Abstract
Human RNA helicase A was recently identified to be a shuttle protein which interacts with the constitutive transport element (CTE) of type D retroviruses. Here we show that a domain of 110 amino acids at the carboxyl terminus of helicase A is both necessary and sufficient for nuclear localization as well as rapid nuclear export of glutathione S-transferase fusion proteins. The import and export activities of this domain overlap but are separable by point mutations. This bidirectional nuclear transport domain (NTD) has no obvious sequence homology to previously identified nuclear import or export signals. However, the Ran-dependent nuclear import of NTD was efficiently competed by excess amounts of the nuclear localization signal (NLS) peptide from simian virus 40 large T antigen, suggesting that import is mediated by the classical NLS pathway. The nuclear export pathway accessed by NTD is insensitive to leptomycin B and thus is distinct from the leucine-rich nuclear export signal pathway mediated by CRM1.
Macromolecular trafficking across the eukaryotic nuclear envelope occurs through the nuclear pore complexes and involves specific signal-receptor interactions. Nuclear localization signals (NLS) are capable of conferring the nuclear import and retention functions to heterologous nonnuclear proteins (3). Both the classical NLS, first identified in simian virus 40 (SV40) large T antigen, and the bipartite NLS, found in a number of Xenopus nuclear proteins, contain stretches of basic amino acid residues. NLS-bearing proteins interact with the NLS receptor, namely, importin α, in the cytoplasm. Subsequent binding to importin β targets the NLS-importin α/β complex to the nuclear pore for translocation across the membrane (12, 31). Another type of nuclear import signal, termed M9, was found in heterogeneous nuclear ribonucleoprotein (hnRNP) A1 and several related hnRNPs (26, 39). M9 specifies both nuclear import and export of hnRNP A1 and bears no sequence resemblance to the classical or bipartite NLS. The only known receptor protein for M9 is a protein distantly related to importin β, named transportin (1, 9, 36). Additional nuclear import sequences have been identified, but their receptors are currently unknown (16, 19, 27). A signal from hnRNP K is believed to bypass the requirement of a soluble receptor and interact directly with the nuclear pore (27).
Nuclear export of RNA and protein is less well understood but also appears to be signal mediated and energy dependent. The best-characterized, leucine-rich nuclear export signals (NES) use CRM1, also a distant relative of importin β, as their functional export receptor (6, 10, 32, 40, 43). Leucine-rich NES complexes with CRM1 and RanGTP in the nucleus prior to translocation through the nuclear pore. This complex formation is sensitive to leptomycin B (LMB). Different classes of cellular RNA were shown to use distinct pathways for nuclear export (18). Recently, the receptor for nuclear export of tRNA, named exportin-t, was shown to be another member of the importin β family (21).
The nuclear export of cellular mRNA is tightly coupled to splicing, such that only completely spliced mRNA is exported into the cytoplasm. However, export of unspliced viral RNA is necessary for the expression and replication of retroviruses. For lentiviruses, a viral regulatory protein named Rev promotes the nuclear export of unspliced and incompletely spliced viral RNA by binding to its cognate RNA sequence, the Rev response element (RRE) (4, 5, 24). Rev contains a leucine-rich NES and uses CRM1 as the export receptor (4, 6). In simple retroviruses, the unspliced RNAs contain a cis-acting, constitutive transport element (CTE) that interacts directly with cellular factors to achieve nuclear export (2, 34, 41, 48). We previously showed that human RNA helicase A specifically binds functional but not mutant CTE in vitro (42) and that antibodies to helicase A blocked CTE function when microinjected into human cells (23). We also showed that helicase A shuttles constantly between the nucleus and cytoplasm despite its apparent steady-state nuclear localization (42). In this paper, we report the identification and characterization of a nuclear transport domain (NTD) in helicase A which directs the bidirectional trafficking of fusion proteins. This domain presumably plays an important role in the posttranscriptional regulation of retroviruses by helicase A.
MATERIALS AND METHODS
Plasmid constructions.
