Abstract
The nucleotide-binding domain, leucine-rich-repeat containing family, pyrin domain-containing 3 (NLRP3) inflammasome is essential in inflammation and inflammatory disorders. Phosphorylation at various sites on NLRP3 differentially regulates inflammasome activation. The Ser725 phosphorylation site on NLRP3 is depicted in multiple inflammasome activation scenarios, but the importance and regulation of this site has not been clarified. The present study revealed that the phosphorylation of Ser725 was an essential step for the priming of the NLRP3 inflammasome in macrophages. We also showed that Ser725 was directly phosphorylated by misshapen (Msn)/NIK-related kinase 1 (MINK1), depending on the direct interaction between MINK1 and the NLRP3 LRR domain. MINK1 deficiency reduced NLRP3 activation and suppressed inflammatory responses in mouse models of acute sepsis and peritonitis. Reactive oxygen species (ROS) upregulated the kinase activity of MINK1 and subsequently promoted inflammasome priming via NLRP3 Ser725 phosphorylation. Eliminating ROS suppressed NLRP3 activation and reduced sepsis and peritonitis symptoms in a MINK1-dependent manner. Altogether, our study reveals a direct regulation of the NLRP3 inflammasome by Msn family kinase MINK1 and suggests that modulation of MINK1 activity is a potential intervention strategy for inflammasome-related diseases.
Keywords: MINK1 kinase, NLRP3 inflammasome, ROS, Phosphorylation
Subject terms: Inflammasome, Monocytes and macrophages
Introduction
The nucleotide-binding domain, leucine-rich-repeat containing family, pyrin domain-containing 3 (NLRP3) inflammasome plays a key role in host defense against pathogens [1, 2]. NLRP3 senses a variety of danger-associated molecular patterns (DAMPs) and pathogen-associated molecular patterns (PAMPs) during infection and tissue damage. It forms a cytosolic multimeric protein complex to process the maturation of caspase-1 and the release of several proinflammatory cytokines, including interleukin-1β (IL-1β) and IL-18 [3–5]. However, dysregulated activation of the NLRP3 inflammasome is closely related to various inflammatory disorders and inflammation-related diseases, including sepsis, type-2 diabetes (T2D), gout and acute peritonitis [1, 4, 6–10]. Classic NLRP3 inflammasome activation follows a 2-step model that requires priming and assembly [11]. PAMPs or cytokines, such as IL-1β and tumor necrosis factor (TNF), regulate the priming step, which is essential for the upregulation of the transcription levels of NLRP3 and other inflammasome components [12]. Numerous PAMPs or DAMPs drive the subsequent activation step, which involves the activation of multiple upstream signaling events [4, 13].
Phosphorylation extensively regulates the activation of NLRP3, which may be phosphorylated during different inflammasome phases, including the unstimulated, priming, activation, and resolution phases. Song et al. demonstrated that JNK1-mediated NLRP3 phosphorylation at the Ser194 residue was a key molecular priming event that poises NLRP3 for self-association and inflammasome assembly [14]. Notably, the phosphorylation of NLRP3 activates or inhibits the activation of NLRP3 [14–18]. Previous research showed that phosphatase 2A-induced inflammasome assembly via dephosphorylation of the NLRP3 pyrin domain, and phosphatase and tensin homolog deleted on chromosome 10 directly interact with and dephosphorylate NLRP3 to enable NLRP3–ASC (apoptosis-associated speck-like protein containing a CARD) interaction, inflammasome assembly, and activation [17, 19]. Therefore, in-depth exploration of the roles of phosphorylation and the responsible kinases in the regulation of NLRP3 inflammasome activation may reveal therapeutic targets of the NLRP3 inflammasome and treatments for NLRP3-related inflammatory diseases. According to the protein phosphorylation site database PhosphoSitePlus (phosphosite.org), Ser725 (the locus corresponding to human NLRP3 S728) is one of the most highly phosphorylated sites in mouse NLRP3, but its function and the kinase responsible for this site is not known [14, 17].
Misshapen (Msn)/NIK-related kinase 1 (MINK1) is a serine-threonine kinase protein in the GCK protein family [20–22]. MINK1 is described as a signaling mediator of the MAP kinase cascades, and it is also named MAP4 kinase 6 (MAP4K6) [20]. MAP4K6 also plays a role in mediating Ras-induced senescence downstream of reactive oxygen species (ROS) in ovarian epithelial cells [23]. MINK1 is involved in various biological processes, such as neurodegeneration, thymocyte selection, and platelet function [24–27]. MINK1 kinases play a role in the Hippo pathway and activate LATS1/2 in parallel to MST1/2, which affects tumor immunity [28, 29]. Our recent research revealed the contribution of MINK1 in regulating Th17 cell differentiation [30]. We found that the knockout of MINK1 in mice resulted in Th17 cell accumulation and increased susceptibility to EAE. However, dysregulated activation of T cells did not result in spontaneous inflammatory symptoms in MINK1 knockout mice, which suggests additional roles of MINK1 in other cell types.
MINK1 expression is highly enriched in myeloid cells and to a lesser extent in dendritic cells. Therefore, we further investigated its function in macrophages. We report that MINK1 deficiency suppressed NLRP3 inflammasome activation and IL-1β secretion in macrophages. We demonstrated that ROS activated MINK1 and promoted NLRP3 activation via direct phosphorylation of the Ser725 residue in the NLRP3 LRR domain, which is as an essential priming event for NLRP3 self-interaction and oligomerization. Disruption of MINK1 kinase activity by ROS scavengers markedly alleviated the activation of NLRP3 in vivo and improved the pathogenesis of inflammatory diseases. Our research reveals a new regulatory function of MINK1 kinase in macrophages and provides a previously unknown mechanistic explanation for ROS activation of the NLRP3 inflammasome.
Results
MINK1 promotes NLRP3-dependent caspase-1 maturation and IL-1β production
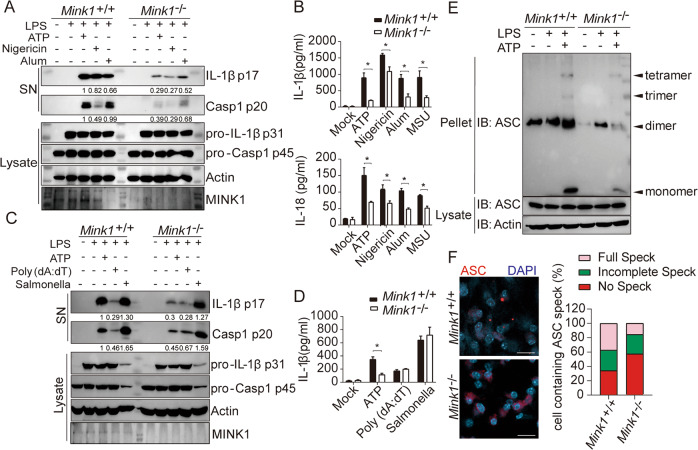
To assess the possible involvement of MINK1 kinase in NLRP3 inflammasome activation, we treated LPS-primed bone marrow-derived macrophages (BMDMs) from WT and Mink1−/− mice with different agonists. Mink1−/− BMDMs showed impaired NLRP3 inflammasome activation triggered by ATP, nigericin, and alum compared to WT BMDMs, which was reflected by reduced levels of IL-1β cleavage and caspase-1 maturation (Fig. 1A) and lower IL-1β and IL-18 secretion (Fig. 1B). A reduction in IL-1β secretion from Mink1−/− BMDMs was observed under optimal and suboptimal ATP concentrations (1–3 mM) (Fig. S1A). NLRP3-dependent ASC oligomerization and ASC speck formation were also decreased in Mink1−/− BMDMs (Fig. 1E, F). These results suggest a requirement of MINK1 for NLRP3 inflammasome activation. Notably, LPS-induced expression of pro-IL-1β and procaspase-1 and the secretion of TNF-α and IL-6 were not changed under different stimulation conditions (Fig. 1A, C, Fig. S1B–D). We further investigated the role of MINK1 during the activation of other inflammasomes. We found that MINK1 deficiency did not affect the activation of AIM2 or NLRC4, triggered by poly (dA:dT) transfection and Salmonella Typhimurium infection, respectively, which indicates the specific function of MINK1 in NLRP3 activation (Fig. 1C, D, Fig. S1E). Together, these results demonstrate the essential role of MINK1 kinase in promoting NLRP3 inflammasome activation in macrophages.
