Abstract
Deregulation of fibroblast growth factor receptor (FGFR) network is common in cancer due to activating mutations, gene amplifications and chromosomal translocations. Currently, various FGFR inhibitors are being developed. In order to optimize their clinical applications, understanding the frequencies and types of FGFR alterations in multiple cancer types appears to be extremely important. This study characterized FGFR1-4 alterations in solid tumors by next-generation sequencing (NGS). Between Jun. 2019 and Aug. 2020, the sequencing data of 5 557 solid tumors of diverse types in the database of Simcere Diagnostics, Inc. (Nanjing, China) were retrospectively analyzed. A panel-based NGS assay was used to detect FGFR1-4 alterations in tumor samples. 9.2% of cancer cases had FGFR1-4 alterations, in which gene amplifications (51.5%) and mutations (40.7%) were frequent, whereas gene rearrangements were less common (10.0%). FGFR1 was involved in 4.6% of 5 557 cases, FGFR2 in 2.1%, FGFR3 in 1.6%, and FGFR4 in 1.4%. Of patients with FGFR1-4 alterations, TP53, MUC16, NSD3, MYC and LRP1B genes were the top 5 mutant genes. FGFR1-4 aberrations occurred in almost every type of solid tumors, with the most common tumor being endometrial carcinoma (22.2%), followed by sarcoma (17.3%), breast cancer (13.2%), gastric cancer (12.2%), and more. 0.6% of cancer cases harbored FGFR1-4 fusions, with the most common fusion partner being TACC3. Two cases of GBM harboring FGFR3-TACC3 fusions were responsive to anlotinib treatment. In conclusion, FGFR1-4 alterations are prevalent in solid tumors of diverse types, with the majority being gene amplifications and mutations. FGFR1-4 fusions only occur in a minority of cancer cases, and those with glioblastoma harboring FGFR3-TACC3 fusions may benefit from anlotinib.
Keywords: Fibroblast growth factor receptors, next-generation sequencing, solid tumors, gene fusion, anlotinib
Introduction
Fibroblast growth factor receptors (FGFRs) pertain to a family of highly-conserved transmembrane tyrosine kinase receptors consisting of extracellular domain, transmembrane domain and intracellular tyrosine kinase domain, and are involved in various aspects of cancer biology, including cell proliferation, differentiation, migration, angiogenesis, and carcinogenesis [1]. There are four typical tyrosine kinase receptors (FGFR1-4) and one without intracellular tyrosine kinase domain (FGFR5 or FGFRL1) in humans [2]. Despite being encoded by different genes, FGFR1-4 share high homology in sequence identify ranging from 56% to 71% [3]. Fibroblast growth factors (FGFs) are the native ligands for FGFRs. When FGFs bind to their cognate receptor, the receptor will dimerize, subsequently resulting in phosphorylated intracellular domain of the receptor, intracellular signaling cascades and gene transcription [4].
Constitutive activation of FGFR genes can be oncogenic due to point mutations, gene amplifications and chromosomal translocations [5-7]. FGFs, ligands of the receptors, could also strengthen the signaling in terms of oncogenesis and neoangiogensis [8]. Specific FGFR alterations are identified in certain cancer types, such as FGFR1 amplification in lung squamous cell carcinoma (LSQCC) and FGFR3 mutations in bladder cancer, and some of them tend to be the driver abnormalities [9,10]. There is evidence suggesting that variations of specific FGFR expression may be associated with the prognosis or sensitivity to cancer treatments [11,12]. Donnem et al. found that co-expression of FGFR-1/platelet-derived growth factor-B (PDGF-B) and FGF2/vascular endothelial growth factor-3 (VEGF-3) was correlated with the poor survival of patients with non-small cell lung cancer (NSCLC) [13]. In contrast to those with wild-type FGFR1, the patients with FGFR1 amplification had a worse progression-free survival (PFS) [14]. Additionally, the tumor FGFR4 level was also found a significant predictor of response to lenvatinib in patients with advanced hepatocellular carcinoma (HCC) [15].
Considering that most FGFR alterations result in gain of function, it is rational to speculate that use of FGFR inhibitors to target these tumors would be beneficial [16]. Although non-selective FGFR inhibitors have been approved for cancer treatments, presence of various complications limits their use in clinic [17]. It seems very necessary to identify FGFR aberrations that may predict the response to treatment in various cancer types and to verify their contributions to the onset of drug-resistant mechanisms. Next-generation sequencing (NGS) with the characteristics of high throughput, automation and low cost makes accurate and rapid identification of FGFR alterations practicable [18]. In this study, we characterized FGFR1-4 alterations involving amplifications, mutations, and rearrangements in 5 557 Chinese patient samples across multiple solid tumors by NGS, aiming at providing a landscape for FGFR alterations in Chinese solid tumors and evidence for more effective use of pharmaceutical agents.
Materials and methods
Between Jun. 2019 and Aug. 2020, the sequencing data of 5 557 solid tumors of diverse types (specific cancer types provided by submitting physicians and their corresponding number of cases are described in Table 1) in the database of Simcere Diagnostics, Co. Ltd. (Nanjing, China) for genomic profiling were retrospectively analyzed. All the patients included in the study were informed consent with respect to genetic testing and research. A panel-based NGS assay was used to detect FGFR1-4 alterations in formalin-fixed, paraffin-embedded (FFPE) tumor samples. First, DNA was extracted from undyed FFPE sections with the proportion of tumor cells more than 20% and whole-blood samples and then purification and library preparation were performed. Second, a probe with 2.29 Mbp in size was used for hybrid capture and enrichment in gene-specific regions where various aberrations of 539 cancer-related genes including single nucleotide variants, copy number variations, small insertions, deletions and gene arrangements were covered. At last, double-ended sequencing on the Illumina NovaSeq 6000 platform was conducted after the captured library was mixed, degenerated and diluted.
Table 1.