The cDNA clone of human RNA helicase A was a kind gift from J. Hurwitz (22). Plasmid pSK-helicase A was first digested with BglII, blunt ended with Klenow polymerase, and then digested with EcoRI. A 3.4-kb fragment that encodes amino acids 1 to 1136 of helicase A was recovered. pEGFP-1 (Clontech) was digested with BamHI, blunt ended, and then digested with NotI. A 0.7-kb fragment encoding the green fluorescent protein (GFP) was recovered. The 3.4- and 0.7-kb fragments were then used in a three-way ligation with the EcoRI-NotI-digested pcDNA3. The appropriate construct encodes amino acids 1 to 1136 of helicase A with GFP fused to the carboxyl terminus and is called GFP/HelA-ΔCTD. The GFP coding sequence from pEGFP-1 was PCR amplified and cloned into the BamHI and EcoRI site of pcDNA3 (Invitrogen) such that the EcoRI site at the 3′ end of the coding sequence can be used for in-frame cloning of GFP fusion proteins. This plasmid was designated pc-GFP. pSK-helicase A was digested with ApaI and EcoRI and then blunt ended with Klenow polymerase, and a 3.9-kb blunt fragment (including the 3′ untranslated region) encoding amino acids 137 to 1269 of helicase A was recovered and cloned into the EcoRI-digested but blunt-ended pc-GFP vector to make GFP/HelA-ApaI. GFP/ApaI-ΔCTD was made in a similar way, but the ApaI-BglII fragment of helicase A was cloned into pc-GFP. To make the GFP-helicase carboxyl-terminal domain fusion proteins, different carboxyl-terminal domain fragments of helicase A were PCR amplified and cloned into the EcoRI site of pc-GFP. Site-directed mutagenesis was carried out by a method similar to that used with the Transformer kit of Clontech. Mutated sequences were used for testing subcellular distributions of the protein domain they encode. Myc-PK plasmid was described by Siomi and Dreyfuss (39). Wild-type and mutant NTDs were PCR amplified as KpnI-NotI fragments and cloned into the KpnI and NotI sites of pcDNA3/Myc-PK to make Myc-PK-NTD and Myc-PK-NTD mutant plasmids. Myc-NPc-TNLS has been described by Michael et al. (26). Wild-type and mutant NTDs were PCR amplified as XhoI-XbaI fragments and cloned into the corresponding sites of pcDNA3/Myc-NPc-TNLS to make NPc-TNLS-NTD, NPc-TNLS-ΔK62, and NPc-TNLS-ΔR65. GFP-TNLS-NTD and mutant constructs were made by cloning the EcoRI-XbaI fragments of the corresponding NPc-TNLS plasmids into EcoRI-XbaI-digested pc-GFP. pGST-NTD was made by cloning the EcoRI fragment of GFP-NTD into yeast vector pGAD10, resulting in pGAD10-NTD. The BamHI-BglII insert of this plasmid was then cloned into the BamHI site of pGEX-2T (Pharmacia) to yield pGST-NTD. pGST-ΔK62 was made in a similar fashion.
Cell cultures, transfections, and Western blotting.
HeLa and Cos-1 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal calf serum. Transfections were done by calcium phosphate precipitation. In experiments where transcription and translation inhibitors were included, they were added 45 h after transfection, 3 h prior to fixation of the cell for immunofluorescence assay. For the Western blot, Myc-PK-NTD plasmids were transfected into Cos-1 cells, and cells were harvested 48 h later and lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis buffer. DNA was sheared by passage through a syringe and boiling. Total proteins were either treated with alkaline phosphatase for 1 h at 37°C or directly loaded onto a 12.5% polyacrylamide gel. The separated proteins were transferred onto a nitrocellulose membrane and Myc-tagged proteins were detected with monoclonal antibody 9E10 (Babco).
Heterokaryon assay.
Interspecies heterokaryons of HeLa and NIH 3T3 cells were formed as described previously (27). For GFP-CTD and GFP-NTD, cells were fixed 1 h after fusion. For the GFP-TNLS-NTD series, NPc-TNLS-NTD series, and Rev, cells were fixed 3 h after fusion. To confirm the inhibitory effect of cycloheximide on new protein synthesis, cells were metabolically labeled with [35S]methionine and [35S]cysteine 30 min after the addition of the translation inhibitor, labeled for 1 h at 37°C, lysed, and assayed for radiation incorporation.
Immunofluorescence analysis.