Fig. 1.
Myeloid MINK1 is essential for NLRP3 inflammasome activation. A, B BMDMs from Mink1+/+ and Mink1−/− mice primed with LPS and stimulated with different secondary signals, including ATP, nigericin, aluminum salts (Alum), and monosodium urate crystals (MSU). Supernatants (SN) and cell extracts (lysate) were analyzed using immunoblotting (A). Supernatant IL-1β and IL-18 were analyzed using ELISA (B). C, D BMDMs from Mink1+/+ and Mink1−/− mice primed with LPS and stimulated with ATP, poly (dA:dT), and Salmonella. SN and lysate were analyzed using immunoblotting (C). SN IL-1β was also analyzed using ELISA (D). E, F BMDMs from Mink1+/+ and Mink1−/− mice primed with LPS and stimulated with ATP followed by immunoblotting for ASC oligomerization analysis in cross-linked cytosolic pellets (E). Representative immunofluorescence images and quantification of ASC speck formation are shown in F. *p < 0.05, two-tailed unpaired Student’s t test was used for B and D. The ELISA and western blot results are representative of three independent experiments
MINK1 ablation ameliorates acute NLRP3-dependent inflammation
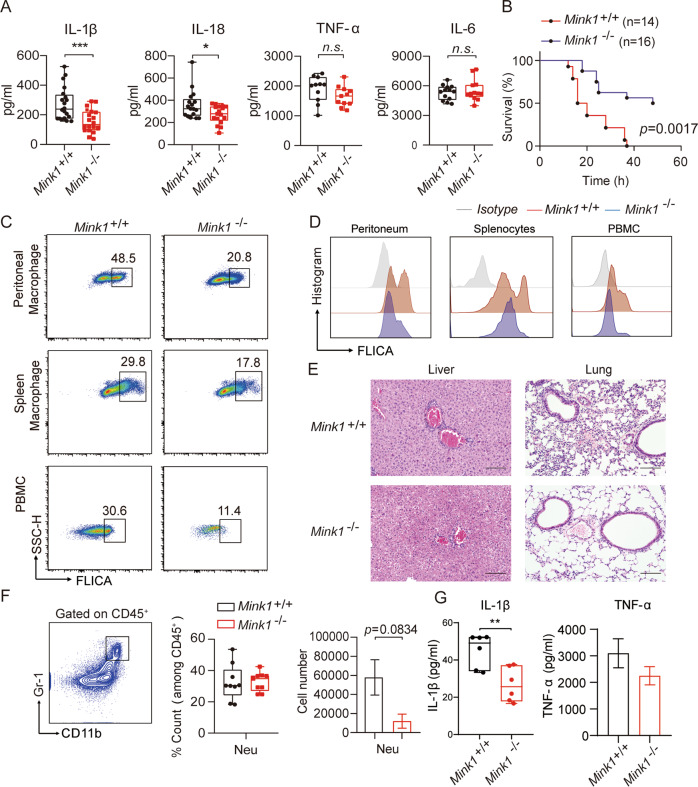
We next used LPS-induced sepsis and alum-induced peritonitis models to evaluate the contribution of MINK1 to the NLRP3-mediated inflammatory response in vivo. A significant decline of IL-1β and IL-18 levels in serum was observed in Mink1−/− mice compared to WT mice in the sepsis model, but the production of TNF-α and IL-6 remained comparable (Fig. 2A). After the injection of LPS, the survival rate of Mink1−/− mice was substantially improved compared to wild-type mice (Fig. 2B). We next performed fluorescent activity-based probe FLICA (FAM-7YVAD-FMK) staining to detect caspase-1 activation. Decreased activation of caspase-1 in peritoneal macrophages (PMs), spleen macrophages, and peripheral blood mononuclear cells (PBMCs) from Mink1−/− mice was observed compared to WT mice (Fig. 2C, D). Further statistical analysis confirmed the reduced frequency and reduced fluorescence intensity of FLICA+ macrophages in Mink1−/− mice (Fig. S2A, B). After LPS injection, Mink1−/− mice also showed reduced inflammatory cell infiltration, edema, interstitial hyperemia, hemorrhage, and alveolar collapse tissue damage in the lung (Fig. 2E, right). Similar results were observed in the liver tissue (Fig. 2E, left). Mink1−/− mice also showed milder inflammatory symptoms than control mice in the alum-induced acute peritonitis experiment, which was evidenced by decreased numbers of neutrophils and IL-1β levels in the peritoneal fluid (Fig. 2F, G). However, the frequency of neutrophils (CD45+ CD11b+ Gr-1+) and TNF-α release in the peritoneal flush fluid showed no difference between the WT and Mink1−/− mice (Fig. 2G). Taken together, our results demonstrate that MINK1 kinase promotes NLRP3-mediated acute inflammatory responses.
Fig. 2.
MINK1 ablation ameliorates acute inflammation. A Mink1+/+ and Mink1−/− mice were intraperitoneally injected with LPS (25 mg/kg), and IL-1β, IL-18, IL-6, and TNF-α in serum were measured using ELISA after 4 h. Data are the means ± SD (n = 20 mice/group). B The survival rates of Mink1+/+ and Mink1−/− mice injected with LPS (20 mg/kg, n = 14 in Mink1+/+ and n = 16 in Mink1−/−). Surface staining of FLICA was performed on macrophages (gated on F4/80+ cells) from the peritoneal cavity, spleen and blood from Mink1+/+ and Mink1−/− mice to determine the proportion of activated caspase-1 macrophages (C) and the intensity of caspase-1 activation (D). Numbers in or adjacent to outlined areas (or in quadrants) indicate the percentages of cells in each throughout. E Representative histology of liver and lung (hematoxylin and eosin stained) from Mink1−/− and WT mice 4 h after LPS (25 mg/kg) injection. Scale bar represents 100 μm. F, G Mink1+/+ and Mink1−/− mice were intraperitoneally injected with alum (2 mg per mouse), and peritoneal fluid was washed out 6 h later for cell surface staining to determine the percentages and cell number of neutrophils between the two groups. F Data are the means ± SD (n = 6 mice/group). Peritoneal IL-1β and TNF-α levels in Mink1−/− and WT mice were also detected using ELISA (G). *p < 0.05, *p < 0.01, ***p < 0.001, two-tailed unpaired Student’s t test for A, F, and G, and Kaplan–Meier method for mouse survival (B). The ELISA and flow cytometry results are representative of three independent experiments
MINK1 relies on its kinase activity to promote NLRP3 activation
We investigated the mechanism of MINK1 promotion of the activation of the NLRP3 inflammasome. We first checked the signaling pathways that were essential for the priming of NLRP3 inflammasome activation [31–33]. No significant difference was found in the upstream events of NLRP3 activation in BMDMs from Mink1−/− and WT mice (Fig. S3A). RT–qPCR analyses also showed that the RNA expression levels of NLRP3 inflammasome components remained unaffected by MINK1 deficiency (Fig. S1F). Considering that the expression of procaspase-1 and pro-IL-1β was also unaltered (Fig. 1A, C), we concluded that the regulation of NLRP3 activation by MINK1 was independent of its effect on the expression of essential NLRP3 components in macrophages.
