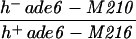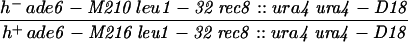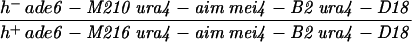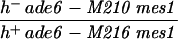Abstract
Our work and that of others defined mitosis-specific (Rad21 subfamily) and meiosis-specific (Rec8 subfamily) proteins involved in sister chromatid cohesion in several eukaryotes, including humans. Mutation of the fission yeast Schizosaccharomyces pombe rec8 gene was previously shown to confer a number of meiotic phenotypes, including strong reduction of recombination frequencies in the central region of chromosome III, absence of linear element polymerization, reduced pairing of homologous chromosomes, reduced sister chromatid cohesion, aberrant chromosome segregation, defects in spore formation, and reduced spore viability. Here we extend the description of recombination reduction to the central regions of chromosomes I and II. We show at the protein level that expression of rec8 is meiosis specific and that Rec8p localizes to approximately 100 foci per prophase nucleus. Rec8p was present in an unphosphorylated form early in meiotic prophase but was phosphorylated prior to meiosis I, as demonstrated by analysis of the mei4 mutant blocked before meiosis I. Evidence for the persistence of Rec8p beyond meiosis I was obtained by analysis of the mutant mes1 blocked before meiosis II. A human gene, which we designate hrec8, showed significant primary sequence similarity to rec8 and was mapped to chromosome 14. High mRNA expression of mouse and human rec8 genes was found only in germ line cells, specifically in testes and, interestingly, in spermatids. hrec8 was also expressed at a low level in the thymus. Sequence similarity and testis-specific expression indicate evolutionarily conserved functions of Rec8p in meiosis. Possible roles of Rec8p in the integration of different meiotic events are discussed.
Meiosis is an essential step in the sexual reproduction of eukaryotes. It serves to reduce the chromosome number from diploidy in the germ line to haploidy in the gametes. This is accomplished by two rounds of chromosome segregation, meiosis I and II, after a single round of DNA replication. During meiotic prophase, the replicated homologous chromosomes pair, recombination occurs between nonsister chromatids, and the resulting crossovers lead to chiasma formation in all bivalents. Chiasmata and sister chromatid cohesion are required for correct segregation of homologous chromosomes at meiosis I (reductional division). Throughout meiotic prophase, numerous events occur at the chromosomes in highly ordered fashion (for reviews, see references 37 and 68).
The events occurring at the DNA level are coordinated with many other cellular processes, and all are integrated into the meiotic cell cycle. Much additional research is required for the elucidation of the regulatory mechanisms governing meiosis. However, it is known that checkpoint proteins involved in the regulation of the mitotic cell cycle are also needed for checkpoint controls in meiosis (47). A number of protein kinases have important functions in meiosis. An example is the kinase encoded by the Drosophila melanogaster mei-41 gene, which is homologous to the human ATM gene (30). Mutation of this gene affects the number and morphology of recombination nodules (12). The CDC28, MEK1/MRE4, ESR2, and IME2 genes, coding for identified or putative protein kinases, have meiosis-specific functions in Saccharomyces cerevisiae: (19, 34, 44, 67, 74). A protein phosphatase was shown to interact with Red1, which is a component of the lateral elements of the S. cerevisiae synaptonemal complex (77, 83). A component of the lateral elements of the rat synaptonemal complex, SCP3, is multiply phosphorylated (43). SCP3 is identical to the Cor1 protein, which relocates from lateral elements to centromeres after anaphase I (20).
An important structural feature of chromosomes during mitotic and meiotic prophase is sister chromatid cohesion. It contributes to meiotic pairing and recombination of the homologous chromosomes. After crossover formation and degradation of the synaptonemal complex, the homologs are kept in alignment by the microscopically visible chiasmata, which do not resolve until anaphase I. If sister chromatid cohesion is resolved prematurely, chromosomes do not segregate properly, resulting in daughter cells with unbalanced genomes (for a review, see reference 52).
In several organisms, genes involved in meiotic sister chromatid cohesion have been identified. The spo76 mutant of Sordaria shows precocious separation of sister chromatids and reduced meiotic recombination levels (55). Mutation of the ord gene of D. melanogaster also leads to premature separation of sister chromatids (7, 51). In mei-S332 mutants of D. melanogaster, meiotic recombination and segregation of the homologs at meiosis I is normal. However, sister chromatids separate precociously during anaphase I, leading to random segregation at meiosis II (35). Mei-S332 protein associates with centromeres in late meiotic prophase and disappears at anaphase II, when sister chromatids separate completely (36). A function similar to Mei-S332 was attributed to mouse Cor1 on the basis of its specific localization to centromeric regions from anaphase I to anaphase II (20, 53).
Loss of mitotic sister chromatid cohesion in S. cerevisiae requires degradation of Pds1p at the metaphase-to-anaphase transition (14). No homologs of Pds1p were identified in other organisms, but Cut2p of the fission yeast S. pombe may have a similar function (24). Recently, additional proteins involved in mitotic sister chromatid cohesion in S. cerevisiae were described (49). One of them, Scc1p, is related to Rad21p of the fission yeast S. pombe (8, 9). Scc1p binds to chromosomes during S phase, dissociates at the metaphase-to-anaphase transition, and is needed for sister chromatid cohesion near centromeres and in the chromosome arms. Binding of Scc1p to chromosomes is Smc1p dependent. The same protein, but named Mcd1p, was also identified in a screen for structural proteins of chromosomes and for proteins interacting with Smc1p, suggesting an additional role for Scc1p/Mcd1p in chromosome condensation (28). In addition to Rad21p, homologs of Scc1p/Mcd1p appear to exist in all eukaryotes, including Homo sapiens (reference 48 and this study).
The rec8 gene of S. pombe was identified by screening for mutants with reduced meiotic recombination in the ade6 gene (63). Point mutations in rec8 reduce meiotic but not mitotic recombination and result in mutants with normal resistance to the DNA-damaging agents UV radiation and methyl methanesulfonate (17). Strong reduction of recombination was reported to be specific for a 2-Mb region around the centromere of chromosome III (18; but see below). Expression of rec8 RNA was concluded to be meiosis specific based on meiosis induction in haploid pat1 mutant cells (46). In mutants with a rec8-110 point mutation, linear-element formation is defective and only amorphous aggregates of linear-element proteins were observed (54). The linear elements appearing in S. pombe prophase resemble axial cores (precursors of the lateral elements in the synaptonemal complex) of other eukaryotes (2). Bouquet formation was normal in rec8-110 mutants, but pairing of chromosomes, studied by fluorescent in situ hybridization (FISH), was reduced. The strongly reduced spore viability was assumed to be a consequence of precocious separation of sister chromatids, which was observed in genetic assays and by FISH. It was concluded that linear-element formation contributes both to sister chromatid cohesion and to high meiotic recombination frequency (54).
Here we expand on the characterization of fission yeast Rec8p by performing an analysis of a new rec8 allele, a gene deletion/disruption likely to create a null phenotype, and using an anti-Rec8p antibody in the study of Rec8p localization and Rec8p phosphorylation. We report the cloning and characterization of a novel human gene, hrec8, and present the results of experiments on complementation of the fission yeast rec8 disruption strain by the human homolog. We describe in detail the rad21/rec8 gene family. Finally, the results are discussed with respect to possible roles of Rec8p in the regulation of meiotic events.
MATERIALS AND METHODS
Strains, plasmids, media, and general methods.
The genotypes of S. pombe strains used in this study are listed in Table 1. All rec8::ura4 mutant strains were constructed from PA32 by genetic crosses. Plasmid pYL3 (46) contains a 3.9-kb rec8 fragment subcloned in pSP2 (16). Plasmid pREP41-hrec8 was prepared by insertion of the full-length human rec8 (hrec8) cDNA into the NdeI site of plasmid pREP41 (5).
TABLE 1.
Description of S. pombe strains used in this study
| Strain | Genotype | Source or reference | |
|---|---|---|---|
| PA3 | h− ade6-52 pro2-1 leu1-32 ura4-D18 | This study | |
| PA4 | h+ ade6-M26 arg3-124 leu1-32 ura4-D18 | This study | |
| PA21 | h− ade6-52 pro2-1 leu1-32 rec8::ura4 ura4-D18 | This study | |
| PA22 | h+ ade6-M26 arg3-124 leu1-32 rec8::ura4 ura4-D18 | This study | |
| PA32 | h− leu1-32 rec8::ura4 ura4-D18 | This study | |
| PA39 |
|
Strain collection, Bern | |
| PA40 |
|
This study | |
| PA41 |
|
This study | |
| PA42 |
|
This study | |
| PA43 | h90 leu1-32 rec8::ura4 ura4-D18 | This study |
General genetic methods and the standard media yeast extract agar (YEA) and malt extract agar (MEA) were as described previously (29). Minimal medium (MMA) consists of 0.67% Difco Nitrogen Base without amino acids, 1% glucose, and 1.8% agar; synthetic growth medium (GMA) consists of 0.17% Difco Nitrogen Base without amino acids, 0.375% sodium glutamate, 1% glucose, and 2% agar. EMM (a modified Edinburgh minimal medium), used for the complementation analysis of rec8::ura4, was prepared with 20 g of agar per liter (71). All growth factors were added to a final concentration of 100 mg/liter. For meiotic time course experiments, the synthetic minimal medium PM (S. pombe minimal) (6) and PM − N (PM without NH4Cl) (86) were used.
rec8 gene disruption.