For GFP-transfected cells, cells were fixed in 4% paraformaldhyde and viewed under a Nikon fluorescence microscope. For Myc-PK or Myc-NPc-TNLS fusion construct-transfected cells, the cells were fixed and stained with monoclonal antibody 9E10 as described previously (26). In all the heterokaryon experiments, 5 μg of Hoechst 33258 (Sigma) per ml was added either in 3% bovine serum albumin (BSA) after permeabilizing the cells with 0.2% Triton X-100 (for GFP fusion proteins) or during the secondary-antibody staining (for NPc-TNLS constructs). MEK-1 was stained with a polyclonal antibody purchased from Santa Cruz Biotechnology Inc. GST-NTD was stained with a monoclonal antibody against GST (Santa Cruz Biotechnology Inc.).
Recombinant protein purification, peptide conjugation, and microinjection.
pGST-NTD was transformed into bacterial strain BL21. Recombinant protein purification was carried out with a commercial GST purification module (Pharmacia). The final protein sample was concentrated to 3 mg/ml for microinjection. NIH 3T3 cells growing on coverslips were injected by using an Eppendorf microinjector. For the LMB experiments, cells were treated with 20 nM LMB 6 h before injection. One group of coverslips was inverted onto 50% polyethylene glycol to form polykaryons; 1 h after cell fusion, GST-NTD was mixed with 1.5 mg of rhodamine-dextran per ml before being injected into selected nuclei of the polykaryon. The cells were fixed for GST staining 30 min after injection. Another group of coverslips was fixed for immunostaining with a polyclonal antibody against MEK-1. BSA conjugates were generated as previously described with either wild-type (CYTPPKKKRKLY) or mutant (CYTPPKTKLV) NLS peptide.
RNA gel shift assays.
Gel mobility shift experiments were carried out as described previously (42). RNA helicase A protein and helicase A protein with the carboxyl terminus deleted were kind gifts from C. Lee and J. Hurwitz. GST-CTD was produced and purified similarly to GST-NTD.
RESULTS
The carboxyl terminus of helicase A harbors an NLS.
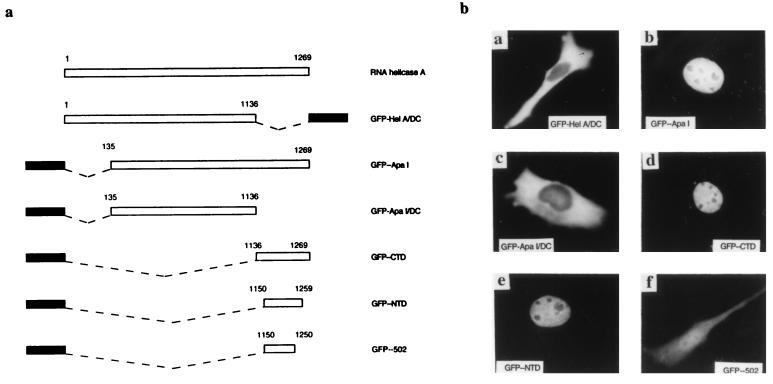
N-terminal and C-terminal deletions of RNA helicase A were generated and fused to GFP (Fig. 1a). The subcellular localization of fusion proteins was compared with that of endogenous helicase A and of GFP alone. We observed that deletion of the N-terminal amino acids (amino acids 1 to 135) did not affect the nuclear localization of the fusion protein (GFP/ApaI). In contrast, deletion of the carboxyl-terminal domain (CTD) (amino acids 1137 to 1269) both in the full-length context (GFP/HelA-ΔC) and together with the N-terminal deletion (GFP/ApaI-ΔC) rendered the fusion protein completely cytoplasmic (Fig. 1b), indicating that the C terminus of helicase A is needed for its nuclear localization. To find out whether the CTD of helicase A is sufficient for nuclear import, we fused the CTD to GFP and examined the distribution of the fusion protein. GFP-CTD also had a nuclear localization similar to that of endogenous helicase A and GFP/ApaI. We therefore conclude that the CTD of helicase A is both necessary and sufficient for nuclear localization.
FIG. 1.
The carboxyl terminus of helicase A is both necessary and sufficient for nuclear localization. (a) Schematic illustration of various domains of RNA helicase A that were fused to GFP to map the NLS of helicase A. Black bar, GFP; dashed line, deleted domain; open bar, domains of helicase A. (b) Subcellular localization of different helicase A domains. The minimal NLS was mapped to amino acids 1150 to 1259.
