dunn J, Forwood J, Tsimbalyuk S, Smith K, Yip D, Malik L, Prasanna T, Milburn P, Rao S. Targeting nuclear LSD1 to reprogram cancer cells and reinvigorate exhausted T cells via a novel LSD1-EOMES switch. Front Immunol. 2020;11:1228. doi: 10.3389/fimmu.2020.01228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inuzuka H, Gao D, Finley LW, Yang W, Wan L, Fukushima H, Chin YR, Zhai B, Shaik S, Lau AW, Wang Z, Gygi SP, Nakayama K, Teruya-Feldstein J, Toker A, Haigis MC, Pandolfi PP, Wei W. Acetylation-dependent regulation of Skp2 function. Cell. 2012;150:179–193. doi: 10.1016/j.cell.2012.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nihira NT, Ogura K, Shimizu K, North BJ, Zhang J, Gao D, Inuzuka H, Wei W. Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function. Sci Signal. 2017;10:eaai8026. doi: 10.1126/scisignal.aai8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Kumar S, Dahiya S, Wang F, Wu J, Newick K, Han R, Samanta A, Beier UH, Akimova T, Bhatti TR, Nicholson B, Kodrasov MP, Agarwal S, Sterner DE, Gu W, Weinstock J, Butt TR, Albelda SM, Hancock WW. Ubiquitin-specific protease-7 inhibition impairs Tip60-dependent Foxp3+ T-regulatory cell function and promotes antitumor immunity. EBioMedicine. 2016;13:99–112. doi: 10.1016/j.ebiom.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song X, Li B, Xiao Y, Chen C, Wang Q, Liu Y, Berezov A, Xu C, Gao Y, Li Z, Wu SL, Cai Z, Zhang H, Karger BL, Hancock WW, Wells AD, Zhou Z, Greene MI. Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep. 2012;1:665–675. doi: 10.1016/j.celrep.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao Y, Nagai Y, Deng G, Ohtani T, Zhu Z, Zhou Z, Zhang H, Ji MQ, Lough JW, Samanta A, Hancock WW, Greene MI. Dynamic interactions between TIP60 and p300 regulate FOXP3 function through a structural switch defined by a single lysine on TIP60. Cell Rep. 2014;7:1471–1480. doi: 10.1016/j.celrep.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Loosdregt J, Vercoulen Y, Guichelaar T, Gent YY, Beekman JM, van Beekum O, Brenkman AB, Hijnen DJ, Mutis T, Kalkhoven E, Prakken BJ, Coffer PJ. Regulation of treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 52.Shen L, Ciesielski M, Ramakrishnan S, Miles KM, Ellis L, Sotomayor P, Shrikant P, Fenstermaker R, Pili R. Class I histone deacetylase inhibitor entinostat suppresses regulatory T cells and enhances immunotherapies in renal and prostate cancer models. PLoS One. 2012;7:e30815. doi: 10.1371/journal.pone.0030815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orillion A, Hashimoto A, Damayanti N, Shen L, Adelaiye-Ogala R, Arisa S, Chintala S, Ordentlich P, Kao C, Elzey B, Gabrilovich D, Pili R. Entinostat neutralizes myeloid-derived suppressor cells and enhances the antitumor effect of PD-1 inhibition in murine models of lung and renal cell carcinoma. Clin Cancer Res. 2017;23:5187–5201. doi: 10.1158/1078-0432.CCR-17-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrews LP, Yano H, Vignali DAA. Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: breakthroughs or backups. Nat Immunol. 2019;20:1425–1434. doi: 10.1038/s41590-019-0512-0. [DOI] [PubMed] [Google Scholar]
- 55.Lopez-Beltran A, Cimadamore A, Blanca A, Massari F, Vau N, Scarpelli M, Cheng L, Montironi R. Immune checkpoint inhibitors for the treatment of bladder cancer. Cancers (Basel) 2021;13:131. doi: 10.3390/cancers13010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Braun DA, Bakouny Z, Hirsch L, Flippot R, Van Allen EM, Wu CJ, Choueiri TK. Beyond conventional immune-checkpoint inhibition - novel immunotherapies for renal cell carcinoma. Nat Rev Clin Oncol. 2021;18:199–214. doi: 10.1038/s41571-020-00455-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan Y, Zhang L, Zuo Y, Qian H, Liu C. Immune checkpoint blockade in cancer immunotherapy: mechanisms, clinical outcomes, and safety profiles of PD-1/PD-L1 inhibitors. Arch Immunol Ther Exp (Warsz) 2020;68:36. doi: 10.1007/s00005-020-00601-6. [DOI] [PubMed] [Google Scholar]