Frequencies of FGFR1-4 alternations and relative distribution of alternation types in solid tumors (%)
| Cancer types | N | All | FGFR1 | FGFR2 | FGFR3 | FGFR4 | Amplification | Mutation | Rearrangement |
|---|---|---|---|---|---|---|---|---|---|
| Endometrial cancer | 36 | 22.2 | 8.3 | 11.1 | 8.3 | 2.8 | 0.0 | 22.2 | 2.8 |
| Sarcoma | 202 | 17.3 | 10.9 | 1.5 | 1.5 | 4.0 | 13.4 | 3.5 | 1.0 |
| Breast cancer | 106 | 13.2 | 8.5 | 2.8 | 0.9 | 0.9 | 8.5 | 3.3 | 0.0 |
| Gastric cancer | 254 | 12.2 | 3.5 | 7.9 | 0.8 | 0.4 | 9.1 | 2.8 | 3.1 |
| Carcinoma of unknown primary | 899 | 11.6 | 7.1 | 1.8 | 1.7 | 1.7 | 6.5 | 2.4 | 1.1 |
| Colorectal cancer | 437 | 11.0 | 6.2 | 1.8 | 3.9 | 0.7 | 5.0 | 6.4 | 0.5 |
| Liver cancer | 625 | 9.6 | 1.9 | 4.0 | 1.9 | 2.1 | 4.3 | 3.5 | 2.2 |
| Biliary tract cancer | 218 | 9.2 | 5.0 | 1.4 | 1.4 | 1.4 | 3.2 | 3.7 | 2.3 |
| Esophageal cancer | 98 | 9.2 | 6.1 | 2.0 | 1.0 | 0.0 | 6.1 | 3.1 | 0.0 |
| Melanoma | 105 | 8.6 | 2.9 | 1.0 | 3.8 | 1.0 | 2.9 | 4.8 | 1.0 |
| Lung cancer | 2262 | 6.9 | 3.8 | 1.0 | 1.1 | 1.4 | 3.1 | 3.8 | 0.3 |
| Pancreatic cancer | 152 | 6.6 | 5.3 | 0.0 | 1.3 | 0.0 | 5.3 | 1.3 | 0.0 |
| Cervical cancer | 67 | 6.0 | 1.5 | 1.5 | 3.0 | 0.0 | 1.5 | 4.5 | 0.0 |
| Brain cancer | 96 | 4.2 | 1.0 | 0.0 | 1.0 | 2.1 | 1.0 | 2.1 | 1.0 |
Results
Frequency of FGFR1-4 alterations in solid tumors
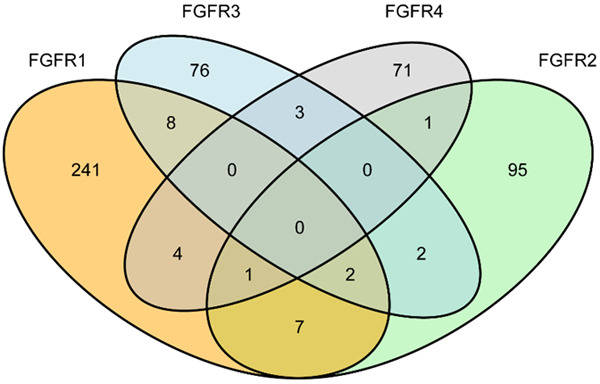
Of the 5 557 tumor samples sequenced, FGFR1-4 alterations occurred in 511 cases, with an overall frequency of 9.2%. It should be noted that 28 cases had more than one alterations (Figure 1). FGFR1 was influenced in 4.6% of 5 557 cases, FGFR2 in 2.1%, FGFR3 in 1.6%, and FGFR4 in 1.4%. In contrast to FGFR2-4 aberrations, FGFR1 alterations were more common (Figure 2A). A great number of FGFR1-4 alterations were gene amplifications (51.5%) and gene mutations (40.7%), with gene rearrangements being less common (10.0%; Figure 2B). The gene amplification was more prevalent in FGFR1 (71.9%), whereas gene mutations were more frequent in FGFR2-4 (50.0%, 68.1%, and 61.3%, respectively; Figure 2C). Among 511 patients with FGFR1-4 aberrations, the top five mutant genes included TP53 (66%), MUC16 (27%), NSD3 (27%), MYC (25%) and LRP1B (22%), among which amplification was more common in NSD3 and MYC genes (Figure 2C). Most concurrently-mutated genes with FGFR1-4 aberrations were found in cell cycle pathways, while those in RAS/MAPK and Wnt/β-catenin pathways were uncommon. Compared with mutations and fusions, FGFR1-4 amplifications were relatively frequent in such pathways as PI3K-ALK, DNA repair, growth factor receptors and transcriptome (Figure 2D; Table 2). The specific forms of FGFR1-4 variations are respectively shown in Figure 3A-D.
Figure 1.
Number of cases with FGFR1-4 aberrations in 5 557 solid tumors.
Figure 2.
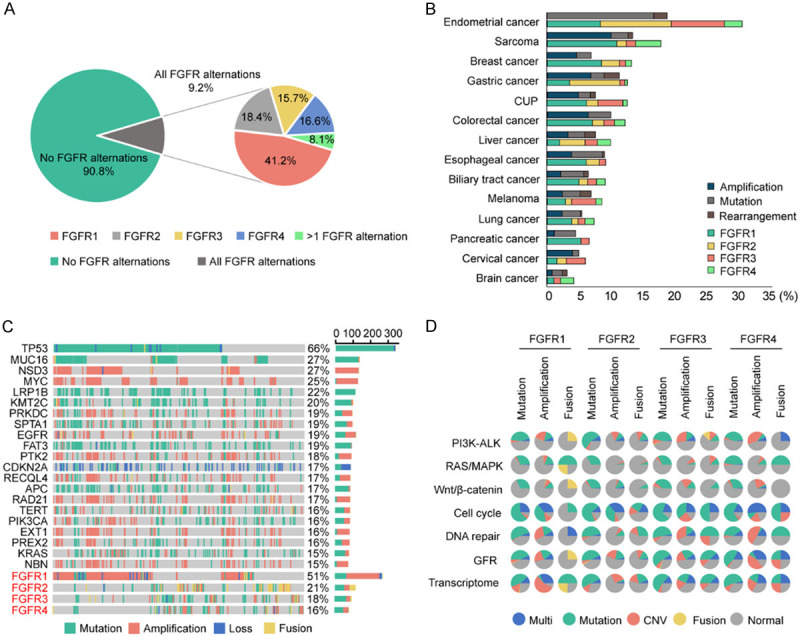
Frequency and distribution of FGFR1-4 variations in solid tumors. A: Frequency of FGFR1-4 alterations in all cases (left); relative proportion of FGFR1-4 alterations in all cases harboring FGFR1-4 aberrations (right). It should be noted that 28 cases had more than one alterations. B: Distribution of FGFR1-4 variations in all cancer types. The horizontal axis represents the percentage of tumors with aberrations. For each cancer type, the frequency of FGFR1-4 variations is described as the percentage of all cancer cases analyzed, and the distribution and type of variations are normalized to 100%. C: Landscape of genomic aberrations of FGFR1-4 in solid tumors. D: The pie chart shows the frequency and types of concurrently mutant genes with FGFR1-4 mutations, amplifications and fusions in seven major pathways.