To map the minimal determinants for nuclear import, we fused a series of 5′ and 3′ deletions of the CTD to GFP and studied their subcellular localization. The minimal functional NLS was mapped between amino acids 1150 and 1259 (Table 1; Fig. 1b). The inability of further CTD deletions to localize in the nucleus was not due to their having insufficient sizes, because M9 of hnRNP A1, which is less than 40 amino acids long, was able to localize GFP in the nucleus in the same experiments (data not shown). Examination of this minimal NLS sequence revealed no homology to either the classical NLS (SV40 T antigen or bipartite NLS), M9 from hnRNP, or the nuclear import-export signal of hnRNP K (27, 39).
TABLE 1.
Mapping the minimal NLS of helicase A
| Hybrid proteina | Helicase domain (aa)b | Subcellular localizationb |
|---|---|---|
| GFP-CTD | 1136–1269 | N |
| GFP-202 | 1136–1259 | N |
| GFP-301 | 1150–1269 | N |
| GFP-NTD | 1150–1259 | N |
| GFP-501 | 1160–1259 | N/C |
| GFP-501.1 | 1165–1259 | N/C |
| GFP-404 | 1170–1259 | N/C |
| GFP-502 | 1150–1250 | N/C |
| GFP-508 | 1150–1220 | N/C |
All the GFP fusion proteins were cloned after a cytomegalovirus promoter and transfected into HeLa cells for expression.
aa, amino acids; N, nuclear; N/C, nuclear and cytoplasmic.
The CTD also contains a functional NES.
We recently demonstrated that helicase A is a shuttle protein (42) and thus probably contains a nuclear export signal. We then tested the CTD for its ability to direct the nuclear export of GFP-CTD. Inhibition of transcription blocks the nuclear import of shuttling proteins, making it possible to detect these steady-state nuclear proteins in the cytoplasm (25, 35). In our previous experiments, actinomycin D treatment caused endogenous helicase A to accumulate in the cytoplasm of HeLa cells. Cytoplasmic accumulation of GFP-CTD was also observed in transfected cells similarly treated with actinomycin D (data not shown), suggesting that CTD is capable of mediating nuclear export.
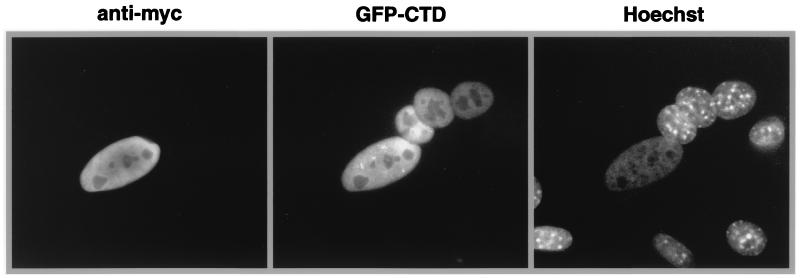
To obtain more definitive evidence for the nuclear export function of CTD, interspecies heterokaryon experiments were carried out with GFP-CTD or the minimal NLS fused to GFP. HeLa cells that were transfected with these constructs were fused to NIH 3T3 cells to form heterokaryons. Cycloheximide was added to inhibit de novo protein synthesis. The appearance of GFP in the mouse nuclei within the heterokaryon indicated that the fusion protein had moved out of the human nuclei and into the mouse nuclei. The mouse nuclei were distinguished from the human nuclei by punctate staining with Hoechst 33258. A Myc-tagged form of NPc-TNLS (26), which contains the core domain of Xenopus nucleoplasmin and an NLS from SV40 T antigen, was cotransfected as a nonshuttling nuclear protein control. As shown in Fig. 2, GFP-CTD was exported from the nucleus of the transfected human cell while Myc-NPc-TNLS stayed in the same nucleus. Similar results were obtained for the minimal NLS domain (data not shown). Therefore, both CTD and the minimal NLS sequence also contain an NES. For this reason, we named the domain between residues 1150 and 1259 the NTD (nuclear transport domain). These results indicate that although GFP-NTD has a steady-state nuclear localization, it actually shuttles constantly between the nucleus and the cytoplasm.
FIG. 2.