To construct a rec8 gene disruption mutant, a 3.9-kb SacI fragment from pYL3 (46) containing the rec8 gene and flanking sequences was subcloned into pBluescript KS (Stratagene). From the resulting plasmid, pBSrec8S-1, a 1.5-kb NsiI-NheI fragment was replaced by a 1.8-kb PstI-XbaI fragment from pB4-3 containing the ura4 marker gene (25) to yield the gene disruption plasmid pBSrec8S-1::ura4. From this plasmid, a 4.2-kb SacI fragment was used to transform the strain h− leu1-32 ura4-D18 by the lithium acetate method (33). Proper integration of the fragment into the genome was verified by Southern blot analysis.
Anti-Rec8p antibody.
A 1.2-kb rec8 DNA fragment covering the originally published open reading frame (ORF) from positions 742 to 1923 (46) was amplified by PCR to introduce a BglII and an EcoRI restriction site at the 5′ and 3′ ends of the fragment, respectively. This PCR product was ligated in frame into the BamHI and EcoRI sites of pGEX-2T (Pharmacia) to obtain a rec8-glutathione S-transferase (rec8-GST) fusion. The construct was verified by DNA sequence analysis.
Fusion protein was produced as described previously (78). Briefly, Escherichia coli DH5α cells were transformed with pGEX-2T-rec8 and selected for the presence of plasmid with ampicillin (50 μg/ml). Synthesis of fusion protein was induced by the addition of isopropyl-β-d-thiogalactoside (IPTG) to a final concentration of 0.1 mM. After further incubation for 3 to 4 h at 37°C, the cells were harvested and stored at −70°C. For preparation of E. coli extracts, approximately 4 g of cells was resuspended in 40 ml of phosphate-buffered saline (PBS) containing 2 mM EDTA, 100 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 1% Triton X-100. The GST fusion protein was isolated from the cleared lysate by the addition of 2 ml of 50% slurry of glutathione-Sepharose 4B (Pharmacia). The fusion protein was eluted in 50 mM Tris-HCl (pH 8.0)–10 mM reduced glutathione, and eluted fractions were concentrated and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The 72-kDa Rec8-GST fusion protein was isolated from the polyacrylamide gel and used for immunization.
Polyclonal antiserum was obtained by injecting a rabbit and three rats three times with approximately 200 μg of Rec8 fusion protein at 2-week intervals. Antisera were affinity purified by three rounds of adsorption to nitrocellulose Western blot strips (65) containing three different proteins (GST, E. coli heat shock protein GroEL, and Rec8 fusion protein). GroEL was a contamination in the preparation of Rec8 fusion protein as identified by protein sequencing.
Meiotic time courses and Rec8p immunofluorescence on nuclear spreads.
The induction of meiosis, the preparation of nuclear spreads, and 4′,6-diamino-2-phenylindole (DAPI) staining of DNA were as described previously (2). Briefly, S. pombe PA39 diploid cells were induced to undergo meiosis by being shifted to nitrogen-free medium. Aliquots collected immediately after the shift (0 h) and at 2-h intervals thereafter were used to prepare spreads of nuclei. For flow-cytometric analysis, 1-ml aliquots of cells were fixed in 70% ethanol and stained with propidium iodide (Sigma) as described elsewhere (6). For immunofluorescence experiments, the wild-type diploid strain PA39 and the control diploid strain PA40 homozygous for the rec8::ura4 deletion were used (Table 1). Slides with nuclear spreads stored at −70°C were soaked in PBS containing 0.1% Photo-Flo (Kodak) to remove the sucrose layer and then washed for 15 min in PBS containing 0.05% Triton X-100 and for 15 min in twofold-diluted blocking buffer (100 mM lysine, 3% nonfat dry milk, 0.05% Triton X-100, and 0.02% NaN3 in PBS [pH 7.3]). The nuclear spreads were then blocked overnight in blocking buffer and subsequently probed with the first antibody (1:10 dilution of rabbit anti-Rec8p in blocking buffer) for 24 h. The slides were sequentially washed in PBS containing 0.1% Photo-Flo and PBS containing 0.05% Triton X-100 for 15 min each prior to incubation with the second antibody for 24 h (1:80 dilution of goat anti-rabbit immunoglobulin G-fluorescein isothiocyanate conjugate in PBS [Sigma]). Before being mounted, the slides were washed once more for 15 min in PBS containing 0.1% Photo-Flo and for 15 min in PBS containing 0.05% Triton X-100, with two additional washes in H2O for 5 min each. The chromatin was counterstained with DAPI in a Vectashield antifade solution (Vecta Laboratories Inc.). The slides were analyzed with an epifluorescence (Zeiss Axiovert) microscope. Foci were quantified with the UTHSCSA Image Tool software (version 1.27).
RNA extraction, Northern blot hybridization, and reverse transcription-PCR.
Total RNA from S. pombe cells was prepared as described previously (26). Hybridization with a 32P-labelled rec8 or byr1 (58) DNA probe was performed as specified by the manufacturer (Bio-Rad) in its standard hybridization protocol. The rec8 probe was the same 1.2-kb DNA fragment used to construct pGEX-2T-rec8. The byr1 probe (a gift from A. M. Schweingruber) was the 0.4-kb PCR fragment from positions 976 to 1387 in the ORF.
A 300-μg portion of total RNA from a meiotic time course (8 h after the shift) was used to prepare meiotic mRNA with the Oligotex poly(A)+ mRNA isolation kit (Qiagen). First-strand rec8 cDNA was synthesized from 1 μg of meiotic mRNA with an oligo(dT)18 primer and Superscript reverse transcriptase (GIBCO). Reverse transcription-PCR was performed with the primers 5′-GGAAAAGGGAGGAATGGGAGTAATTTGG-3′ (positions 186 to 213 of rec8) and 5′-GTGAAAAGTTTCAAATGGCATCGGTGC-3′ (positions 2013 to 2039 of rec8). The resulting PCR fragment was subcloned into pGEM-T (Promega) and subjected to DNA sequencing.
Northern blot analysis of the human rec8 mRNA was performed as described previously (48). A multiple human tissue blot was obtained from Clontech (no. 7754-1); hybridization and wash conditions were as specified by the manufacturer. The blots were hybridized with 32P-labelled cDNA probes.
Immunoblotting, immunoprecipitation, and phosphatase treatment.
For the preparation of S. pombe extracts, approximately 1 g of cells from different time points of a meiotic time course was suspended in disruption buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 5 mM benzamidine, 13 mM β-mercaptoethanol), mixed with an equal volume of glass beads, and disrupted in a mini bead-beater. Following SDS-PAGE, the proteins were transferred to nitrocellulose and the filter was blocked overnight in TBST (150 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.05% Tween 20, 0.01% NaN3) containing 5% nonfat dry milk. The filter was then incubated with the primary rat anti-Rec8 antibody (1:20 in TBST containing 5% nonfat dry milk) for 2 h. After three washes in TBST, the second antibody (1:500 dilution of rabbit anti-rat IgG peroxidase conjugate [DAKO] in TBST) was added for 2 h, and the mixture was washed three times in TBST and developed with the ECL chemiluminescent detection kit (Amersham).
For immunoprecipitation, Dynabeads M-280 (Dynal) were coated with affinity-purified rabbit anti-Rec8p antibodies and used for magnetic Rec8p purification from freshly prepared crude extracts of wild-type or mei4 diploid cells, respectively, as described by the manufacturer.
Lambda protein phosphatase (New England BioLabs) was used in dephosphorylation experiments. Crude extracts from 1 g of meiotic wild-type cells (8 h after induction of meiosis) were prepared and used to immunoprecipitate Rec8p as described above. Beads containing Rec8p were washed twice in reaction buffer (50 mM Tris-HCl, 0.1 mM disodium EDTA, 5 mM dithiothreitol, 0.01% Brij 35 [pH 7.5]) and split into three portions. The first was used as control for immunoprecipitation and used directly for SDS-PAGE, the second was incubated in reaction buffer with 2 mM MnCl2 for 1 h at 30°C (no phosphatase), and the third was incubated with 800 U of lambda protein phosphatase for 1 h at 30°C.
Cloning and sequence analysis of hrec8.
General molecular biology procedures were essentially as described previously (69). Two 20-mer primers were designed based on the nucleotide sequence of EST T33286, whose translated product was homologous to the C-terminal end of hHR21sp (48) (see Results). The primers had their 5′ ends at 102 nucleotides before and 140 nucleotides after the terminal nucleotide of the hrec8 ORF and were used to amplify a 241-bp fragment of the hrec8 gene from a human T-cell leukemia cell line cDNA (cDNA kindly supplied by Karin van Gool). This PCR fragment was used as a probe to isolate hrec8 cDNA clones from a normalized, gridded human infant-brain plasmid library (79), kindly supplied by P. Heutink. These hrec8 clones were sequenced on both strands. Amino acid sequences were aligned with the ESEE program (version 1.09e; supplied by Eric Cabot). An unrooted phylogenic tree (see Fig. 5C) was constructed based on the protein sequence alignments of the rec8/rad21 homologs shown in Fig. 5B; a tree based on alignment of all full-length rec8/rad21 proteins with the DNA Man program (version 3.2; Lynnon Biosoft, Quebec, Canada) gave very similar results (not shown). Trees were drawn with the PHYLIP software package (22); pairwise distances were calculated with the Dayhoff PAM 001 matrix. PEST sequences (66) were identified by using PESTfind at IMB Jena.