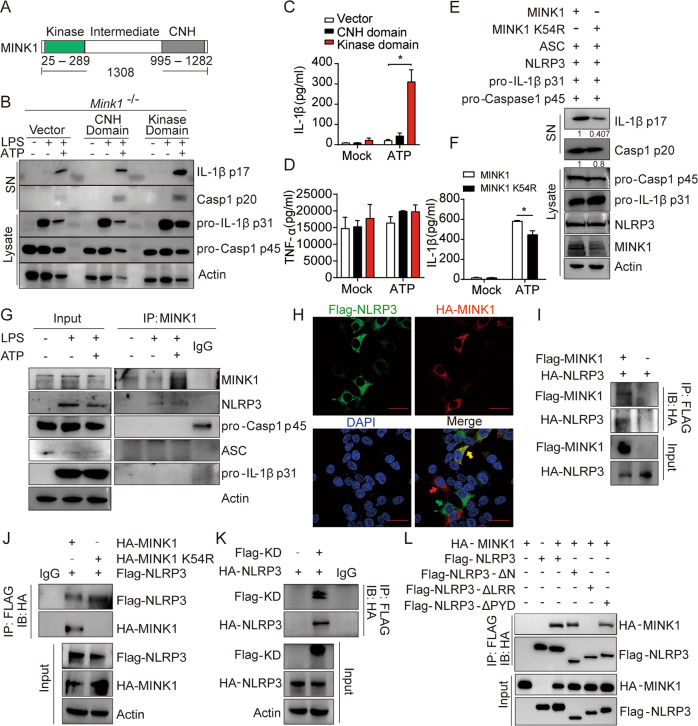
MINK1 is a member of the mammalian GCK family of kinases, which exhibit multiple functions that are highly dependent on kinase activity [34, 35]. Protein phosphorylation regulates NLRP3 activation [19, 36]. Therefore, we next investigated whether the kinase activity of MINK1 was essential for promoting NLRP3 inflammasome activation. MINK1 is composed of an N-terminal catalytic domain (kinase domain), a C-terminal citron homology (CNH) domain (regulatory domain), and an intermediate region (Fig. 3A and [34]). We reconstituted Mink1−/− BMDMs with the MINK1 kinase domain or regulatory domain. We found that the overexpression of the kinase domain, but not the regulatory domain, restored ATP-induced IL-1β secretion and caspase-1 activation (Fig. 3B, C), but TNF-α release was not affected (Fig. 3D). To further corroborate this result, we reconstituted the NLRP3 inflammasome in HEK293T cells with WT MINK1 and a MINK1K54R mutant that abrogated MINK1 kinase activity. We found that the inactive MINK1K54R mutant did not induce caspase-1 maturation or IL-1β secretion compared to the WT protein (Fig. 3E, F, Fig. S3B). Collectively, these results suggest that the kinase activity of MINK1 is required for NLRP3 inflammasome activation.
Fig. 3.
MINK1 directly interacts with NLRP3 and regulates NLRP3 activation via its kinase activity. A Schematic diagram of the structure of the MINK1 protein. B–D BMDMs from Mink1−/− mice primed with LPS retrovirally transduced with pHAGE empty vector, pHAGE-CNH domain, or pHAGE-Kinase domain stimulated with ATP. Supernatants (SN) and cell extracts (lysate) were examined using immunoblotting (B). IL-1β and TNF-α secretion was determined using ELISA (C, D). E, F Reconstruction of NLRP3 activation in HEK293T cells by overexpressing NLRP3 inflammasome activation-related components with HA-MINK1 or HA-MINK1-K54R. Supernatants (SN) and cell extracts (lysate) were determined using immunoblotting (E). IL-1β secretion was determined using ELISA (F). G IP and immunoblot analysis of the interaction between endogenous MINK1 and NLRP3 inflammasome activation-related components in BMDMs primed with LPS then stimulated with ATP. H Representative confocal images of HEK293T cells overexpressing HA-MINK1 (red) and Flag-NLRP3 (green). Scale bar represents 50 μm. I IP and immunoblot analysis of the interaction between Flag-MINK1 and HA-NLRP3. J IP and immunoblot analysis of the interaction between Flag-NLRP3 and HA-MINK1 or HA-MINK1-K54R. K IP and immunoblot analysis of the interaction between HA-NLRP3 and the Flag-kinase domain. L IP and immunoblot analysis of the interaction between HA-MINK1 and full-length or truncated NLRP3. *p < 0.05, one-way ANOVA was used for C and D, and two-tailed unpaired Student’s t test was used for F. The ELISA, IF, and western blot results are representative of three independent experiments
MINK1 directly interacts with NLRP3
We next investigated whether MINK1 directly interacted with key components of the NLRP3 inflammasome. Immunoprecipitation (IP) assays of BMDMs and PMs found that ATP stimulation promoted the specific binding of endogenous MINK1 kinase to NLRP3 but not to other components, including ASC, procaspase-1, pro-IL-1β, or NEK7 (Fig. 3G, Fig. S3D–G). Immunofluorescence results also revealed substantial colocalization of MINK1 and NLRP3 (Fig. 3H and Fig. S3C). When coexpressed in HEK293T cells, an interaction of MINK1 kinase and NLRP3 was also found (Fig. 3I). We further mapped the physical domains responsible for the MINK1-NLRP3 interaction in HEK293T cells. We found that the MINK1 kinase domain directly bound to NLRP3, but mutant MINK1K54R (which had ablated kinase activity) lost the ability to bind to NLRP3 (Fig. 3J, K). These results further confirmed the function of MINK1 kinase and its kinase activity in NLRP3 inflammasome activation. When different NLRP3 truncated mutants were expressed with MINK1 in HEK293T cells, only the LRR domain of NLRP3 was capable of binding with MINK1 (Fig. 3L). These results suggest that MINK1 promotes NLRP3 inflammasome activation via direct interaction with NLRP3.
MINK1-mediated NLRP3 phosphorylation is essential for inflammasome activation
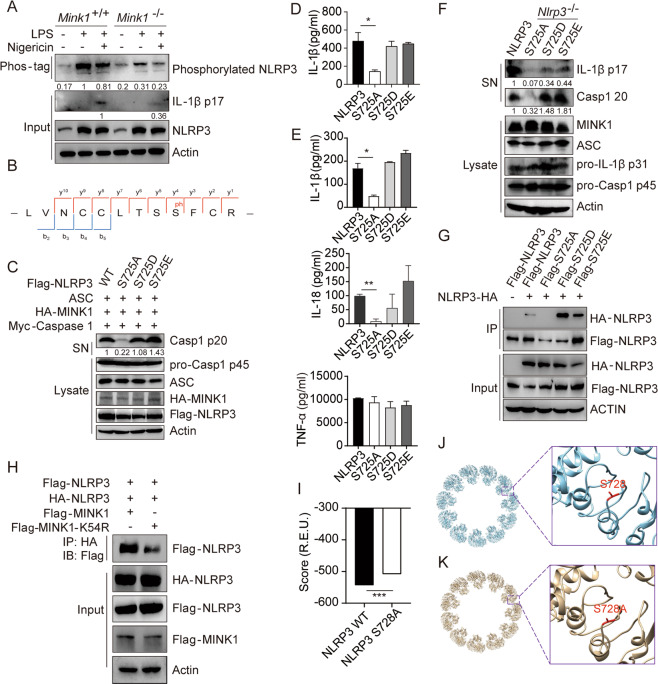
To determine whether MINK1 phosphorylated NLRP3 directly, we used Phos-tagTM SDS–PAGE to detect the phosphorylation of NLRP3. Significantly decreased phosphorylation of NLRP3 was observed in Mink1−/− BMDMs (Fig. 4A, Fig. S4A). We also performed mass spectrometry (MS) analysis of NLRP3 in MINK1-overexpressing cells, which led to the detection of Ser725 phosphorylation (Fig. 4B, Fig. S4B). To further investigate how the phosphorylation of this residue affected NLRP3 activation, we reconstituted the NLRP3 inflammasome in HEK293T cells with WT NLRP3, an NLRP3S725A mutant (to mimic dephosphorylation), or the NLRP3S725D and NLRP3S725E mutants (to mimic phosphorylation). We found that ATP-induced caspase-1 activation and IL-1β production were abrogated in NLRP3S725A cells but remained intact in cells that were reconstituted with WT, NLRP3S725D, or NLRP3S725E (Fig. 4C, D). When these mutants were overexpressed in BMDMs from Nlrp3−/− mice, NLRP3S725D and NLRP3S725E restored ATP-induced IL-1β and IL-18 secretion and caspase-1 activation to the level of WT cells, but NLRP3S725A did not induce these effects (Fig. 4E, F). None of these manipulations affected the production of TNF-α (Fig. 4E). These results suggest that MINK1-mediated phosphorylation at Ser725 is essential for NLRP3 activation.