Table 2.
List of common genes in different pathways
| Pathways | Genes |
|---|---|
| PI3K-AKT | AKT3, TSC2, PTEN, mTOR, PIK3CA, RPTOR, TSC1, TCS2, RICOTR, PI3K3R2, PI3K3CB, INPP4B, STK11, PPP2R1A, PIK3R1, AKT2, RHEB, PDK1, AKT1 |
| RAS/MAPK | KRAS, MAP2K1, NF1, NF2 |
| Wnt/β-catenin | AXIN1, AXIN2, RNF43, WNT7B, WNT108, AMER1, CHD4, TCF7L2, WNT10A |
| Cell cycle | TP53, TERT, CTNNB1, ATM, APC, CDK4, CCND1, CCND3, CCND2, CDKN2A, RB1, CCNE1, CDK6, CDK8, CCKN2A/B, CDKN1B CDKN2B |
| DNA repair | ATR, ATM, BACA1, BRCA2, MLH1, MSH2, MSH6, FANCA, BRIP1, BAP1, CHEK2, ERCC2, FANCA/D2/E/L, MUTYH, NBN, PMS2, RAD50/51B, WRN |
| Transcriptome | IGF1R, RET, ERBB2, EGFR, KIT, PDGFRA, KDR, ERBB4, FLT4, PDGFRB, MET, FLT3, ERBB3 |
| Growth factor receptors | MYC, SOX2, KDM6A, SMARCA4, RUNX1, MYCN, EZH2, NKX2, KMT2D, SMARCB1, DNMT3A, MYCL1, SOX10, ARID2, ASXL1, DNMT3A, EP300, KMT2C, KMT2D, NSD3, PBRM1, SMARCB1, SETD2, BAP1, IDH1, IDH2 |
Figure 3.
Specific types of FGFR1-4 variations. A: FGFR1; B: FGFR2; C: FGFR3; D: FGFR4.
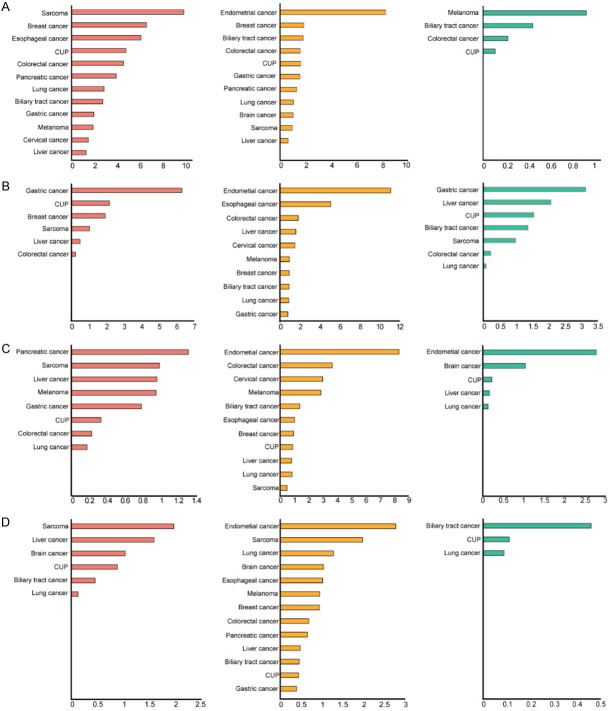
In Table 1, it could be observed that the frequencies of FGFR1-4 alterations were different in solid tumors of diverse types, ranging from 4.2% to 22.2%. Endometrial carcinoma had the highest frequency of aberrations (22.2%), followed by sarcoma (17.3%), breast cancer (13.2%), gastric cancer (12.2%), carcinoma of unknown primary (11.6%), colorectal cancer (11.0%), liver cancer (9.6%), biliary tract cancer (9.2%), esophageal cancer (9.2%), melanoma (8.6%), lung cancer (6.9%), pancreatic cancer (6.6%), cervical cancer (6.0%) and brain cancer (4.2%). The proportions of FGFR1-4 alterations in various cancer types are shown in Figure 4A-D.
Figure 4.
Proportions of FGFR1-4 alterations in various cancer types. The horizontal axis represents the percentage of tumors with aberrations. A: FGFR1; B: FGFR2; C: FGFR3; D: FGFR4. Orange: gene amplification; Yellow: gene mutation; Green: gene rearrangement.
Endometrial carcinoma
There were 36 cases of endometrial carcinoma in the dataset, with the overall frequency of 22.2% in FGFR1-4 aberrations. A great number of aberrations were gene mutations, with gene rearrangements being less common and without gene amplifications (Figure 5A). FGFR2 mutations were relatively common, including S252W (2 cases), N549K (1 case), D794Y (1 case) and E806K (1 case), and all of these were gain-of-function mutations.
Figure 5.
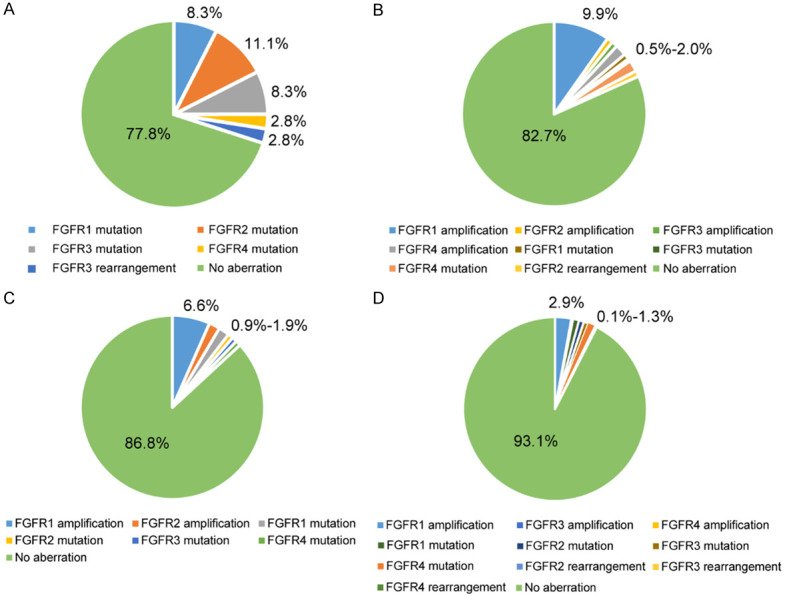
Frequency of FGFR1-4 aberrations in specific cancer types. A: Endometrial cancer; B: Sarcoma; C: Breast cancer; D: Lung cancer.