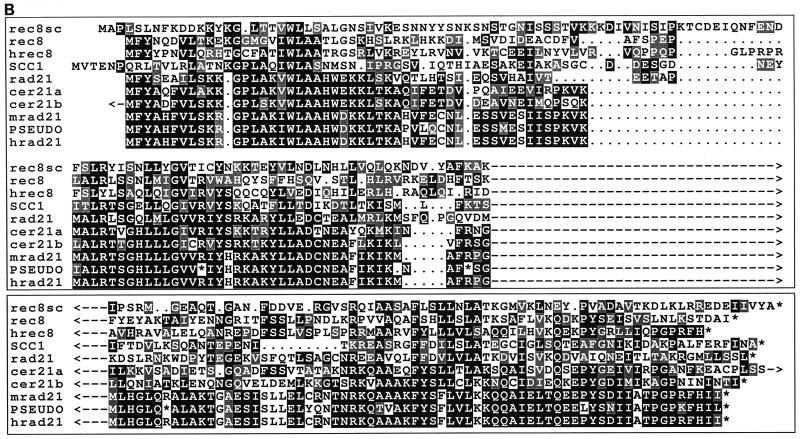
FIG. 5.
rad21/rec8 gene family. (A) Alignment of the predicted amino acid sequence of S. pombe Rec8p with human Rec8p, an S. cerevisiae Rec8p homolog (Rec8psc), and the founding family member, Rad21p. Letters in black boxes represent identical amino acids in at least two species, whereas those in gray boxes represent similar (P, A, G, S, and T; E, D, N, and Q; V, I, L, and M; F, W, and Y; R, K, and H) amino acids. Gaps introduced into the sequences for alignment optimization are shown as dots. Numbers denote amino acid numbers in the sequence. (B) Alignment of the conserved N- and C-terminal amino acid regions (top and bottom, respectively) of rad21/rec8 gene family members from different species. Sequences shown here are as follows: rec8sc, S. cerevisiae Rec8p homolog (GenBank accession no. U31900); rec8 (reference 46 and this study); hrec8 (present study); SCC1, S. cerevisiae Rad21p homolog (28, 49) (GenBank U23759); rad21, S. pombe Rad21p (8); cer21a and cer21b, first and second C. elegans Rad21p homologs (reference 48 and this study) (GenBank U40029 and U38377, respectively); mrad21 and hrad21, mHR21sp and hHR21sp (mouse and human Rad21p homologs) (48) (GenBank X98293 and X98294, respectively); and PSEUDO, hHR21sp pseudogene on chromosome X (GenBank HSU85A3). Rec8sc, Rec8p, and hRec8p are the S. cerevisiae, S. pombe, and human Rec8 proteins that are the subject of this publication. Amino acid shading and general features of the alignment are as in panel A. Regions of less highly conserved sequence exist between the blocks and are indicated by dashes and arrowheads. The arrowhead at the end of cer21a represents 22 nonconserved amino acids at its C terminus. The arrowhead at the beginning of cer21b represents 55 nonconserved amino acids at its N terminus. (C) Unrooted phylogenetic tree of Rad21 and Rec8 proteins from different species. Evolutionary distances between the proteins from different species are indicated by the lengths of the lines in the figure. The sequences represented here are the same as in panel B, except that for reasons of clarity, the mHR21sp protein (and the pseudogene) are not on the tree because of their evolutionary proximity to hHR21spp.
Complementation of rec8::ura4 by the human hrec8 gene.
S. pombe PA43 was transformed with either pREP41 or pREP41-hrec8 by the lithium acetate method (33). Transformants were selected on EMM plates lacking leucine. Transformants were plated onto MEA and allowed to sporulate for 72 h at 25°C. Cell-free spore suspensions were prepared by incubation of cell material with β-glucuronidase (Sigma) at 22°C for 16 h. The spore titer was determined by counting. The percentage of viable spores was determined by plating on YEA and counting of the forming colonies.
S. pombe PA21 and PA22 were transformed with either pREP41 or pREP41-hrec8 by the lithium acetate method (33). Transformants were selected following growth at 30°C for 5 days on EMM plates containing adenine, proline, and arginine but lacking leucine. Individual colonies were streaked on the same medium and grown for 24 h at 30°C. Crosses were conducted at 25°C for 3 days on EMM plates containing adenine, proline, and arginine. Cell-free spore suspensions were prepared by overnight treatment of aliquots of the crossing material with snail digestive juice. Measurement of spore viability and intragenic and intergenic recombination were done by standard genetic methods. The mean values of three experiments were determined.
Chromosomal localization.
FISH experiments were performed with biotinylated hrec8 cDNA probes, hybridized to metaphase spreads of normal human lymphocytes, as described previously (62, 84). Chromosomes were banded with DAPI and actinomycin D and counterstained with propidium iodide in antifade solution.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the fission yeast and human rec8 DNA sequences are AJ223299 and AF006264, respectively.
RESULTS
The phenotypes of a rec8 gene disruption and reevaluation of the region specific reduction of recombination.
A renewed analysis of the rec8 gene led to extension of the ORF at both ends (see the legend to Fig. 1). This result led to the discovery of homology to other genes in the data banks (see below). A gene disruption strain was constructed by replacement of a large part of the rec8 ORF by the ura4 marker gene (Fig. 1) for comparison of its phenotypes with those of the rec8-110 point mutation studied previously (54). Spore viabilities measured by random spore analysis were 12 and 20% in the disruption and point mutants, respectively. This difference is statistically significant but of doubtful relevance. The frequencies of intragenic and intergenic recombination were clearly reduced in comparison to those of the wild type, but no significant differences between disruption and point mutation crosses were found. At lys7 (lys7-1 × lys7-2) and at ade6 (ade6-M26 × ade6-52), the differences were at most threefold. In the three intervals pro2-arg3, leu2-lys7, and ade6-arg1, the values were two- to threefold lower in the rec8::ura4 than in the point mutation crosses (data not shown). The cytological analysis revealed an absence of linear elements and a shortened meiotic prophase in the rec8-110 strain (54). Linear-element formation in rec8::ura4 was impaired to a similar degree: no filaments and amorphous complexes of linear-element material were found. In contrast to the point mutation, the rec8::ura4 strain showed no shortening of prophase in three independent time courses (data not shown). Shortening of the prophase in the point mutation strain may indicate a role of Rec8p in meiosis regulation. Alternative explanations, like shortening of prophase by an additional mutation, were not excluded.
FIG. 1.
Schematic diagram of a 2.3-kb fragment carrying the rec8 gene and of the rec8::ura4 insertion mutation. The open box indicates the ORF of the rec8 gene. Solid boxes indicate the four introns at the following nucleotide positions: I at 213 to 255, II at 444 to 495, III at 593 to 644, and IV at 1895 to 1934. The putative TATA box (T), translation start and stop sites, and three potential polyadenylation signals (A1 to A3) are also indicated. In contrast to the originally published rec8 sequence (742 to 1923) (46), the actual gene structure contains four introns that were first identified on the basis of the published splice consensus sequences for S. pombe (64) (reference 45 and data not shown). The existence of these introns was confirmed as described in Materials and Methods. The A of the ATG is position 157, and the A of the TGA is position 2029. To produce the rec8::ura4 deletion mutant, the ura4 marker gene was used to replace the fragment between the 5′ NsiI and 3′ NheI restriction sites.
It was previously reported that a study of the rec8-110 point mutation showed that Rec8p is strongly required for activation of recombination in a 2-Mb region surrounding the ade6 gene on chromosome III but only marginally involved at other regions of the genome (18). Here, we extend this recombination analysis both by using the rec8 gene disruption mutant (rec8::ura4) and by testing new regions of chromosomes I and II. All data on intergenic recombination obtained with the point mutant (18) and our results from the gene disruption mutant are summarized in Fig. 2. The recombination frequencies obtained with the rec8 gene disruption, the genotypes of the strains used, and the specific methods applied are available upon request.
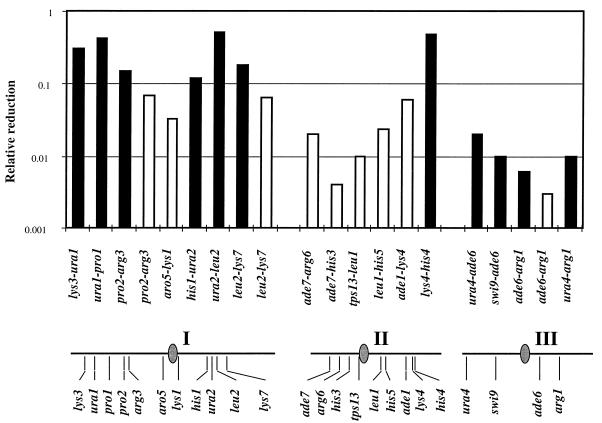
FIG. 2.
Meiotic recombinant frequencies in rec8 mutants. For each interval, the frequency is expressed relative to the one measured in the rec+ cross. The solid bars represent the data from reference 18, and the open bars represent those obtained from rec8::ura4 crosses. Recombination frequencies were measured twice in every interval in the rec+ and the rec8::ura4 backgrounds. The approximate positions of the genes on the chromosomes are shown below the histogram.
We observed the strongest reduction of recombination (approximately 300-fold) in crosses with rec8::ura4 at the ade6–arg1 interval on chromosome III, confirming the published results (18). On chromosomes I and II, the strongest reductions of recombination were also observed in the vicinity of the centromeres. The centromere-spanning interval tps13–leu1 and the centromere-proximal interval ade7–his3 on the short arm of chromosome II showed reductions of 100- and 300-fold, respectively. These values are comparable to those obtained for chromosome III. On chromosome I, the longest chromosome, the centromeric interval aro5–lys1 showed a 30-fold reduction. Moderate but significant reductions were found in the arms of chromosome I and close to the telomeres of chromosomes I and II.