Fig. 4.
MINK1-mediated NLRP3 phosphorylation at Ser725 is critical for inflammasome activation. A Phos-tagTM SDS–PAGE was used to detect phosphorylated NLRP3 in BMDMs primed with LPS and stimulated with nigericin. B IP was performed after overexpressing MINK1 and NLRP3 in HEK293T cells, followed by phosphorylation mass spectrometry to show the phosphorylated residue. C, D Reconstruction of NLRP3 activation in HEK293T cells, supernatants (SN) and cell extracts (lysate) was determined using immunoblotting (C). IL-1β release was determined using ELISA (D). ELISA analyses of IL-1β, IL-18, and TNF-α in culture supernatants (E) or immunoblot analysis of p17 and p20 (F) from Nlrp3−/− BMDMs transduced with mouse WT or mutant NLRP3-expressing lentivirus and primed with LPS and stimulated with ATP. G IP and immunoblot analysis of the aggregation of NLRP3 after overexpressing HA-NLRP3 with different Flag-NLRP3 in HEK293T cells. H IP and immunoblot analysis of the aggregation of NLRP3 after overexpressing Flag-MINK1, Flag-MINK1-K54R, and different tagged NLRP3 in HEK293T cells. I–K The structural model of WT NLRP3-NEK is shown in blue, and serine 728 is shown in red. The inset shows the detailed conformation (J), and the S728A mutant is shown in K. The inset shows the detailed conformation. The score (in the unit of R.E.U.) reflecting the structural stability was calculated by Rosetta for WT NLRP3-NEK and the S728A mutant (I). *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA was used for D and E, two-tailed unpaired Student’s t test was used for I. The ELISA and western blot results are representative of three independent experiments
MINK1 promotes NLRP3 self-association
NLRP3 self-association is a critical step in inflammasome assembly and activation [14]. Therefore, we investigated whether phosphorylation at the S725 site of NLRP3 mediated by MINK1 affected the assembly of the NLRP3 inflammasome. Flag-tagged NLRP3, with or without the S725 mutation, was cotransfected with HA-tagged NLRP3 in HEK293T cells. We found that the S725A mutation limited NLRP3 self-interaction but the S725D and S725E mutations significantly enhanced the interaction (Fig. 4G). Similarly, a reduction in NLRP3 self-interaction was also observed in cells overexpressing the kinase-dead MINK1 protein (Fig. 4H, Fig. S4D). To investigate potential structural mechanisms underlying phosphorylation-induced changes in NLRP3, we performed symmetric docking of human NLRP3 and NEK7 protein subunits. Because this site is well conserved in different organisms (Fig. S4C), we used the structure of human NLRP3 in the database as a template. The calculated score of the complex of the S728A (dephosphorylation-mimicking) mutant was significantly decreased compared to the WT (Fig. 4I–K), but S728E showed no significant difference (Fig. S4E), which suggests that the dephosphorylation-mimicking mutant complex is less stable than the WT. These results revealed a regulatory role for S725 phosphorylation in NLRP3-NLRP3 interactions.
ROS activates MINK1 kinase to prime the activation of NLRP3
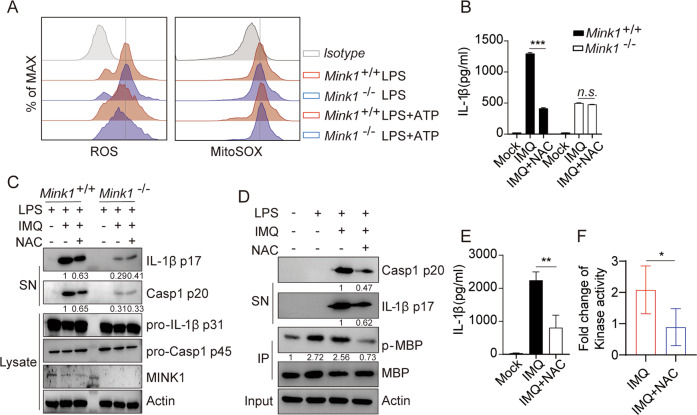
ROS activated MINK1 and our previous study showed that NAC (ROS scavenger) treatment profoundly reduced the kinase activity of MINK1 [23, 37]. ROS also serves as a priming factor for the NLRP3 inflammasome [38, 39]. We next asked whether MINK1-mediated NLRP3 phosphorylation and activation were downstream events following ROS production. We found that the levels of total ROS and mitochondrial ROS production in LPS-primed BMDMs stimulated with ATP were comparable between Mink1−/− and the control (Fig. 5A), which indicates that MINK1 ablation does not affect mitochondrial damage in NLRP3 activation. We next treated BMDMs with the ROS scavenger NAC or Mito-TEMPO (a mitochondria-targeted superoxide dismutase mimetic with superoxide and alkyl radical scavenging properties) under different stimuli [30, 40, 41]. We found that IL-1β and caspase-1 maturation and IL-1β secretion were critically compromised by these treatments (Fig. 5B, C, Fig. S5B–E), but TNF-α release was not affected by NAC or Mito-TEMPO treatment (Fig. S5A, E). More importantly, NAC or Mito-TEMPO treatment failed to further suppress IL-1β and caspase-1 maturation in Mink1−/− cells (Fig. 5B, C, Fig. S5B–E). In vitro kinase activity assays further confirmed that the activation of MINK1 was a direct priming event after LPS treatment, which was significantly reduced by the removal of ROS (Fig. 5D, F), in accordance with the suppressed IL-1β and caspase-1 maturation and IL-1β secretion after treatment with NAC or Mito-TEMPO (Fig. 5D, E, Fig. S5F, G). These results indicate that ROS scavengers reduced NLRP3 inflammasome activation in vitro in a MINK1-dependent manner at the priming stage.
Fig. 5.
ROS activates MINK1 kinase to prime the activation of NLRP3. A LPS-primed BMDMs from Mink1−/− and WT mice were stimulated with ATP, and total ROS and mitochondrial ROS levels were measured by ROS and MitoSOX incorporation using flow cytometry. B, C LPS-primed BMDMs from Mink1−/− and WT mice were treated with NAC (3 mM) 30 min before imiquimod (a small-molecule ligand of Toll-like receptor-7 (TLR7)) stimulation. Supernatants (SN) and cell extracts (lysate) were analyzed using immunoblotting (C), and supernatants were analyzed using ELISA for IL-1β (B). D–F LPS-primed BMDMs were treated with NAC 30 min before imiquimod stimulation, MINK1 was immunoprecipitated, and in vitro kinase assays were performed with myelin basic protein as the substrate in the presence of ATP-γ-S. The reaction products were immunoblotted with an antithiophosphate ester antibody (D). Supernatants were analyzed using ELISA for IL-1β (E), and the fold change of kinase activity was quantified and is presented in F. *p < 0.05, **p < 0.01, ***p < 0.001, two-tailed unpaired Student’s t test was used for F, and ANOVA was used for B and E. The flow cytometry, ELISA, and western blot results are representative of three independent experiments
NAC alleviates the NLRP3-mediated acute inflammatory response in a mouse model and human cells
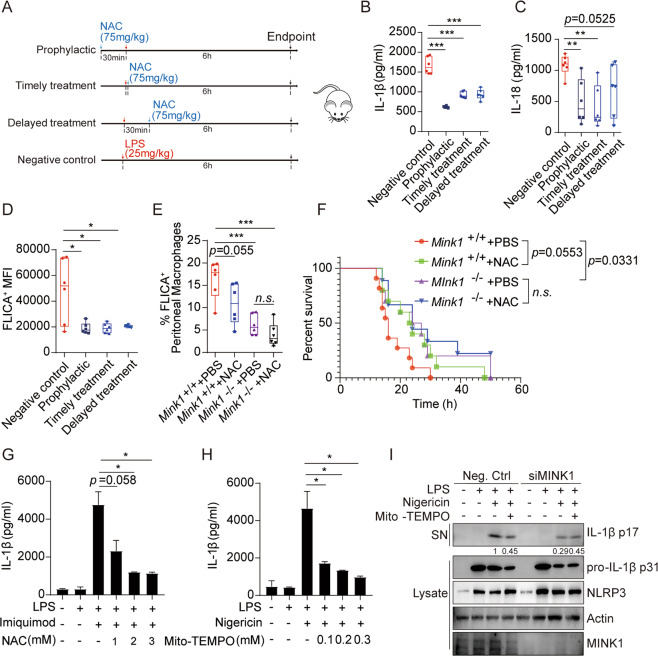
To determine whether reduced NLRP3 inflammasome activation upon NAC treatment resulted in pathological improvements in acute inflammation, we again performed in vivo LPS-induced sepsis with or without NAC treatment at different time points. We found that serum levels of IL-1β and IL-18 were consistently lower in the NAC-treated groups, although the reduction rates varied between groups (Fig. 6A–C). The secretion of TNF-α and IL-6 was similar (Fig. S6A, B). Prophylactic treatment with NAC showed the most beneficial effects, and NAC treatment afterward was the least effective. FLICA staining assays revealed blunted LPS-induced caspase-1 activation in macrophages from NAC-treated mice (Fig. 6D). NAC supplementation significantly improved the survival of mice challenged with LPS (Fig. 6F). These results confirmed the protective role of NAC supplementation in acute inflammation. Notably, we found that NAC treatment in Mink1−/− mice produced limited improvements in inflammatory symptoms, as evaluated by the survival rate and the proportion of FLICA+ macrophages, which was consistent with our in vitro results (Fig. 6E, F). These results demonstrate that ROS are the major upstream activator of MINK1 in promoting NLRP3 inflammasome activation.