The carboxyl terminus of helicase A harbors an NES. An interspecies heterokaryon assay confirmed the shuttling ability of NTD. Mouse nuclei were identified by the punctate staining by Hoechst 33258. Anti-Myc antibody detected the cotransfected Myc-NPc-TNLS. GFP was found only in transfected HeLa cell nuclei and mouse nuclei within the heterokaryons that contained transfected human cells.
The nuclear import and export activities of NTD are separable by point mutations.
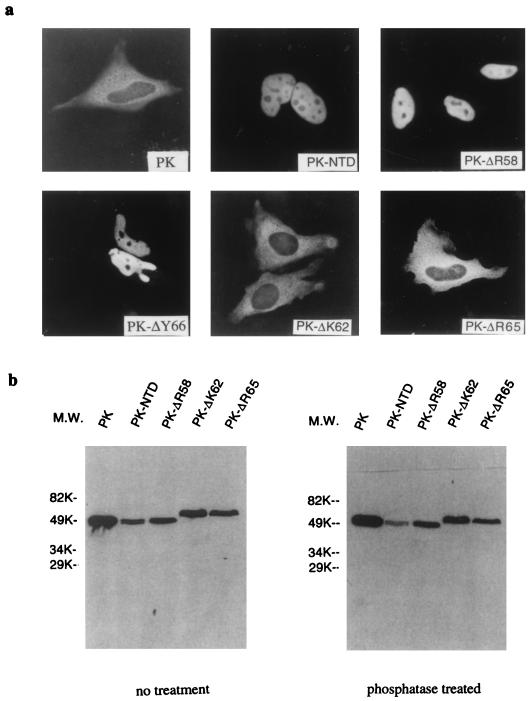
NTD, which contains part of the RGG box of helicase A (22, 46), is rich in glycine and serine residues. The N terminus of the domain contains several closely spaced basic residues (Fig. 3). Site-directed mutagenesis was used to identify specific amino acid residues that are critical for the import and export functions of NTD. Point mutations were introduced across the domain, and the mutants were tested for their ability to direct the GFP fusion protein in and out of the nucleus (Table 2). All the point mutants that retained the nuclear import function also maintained their nuclear export activity. Mutations in two basic residues, lysine 1162 and arginine 1165 (ΔK62, ΔR65, or R65L) abolished the nuclear localization of GFP-NTD, but deletion of arginine 1158 (ΔR58) or a neighboring tyrosine 1166 (ΔY66) did not have any effect. The loss of import function of the K62 and R65 mutants was confirmed in another experiment. NTD, ΔR58, ΔK62, ΔR65, and ΔY66 were fused to the carboxyl terminus of a Myc-tagged pyruvate kinase (39) and transiently expressed in HeLa cells. The subcellular localization of the fusion proteins was determined by immunostaining with a monoclonal antibody against Myc. Myc-PK localized exclusively in the cytoplasm, as previously reported (39). PK-NTD, PK-ΔR58, and PK-ΔY66 were all localized in the nucleus, while the PK-ΔK62 and PK-ΔR65 derivatives were localized in the cytoplasm (Fig. 4a).
FIG. 3.
Sequence of the RNA helicase A NTD. (a) Protein sequence of helicase A NTD. Residues that were mutated in this study are underlined. Positions of residues are those of Lee and Hurwitz (22) but adjusted to the carboxyl-terminal sequence of nuclear DNA helicase II as given by Zhang and Grosse (46). We independently sequenced the carboxyl terminus of RNA helicase A clone 1 (22), from which we identified NTD; we found that it is identical to that of nuclear DNA helicase II. (b) Positive residues are important for NTD and other nuclear import signals. Arginines and lysines are underlined; residues in boldface type are essential for the NLS activity of the protein. Only part of the NTD sequence is shown in panel b.
TABLE 2.
Summary of point mutations of helicase A NTD and their ability for nuclear import and export
| Mutation | Ability for:
|
|
|---|---|---|
| Nuclear importa | Nuclear exportb | |
| Wild type | + | + |
| P1160A | + | + |
| Y1179A | + | + |
| Y1199A | + | + |
| R1219G | + | + |
| G1220A | + | + |
| F1125L | + | + |
| ΔR1158 | + | + |
| ΔK1162 | − | + |
| ΔR1165 | − | + |
| ΔY1166 | + | + |
| R1165L | − | NDc |
The subcellular localization of all mutants was examined with the mutants as GFP fusion proteins. The wild type, ΔR1158, ΔK1162, and ΔR1165 were also studied as Myc-PK fusions.