Rec8p is localized in foci in the nucleus.
When intact wild-type cells from vegetative or meiotic cultures were fixed and treated with affinity-purified anti-Rec8p antibody, no signal was detected unless Rec8p was overexpressed from a plasmid, in which case the signal was confined to the nucleus (data not shown).
Gentle lysis of cells and spreading of nuclei on surfaces were used previously for the analysis of linear-element formation and chromosome pairing (2, 54, 70). Thus, spread meiotic nuclei from wild-type diploids were treated with affinity-purified anti-Rec8p antibody (see Materials and Methods). Fluorescence microscopy revealed that Rec8p is localized in foci throughout the spread nuclei. Meiotic time course experiments were then performed with the wild-type diploid strain PA39 and with the rec8::ura4 disruption diploid strain PA40. An example of the analysis of meiotic landmarks, the presence of rec8 mRNA, and the subnuclear localization of Rec8p (quantitation of Rec8p foci) are presented in Fig. 3.
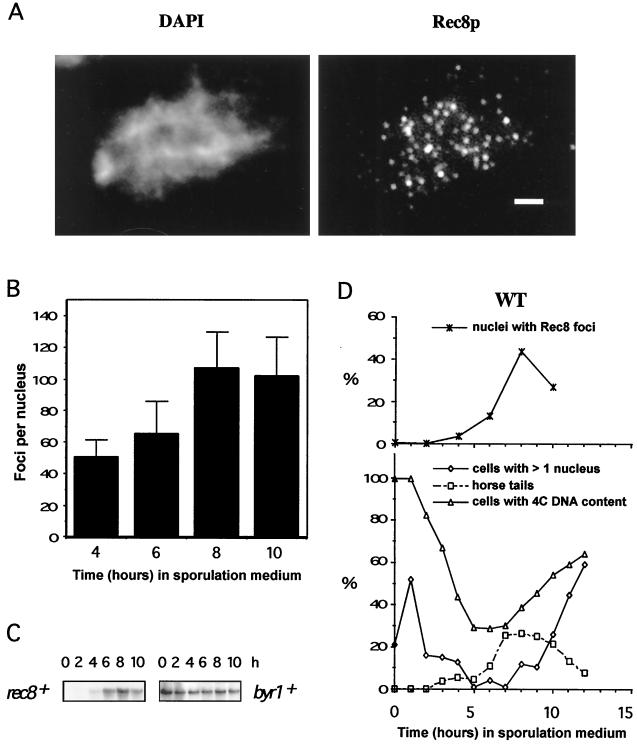
FIG. 3.
Immunolocalization of Rec8p-staining foci and rec8 mRNA expression in relation to a time course of cytological events during wild-type meiosis. Nuclear spreads were prepared from wild-type (PA39) diploid cells 0, 2, 4, 6, 8, and 10 h after transfer to meiosis medium, and all data are from the same meiosis time course experiment. (A) Example of a spread wild-type nucleus 8 h after induction of meiosis. Rec8p was visualized with an affinity purified polyclonal anti-Rec8p antibody (right), and the DNA was counterstained with DAPI (left). Bar, 2 μm. (B) Quantitation of Rec8p-staining foci during meiosis. The number of foci was determined in samples of 30 well-spread nuclei at the indicated time points. The error bars indicate 1 standard deviation. (C) Induction of rec8 mRNA expression during wild-type meiosis. Northern blots of 20 μg of total RNA at the indicated time points were hybridized with a rec8-specific probe (left) and later hybridized to a byr1-specific cDNA probe to verify loading (right). (D) Timing of cytological events during wild-type (WT) meiosis. The number of horse-tail nuclei, which are markers of meiotic prophase, was determined by DAPI staining, and the percentage of cells with 4C DNA content was measured by flow cytometry. Cells with more than one nucleus represent the percentage of the cells that have completed the first meiotic division. The increase in the number of cells containing two nuclei 1 h after induction of meiosis is due to the mitotic division of cells in G2 phase. This last mitotic division must occur before cells can enter meiosis from the G1 phase (21).
Rec8p was localized to distinct foci of different intensities within the nucleus (Fig. 3A). The intensity of Rec8p foci was dependent on the focal plane. No staining above background was observed on spreads of a time course of the diploid PA40 (rec8::ura4 [data not shown]). In general, the foci were round. The few elongated foci may represent two or more unresolved structures. Some adjacent foci (three to seven foci) resembled pearls on a string, while other foci were more widely separated and scattered in the same nucleus. We quantified the Rec8p foci per individual nucleus during a meiotic time course experiment (Fig. 3B). The average number of foci per nucleus increased from about 50 at early prophase (4 h after the shift to meiotic conditions) to more than 100 at late prophase (8 and 10 h). Similar results were obtained in a second independent time course. Foci first appeared after 4 h, when premeiotic DNA replication had started and when cells with elongated and deformed horse-tail nuclei (typical of meiotic prophase) started to accumulate (Fig. 3D). At 8 h, the fraction of nuclei with foci reached its maximum (42%). After 10 h, reliable scoring of nuclei with foci and quantitation of foci within single nuclei were no longer possible due to poor spreading properties of the nuclei (spore formation).
The number of nuclei carrying Rec8p foci correlated with the relative amount of rec8 mRNA at the different time points (Fig. 3C and D). Rec8p mRNA expression was previously shown to be specifically induced in haploid pat1 meiosis (46). The size of the transcript (2.0 kb) is consistent with the rec8 ORF (1,683 nucleotides) shown in Fig. 1.
The percentage of nuclei with foci was brought into the context of classical landmarks of fission yeast meiosis (Fig. 3D). The transition from mitotic G2 to G1 cells followed by premeiotic S phase was visualized by flow cytometry for DNA content. The large number of cells with two nuclei 1 h after induction of meiosis represents the final mitotic division before cells entered the meiotic prophase. The number of cells with more than one nucleus was smallest from 5 to 7 h and then increased again, indicating the onset of meiosis I. At 12 h after induction of meiosis, roughly 60% of the cells had completed the first meiotic division. The abundance of nuclei with foci coincided fully with the presence of nuclei with an extended shape (horse-tail nuclei), which appear to be due to the vigorous movement of prophase nuclei (13, 81).
Rec8p is phosphorylated during prophase and persists beyond meiosis I.
To further examine Rec8p expression during meiosis, time course Western analysis and immunoprecipitation experiments were performed with both wild-type diploid PA39 cells and diploid cells homozygous for mutations that block meiosis at specific stages. The particular PA39 culture used for the experiment in Fig. 4A was somewhat delayed compared to the cultures used for the study of Rec8p foci (Fig. 3). Rec8p first appeared 6 h after induction of meiosis, when the first horse-tail nuclei also became visible (data not shown), and grew more abundant at the later time points. At 6 h, Rec8p was visible as a single band of 87 kDa. At 8 h, an additional band migrating at 95 kDa became apparent and its intensity increased (10 and 12 h). After 12 h, protein extraction became difficult due to the presence of high percentages of spores. At 12 h, 40% of cells had performed the first meiotic division and fewer than 10% of the cells were still in meiotic prophase (horse-tail nuclei [data not shown]). The apparently large amount of Rec8p in the extract from cells harvested at 12 h indicated to us that Rec8p may persist beyond meiosis I (see below).
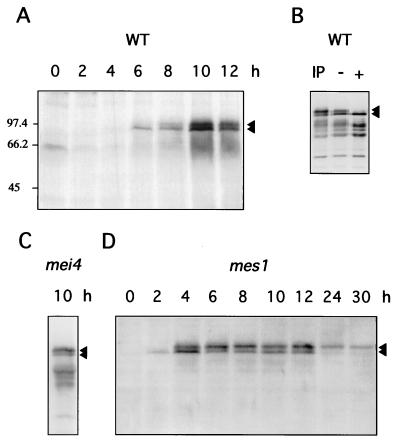
FIG. 4.
Expression and phosphorylation of Rec8p during meiosis. (A) Protein extracts were prepared at different time points of a meiosis time course of the wild-type (PA39; WT) diploid. Aliquots of 300 μg were subjected to SDS-PAGE and Western blot analysis with affinity-purified anti-Rec8p antiserum. Arrowheads indicate the two forms of Rec8p. (B) Phosphatase treatment of Rec8p from wild-type (WT) cells (strain PA39) 8 h after induction of meiosis. IP, immunoprecipitate; −, no phosphatase treatment; +, phosphatase treatment. (C) Immunoprecipitation of Rec8p from mei4 (PA41) cell extracts 10 h after a shift to sporulation medium. (D) Meiotic time course of the mes1 diploid strain PA42. Protein extracts were prepared at the indicated time points, and aliquots of 150 μg were subjected to SDS-PAGE and Western blot analysis with affinity-purified rat anti-Rec8p antiserum.
The appearance of an additional protein band suggested that Rec8p is modified during meiotic prophase. To test whether the additional Rec8p band was due to phosphorylation, Rec8p was immunoprecipitated from extracts of wild-type cells and treated with phosphatase. Figure 4B shows that the low-mobility Rec8p band was removed by this treatment, suggesting that Rec8p is phosphorylated during meiotic prophase. The high-mobility form, appearing first as a single band, did not change upon phosphatase treatment (control experiment; results not shown).