Fig. 6.
NAC significantly alleviates the acute inflammatory response mediated by NLRP3 activation in human and mouse cells. A Schematic diagram of NAC treatment model. B, C Eight-week-old mice were intraperitoneally injected with LPS (25 mg/kg), followed by different NAC treatments at different time points. Serum was collected to detect IL-1β and IL-18. Data are the means ± SD (n= 6 mice/group). D Surface staining of F4/80 and FLICA on macrophages in spleens from C57BL/6 (B6) mice to determine the intensity of caspase-1 activation. E Surface staining of F4/80 and FLICA on peritoneal macrophages from Mink1+/+ and Mink1−/− mice to determine the proportion of activated caspase-1 macrophages. F Survival rate of Mink1+/+ and Mink1−/− mice injected with 20 mg/kg LPS. G, H PBMCs were freshly isolated from healthy donors and LPS-primed PBMCs with NAC or Mito-TEMPO before imiquimod or nigericin stimulation. Supernatant IL-1β was analyzed using ELISA. I LPS-primed THP-1 cells were treated with Mito-TEMPO before nigericin stimulation. Supernatants (SN) and cell extracts (lysate) were analyzed using immunoblotting. *p < 0.05, **p < 0.01, ***p < 0.001. One-way ANOVA was used for B–E, G, and H, and the Kaplan–Meier method was used for mouse survival F. The ELISA and western blot results are representative of three independent experiments
We examined whether similar mechanisms existed in humans. We performed similar treatments in PBMCs freshly isolated from healthy donors. The results showed that NAC and Mito-TEMPO suppressed IL-1β production in a dose-dependent manner (Fig. 6G, H). We used siRNA to knockdown MINK1 in the human THP-1 cell line (Fig. S6C) and found that ROS scavenging reduced NLRP3 activation only in the control group but not in MINK1 knockdown cells (Fig. 6I). Taken together, these results indicate that MINK1 in macrophages contributes to the progression of NLRP3-dependent inflammation in mice and humans, which suggests that MINK1 is a potential target for treating NLRP3-related inflammatory diseases in human patients.
Discussion
The phosphorylation of NLRP3 at different residues contributes to NLRP3 activation [14, 17, 19]. However, contradictions between these findings and the mechanisms remain. The present study found that the phosphorylation of NLRP3 at Ser725 was a critical priming step in the upregulated expression for inflammasome activation. This single amino acid phosphorylation in the LRR domain of NLRP3 facilitates its self-association. We also demonstrated that MINK1 kinase activity triggered by ROS following LPS stimulation mediated this phosphorylation. Therefore, we revealed an undiscovered regulatory pathway associated with NLRP3 priming.
Previous studies in human macrophage studies reported that NLRP3 S728 (equivalent to the murine NLRP3 S725 residue) was a phosphorylation site. However, Song et al. and Stutz et al. considered that phosphorylation of Ser728 was not essential for human NLRP3 activation [14, 17], which contradicts our findings. To clarify these results, we used human macrophage cells and confirmed that the S728 mutation in human NLRP3 impaired inflammasome activation. The reason for this inconsistency may be the different experimental settings. However, a nonphosphorylation mutation at the S728 residue did reduce NLRP3 activation in these previous reports to a lesser extent than in the present study [14, 17]. Therefore, the importance of this residue, and its phosphorylation status, must be reevaluated.
We previously showed that MINK1 knockout in mice increased Th17 cell polarization and the susceptibility to EAE [30]. However, we did not observe a spontaneous autoimmune phenotype in MINK1-deficient mice. MINK1 is widely expressed in different immune cells, and it may exert immune regulatory roles in other immune cells. We investigated its possible role in macrophages because macrophages are a major cell type mediating inflammatory responses. We demonstrated a regulatory role of MINK1 in NLRP3 priming and inflammasome activation. Therefore, the insufficient inflammasome activation in MINK1 KO mice may counterbalance the proinflammatory effect of increased Th17 differentiation and avert spontaneous inflammation.
Many infectious and stress signals activate the NLRP3 inflammasome to elicit inflammatory responses. ROS are a major intermediate activator of the NLRP3 inflammasome in the priming and activation stages [42, 43]. The function of ROS in NLRP3 activation is well confirmed, and ROS elimination and blockade of cellular antioxidant responses inhibit NLRP3 inflammasome activation [12, 44–46]. However, the mechanisms or pathways responsible for this process remain elusive. Thioredoxin-interacting protein (TXNIP) and the transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2), which are the two major responders to oxidative stress, are links between ROS and NLRP3 activation. Zhou et al. proposed a model in which TXNIP released from TRX during oxidative stress bound to NLRP3 directly to activate the inflammasome [47]. Park et al. demonstrated that TXNIP promoted the activation of inflammasomes indirectly, and TXNIP inhibited the transcriptional activity of NF-κB to attenuate the LPS-induced upregulation of inducible nitric oxide synthase (iNOS) [48]. However, these mechanisms are not supported by genetic evidence, because wild-type and Txnip−/− macrophages secrete similar levels of IL-1β upon exposure to different stimuli [49]. The transcription factor Nrf2 is essential for antioxidant responses. Previous studies demonstrated that macrophages from Nrf2−/− mice secreted less IL-1β than wild-type cells in response to NLRP3 activators, and several different mechanisms were proposed. Nrf2 is in a complex comprised of Keap1 and the mitochondrial outer-membrane protein PGAM5 in its resting state. ROS causes dissociation of Nrf2 from the complex, and it translocates to the cytosol and interacts with ASCs to activate the inflammasome [50]. Some of the Nrf2 translocates to the nucleus and upregulates heme oxygenase-1 expression, which activates the inflammasome at the priming stage [51]. Nrf2 also upregulates the expression of antioxidant genes and downregulates the expression of TXNIP to mediate the activation of NLRP3 [42]. However, Nrf2 does not appear to play a direct role in priming, and all of the above-proposed mechanisms are indirect processes that involve transcriptional regulation [52]. Consistent with previous findings that ROS activates the kinase activity of MINK1, our findings offer a more attractive mechanism by which PAMP- and DAMP-induced ROS activate MINK1 to prime NLRP3 activation via direct phosphorylation. This posttranslational mechanism may be fast and efficient, which is consistent with the general characteristics of rapid NLRP3 inflammasome activation.