Sarcoma
Totally 202 cases of sarcoma were involved in the dataset, and 35 cases of them harbored FGFR1-4 variations. The gene amplifications were the most common alterations, especially FGFR1 amplification (9.9%). FGFR1, FGFR3 and FGFR4 mutations varied from 0.5%-2.0% in sarcoma (Figure 5B).
Breast cancer
In the dataset, there were 106 breast cancer cases in total, mainly including invasive lobular carcinoma, invasive ductal carcinoma and invasive metaplastic carcinoma. About 13.2% of breast cancer cases harbored FGFR1-4 alterations, with FGFR1 amplification being the most frequent (6.6%), while FGFR2 amplification and FGFR1-4 mutations less common (0.9%-1.9%; Figure 5C). PIK3CA aberrations were extraordinarily prevalent in breast cancer [19]. In our dataset, 2 out of 5 cases with FGFR mutations (40.0%) had PIK3CA mutations, including PIK3CA-H1047R, PIK3CA-H1048R, and PIK3CA-C378Y. It should be noted that these 2 cases both suffered from PIK3CA-H1047R.
Lung cancer
A total of 2 262 lung cancer cases were involved in the dataset, including adenocarcinoma, squamous cell carcinoma, adenosquamous carcinoma and small cell carcinoma. Approximately 6.9% of lung cancer cases analyzed harbored FGFR1-4 alterations, the most frequent of which was FGFR1 amplification (2.9%), while FGFR1-4 mutations were less common (0.8%-1.3%), and FGFR3-4 amplifications and FGFR2-4 rearrangements were rare (0.1%-0.2%) (Figure 5D). In our previous CHOICE study including 245 cases of NSCLC [20], the frequencies of FGFR1, FGFR2, FGFR3 and FGFR4 mutations were 2.45%, 0.41%, 2.04%, and 0.41%, respectively, further suggesting that the frequencies of FGFR1-4 mutations were low in Chinese lung cancer patients.
Gene fusions
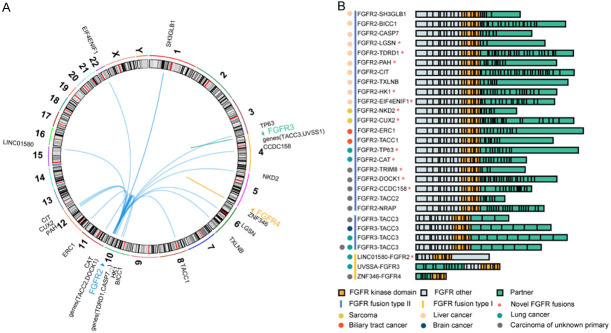
The genes fusing with FGFR1-4 in our dataset are summarized in Table 3 and Figure 6. It could be observed that 31 cases (0.6%, 31/5 557) harbored fusions of FGFR genes with other genes, a majority of which were with FGFR2 (24 cases). Transforming acidic coiled-coil containing protein 3 (TACC3) was the most common fusion partner (5 cases), followed by TACC2 (2 cases) and SH3GLB1 (2 cases). The other fusion partners, including BICC1, CAT, TDRD1, CCDC158, CIT, TXLNB, NKD2, CUX2, HK1, DOCK1, TACC1 and more, were found in single cases. Of 31 cases, gene fusions occurred in 10 cases of liver cancer, 8 of carcinoma of unknown primary, 5 of lung cancer, 2 of gastric cancer, 2 of sarcoma, 2 of biliary tract cancer, 1 of renal cancer and 1 of brain cancer.
Table 3.
The genes fusing with FGFR1-4 in the dataset (31 cases)
| Gene partner, chromosome location | Specific fusion (n) | Tumor types (n) |
|---|---|---|
| SH3GLB1, chromosome 1: 87191379 | FGFR2-SH3GLB1 (2) | Liver cancer (2) |
| BICC1, chromosome 10: 60426432 | FGFR2-BICC1 (1) | Liver cancer (1) |
| TACC2, chromosome 10: 123996320/123991849 | FGFR2-TACC2 (2) | Renal cancer (1), gastric cancer (1) |
| TACC3, chromosome 4: 1743533/1737371/1739077/1740504/1739733 | FGFR3-TACC3 (5) | CUP (3), lung cancer (1), brain cancer (1) |
| CAT, chromosome 11: 34463828 | FGFR2-CAT (1) | Lung cancer (1) |
| TDRD1, chromosome 10: 115939394 | FGFR2-TDRD1 (1) | Liver cancer (1) |
| CCDC158, chromosome 4: 77292242 | FGFR2-CCDC158 (1) | CUP (1) |
| CIT, chromosome 12: 120186475 | FGFR2-CIT (1) | Liver cancer (1) |
| TXLNB, chromosome 6: 139595138 | FGFR2-TXLNB (1) | Liver cancer (1) |
| NKD2, chromosome 5: 1011532 | FGFR2-NKD2 (1) | Sarcoma (1) |
| CUX2, chromosome 12: 111570060 | FGFR2-CUX2 (1) | Sarcoma (1) |
| HK1, chromosome 10: 71051284 | FGFR2-HK1 (1) | Liver cancer (1) |
| EIF4ENIF1, chromosome 22: 31884504 | FGFR2-EIF4ENIF1 (1) | CUP (1) |
| DOCK1, chromosome 10: 129093385 | FGFR2-DOCK1 (1) | Gastric cancer (1) |
| TACC1, chromosome 8: 38683244 | FGFR2-TACC1 (1) | Biliary tract cancer (1) |
| TP63, chromosome 3: 189457592 | FGFR2-TP63 (1) | Lung cancer (1) |
| PAH, chromosome 12: 103300452 | FGFR2-PAH (1) | Liver cancer (1) |
| TRIM8, chromosome 10: 104408969 | FGFR2-TRIM8 (1) | CUP (1) |
| ERC1, chromosome 12: 1241041 | FGFR2-ERC1 (1) | Biliary tract cancer (1) |
| CASP7, chromosome 10: 115441363 | FGFR2-CASP7 (1) | Liver cancer (1) |
| LGSN, chromosome 6: 64006372 | FGFR2-LGSN (1) | Liver cancer (1) |
| ZNF346, chromosome 5: 176472292 | ZNF346-FGFR4 (1) | Lung cancer (1) |
| UVSSA, chromosome 4: 1377083 | UVSSA-FGFR3 (1) | Lung cancer (1) |
| LINC01580, chromosome 15: 94446896 | LINC01580-FGFR2 (1) | CUP (1) |
| NRAP, chromosome 10: 115383150 | FGFR2-NRAP (1) | CUP (1) |
Notes: CUP, carcinoma of unknown primary.