The data in Fig. 4A indicate that Rec8p phosphorylation occurs prior to the first meiotic division. If this is the case, mutant cells blocked before meiosis I are expected to contain phosphorylated Rec8p. Thus, the protein was immunoprecipitated from extracts of mei4 diploid cells, which are blocked in meiotic prophase at the stage with fully developed linear elements (2). In contrast to wild-type meiosis, no cells in the mei4 culture had two nuclei (indicated by DAPI staining) 10 h after induction of meiosis, confirming that the mei4 cells were blocked in prophase. Both versions of Rec8p were apparent as distinct bands; in addition, smaller proteins which may be degradation products were present (Fig. 4C). This result is consistent with Rec8p becoming phosphorylated prior to meiosis I.
Persistence of Rec8p beyond meiosis I is expected to lead to the presence of the protein in cells blocked after meiosis I and before meiosis II. Diploids homozygous for a mes1 mutation are blocked after the first and before the second meiotic division (11, 74). The same Rec8p double-band pattern was observed in wild-type cells (Fig. 4A) and mes1 cells (Fig. 4D) up to 12 h after the medium shift. In the mes1 time course, horse-tail nuclei were already visible 2 h after the shift and disappeared completely after 12 h. At 24 h, all of the cells which had entered meiosis (32%) had performed the first meiotic division, as determined by DAPI staining. At 30 h, the percentage of cells with two nuclei was also 32%. This shows that in mes1 mutants, by 24 h the cells were fully blocked after the first meiotic division. At these time points, the low-mobility Rec8p predominated in Western blots of crude extracts (Fig. 4D). Upon immunoprecipitation, both protein forms were detectable, but for unknown reasons the low-mobility form could not be abolished by phosphatase treatment (data not shown). The observations made with the mes1 mutant are consistent with persistence of Rec8p after meiosis I.
Cloning and sequence analysis of a human rec8 gene.
Since the conservation of the rad21 class of genes extended from yeasts to mammals and rec8 genes existed in both budding and fission yeast species (see below), it was likely that rec8 was also conserved in mammals. A human expressed sequence tag, T33286, was identified by its homology to the human rad21 homolog, hHR21sp (for human homolog of rad21, S. pombe [48]) on BLAST (1) database searching. A cDNA fragment corresponding to the C-terminal region of this new human gene, which we denote hrec8, was amplified from a human T-cell leukemia cDNA pool and used to screen a cDNA library from which full-length hrec8 cDNA clones were obtained.
hrec8 corresponded to an ORF of 1,641 nucleotides. Consistent with the generally low mRNA expression of this gene (see below), the sequence around the initiation codon did not conform strongly to the Kozak consensus (41); however, its translation start site was likely to be the correct one because of its highly conserved nature (below) and the presence of stop codons in all three reading frames in the 5′ untranslated region (5′ UTR) (data not shown). The 5′ UTR was at least 477 nucleotides long, while the 3′ UTR was 160 nucleotides (data not shown). The translation of hrec8 is shown in Fig. 5A, aligned with the full-length fission yeast Rec8p, a putative Rec8p homolog in budding yeast (Rec8sc; see below), and the founding member of the rad21/rec8 gene family, Rad21p.
Rec8p and its homologs are clearly sequence related to Rad21p and its homologs (Fig. 5B). To distinguish between previously reported Rad21-related proteins and those described in this paper, the new proteins described here are referred to as the rec8 members of the rad21/rec8 gene family. Sequence homology is higher at the N- and C-terminal ends of the proteins, where all proteins of the family are around 30% identical. Overall, Hrec8p had 49% similarity and 26% identity to hHR21sp (data not shown).
The PROSITE protein motif library failed to reveal major structural motifs, indicating a particular biological function for Hrec8p or related proteins. The unusual alternating basic-acidic region in the C-terminal region of the hHR21sp protein (see Fig. 2A of reference 48) was absent from the hrec8 product. Overall, Hrec8p is, like hHR21sp, an acidic protein (pI 5.0), but there were dramatic variations in charge and pI across Hrec8p. Another notable feature of Hrec8p was its unusually high proline content of 14.3%. Like fission yeast Rec8p, which had conserved basic residues at amino acids 284 to 290, Hrec8p contained a stretch of 6 arginine residues preceded by a long stretch of prolines (amino acids 298 to 304; Fig. 5A). This basic region, conserved in rad21/rec8 family homologs from other species (see Fig. 2 of reference 48), could represent a nuclear localization signal. Consistent with cell cycle regulation of these proteins by proteolysis (28, 49), potential PEST sequences (66) were identified in the central, less highly conserved regions of all the Rad21/Rec8 family proteins (data not shown).
Phylogenetic analysis of rad21/rec8 gene family.
A phylogenetic comparison of the Rec8 and Rad21 proteins from the different eukaryotes was performed. Overall, the estimated phylogenetic relationships between the Rec8/Rad21 proteins from the different species (indicated by the lengths of the lines in Fig. 5C) are consistent with those observed for other groups of orthologous and paralogous genes; i.e., they are congruent with the evolutionary relationship of the various species (32, 76, 82). There is no clear distinction between the rad21 and rec8 members of the gene family based on phylogeny. The common root of the two nematode proteins and their proximity to human rad21 indicates that unlike in the other species examined to date, there are two rad21 genes in Caenorhabditis elegans. However, no genes with sequence homology closer to rec8 than to rad21 have yet been identified in that species, although its sequence analysis has been completed (82).
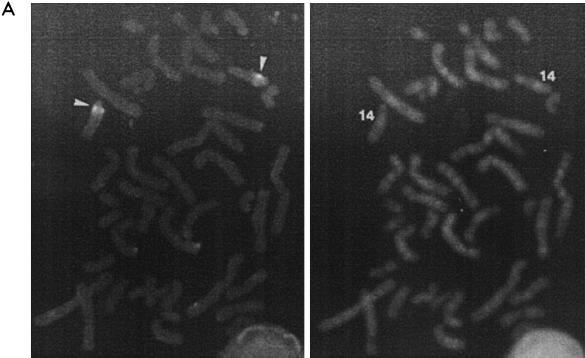
Chromosomal localization and mRNA expression of hrec8.
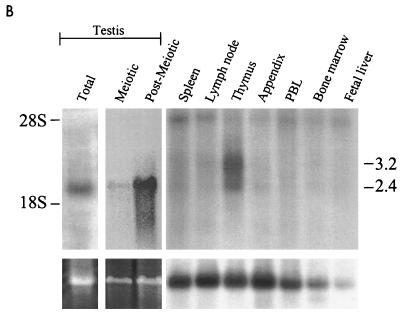
By using FISH, hrec8 was mapped to chromosome 14q11.2-12 (Fig. 6A). This locus is not apparently implicated in human disease syndromes. The mRNA expression of hrec8 was examined in different mammalian tissues. On blots prepared with total RNA, a 2.4-kb mouse rec8 mRNA species, consistent with the size of the hrec8 ORF, was detected only in testis cells (Fig. 6B, top left) and not in other tissues. Mouse testis tissues were fractionated into meiotic and postmeiotic compartments (spermatocytes and spermatids, respectively [27]). In contrast to the pattern seen for the related mHR21sp gene (48), weak mrec8 expression was detected in meiotic cells and greater expression was detected in postmeiotic spermatids (Fig. 6B, top middle). In addition, a multiple-human-tissue poly(A)+ RNA blot was hybridized with an hrec8 probe. Although not detected on the total-RNA blot, the hrec8 mRNA level was increased in thymus tissue and, unlike in testis tissue, there was an additional 3.2-kb mRNA species (Fig. 6B, top right). Expression was not detectable in other tissues, including some with high cellular proliferation. This indicates that increased thymic and testicular mrec8 expression was unlikely to simply reflect cellular proliferation in these tissues. We conclude that hrec8 mRNA expression is increased in meiotic as well as postmeiotic testis cells and is also detectable in thymus cells.
FIG. 6.
Chromosomal assignment and tissue specificity of mammalian rec8 mRNA expression. (A) FISH was used for hrec8 chromosomal assignment; biotinylated full-length hrec8 cDNA was hybridized with human metaphase spreads. (Left) Specific double-hybridization signals on chromosome 14q11.2-12 (arrowheads). (Right) DAPI staining of the same metaphase spread. (B) To examine mammalian rec8 mRNA expression, Northern blots of RNA from various mouse and human tissues and different testis fractions were hybridized with an hrec8 cDNA probe (top). (Top left) Single lane from a previously reported total RNA blot of mouse testis (Fig. 4 of reference 48); since no transcripts were detected in other tissues (thymus, brain, muscle, kidney, heart, liver, spleen, and ovary), these lanes are not shown here. (Top middle) Murine testis fractions representing meiotic (spermatocytes) and postmeiotic (a mixture of round and elongating spermatids) fractions. (Top right) Clontech multiple-human-tissue blot (no. 7754-1). There was some cross-hybridization of the hrec8 probe with trace amounts of 28S rRNA, as previously observed by us (data not shown). (Bottom left and middle) Ethidium bromide staining of rRNA indicating lane loading. (Bottom right) The multiple tissue blot was rehybridized with a β-actin control for lane loading. The positions of the 28S and 18S rRNA bands are shown on the left, while the transcript sizes (in kilobases) are indicated on the right.
Experiments on complementation of fission yeast rec8::ura4 by human hrec8 cDNA.
Sequence similarity was demonstrated for fission yeast and human Rec8p, and both genes were expressed in meiosis. If the two proteins have common functions, hRec8p might be able to substitute for Rec8p function in S. pombe. Two sets of experiments were performed. The homothallic strain PA43 with the rec8::ura4 disruption was transformed with pREP41 (control) and pREP41-hrec8. Individual transformants were brought to conjugation and sporulation, and the resulting spores were checked for viability. Untransformed PA43 showed a low spore viability of 9% compared to that of a rec8 wild-type strain. The vector alone had no effect after transformation into PA43, but pREP41-hrec8 increased spore viability to 48% (normalized to that of the rec8 wild-type strain [data not shown]).