Abnormal activation of the NLRP3 inflammasome is closely related to many human diseases [1, 4, 16]. NLRP3 activation and IL-1β accumulation mediate inflammation during viral infection in the lung and the pathogenesis of ARDS, such as in severe COVID-19 [53]. Therefore, targeting NLRP3 may have great potential in the treatment of these diseases. The delineation of MINK1 function in inflammasome activation shown in the present study adds MINK1 as a promising candidate in the treatment of NLRP3-related diseases because the targeting of kinases is a highly practical approach.
Materials and methods
Mice
Mink1−/− mice (on a C57BL/6J background) were generated as described previously [27, 30]. Nlrp3−/− mice (on a C57BL/6J background) were kindly provided by Prof. Di Wang (Zhejiang University). C57BL/6 (B6) mice were purchased from the Model Animal Research Center of Nanjing University. All mice were housed in a specific pathogen-free facility in the Laboratory Animal Center of Zhejiang University in accordance with the National Institute of Health Guide for Care and Use of Laboratory Animals. The animal experimental protocols were approved by the Review Committee of Zhejiang University School of Medicine (Approval No. ZJU20210015) and were performed in compliance with institutional guidelines. The mice for the experiment were aged 6–8 weeks and weighed 20–25 g.
Human samples
PBMCs were isolated from peripheral blood from six healthy volunteers aged 20–30 years (three men and three women) who provided informed consent. Briefly, human peripheral blood samples were centrifuged, and the serum was discarded. The samples were resuspended in PBS and added to Lympholyte-H (Cedarlane). The mesospheric cells were collected and spread into six-well plates after centrifugation. Cells were treated after 24 h. Prior written informed consent was given by the donors, and approval was obtained from the Zhejiang University School of Medicine (Approval No. 2021–076).
Cells
Mouse bone marrow cells (BMDMs) were flushed from tibias and femurs with chilled Dulbecco’s modified Eagle’s medium (DMEM) and cultured in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 10 ng/mL macrophage colony stimulating factor (PeproTech) to generate BMDMs. Mouse PMs were harvested 4 days after thioglycolate (Merck) injection. Stimulation conditions were performed as described previously [16]. Briefly, cells were primed with LPS (500 ng/mL) for 3.5 h in Opti-MEM, followed by stimulation with various NLRP3 inflammasome activators: 2 mM ATP (30 min); 10 µM nigericin (30 min); 200 µg/mL MSU (5 h); and 300 µg/mL aluminum salts (5 h). For AIM2 inflammasome activation, poly(dA:dT) (1 µg/mL) was transfected using Lipofectamine 2000 (3 mL/mg DNA) (Invitrogen). For NLRC4 inflammasome activation, S. Typhimurium was grown overnight in Luria-Bertani broth, and BMDMs were infected for 1 h with Salmonella culture (1:100) then incubated for another 1 h in the presence of gentamycin. For reconstruction of the NLRP3 inflammasome in HEK293T cells, cells were seeded into six-well plates at 5 × 105 per well in complete cell culture medium. After 24 h, the cells were transfected with plasmids expressing pro-IL-1β (1 µg), Myc-procaspase-1 (60 ng), HA-ASC (150 ng), HA-NLRP3 (200 ng), and MINK1 or its mutants (200 ng) using Lipofectamine 2000. Forty-eight hours later, the cells were stimulated with NLRP3 inflammasome activators, and cell lysates and precipitated supernatants were further analyzed using western blotting and ELISA.
Reagents
Primary antibodies against NLRP3 (AdipoGen), mouse IL-1β (R&D Systems), mouse caspase-1 (p20) (AdipoGen), NEK7 (Santa Cruz Biotechnology), ASC (Santa Cruz Biotechnology), and MINK1 (NOVUS) were used for western blotting. For stimulation, MSU, nigericin, ATP and poly(dA:dT) were purchased from Sigma-Aldrich, and LPS, MitoTracker, and MitoSOX were obtained from Invitrogen. The Salmonella was a gift from Prof. Di Wang. The FAM-FLICA-caspase-1 assay kit was obtained from Immunochemistry. The cytoTox 96 nonradioactive cytotoxicity assay kit for LDH release was obtained from Promega. Phos-tagTM SDS–PAGE was obtained from Wako.
Protein precipitation from supernatant
Briefly, supernatants from different stimulation conditions were collected and centrifuged to remove the precipitate, then 1/4 volume of chloroform and 1 volume of methanol were added to the supernatant. The samples were centrifuged at room temperature, 12000 rpm for 5 min. The methanol layer was aspirated, and 1 volume of methanol was added. The samples were centrifuged at room temperature, 12000 rpm for 5 min again. The samples were air dried or dried on a heat block at 50 °C for 5 min. The dried samples were lysed in cell lysis buffer.
Flow cytometry and cell sorting
The antibodies and reagents used for surface staining were fixed viability dye, anti-CD45 (APC-Cyanine7, 30-F11), anti-CD11b (PE, N418), anti-Gr-1 (FITC, RB6–8C5), and anti-F4/80 (APC-F4/80), all from BioLegend. Samples were run on an LSR Fortessa (BD) or NovoCyte (ACEA). Cell sorting was performed by Beckman Moflo Astrios EQ. Data were analyzed using FlowJo v10 software.
ROS and mitochondrial ROS measurements
Total ROS was measured using a Reactive Oxygen Species Assay Kit from Beyotime (Cat. S0033). Cells were incubated with antibody after stimulation in the incubator for 30 min, rinsed in PBS, and analyzed using NovoCyte flow cytometers. Mitochondrial ROS was measured following incubation with 5 mM MitoSOX in PBS for 30 min at 37 °C. The cells were rinsed in cold PBS and analyzed using NovoCyte flow cytometers. Data were processed using FlowJo v10 software.
Western blotting and IP
Cells were lysed in cell lysis buffer (100 mM Tris-HCl, 4% SDS, 20% glycerol, 2% 2-mercaptoethanol, and 0.05% bromophenol blue, 1 mM PMSF, and protease inhibitors). The samples were boiled and separated on 10% or 12% SDS–PAGE gels and transferred to nitrocellulose membranes (Millipore). The membranes were blocked with 5% BSA in TBST (0.5 M NaCl, Tris-HCl, pH 7.5, and 0.05% [vol/vol] Tween 20) for 1 h at room temperature. After incubation with primary and secondary antibodies, ECL blotting reagents (Thermo Fisher) were used for immunoblot detection. For transfection and coimmunoprecipitation, constructs were transfected into HEK293T cells using polyethyleneimine (PEI) or Lipofectamine 2000. After 24 h, the cells were collected and resuspended in cell lysis buffer. Cell lysates were immunoprecipitated by incubation of lysates with 1–10 µg antibody overnight at 4 °C, and antibody–protein precipitates were pulled down with SureBeads Protein A (or protein G) magnetic beads (Bio-Rad Laboratories). The presence of immunocomplex proteins was determined using western blot analysis.
ASC oligomerization and ASC speck formation
ASC oligomerization and speck formation conditions were established as previously described [15]. After stimulation, BMDMs were rinsed in PBS, and 500 µL ice-cold buffer (50 mM Tris-HCl pH 7.6, 0.5% Triton X-100, 0.1 mM PMSF, and a protease inhibitor cocktail) was added. Cells were scraped, lysed, and centrifuged at 330 × g for 10 min, and the pellets were washed twice and resuspended in 500 µL PBS. Disuccinimidyl suberate (2 mM) was added to the resuspended pellets and incubated at room temperature for 30 min with rotation. Samples were centrifuged and resuspended in SDS loading buffer for western blotting.
For ASC speck formation, BMDMs were seeded and cultured overnight on glass coverslips. The following day, the cells were primed with LPS and treated with ATP in the presence or absence of the indicated inhibitors. The cells were fixed in 4% paraformaldehyde followed by ASC and DAPI staining.