Figure 6.
Distribution of the genes fusing with FGFR1-4 in all cancer cases. A: The Circos plot graphically depicts FGFR and their fusion partners, showing specific chromosomal locations of fusion partners. B: Schematic of all FGFR fusion proteins identified, at scale with exons represented by individual boxes. The upstream partner is colored grey, with FGFR kinase domain colored orange and other FGFR exons colored green.
Chromosomal translocations that can result in fusion proteins play their oncogenic roles in cancer by over-expressing another normal gene or creating a chimeric gene where parts of two genes are fused. Take FGFR3-TACC3 for example, the TACC domain mediating microtubule binding is fused with the entire FGFR3 kinase domain. The MAPK pathway can be activated by the fusion genes transfected into the normal urothelial cells in humans, highlighting the presence of active signaling in fusion genes. Moreover, the cell lines with fusion genes were found extremely sensitive to FGFR-selective agents, suggesting that the presence of fusion genes may be a potential therapeutic target in cancer cells [21]. Studies have shown that FGFR-TACC fusions may be conductive to identifying a subset of patients with glioblastoma multiforme (GBM) who would get benefits from targeted FGFR kinase inhibition [22,23]. Here, we shared the treatment experience of two cases of GBM with FGFR3-TACC3 fusions who both achieved partial responses (PR) after use of anlotinib.
Case presentation
The first patient with FGFR3-TACC3 fusion was a 65-year-old women diagnosed as GBM in the right splenium of corpus callosum. She underwent GBM resection on Feb. 13, 2020, recovered well and left hospital after radiotherapy concurrently with temozolomide chemotherapy. Later, she successively visited our hospital and 5 cycles of temozolomide chemotherapy were given. On May 4, 2020, she began to use anlotinib (12 mg) every other day, and achieved PR after use for 3 months. Until now, the patient is alive, and still on the treatment of anlotinib. Magnetic resonance imaging (MRI) images of this patient at different time points after use of anlotinib are shown in Figure 7A.
Figure 7.
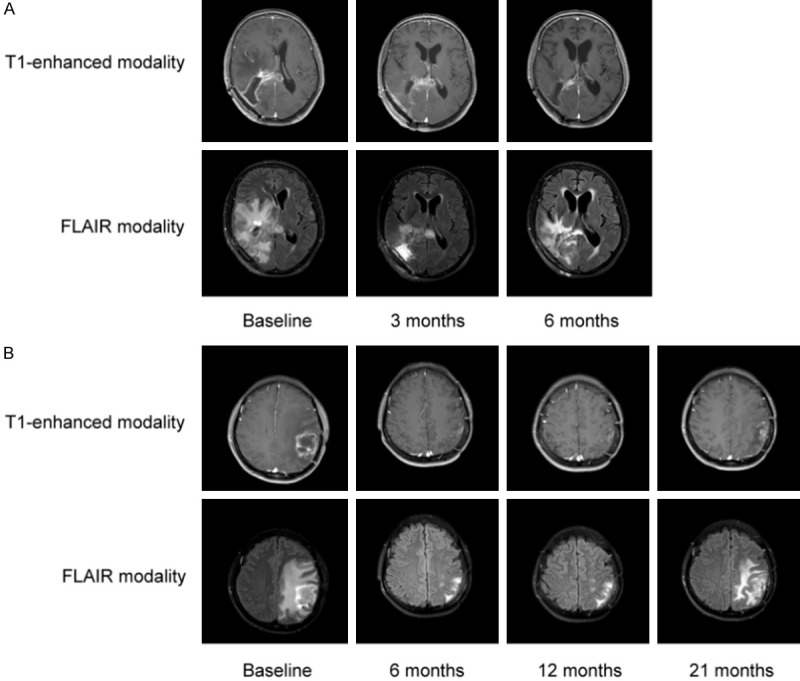
MRI images of two cases with FGFR3-TACC3 fusions at different time points after anlotinib use. A: A 65-year-old women diagnosed as glioblastoma multiforme in the right splenium of corpus callosum; B: A 46-year-old women with glioma in the left thalamus and parietal lobe.
The second patient with FGFR3-TACC3 fusion and FGFR3 amplification was a 46-year-old woman with GBM in the left thalamus and parietal lobe. She visited the hospital on October 1, 2017, with the chief complaint of 3 sudden onsets of epilepsy, and underwent GBM resection on October 10, 2017. The patient first came to our hospital on November 29, 2017 and underwent intensity modulated radiation therapy concurrently with temozolomide chemotherapy. Later, she was successively treated with temozolomide, nedaplatin and bevacizumab, but residual recurrence occurred on June 5, 2018, then 1 cycle of temozolomide chemotherapy was administrated. On July 4, 2018, the patient received 1 cycle of temozolomide chemotherapy again, and then used anlotinib (12 mg, every other day) for 21 months and maintenance chemotherapy of temozolomide to date. She got PR after using anlotinib for 2 months. This patient is still alive now. Her MRI images at different time points after use of anlotinib are shown in Figure 7B.