Complementation of spore viability is expected to be due to restoration of high recombination frequency. Therefore, the heterothallic strains PA21 and PA22, suitable for recombination analysis, were transformed by pREP41 (control) and pREP41-hrec8. In this experiment, crossing the two untransformed strains resulted in low (18%) spore viability while transformation with the vector yielded 24% spore viability. Transformation with pREP41-hrec8 increased the spore viability to 38%. These results are qualitatively similar to those described above and may indicate partial complementation of the reduced spore viability phenotype by Hrec8p (data not shown). Neither intragenic recombination measured between the ade6-M26 and ade6-52 alleles nor intergenic recombination examined in the pro2-arg3 interval was significantly different from that in crosses of the untransformed strains (data not shown). These results show that the partial complementation of spore viability may be of doubtful significance.
DISCUSSION
In this study, we have further elaborated the function of the fission yeast rec8 gene, whose product is a phosphoprotein critical for meiotic sister chromatid cohesion and correct chromosome segregation. We describe the sequence conservation of rec8 in humans and present evidence that hrec8 is the human homolog of fission yeast rec8. We also demonstrate that the rec8 class of genes shows sequence homology to the rad21 class of mitotic, cell cycle-regulated phosphoproteins involved in sister chromatid cohesion, chromosome condensation, and DNA double-strand break repair. The findings suggest that rad21 and rec8 are the mitotic and meiotic members, respectively, of a new gene family involved in multiple aspects of DNA metabolism.
Here we present a unifying hypothesis to explain the observations on Rec8p. In this context, some special features of fission yeast meiosis need to be kept in mind. S. pombe maintains the bouquet structure of chromosomes throughout meiotic prophase, with concomitant absence of a fully tripartite synaptonemal complex and of crossover interference (for reviews, see references 37, 39, and 68). The retention of linear elements resembling the axial cores of synaptonemal complex indicates that these structures are of fundamental importance for meiotic recombination and chromosome segregation. During prophase of fission yeast meiosis the horse-tail nuclei move continuously from end to end of the cylindrical cells. At the leading end of the elongated nuclei, all telomeres are clustered to form and maintain the bouquet structure of chromosomes (13, 81). Disruption of the integrity of the spindle-pole body and/or telomere clustering by mutation leads to reduction of meiotic intra- and intergenic recombination frequencies (15, 59, 73). When nuclear movement was abolished by mutation of a motor protein required exclusively for horse-tail nucleus movement, recombination was also reduced (31). In all cases, about a fivefold reduction of recombination frequencies resulted.
The primary functions of Rec8p: a working hypothesis.
We propose that freshly synthesized Rec8p binds to the sites of initiation of recombination that are a subset of the sites of early pairing between related DNA sequences (re)established immediately after premeiotic DNA replication. It is likely that at this early stage, ectopic interactions between repeated sequences on different chromosomes occur as well as truly homologous DNA contacts. Little is known about the mechanisms involved in the formation of the first contacts between homologous chromosomes and the resolution of ectopic interactions. In S. cerevisiae, early contacts between homologous chromosomes occur before double-strand breaks initiate recombination, and it was proposed that assembly of the recombination initiation complex occurs in a succession of events at sites of sister chromatid cohesion (for reviews, see references 37 and 68).
Rec8p binding to the developing recombination initiation complex is proposed to be required for the start of linear-element polymerization. Linear elements, in turn, are proposed to contribute to extension and stabilization of homologous chromosome pairing and to maintenance of recombination intermediates once initiation has occurred. In particular, we suggest that linear-element formation is important in chromosome regions far from the telomeres. Rec8p binding to early homologous contacts is also proposed to enhance sister chromatid cohesion directly, or indirectly by promotion of linear-element polymerization.
The phosphorylation of Rec8p may play a structural role in organizing the chromosomes during prophase (e.g., repulsion of DNA backbones). Alternatively, or in addition, phosphate groups may be introduced in response to completion of specific steps of the pairing and recombination pathway, in order to signal their occurrence to other components of the nucleus. Another regulatory function of the kinase(s) phosphorylating Rec8p may be the promotion of events at the chromosomes after completion of processes elsewhere in the cell, in particular, resolution of recombination intermediates and relaxation of sister chromatid cohesion in the chromosome arms at meiosis I.
The persistence of phosphorylated Rec8p after meiosis I may be required for cohesion of the centromeres, as proposed for the phosphoprotein Cor1/SCP3 of rats. Cor1 relocates from the dissolving lateral elements to the centromeric regions of chromosomes and persists there until anaphase II (20, 53). A role in centromere cohesion before anaphase II has been established for the Mei-S332 protein of Drosophila (36).
Evaluation of the hypothesis on primary Rec8p functions.
First, we consider early Rec8p functions. An alternative to the model presented above may propose binding of Rec8p to specific sites on replicated chromosomes and subsequent formation of homologous contacts, leading to linear-element formation and recombination initiation. If this equivalent model applies, it may also be proposed that Rec8p binds to sites of sister chromatid cohesion formed during premeiotic DNA replication by the same mechanism as in mitotic DNA replication and thus involving mitotic cohesins. Scc1p/Mcd1p, the S. cerevisiae homolog of Rad21p, was reported to bind to chromosomes during mitotic S phase (28, 49).
A number of proteins involved in recombination were shown to form foci on the chromosomes during meiotic prophase (see, e.g., references 10 and 72). It remains to be demonstrated whether Rec8p foci colocalize with recombination enzymes loading onto the initiation sites of recombination. We propose loading of Rec8p onto emerging recombination initiation complexes as a parsimonious way to ensure that linear-element formation occurs at the sites where stabilization of recombination intermediates is required. Ultimately, sister chromatid cohesion in the chromosome arms is required after crossover formation and recombination complex (nodule) dissolution, and it must then be maintained up to anaphase I. However, sister chromatid cohesion may also have a function in early prophase of fission yeast meiosis. Its presence in wild-type early prophase and its absence in rec8 mutant prophase have been demonstrated (54, 70).
With Rec8p antisera, the protein was localized to about 100 foci in prophase nuclei (Fig. 3). The average number of crossovers per fission yeast meiosis is 45 (56). Thus, sufficient Rec8p foci form during prophase to account also for conversion events not associated with crossing over. A roughly 1:2 relation between crossover and conversion events was proposed early in the analysis of intragenic recombination of S. cerevisiae (for a review, see reference 61) and is consistent with the existing data on S. pombe (88). Smith and Roeder (77) proposed that Red1 protein nucleates the formation of axial elements in S. cerevisiae and plays a role in meiotic sister chromatid cohesion. Red1 also interacts with a protein phosphatase (83). The respective roles of Red1 and the S. cerevisiae Rec8p homolog (see below) remain to be investigated.
As reviewed by Kleckner (37), the lateral elements may stabilize the initial homologous contacts by firmly organizing the two sister chromatids attached to them. DNA regions close to the sites of attachment of chromatin loops to the lateral elements may then be protected for maintenance of their interactions with one of the homologous chromatids throughout prophase, until the recombination intermediates are resolved. Genetic and cytological data collected on rec8 mutants clearly demonstrated the involvement of Rec8p in sister chromatid cohesion and consequently in correct chromosome segregation in meiosis I (54). Rec8p may also be involved in late sister chromatid cohesion that is required for maintenance of chiasmata until transition from metaphase to anaphase I (for a review, see reference 52). Late sister chromatid cohesion must be independent of linear elements, since degradation of linear elements clearly occurs before meiosis I (2). Concurrently, nuclei approaching meiosis I also stop nuclear movement (13, 81).
Rec8p is phosphorylated during meiotic prophase. Its mitotic homolog, Rad21p, is multiply phosphorylated on serine and threonine residues, and its phosphorylation status varied through the cell cycle (9). The transition from unphosphorylated to phosphorylated Rec8p occurs early in prophase (Fig. 4), perhaps in connection with linear-element polymerization. Additional (de)phosphorylation steps may occur at the transitions to completion of recombination and linear-element degradation. The presence of Rec8p even after meiosis I suggests a further role for the protein that may be similar to the one proposed for the rat SCP3/Cor1 lateral-element component. SCP3/Cor1 is a phosphoprotein whose phosphorylation state changes during pachytene (43). Relocation of Rec8p to the centromeres of chromosomes, as demonstrated for Cor1 in rat meiosis (20, 53), is consistent with our results in Fig. 4. Recently, Watanabe and Nurse (87) obtained evidence for binding of Rec8p to the centromeric regions of chromosomes before meiosis I and persistence of this binding up to meiosis II. Their work emphasizes the function of Rec8p at the centromeric regions of the chromosomes, but they also observed localization of Rec8p throughout the nucleus during meiotic prophase.
Region specificity of recombination reduction in rec8 mutants.
De Veaux and Smith (18) showed that rec8, rec10, and rec11 point mutations reduce the recombination frequency more than 100-fold in the central part of chromosome III but not more than 10-fold in chromosomes I and II. They proposed that the respective wild-type proteins activate recombination in some chromosome regions but not in others. Our additional data showing strong reduction also in the central regions of chromosomes II and I (Fig. 2), as well as similar results obtained independently (42), do not exclude this hypothesis. However, the results are at odds with our hypothesis proposing that Rec8p binds to all early homologous contacts that will be resolved as recombination events. We suggest an alternative explanation, assuming that several factors contribute to the high level of meiotic recombination and that they are of variable importance in different regions of the genome.