- 58.Huang Z, Su W, Lu T, Wang Y, Dong Y, Qin Y, Liu D, Sun L, Jiao W. First-line immune-checkpoint inhibitors in non-small cell lung cancer: current landscape and future progress. Front Pharmacol. 2020;11:578091. doi: 10.3389/fphar.2020.578091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Federico P, Petrillo A, Giordano P, Bosso D, Fabbrocini A, Ottaviano M, Rosanova M, Silvestri A, Tufo A, Cozzolino A, Daniele B. Immune checkpoint inhibitors in hepatocellular carcinoma: current status and novel perspectives. Cancers (Basel) 2020;12:3025. doi: 10.3390/cancers12103025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Silva P, Aiello M, Gu-Trantien C, Migliori E, Willard-Gallo K, Solinas C. Targeting CTLA-4 in cancer: is it the ideal companion for PD-1 blockade immunotherapy combinations? Int J Cancer. 2021;149:31–41. doi: 10.1002/ijc.33415. [DOI] [PubMed] [Google Scholar]
- 61.Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: clinical impact and mechanisms of response and resistance. Annu Rev Pathol. 2021;16:223–249. doi: 10.1146/annurev-pathol-042020-042741. [DOI] [PubMed] [Google Scholar]
- 62.Brigida M, Perricelli A, Sposato F, Spadafora MG, Pomillo A, Sisto M. Management of immunotherapy adverse events in oncological patients: anti-CTLA-4, anti-PD-1/PD-L1. Rev Recent Clin Trials. 2020;15:339–346. doi: 10.2174/1574887115666200622161418. [DOI] [PubMed] [Google Scholar]
- 63.Restifo NP, Smyth MJ, Snyder A. Acquired resistance to immunotherapy and future challenges. Nat Rev Cancer. 2016;16:121–126. doi: 10.1038/nrc.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu Y, Liu T, Li J, Mai F, Chen Y, Jing Y, Dong X, Lin L, He J, Xu Y, Shan C, Hao J, Yin Z, Chen T, Wu Y. Selenium nanoparticles as new strategy to potentiate gammadelta T cell anti-tumor cytotoxicity through upregulation of tubulin-alpha acetylation. Biomaterials. 2019;222:119397. doi: 10.1016/j.biomaterials.2019.119397. [DOI] [PubMed] [Google Scholar]
- 66.Lienlaf M, Perez-Villarroel P, Knox T, Pabon M, Sahakian E, Powers J, Woan KV, Lee C, Cheng F, Deng S, Smalley KSM, Montecino M, Kozikowski A, Pinilla-Ibarz J, Sarnaik A, Seto E, Weber J, Sotomayor EM, Villagra A. Essential role of HDAC6 in the regulation of PD-L1 in melanoma. Mol Oncol. 2016;10:735–750. doi: 10.1016/j.molonc.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen MC, Lin YC, Liao YH, Liou JP, Chen CH. MPT0G612, a novel HDAC6 inhibitor, induces apoptosis and suppresses IFN-gamma-induced programmed death-ligand 1 in human colorectal carcinoma cells. Cancers (Basel) 2019;11:1617. doi: 10.3390/cancers11101617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knox T, Sahakian E, Banik D, Hadley M, Palmer E, Noonepalle S, Kim J, Powers J, Gracia-Hernandez M, Oliveira V, Cheng F, Chen J, Barinka C, Pinilla-Ibarz J, Lee NH, Kozikowski A, Villagra A. Selective HDAC6 inhibitors improve anti-PD-1 immune checkpoint blockade therapy by decreasing the anti-inflammatory phenotype of macrophages and down-regulation of immunosuppressive proteins in tumor cells. Sci Rep. 2019;9:6136. doi: 10.1038/s41598-019-42237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adeshakin AO, Yan D, Zhang M, Wang L, Adeshakin FO, Liu W, Wan X. Blockade of myeloid-derived suppressor cell function by valproic acid enhanced anti-PD-L1 tumor immunotherapy. Biochem Biophys Res Commun. 2020;522:604–611. doi: 10.1016/j.bbrc.2019.11.155. [DOI] [PubMed] [Google Scholar]
- 70.Inaba K, Yashiro T, Hiroki I, Watanabe R, Kasakura K, Nishiyama C. Dual roles of PU.1 in the expression of PD-L2: direct transactivation with IRF4 and indirect epigenetic regulation. J Immunol. 2020;205:822–829. doi: 10.4049/jimmunol.1901008. [DOI] [PubMed] [Google Scholar]