Discussion
In this study, overall 9.2% of patients with solid tumors were found to harbor FGFR1-4 alterations, with the majority being gene amplifications (mostly FGFR1 amplification) and mutations. FGFR1 was influenced in 4.6% of 5 557 cases, FGFR2 in 2.1%, FGFR3 in 1.6%, and FGFR4 in 1.4%. The frequencies of TP53, MUC16, NSD3, MYC and LRP1B mutations were relatively high in patients with FGFR1-4 aberrations, and most concurrently-mutated genes were identified in the cell cycle pathway. In solid tumors, the frequencies of FGFR1-4 aberrations varied from 4.2% to 22.2%. In addition, FGFR fusions were also identified in 0.6% of cancer cases, among which liver cancer showed the highest fusion frequency. The subsequent case studies suggested that GBM patients with FGFR3-TACC3 fusions may derive benefits from anlotinib.
Up to 22.2% of FGFR1-4 aberrations occurred in cases with endometrial cancer in our dataset, mostly being activating mutations in FGFR2, such as S252W and N549K. FGFR2-S252W mutation can increase the affinity to FGF ligands, especially FGF9, which may be extremely crucial for endometrial cancer due to its extensive presence in endometrial stroma [24]. Interestingly, in endometrial cancer, it appears to be mutually exclusive in FGFR2 and KRAS mutations, indicating redundancy with respect to activation of MAPK pathway [25].
There were 13.2% of breast cancer cases with FGFR1-4 alterations in our dataset, with the majority being FGFR1 amplification (6.6%) which had been identified to be a significant prognostic predictor for the patients with breast cancer and were associated with endocrine therapy resistance [26-28]. Additionally, the prognosis of patients with FGFR1 and human epidermal growth factor receptor 1 or 2 (HER1/2) co-amplification may be worse than those with FGFR1 or HER1/2 amplification alone or no amplification [29]. These findings all highlighted the importance of evaluating FGFR1 status. Lucitanib, an inhibitor of VEGF1-3, FGFR1-3, and platelet-derived growth factor receptor (PDGFR) α/β, exerts a certain effect on FGFR1-amplified breast cancer, but its antitumor activity is modest in HR+/HER2-metastatic breast cancer, with conspicuous hypertension-related toxicity [30]. Therefore, it still needs more studies to explore the potential role of FGFR inhibition in reversing endocrine therapy resistance.
Although FGFR variations did not seem to segregate well with histological changes, it was observed that some certain cancers harbored more frequent alterations. In our study, FGFR1 amplification was not found in endometrial cancer, but more commonly in sarcoma, breast cancer and lung cancer. Of 2 262 lung cancer cases sequenced, 6.9% of them was detected to have FGFR1-4 aberrations, especially FGFR1 amplification. Previous studies also showed the high frequency of FGFR1 amplification in LSQCC [31,32], but it was not analyzed in details in our study due to insufficient cases with LSQCC. Currently, except for erdafitinib approved by Food and Drug Administration, a selective pan-FGFR tyrosine kinase inhibitor, other selective FGFR inhibitors for various cancer types are under way, such as AZD4547, Ly2874455, CH5183284, NVP-BGJ398, and more [3].
In our study, it was also observed FGFR gene fusions (especially FGFR2 and FGFR3) in a small number of the cases, with the most common fusion partner being TACC3, which might be attributed to the fact that FGFR3 was close to TACC3 on chromosome 4p16 [33]. The emergence of the TACC coiled-coil domain can enhance FGFR3 activity by phosphorylating more miscellaneously constituted tyrosine kinase residues in FGFR3 protein and activating MAPK kinase [34]. FGFR3-TACC3 fusions can result in loss of an miR-99a binding site within the 3’-untranslated region of FGFR3, leading to the release of FGFR3 signaling from miR-99a-dependent inhibition and reinforcement of tumor progression in contrast to wild-type FGFR3 [35]. Emerging evidence suggests that the constitutional kinase activity of FGFR3-TACC3 fusion protein can be suppressed by FGFR inhibitors [22,36,37]. Anlotinib is an orally administrated, small-molecule receptor tyrosine kinase inhibitor that can target VEGFR, FGFR, PDGFR and c-kit, with inhibitory effects on tumor cell proliferation and tumor angiogenesis. It can suppress the activation of FGFRs by inhibiting the phosphorylation of FGFR1 with the inhibition rate of 45.0% (p-FGFR1/FGFR1) at 1 μM, and an IC50 value of 25 nM in AN3Ca cells overexpressing a FGFR2 mutant protein has been reported [38]. In our study, two GBM patients harboring FGFR3-TACC3 fusions both achieved PR after using anlotinib, suggesting that FGFR3-TACC3 fusions might be a potential biomarker for administration of anlotinib to treat GBM, which was confirmed by our previous study [39]. Moreover, there are also evidences showing that GBM patients harboring FGFR3-TACC3 fusions may benefit from targeted FGFR kinase inhibition [40,41].
A major strength of this study was that FGFR aberrations were comprehensively analyzed in a large group of Chinese patients with diverse cancer types, which may provide a better landscape for FGFR alterations in solid tumors. Additionally, other FGFR fusion partners (SH3GLB1, BICC1, CAT, TDRD1, CCDC158, CIT, TXLNB, NKD2, CUX2, HK1, and DOCK1) were also identified except for TACC, particularly the most common fusion partner TACC3. However, some limitations should also be interpreted cautiously. First, the number of patients with each cancer type was based on the number of tumor samples presented by physicians for genomic profiling, which may lead to the bias in sample size. Second, we did not evaluate whether some novel alterations of FGFRs had functional effects in in vitro or in vivo models. Further clinical relevance of these alterations needs investigation.
Conclusions
FGFR1-4 aberrations are common in solid tumors of diverse types, approximately accounting for 4.2%-22.2%. Of FGFR1-4 variations, gene amplifications and gene mutations are more frequent, especially FGFR1 amplifications. The frequencies of TP53, MUC16, NSD3, MYC and LRP1B mutations are relatively high in patients with FGFR1-4 aberrations. FGFR fusions (mostly FGFR2 and FGFR3) occur in 0.6% of cancer cases, and its relevant case studies suggest that GBM patients with FGFR3-TACC3 fusions may derive benefits from anlotinib.
Acknowledgements
This work was supported by following grants: Guangdong Provincial Natural Science Program (No. 2019A1515010900, X Zhang); GDPH Dengfeng Program (No. DFJH201903 & KJ012019444 & 8197103306, X Zhang); General Research Project of Guangzhou Science and Technology Bureau (No. 201607010391, X Zhang).