Immunofluorescence staining and confocal microscopy
Briefly, 2 × 105 BMBMs were plated on coverslips overnight. After blocking with 5% BSA (Sigma), cells were incubated with primary antibodies overnight at 4 °C in PBS containing 1% BSA. After three washes in PBS with Tween 20 (PBST), the cells were incubated with secondary antibodies in PBS for 1 h at room temperature, rinsed in PBST, and mounted with mounting media containing DAPI (Sigma). Confocal microscopy analyses were performed using a Zeiss LSM880/800 or Nikon A1R.
ELISA
Supernatants from cell cultures and sera were collected, and the concentrations of IL-1β, IL-18, IL-6, and TNF-α (all from Thermo Fisher) were determined according to the manufacturer’s instructions.
Quantitative PCR (qPCR) with reverse transcription
RNA was extracted using a total RNA extraction reagent (Vazyme Biotech). Complementary DNA was synthesized using HiScript® II Reverse Transcriptase (Vazyme Biotech) according to the manufacturer’s instructions. qPCR was performed using SYBR Green (Vazyme Biotech) on a CFX96 Touch Real-Time PCR (Bio-Rad). All samples were individually normalized to β-actin. The primers are shown in Supplementary Table 1.
MS analysis
MINK1 and NLRP3 were cotransfected into HEK293T cells using PEI. After 24 h, cells were collected and resuspended in cold cell lysis buffer. Extracts were immunoprecipitated with Flag-M2 monoclonal antibody–agarose beads and dissolved in sample buffer. Cell lysates were separated on SDS–PAGE, followed by in-gel digestion, desalting, and analysis with the assistance of the Tsinghua University Protein Research Technology Center. Data analysis was performed with Maxquant (v1.6.8.0).
In vivo animal model
Mink1−/− and WT mice were injected intraperitoneally with LPS or aluminum salts. For the sepsis model, mice were sacrificed 4 h after LPS challenge (25 mg/kg body weight), and the serum levels of IL-1β, IL-18, and TNF-α were measured using ELISA (Thermo Fisher). For the acute peritonitis model, mice were sacrificed 6 h after alum (2 mg per mouse) challenge. Flow cytometry was used to detect the proportion of neutrophils in the peritoneal flushing fluid, and the release of IL-1β and TNF-α was also detected using ELISA. For the survival model, mice were challenged with LPS (20 mg/kg body weight) and observed up to 30 h.
In vitro kinase activity assay
MINK1 kinase was immunoprecipitated in cell lysates of LPS-stimulated macrophages and MEFs using a MINK1 antibody and protein A/G-Sepharose. Beads were washed three times with lysis buffer and two times with 1× kinase assay buffer. For kinase reaction in vitro, beads containing MINK1 were incubated with 20 µg purified substrates myelin basic protein, 50 μM ATP, 10× kinase assay buffer, and 1× protease inhibitor cocktail at 30 °C for 30 min with flicking. All kinase assay reactions were stopped with 5× SDS loading buffer, boiled at 100 °C for 5 min, and subjected to immunoblot analysis with an antithiophosphate antibody.
Human NLRP3-NEK7 symmetric docking
The input model was the cryo-EM structure of NLRP3 bound to NEK7 [Protein Data Bank (PDB) ID:6npy] [54, 55]. A total of 100 models were relaxed using the relax application [56] in the Rosetta suite. The model with the lowest energy was used for symmetric docking. Point mutation S728A was introduced by the backrub application [57] in Rosetta. The models of mutants were again relaxed by Rosetta before docking.
The symmetric docking application [58] was used to dock. A total of 10,000 models were generated for each docking trial. After docking, the top ten models with the lowest scores were chosen as candidates. The model with the lowest score among the largest cluster of the top ten models was used as the representative model.
Statistical analysis
All results are presented as the means ± SEM or SD. All the data were tested for normal distributions. Statistical analyses were performed using Student’s t test (two-tailed unpaired) for two groups, ANOVA for multigroup comparisons and the Kaplan–Meier method for mouse survival were performed as indicated using GraphPad Prism 8, unless otherwise noted. Differences were considered significant when p < 0.05.
Supplementary information
Supplementary figures and figure legends
All the blots and the whole membrane (not cut)
Acknowledgements
The authors thank Xinhui Song, Chun Guo, Jiajia Wang, and Yingying Huang from the core facilities (Zhejiang University School of Medicine) for technical assistance in histology and FACS analysis. The authors are grateful to Prof. Fan Yang and Wenxuan Zhen for human NLRP3-NEK7 symmetric docking. The authors thank the Key Laboratory of Immunity and Inflammatory Diseases of Zhejiang Province for their support. This work was supported by grants from the National Natural Science Foundation of China (31930038, 31770954, and 31530019 to L.L.).
Author contributions
K.Z., X.J., D.W., X.L. and L.L. designed the research. K.Z., X.J., Z.C., S.C. and S.W. performed the research. K.Z., X.J. and L.L. analyzed the data. K.Z., X.J. and L.L. wrote the paper. R.D.S., H.H. and D.N. edited the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Kaixiang Zhu, Xuexiao Jin.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00761-1.
References
- 1.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–35. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evavold CL, Kagan JC. Inflammasomes: threat-assessment organelles of the innate immune system. Immunity. 2019;51:609–24. doi: 10.1016/j.immuni.2019.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y, Hara H, Nunez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem Sci. 2016;41:1012–21. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broz P. Inflammasomes in host defense and autoimmunity. Chimia. 2016;70:853–5. doi: 10.2533/chimia.2016.853. [DOI] [PubMed] [Google Scholar]
- 5.Broz P. Inflammasomes: intracellular detection of extracellular bacteria. Cell Res. 2016;26:859–60. doi: 10.1038/cr.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baroja-Mazo A, Martin-Sanchez F, Gomez AI, Martinez CM, Amores-Iniesta J, Compan V, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15:738–48. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 7.Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–41. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 8.Schroder K, Zhou R, Tschopp J. The NLRP3 inflammasome: a sensor for metabolic danger? Science. 2010;327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 9.Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, et al. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol. 2011;12:408–15. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kummer JA, Broekhuizen R, Everett H, Agostini L, Kuijk L, Martinon F, et al. Inflammasome components NALP 1 and 3 show distinct but separate expression profiles in human tissues suggesting a site-specific role in the inflammatory response. J Histochem Cytochem. 2007;55:443–52. doi: 10.1369/jhc.6A7101.2006. [DOI] [PubMed] [Google Scholar]
- 11.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–20. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 12.Carta S, Tassi S, Pettinati I, Delfino L, Dinarello CA, Rubartelli A. The rate of interleukin-1beta secretion in different myeloid cells varies with the extent of redox response to Toll-like receptor triggering. J Biol Chem. 2011;286:27069–80. doi: 10.1074/jbc.M110.203398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol. 2019;19:477–89. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song N, Liu ZS, Xue W, Bai ZF, Wang QY, Dai J, et al. NLRP3 phosphorylation is an essential priming event for inflammasome activation. Mol Cell. 2017;68:185–97. doi: 10.1016/j.molcel.2017.08.017. [DOI] [PubMed] [Google Scholar]
- 15.Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, et al. Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity. 2016;45:802–16. doi: 10.1016/j.immuni.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Chi Z, Chen S, Xu T, Zhen W, Yu W, Jiang D, et al. Histone deacetylase 3 couples mitochondria to drive IL-1beta-dependent inflammation by configuring fatty acid oxidation. Mol Cell. 2020;80:43–58. doi: 10.1016/j.molcel.2020.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Stutz A, Kolbe CC, Stahl R, Horvath GL, Franklin BS, van Ray O, et al. NLRP3 inflammasome assembly is regulated by phosphorylation of the pyrin domain. J Exp Med. 2017;214:1725–36. doi: 10.1084/jem.20160933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spalinger MR, Kasper S, Gottier C, Lang S, Atrott K, Vavricka SR, et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Investig. 2016;126:4388. doi: 10.1172/JCI90897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Wang H, Hao Y, Lin H, Dong M, Ye J, et al. Myeloid PTEN promotes chemotherapy-induced NLRP3-inflammasome activation and antitumour immunity. Nat Cell Biol. 2020;22:716–27. doi: 10.1038/s41556-020-0510-3. [DOI] [PubMed] [Google Scholar]