Meiotic recombination frequencies are 2 to 3 orders of magnitude above the mitotic recombination frequencies in fission yeast (50), as in other organisms (3). Disruption of spindle pole integrity and telomere clustering leads to about a fivefold reduction of recombination frequencies (15, 59, 73). A similar result was obtained when nuclear movement was abolished (31). Thus, formation and maintenance of the bouquet and nuclear movement are two factors contributing to high levels of meiotic recombination. However, this contribution is minor compared to other factors. We suggest that the vigorous nuclear movement may also act negatively, leading to disruption of homologous pairing before recombination intermediates are resolved. This would reduce recombination frequencies most strongly in regions that are under greater mechanical stress.
Our model for primary Rec8p function proposes that linear elements contribute to the stabilization of recombination intermediates. The small reduction of recombination in the vicinity of the telomeres (Fig. 2) may then result from maintenance of pairing due to telomere clustering and little disruption by chromosome movement. In contrast, in the regions far from the telomeres, the loss of stabilization by linear elements may frequently lead to disruption of recombination intermediates due to mechanical stress. While recombination reduction is larger than 2 orders of magnitude in the centromeric regions of chromosomes II and III, it is less pronounced in the centromeric region of chromosome I. Interestingly, both arms of chromosome III and one arm of chromosome II are short, while both arms of chromosome I are longer. Long arms may experience less mechanical stress than short arms.
An alternative explanation for the strong region-specific reduction of recombination frequency in rec8 mutant strains may be the involvement of another protein, fulfilling similar functions to Rec8p only at the ends of chromosomes. However, the slight reduction of recombination frequencies at the telomere regions in rec8 mutant strains indicates that Rec8p also has a function at the ends of chromosomes.
The rad21/rec8 gene family and cloning of a human rec8 homolog.
On the basis of amino acid similarity to Rec8p, we identified a human homolog of fission yeast rec8. hrec8 was homologous both to rec8 and rad21 of fission yeast and to the previously described human rad21 homolog (48) (see above). The degree of homology is well within the range previously found for orthologous yeast-mammalian DNA repair and metabolism genes (23).
We also identified a rec8 homolog, rec8sc, within the published budding-yeast genome. Interestingly, disruption of the S. cerevisiae rec8sc gene resulted in no detectable phenotype under vegetative growth (spore germination, growth on different media, and resistance to UV radiation and methyl methanesulfonate). However, homozygous deletion of rec8sc resulted in loss of sporulation (4).
Given the sequence homology between rad21 and rec8 family members, which genes are true homologs in the different species? For a number of reasons, it is likely that the designation we use here is correct. First, in the regions of conserved sequence, there are key residues conserved between the rec8 family members which are not conserved in the rad21 group, and vice versa. For example, with reference to human Hrec8p (Fig. 5A and B), the residues at position 17 are T or V in rec8 genes but mostly K in rad21 genes; at position 25, rec8 genes have all G residues whereas the others have different amino acids; at position 94, H and S are present in rec8 genes and mostly E is present in rad21 genes. Likewise, T23 and S26 are conserved between Rec8p and Hrec8p, but the rad21 genes carry a chemically different amino acid. Second, deletion of the rad21 and SCC1/MCD1 genes in fission and budding yeasts revealed that they are essential for mitotic growth. Hypomorphic alleles show similar phenotypes in mitotic cells (8, 28, 49). In contrast, deletion of the rec8 genes in both yeast species resulted in nonvital phenotypes restricted to meiosis and sporulation (reference 4 and our results). Third, Hrec8p may partially complement the low spore viability of a fission yeast rec8 deletion strain but fails to complement recombination deficiency.
We did not find a clear separation of rad21 from rec8 classes on phylogenetic analysis of the rad21/rec8 gene family. For some duplicated genes involved in DNA metabolism, e.g., RAD23 and RAD6 of S. cerevisiae (40, 84), gene duplication apparently occurred during the one billion or more years of evolution from yeasts to mammals (38). For the rad21/rec8 family, however, it appears that gene duplication occurred earlier, before differentiation of yeasts and other eukaryotes, probably with the evolution of meiosis itself. Further, as evidenced by the lengths of the lines in Fig. 5C, the rec8 genes appear to have evolved at a higher rate than the corresponding rad21 genes in the respective species. The common root between the two C. elegans proteins and hHR21sp (Fig. 5C) also suggests that these sequences are derived from the same ancestral gene. Since rad21 and rec8 genes exist in budding yeast, fission yeast, Drosophila (85), and mammals, they are likely to be present in intermediate species as well. Thus, a rec8 homolog may eventually be identified in C. elegans.
hrec8 was mapped by FISH to chromosome 14q11.2-12 (Fig. 6A). This locus does not correspond to any known human disease. Previously, sequence-tagged sites corresponding to hrec8 were mapped to chromosomes 1, 5, 14, and X (Genethon). The mapping to X may represent cross-hybridization to the hHR21sp intronless pseudogene located on the X chromosome (GenBank accession no. HSU85A3). Apparently this pseudogene originated recently in evolution, given the high degree of identity to hHR21sp (data not shown).
As with the mouse rad21 homolog, mHR21sp, mRNA formation from mrec8 was high in testis tissue and detectable above baseline in thymus tissue. Recombination occurs in both these tissues. However, unlike mHR21sp, mrec8 was highly expressed in postmeiotic testis tissue (Fig. 6B). The transcript seen in postmeiotic cells did not represent cross-hybridization of mrec8 with the mHR21sp gene, since mHR21sp mRNAs differ in size and since nucleotide sequence similarity between these genes is poor in the region of the probe used for hybridization (data not shown). The significance of mrec8 expression in postmeiotic testis cells is unknown. However, in view of the role for a rad21/rec8 family member in chromatin condensation (28), mrec8 may be involved in the marked chromatin repackaging accompanying spermiogenesis.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank the following persons for communication of unpublished results: Y. Lin, D. Evans, and G. Smith; W.-D. Heyer and V. Bashkirov; Y. Hiraoka; M. D. Krawchuk and W. P. Wahls; Y. Watanabe and P. Nurse; K. Nasmyth and F. Klein; and W. Warren. We thank H. Scherthan for technical advice, S. Verschoor for technical assistance, J. Hoogerbrugge and A. Grootegoed for fractionated mouse testis cells, and F. Fabre for supplying basic budding yeast strains. M.J.M. thanks colleagues in the Bootsma/Hoeijmakers laboratory for general advice and discussion.
R.K. is a Fellow of the Royal Netherlands Academy of Arts and Sciences. This work was supported by the Swiss National Science Foundation, a grant of the Human Frontier Science Program, the Royal Australasian College of Radiologists, the Netherlands Organization for Scientific Research (NWO; 901-01-097), and International Human Frontier Science Program postdoctoral fellowship LT-506/94 to M.J.M.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bähler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker B S, Carpenter A T C, Esposito M S, Esposito R E, Sandler L. The genetic control of meiosis. Annu Rev Genet. 1976;10:53–134. doi: 10.1146/annurev.ge.10.120176.000413. [DOI] [PubMed] [Google Scholar]
- 4.Bashkirov, V., and W.-D. Heyer. Personal communication.
- 5.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 6.Beach D, Rodgers L, Gould J. RAN1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- 7.Bickel S E, Wyman D W, Orr-Weaver T L. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics. 1997;146:1319–1331. doi: 10.1093/genetics/146.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenbihl R P, Subramani S. Cloning and characterization of rad21 an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 1992;20:6605–6611. doi: 10.1093/nar/20.24.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birkenbihl R P, Subramani S. The rad21 gene product of Schizosaccharomyces pombe is a nuclear, cell-cycle regulated phosphoprotein. J Biol Chem. 1995;270:7703–7711. doi: 10.1074/jbc.270.13.7703. [DOI] [PubMed] [Google Scholar]
- 10.Bishop D K. RecA homologs Rad51 and Dmc1 interact to form discrete nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 11.Bresch C, Müller G, Egel R. Genes involved in meiosis and sporulation of a yeast. Mol Gen Genet. 1968;102:301–306. doi: 10.1007/BF00433721. [DOI] [PubMed] [Google Scholar]
- 12.Carpenter A T C. Recombination nodules and synaptonemal complex in recombination-defective females of Drosophila melanogaster. Chromosoma. 1979;75:259–292. doi: 10.1007/BF00293472. [DOI] [PubMed] [Google Scholar]
- 13.Chikashige Y, Ding D Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- 14.Cohen-Fix O, Peters J M, Kirschner M W, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 15.Cooper J P, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- 16.Cottarel G, Beach D, Deuschle U. Two new multipurpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr Genet. 1993;23:547–548. doi: 10.1007/BF00312650. [DOI] [PubMed] [Google Scholar]
- 17.De Veaux L C, Hoagland N A, Smith G R. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–261. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Veaux L C, Smith G R. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- 19.Dirick L, Goetsch L, Ammerer G, Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science. 1998;281:1854–1857. doi: 10.1126/science.281.5384.1854. [DOI] [PubMed] [Google Scholar]
- 20.Dobson M J, Pearlman R E, Karaiskakis A, Spyropoulos B, Moens P B. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- 21.Egel R, Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp Cell Res. 1974;88:127–134. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. PHYLIP, version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 23.Friedberg E C, Bardwell A J, Bardwell L, Feaver W J, Kornberg R D, Svejstrup J Q, Tomkinson A E, Wang Z. Nucleotide excision repair in the yeast Saccharomyces cerevisiae: its relationship to specialized mitotic recombination and RNA polymerase II basal transcription. Philos Trans R Soc London Ser B. 1995;347:63–68. doi: 10.1098/rstb.1995.0010. [DOI] [PubMed] [Google Scholar]
- 24.Funabiki H, Yamano H, Kumada K, Nagao K, Hunt T, Yanagida M. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature. 1996;381:438–441. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 25.Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe. A system for gene disruption and replacement using the ura4 gene as selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- 26.Grimm C, Schär P, Munz P, Kohli J. The strong adh promoter stimulates mitotic and meiotic recombination at the ade6 gene of Schizosaccharomyces pombe. Mol Cell Biol. 1991;11:289–298. doi: 10.1128/mcb.11.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grootegoed J A, Janson R, van der Molen H J. Effect of glucose on ATP dephosphorylation in rat spermatids. J Reprod Fertil. 1986;77:99–107. doi: 10.1530/jrf.0.0770099. [DOI] [PubMed] [Google Scholar]
- 28.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutz H, Heslot H, Leupold U, Loprieno N. Schizosaccharomyces pombe. In: King R C, editor. Handbook of genetics. Vol. 1. New York, N.Y: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- 30.Hari K L, Santerre A, Sekelsky J J, McKim K S, Boyd J B, Hawley R S. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 31.Hiraoka, Y. Personal communication.