Two cases of GBM with FGFR3-TACC3 fusions voluntarily participated in the study and signed the informed consent form. The study was in accordance with regulations of the Declaration of Helsinki.
Disclosure of conflict of interest
None.
References
- 1.Katoh M, Nakagama H. FGF receptors: cancer biology and therapeutics. Med Res Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 2.Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer. 2017;17:318–332. doi: 10.1038/nrc.2017.8. [DOI] [PubMed] [Google Scholar]
- 3.Dai S, Zhou Z, Chen Z, Xu G, Chen Y. Fibroblast growth factor receptors (FGFRs): structures and small molecule inhibitors. Cells. 2019;8:614. doi: 10.3390/cells8060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Porta R, Borea R, Coelho A, Khan S, Araujo A, Reclusa P, Franchina T, Van Der Steen N, Van Dam P, Ferri J, Sirera R, Naing A, Hong D, Rolfo C. FGFR a promising druggable target in cancer: molecular biology and new drugs. Crit Rev Oncol Hematol. 2017;113:256–267. doi: 10.1016/j.critrevonc.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 6.Brooks AN, Kilgour E, Smith PD. Molecular pathways: fibroblast growth factor signaling: a new therapeutic opportunity in cancer. Clin Cancer Res. 2012;18:1855–1862. doi: 10.1158/1078-0432.CCR-11-0699. [DOI] [PubMed] [Google Scholar]
- 7.Greulich H, Pollock PM. Targeting mutant fibroblast growth factor receptors in cancer. Trends Mol Med. 2011;17:283–292. doi: 10.1016/j.molmed.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jing Q, Wang Y, Liu H, Deng X, Jiang L, Liu R, Song H, Li J. FGFs: crucial factors that regulate tumour initiation and progression. Cell Prolif. 2016;49:438–447. doi: 10.1111/cpr.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Zhang W, Geng D, He J, Zhao Y, Yu L. Clinical significance of fibroblast growth factor receptor-3 mutations in bladder cancer: a systematic review and meta-analysis. Genet Mol Res. 2014;13:1109–1120. doi: 10.4238/2014.February.20.12. [DOI] [PubMed] [Google Scholar]
- 10.Heist RS, Mino-Kenudson M, Sequist LV, Tammireddy S, Morrissey L, Christiani DC, Engelman JA, Iafrate AJ. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7:1775–1780. doi: 10.1097/JTO.0b013e31826aed28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, Cummings CT, Graham DK, Astling D, Tan AC, Heasley LE. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis. 2013;2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katoh M. Genetic alterations of FGF receptors: an emerging field in clinical cancer diagnostics and therapeutics. Expert Rev Anticancer Ther. 2010;10:1375–1379. doi: 10.1586/era.10.128. [DOI] [PubMed] [Google Scholar]
- 13.Donnem T, Al-Shibli K, Al-Saad S, Busund LT, Bremnes RM. Prognostic impact of fibroblast growth factor 2 in non-small cell lung cancer: coexpression with VEGFR-3 and PDGF-B predicts poor survival. J Thorac Oncol. 2009;4:578–585. doi: 10.1097/JTO.0b013e31819f2e38. [DOI] [PubMed] [Google Scholar]
- 14.Formisano L, Lu Y, Servetto A, Hanker AB, Jansen VM, Bauer JA, Sudhan DR, Guerrero-Zotano AL, Croessmann S, Guo Y, Ericsson PG, Lee KM, Nixon MJ, Schwarz LJ, Sanders ME, Dugger TC, Cruz MR, Behdad A, Cristofanilli M, Bardia A, O’Shaughnessy J, Nagy RJ, Lanman RB, Solovieff N, He W, Miller M, Su F, Shyr Y, Mayer IA, Balko JM, Arteaga CL. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer. Nat Commun. 2019;10:1373. doi: 10.1038/s41467-019-09068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamauchi M, Ono A, Ishikawa A, Kodama K, Uchikawa S, Hatooka H, Zhang P, Teraoka Y, Morio K, Fujino H, Nakahara T, Murakami E, Miki D, Kawaoka T, Tsuge M, Hiramatsu A, Imamura M, Hayes CN, Fujita M, Nakagawa H, Yasui W, Aikata H, Chayama K. Tumor fibroblast growth factor receptor 4 level predicts the efficacy of lenvatinib in patients with advanced hepatocellular carcinoma. Clin Transl Gastroenterol. 2020;11:e00179. doi: 10.14309/ctg.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dienstmann R, Rodon J, Prat A, Perez-Garcia J, Adamo B, Felip E, Cortes J, Iafrate AJ, Nuciforo P, Tabernero J. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann Oncol. 2014;25:552–563. doi: 10.1093/annonc/mdt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghedini GC, Ronca R, Presta M, Giacomini A. Future applications of FGF/FGFR inhibitors in cancer. Expert Rev Anticancer Ther. 2018;18:861–872. doi: 10.1080/14737140.2018.1491795. [DOI] [PubMed] [Google Scholar]
- 18.Helsten T, Elkin S, Arthur E, Tomson BN, Carter J, Kurzrock R. The FGFR landscape in cancer: analysis of 4,853 tumors by next-generation sequencing. Clin Cancer Res. 2016;22:259–267. doi: 10.1158/1078-0432.CCR-14-3212. [DOI] [PubMed] [Google Scholar]