- 20.Dan I, Watanabe NM, Kobayashi T, Yamashita-Suzuki K, Fukagaya Y, Kajikawa E, et al. Molecular cloning of MINK, a novel member of mammalian GCK family kinases, which is up-regulated during postnatal mouse cerebral development. FEBS Lett. 2000;469:19–23. doi: 10.1016/S0014-5793(00)01247-3. [DOI] [PubMed] [Google Scholar]
- 21.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–30. doi: 10.1016/S0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 22.Leong SY, Ong BK, Chu JJ. The role of misshapen NCK-related kinase (MINK), a novel Ste20 family kinase, in the IRES-mediated protein translation of human enterovirus 71. PLoS Pathog. 2015;11:e1004686. doi: 10.1371/journal.ppat.1004686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicke B, Bastien J, Khanna SJ, Warne PH, Cowling V, Cook SJ, et al. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol Cell. 2005;20:673–85. doi: 10.1016/j.molcel.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 24.Larhammar M, Huntwork-Rodriguez S, Rudhard Y, Sengupta-Ghosh A, Lewcock JW. The Ste20 family kinases MAP4K4, MINK1, and TNIK converge to regulate stress-induced JNK signaling in neurons. J Neurosci. 2017;37:11074–84. doi: 10.1523/JNEUROSCI.0905-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hussain NK, Hsin H, Huganir RL, Sheng M. MINK and TNIK differentially act on Rap2-mediated signal transduction to regulate neuronal structure and AMPA receptor function. J Neurosci. 2010;30:14786–94. doi: 10.1523/JNEUROSCI.4124-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarty N, Paust S, Ikizawa K, Dan I, Li X, Cantor H. Signaling by the kinase MINK is essential in the negative selection of autoreactive thymocytes. Nat Immunol. 2005;6:65–72. doi: 10.1038/ni1145. [DOI] [PubMed] [Google Scholar]
- 27.Yue M, Luo D, Yu S, Liu P, Zhou Q, Hu M, et al. Misshapen/NIK-related kinase (MINK1) is involved in platelet function, hemostasis, and thrombus formation. Blood. 2016;127:927–37. doi: 10.1182/blood-2015-07-659185. [DOI] [PubMed] [Google Scholar]
- 28.Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moroishi T, Hayashi T, Pan WW, Fujita Y, Holt MV, Qin J, et al. The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell. 2016;167:1525–39. doi: 10.1016/j.cell.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu G, Xu Q, Qiu Y, Jin X, Xu T, Dong S, et al. Suppression of Th17 cell differentiation by misshapen/NIK-related kinase MINK1. J Exp Med. 2017;214:1453–69. doi: 10.1084/jem.20161120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J Immunol. 2009;183:787–91. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sutterwala FS, Ogura Y, Zamboni DS, Roy CR, Flavell RA. NALP3: a key player in caspase-1 activation. J Endotoxin Res. 2006;12:251–6. doi: 10.1177/09680519060120040701. [DOI] [PubMed] [Google Scholar]
- 33.Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, et al. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–56. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyodo T, Ito S, Hasegawa H, Asano E, Maeda M, Urano T, et al. Misshapen-like kinase 1 (MINK1) is a novel component of striatin-interacting phosphatase and kinase (STRIPAK) and is required for the completion of cytokinesis. J Biol Chem. 2012;287:25019–29. doi: 10.1074/jbc.M112.372342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaneko S, Chen X, Lu P, Yao X, Wright TG, Rajurkar M, et al. Smad inhibition by the Ste20 kinase misshapen. Proc Natl Acad Sci USA. 2011;108:11127–32. doi: 10.1073/pnas.1104128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Pichulik T, Wolz OO, Dang TM, Stutz A, Dillen C, et al. Human NACHT, LRR, and PYD domain-containing protein 3 (NLRP3) inflammasome activity is regulated by and potentially targetable through Bruton tyrosine kinase. J Allergy Clin Immunol. 2017;140:1054–67. doi: 10.1016/j.jaci.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Martinez GJ. MINK1: the missing link between ROS and its inhibition of Th17 cells. J Exp Med. 2017;214:1205–6. doi: 10.1084/jem.20170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 39.Tschopp J, Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–5. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 40.Du K, Ramachandran A, Weemhoff JL, Woolbright BL, Jaeschke AH, Chao X, et al. Mito-tempo protects against acute liver injury but induces limited secondary apoptosis during the late phase of acetaminophen hepatotoxicity. Arch Toxicol. 2019;93:163–78. doi: 10.1007/s00204-018-2331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, et al. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity. 2013;39:311–23. doi: 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dan Dunn J, Alvarez LA, Zhang X, Soldati T. Reactive oxygen species and mitochondria: a nexus of cellular homeostasis. Redox Biol. 2015;6:472–85. doi: 10.1016/j.redox.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu T, Wang L, Liang P, Wang X, Liu Y, Cai J, et al. USP19 suppresses inflammation and promotes M2-like macrophage polarization by manipulating NLRP3 function via autophagy. Cell Mol Immunol. 2020. 10.1038/s41423-020-00567-7. [DOI] [PMC free article] [PubMed]
- 44.Bauernfeind F, Bartok E, Rieger A, Franchi L, Nunez G, Hornung V. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J Immunol. 2011;187:613–7. doi: 10.4049/jimmunol.1100613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tassi S, Carta S, Delfino L, Caorsi R, Martini A, Gattorno M, et al. Altered redox state of monocytes from cryopyrin-associated periodic syndromes causes accelerated IL-1beta secretion. Proc Natl Acad Sci USA. 2010;107:9789–94. doi: 10.1073/pnas.1000779107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez-Rodriguez R, Munari F, Angioni R, Venegas F, Agnellini A, Castro-Gil MP, et al. Targeting monoamine oxidase to dampen NLRP3 inflammasome activation in inflammation. Cell Mol Immunol. 2021;18:1311–3. [DOI] [PMC free article] [PubMed]
- 47.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–40. doi: 10.1038/ni.1831. [DOI] [PubMed] [Google Scholar]
- 48.Park YJ, Yoon SJ, Suh HW, Kim DO, Park JR, Jung H, et al. TXNIP deficiency exacerbates endotoxic shock via the induction of excessive nitric oxide synthesis. PLoS Pathog. 2013;9:e1003646. doi: 10.1371/journal.ppat.1003646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masters SL, Dunne A, Subramanian SL, Hull RL, Tannahill GM, Sharp FA, et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1beta in type 2 diabetes. Nat Immunol. 2010;11:897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao C, Gillette DD, Li X, Zhang Z, Wen H. Nuclear factor E2-related factor-2 (Nrf2) is required for NLRP3 and AIM2 inflammasome activation. J Biol Chem. 2014;289:17020–9. doi: 10.1074/jbc.M114.563114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wegiel B, Larsen R, Gallo D, Chin BY, Harris C, Mannam P, et al. Macrophages sense and kill bacteria through carbon monoxide-dependent inflammasome activation. J Clin Investig. 2014;124:4926–40. doi: 10.1172/JCI72853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jhang JJ, Yen GC. The role of Nrf2 in NLRP3 inflammasome activation. Cell Mol Immunol. 2017;14:1011–2. doi: 10.1038/cmi.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Freeman TL, Swartz TH. Targeting the NLRP3 inflammasome in severe COVID-19. Front Immunol. 2020;11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharif H, Wang L, Wang WL, Magupalli VG, Andreeva L, Qiao Q, et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 2019;570:338–43. doi: 10.1038/s41586-019-1295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Z, Yan C, Liu P, Huang Z, Ma R, Zhang C, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–5. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 56.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci. 2014;23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis IW, Arendall WB, 3rd, Richardson DC, Richardson JS. The backrub motion: how protein backbone shrugs when a sidechain dances. Structure. 2006;14:265–74. doi: 10.1016/j.str.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 58.Andre I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc Natl Acad Sci USA. 2007;104:17656–61. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and figure legends
All the blots and the whole membrane (not cut)