- 32.Huysmans E, Dams E, Vandenberghe A, De Wachter R. The nucleotide sequence of the 5S rRNAs of four mushrooms and their use in studying the phylogenetic position of basidiomycetes among the eukaryotes. Nucleic Acids Res. 1983;11:2871–2880. doi: 10.1093/nar/11.9.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact cells treated with alkali cations. J Bacteriol. 1983;153:487–493. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerrebrock A W, Miyazaki W Y, Birnby D, Orr-Weaver T L. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130:827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerrebrock A W, Moore D P, Wu J S, Orr-Weaver T L. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- 37.Kleckner N. Meiosis: how could it work? Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knoll A H. The early evolution of eukaryotes: a geological perspective. Science. 1992;256:622–627. doi: 10.1126/science.1585174. [DOI] [PubMed] [Google Scholar]
- 39.Kohli J. Telomeres lead chromosome movement. Curr Biol. 1994;4:724–727. doi: 10.1016/s0960-9822(00)00160-3. [DOI] [PubMed] [Google Scholar]
- 40.Koken M H, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers J H. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc Natl Acad Sci USA. 1991;88:8865–8869. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981;9:5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krawchuk, M. D., and W. P. Wahls. Personal communication.
- 43.Lammers J H M, van Alderen M, Peters A H F M, van Pelt A A M, Gaemmers I C, de Rooij D G, de Boer P, Offenberg H H, Dietrich A J J, Heyting C. A change in the phosphorylation pattern of the 30000 to 33000 Mr synaptonemal complex proteins of the rat between early and mid-pachytene. Chromosoma. 1995;104:154–163. doi: 10.1007/BF00352179. [DOI] [PubMed] [Google Scholar]
- 44.Leem S-H, Ogawa H. The MER4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:449–457. doi: 10.1093/nar/20.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, Y., D. Evans, and G. R. Smith. Personal communication.
- 46.Lin Y, Larson K L, Dorer R, Smith G R. Meiotically induced rec7 and rec8 genes of Schizosaccharomyces pombe. Genetics. 1992;132:75–85. doi: 10.1093/genetics/132.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lydall D, Nikolsky Y, Bishop D K, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 48.McKay M J, Troelstra C, van der Spek P, Kanaar R, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers J H. Sequence conservation of the rad21 Schizosaccharomyces pombe DNA double-strand break repair gene in human and mouse. Genomics. 1996;36:305–315. doi: 10.1006/geno.1996.0466. [DOI] [PubMed] [Google Scholar]
- 49.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 50.Minet M, Grossenbacher-Grunder A-M, Thuriaux P. The origin of a centromere effect on mitotic recombination. Curr Genet. 1980;2:53–60. doi: 10.1007/BF00445694. [DOI] [PubMed] [Google Scholar]
- 51.Miyazaki W Y, Orr-Weaver T L. Sister-chromatid misbehavior in Drosophila ord mutants. Genetics. 1992;132:1047–1061. doi: 10.1093/genetics/132.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miyazaki W Y, Orr-Weaver T L. Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet. 1994;28:167–187. doi: 10.1146/annurev.ge.28.120194.001123. [DOI] [PubMed] [Google Scholar]
- 53.Moens P B, Spyropoulos B. Immunocytology of chiasmata and chromosomal disjunction at mouse meiosis. Chromosoma. 1995;104:175–182. doi: 10.1007/BF00352182. [DOI] [PubMed] [Google Scholar]
- 54.Molnar M, Bähler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreau P J F, Zickler D, Leblon G. One class of mutants with disturbed centromere cleavage and chromosome pairing in Sordaria macrospora. Mol Gen Genet. 1985;198:189–197. [Google Scholar]
- 56.Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Munz P, Leupold U. Gene conversion in nonsense suppressors of Schizosaccharomyces pombe. I. The influence of the genetic background and of three mutant genes (rad2, mut1 and mut2) on the frequency of postmeiotic segregation. Mol Gen Genet. 1979;170:145–148. doi: 10.1007/BF00337789. [DOI] [PubMed] [Google Scholar]
- 58.Nadin-Davis S A, Nasim A. Schizosaccharomyces pombe ras1 and byr1 are functionally related genes of the ste family that affect starvation-induced transcription of mating-type genes. Mol Cell Biol. 1990;10:549–560. doi: 10.1128/mcb.10.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nimmo E R, Pidoux A L, Perry P E, Allshire R C. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature. 1998;392:825–828. doi: 10.1038/33941. [DOI] [PubMed] [Google Scholar]
- 60.Pawson T, Schlessinger J. SH2 and SH3 domains. Curr Biol. 1993;3:434–442. doi: 10.1016/0960-9822(93)90350-w. [DOI] [PubMed] [Google Scholar]
- 61.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E E, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae: genome dynamics, protein synthesis and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- 62.Pinkel D, Straume T, Gray J W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ponticelli A S, Smith G R. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prabhala G, Rosenberg G H, Käufer N F. Architectural features of pre-mRNA introns in the fission yeast Schizosaccharomyces pombe. Yeast. 1992;8:171–182. doi: 10.1002/yea.320080303. [DOI] [PubMed] [Google Scholar]
- 65.Pringle J R, Adams A E M, Drubin D G, Haarer B K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 66.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 67.Rockmill B, Roeder G S. A meiosis-specific protein kinase homolog required for chromosome synapsis and recombination. Genes Dev. 1991;5:2392–2404. doi: 10.1101/gad.5.12b.2392. [DOI] [PubMed] [Google Scholar]
- 68.Roeder G S. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 69.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 70.Scherthan H, Bähler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schweingruber M E, Edenharter E. Thiamin regulates agglutination and zygote formation in Schizosaccharomyces pombe. Curr Genet. 1990;17:191–194. doi: 10.1007/BF00312609. [DOI] [PubMed] [Google Scholar]
- 72.Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Asley T, Livingston D M. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 73.Shimanuki M, Miki F, Ding D-Q, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- 74.Shimoda C, Hirata A, Kishida M, Hashida T, Tanaka K. Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1985;200:252–257. doi: 10.1007/BF00425432. [DOI] [PubMed] [Google Scholar]
- 75.Shuster E O, Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sipiczki M. Taxonomy and phylogenesis. In: Nasim A, Young P, Johnson B F, editors. Molecular biology of the fission yeast. New York, N.Y: Academic Press, Inc.; 1989. pp. 431–452. [Google Scholar]
- 77.Smith A V, Roeder G S. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith D B, Johnson K S. Single step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 79.Soares M B, Bonaldo M F, Jelene P, Su L, Lawton L, Efstratiadis A. Construction and characterization of a normalized cDNA library. Proc Natl Acad Sci USA. 1994;91:9228–9232. doi: 10.1073/pnas.91.20.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Strauss A. Isolation and genetic classification of aromatic amino acid auxotrophic mutants in Schizosaccharomyces pombe. J Gen Microbiol. 1979;113:173–176. [Google Scholar]
- 81.Svoboda A, Bähler J, Kohli J. Microtubule-driven nuclear movement and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma. 1995;104:203–214. doi: 10.1007/BF00352185. [DOI] [PubMed] [Google Scholar]
- 82.The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 83.Tu J, Song W, Carlson M. Protein phosphatase type I interacts with proteins required for meiosis and other cellular processes in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4199–4206. doi: 10.1128/mcb.16.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Spek P J, Visser C E, Hanaoka F, Smit B, Hagemeijer A, Bootsma D, Hoeijmakers J H. Cloning, comparative mapping, and RNA expression of the mouse homologues of the Saccharomyces cerevisiae nucleotide excision repair gene RAD23. Genomics. 1996;31:20–27. doi: 10.1006/geno.1996.0004. [DOI] [PubMed] [Google Scholar]
- 85.Warren, W. Personal communication.
- 86.Watanabe Y, Iino Y, Furuhata K, Shimoda C, Yamamoto M. The S. pombe mei2 gene encoding a crucial molecule for commitment to meiosis is under the regulation of cAMP. EMBO J. 1988;7:761–767. doi: 10.1002/j.1460-2075.1988.tb02873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watanabe, Y., and P. Nurse. Personal communication.
- 88.Zahn-Zabal M, Lehmann E, Kohli J. Hot spots of recombination in fission yeast: inactivation of the M26 hot spot by deletion of the ade6 promoter and the novel hotspot ura4-aim. Genetics. 1995;140:469–478. doi: 10.1093/genetics/140.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]