- 19.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L, Nolden LK, Horlings H, Berns K, Hung MC, van de Vijver MJ, Valero V, Gray JW, Bernards R, Mills GB, Hennessy BT. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–6091. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XC, Wang J, Shao GG, Wang Q, Qu X, Wang B, Moy C, Fan Y, Albertyn Z, Huang X, Zhang J, Qiu Y, Platero S, Lorenzi MV, Zudaire E, Yang J, Cheng Y, Xu L, Wu YL. Comprehensive genomic and immunological characterization of Chinese non-small cell lung cancer patients. Nat Commun. 2019;10:1772. doi: 10.1038/s41467-019-09762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, Liu EM, Reichel J, Porrati P, Pellegatta S, Qiu K, Gao Z, Ceccarelli M, Riccardi R, Brat DJ, Guha A, Aldape K, Golfinos JG, Zagzag D, Mikkelsen T, Finocchiaro G, Lasorella A, Rabadan R, Iavarone A. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamont FR, Tomlinson DC, Cooper PA, Shnyder SD, Chester JD, Knowles MA. Small molecule FGF receptor inhibitors block FGFR-dependent urothelial carcinoma growth in vitro and in vivo. Br J Cancer. 2011;104:75–82. doi: 10.1038/sj.bjc.6606016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai SJ, Wu MH, Chen HM, Chuang PC, Wing LY. Fibroblast growth factor-9 is an endometrial stromal growth factor. Endocrinology. 2002;143:2715–2721. doi: 10.1210/endo.143.7.8900. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad I, Iwata T, Leung HY. Mechanisms of FGFR-mediated carcinogenesis. Biochim Biophys Acta. 2012;1823:850–860. doi: 10.1016/j.bbamcr.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Jang M, Kim E, Choi Y, Lee H, Kim Y, Kim J, Kang E, Kim SW, Kim I, Park S. FGFR1 is amplified during the progression of in situ to invasive breast carcinoma. Breast Cancer Res. 2012;14:R115. doi: 10.1186/bcr3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, Natrajan R, Marchio C, Iorns E, Mackay A, Gillett C, Grigoriadis A, Tutt A, Reis-Filho JS, Ashworth A. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–2094. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas: a chromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Qiu Y, Guo P, Pu T, Feng Y, Bu H. FGFR1 and HER1 or HER2 co-amplification in breast cancer indicate poor prognosis. Oncol Lett. 2018;15:8206–8214. doi: 10.3892/ol.2018.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hui R, Pearson A, Cortes J, Campbell C, Poirot C, Azim HA Jr, Fumagalli D, Lambertini M, Daly F, Arahmani A, Perez-Garcia J, Aftimos P, Bedard PL, Xuereb L, Scheepers ED, Vicente M, Goulioti T, Loibl S, Loi S, Pierrat MJ, Turner NC, Andre F, Curigliano G. Lucitanib for the treatment of HR(+)/HER2(-) metastatic breast cancer: results from the multicohort phase II FINESSE study. Clin Cancer Res. 2020;26:354–363. doi: 10.1158/1078-0432.CCR-19-1164. [DOI] [PubMed] [Google Scholar]
- 31.Kim HR, Kim DJ, Kang DR, Lee JG, Lim SM, Lee CY, Rha SY, Bae MK, Lee YJ, Kim SH, Ha SJ, Soo RA, Chung KY, Kim JH, Lee JH, Shim HS, Cho BC. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J. Clin. Oncol. 2013;31:731–737. doi: 10.1200/JCO.2012.43.8622. [DOI] [PubMed] [Google Scholar]
- 32.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A, Moch H, Wagener P, Fischer F, Heynck S, Koker M, Schottle J, Leenders F, Gabler F, Dabow I, Querings S, Heukamp LC, Balke-Want H, Ansen S, Rauh D, Baessmann I, Altmuller J, Wainer Z, Conron M, Wright G, Russell P, Solomon B, Brambilla E, Brambilla C, Lorimier P, Sollberg S, Brustugun OT, Engel-Riedel W, Ludwig C, Petersen I, Sanger J, Clement J, Groen H, Timens W, Sietsma H, Thunnissen E, Smit E, Heideman D, Cappuzzo F, Ligorio C, Damiani S, Hallek M, Beroukhim R, Pao W, Klebl B, Baumann M, Buettner R, Ernestus K, Stoelben E, Wolf J, Nurnberg P, Perner S, Thomas RK. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lasorella A, Sanson M, Iavarone A. FGFR-TACC gene fusions in human glioma. Neuro Oncol. 2017;19:475–483. doi: 10.1093/neuonc/now240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson KN, Meyer AN, Siari A, Campos AR, Motamedchaboki K, Donoghue DJ. Oncogenic gene fusion FGFR3-TACC3 is regulated by tyrosine phosphorylation. Mol Cancer Res. 2016;14:458–469. doi: 10.1158/1541-7786.MCR-15-0497. [DOI] [PubMed] [Google Scholar]
- 35.Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, Li X, Gumin J, Zheng H, Hu L, Yli-Harja O, Haapasalo H, Visakorpi T, Liu X, Liu CG, Sawaya R, Fuller GN, Chen K, Lang FF, Nykter M, Zhang W. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123:855–865. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Costa R, Carneiro BA, Taxter T, Tavora FA, Kalyan A, Pai SA, Chae YK, Giles FJ. FGFR3-TACC3 fusion in solid tumors: mini review. Oncotarget. 2016;7:55924–55938. doi: 10.18632/oncotarget.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan L, Liu ZH, Lin ZR, Xu LH, Zhong Q, Zeng MS. Recurrent FGFR3-TACC3 fusion gene in nasopharyngeal carcinoma. Cancer Biol Ther. 2014;15:1613–1621. doi: 10.4161/15384047.2014.961874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen G, Zheng F, Ren D, Du F, Dong Q, Wang Z, Zhao F, Ahmad R, Zhao J. Anlotinib: a novel multi-targeting tyrosine kinase inhibitor in clinical development. J Hematol Oncol. 2018;11:120. doi: 10.1186/s13045-018-0664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Liang D, Chen J, Chen H, Fan R, Gao Y, Gao Y, Tao R, Zhang H. Targeted therapy with anlotinib for a patient with an oncogenic FGFR3-TACC3 fusion and recurrent glioblastoma. Oncologist. 2021;26:173–177. doi: 10.1002/onco.13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Stefano AL, Fucci A, Frattini V, Labussiere M, Mokhtari K, Zoppoli P, Marie Y, Bruno A, Boisselier B, Giry M, Savatovsky J, Touat M, Belaid H, Kamoun A, Idbaih A, Houillier C, Luo FR, Soria JC, Tabernero J, Eoli M, Paterra R, Yip S, Petrecca K, Chan JA, Finocchiaro G, Lasorella A, Sanson M, Iavarone A. Detection, characterization, and inhibition of FGFR-TACC fusions in IDH wild-type glioma. Clin Cancer Res. 2015;21:3307–3317. doi: 10.1158/1078-0432.CCR-14-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang G, Liu Z, Wu J, Cai Y, Li X. Anticancer molecules targeting fibroblast growth factor receptors. Trends Pharmacol Sci. 2012;33:531–541. doi: 10.1016/j.tips.2012.07.001. [DOI] [PubMed] [Google Scholar]