For all the mutants that retained NLS activity, nuclear export was assayed by accumulation of the GFP fusion proteins in the cytoplasm after treatment with 4 μg of actinomycin D per ml. For ΔK1162 and ΔR1165, nuclear export was assayed in interspecies heterokaryons as both NPc-TNLS fusions and GFP-TNLS fusions.
ND, not determined.
FIG. 4.
Subcellular localization and expression of NTD mutants in HeLa cells. (a) Subcellular localization of NLS mutants. Myc-PK localized in the cytoplasm (39); NTD and ΔR58 targeted this protein into the nucleus, while ΔK62 and ΔR65 did not. (b) Immunoblotting analysis of PK-NTD and mutant fusion proteins. PK was truncated by a KpnI site at amino acid 443 during cloning so that PK-NTD is approximately the same size as PK. PK-ΔK62 and PK-ΔR65 had a lower gel mobility than expected, possibly because NTD is modified in the cytoplasm. Phosphatase treatment did not alter the gel mobility of PK-ΔK62 and PK-ΔR65. M.W., molecular weight.
The efficient expression of the PK-NTD fusion proteins in mammalian cells was verified by Western blotting with the antibody against the Myc tag (Fig. 4b). PK was truncated at the carboxyl terminus when fused to the NTD series, and so Myc-PK-NTD is about the same size as Myc-PK. This analysis revealed that the PK-ΔK62 and PK-ΔR65 derivatives had a lower mobility than PK-NTD and PK-ΔR58. This reduced electrophoretic mobility was probably due to posttranslational modifications of NTD in the cytoplasm of eukaryotic cells, since GST fusion proteins of NTD and ΔK62 expressed in Escherichia coli exhibited similar electrophoretic mobility (data not shown). Treatment of the transfected cell lysates with alkaline phosphatase did not alter the mobility of PK-ΔK62 and PK-ΔR65 (Fig. 4b), suggesting that the shift is not due to phosphorylation.
ΔK62 and ΔR65 were further studied for their ability to mediate export. Wild-type NTD, ΔK62, and ΔR65 were fused to the carboxyl terminus of Myc-NPc-TNLS, and the resultant hybrid proteins were tested for their abilities to shuttle between the cytoplasm and nucleus by interspecies heterokaryon assays. As shown in Fig. 5a, both mutants still shuttled. The ΔK62 and ΔR65 NTD mutations were also able to facilitate the shuttling of GFP-TNLS in heterokaryon assays (data not shown).
FIG. 5.
NTD import mutants retain nuclear export function. (a) NPc-TNLS had a nuclear localization and did not shuttle in heterokaryon assays; both import mutants (ΔK62 and ΔR65) shuttled as well as the wild-type NTD when fused to carboxyl terminus of NPc-TNLS. Arrows indicate the mouse nuclei in the heterokaryons. (b) ΔK62 directed the rapid export of GST fusion protein when injected in to nucleus. GST-ΔK62 was purified from E. coli as a recombinant protein and injected into either the nuclei or the cytoplasm of NIH 3T3 cells. The cells were then fixed for an immunofluorescence assay with a GST monoclonal antibody after a 30-min incubation at 37°C. The injection site was identified by coinjecting 1.5 mg of rhodamine-dextran per ml with GST-ΔK62. Hoechst staining identifies the nuclei.
The effects of the ΔK62 NTD mutation on import and export function were further studied by microinjection analysis of proteins. A fragment encompassing ΔK62 NTD was fused to GST (GST-ΔK62). The expressed recombinant protein was purified and microinjected into either the cytoplasm or the nuclei of NIH 3T3 cells. When injected into the cytoplasm, the fusion protein stayed in the cytoplasm (Fig. 5b), but when injected into the nuclei, it became cytoplasmic after 30 min of incubation at 37°C (Fig. 5b). These results confirm the observations obtained in the transfection studies of the ΔK62 NTD mutation. Again, the ΔK62 mutation disrupted the nuclear import function of NTD while having no effect on its nuclear export. These results demonstrate that the import and export functions of the NTD are separable activities of the NTD.
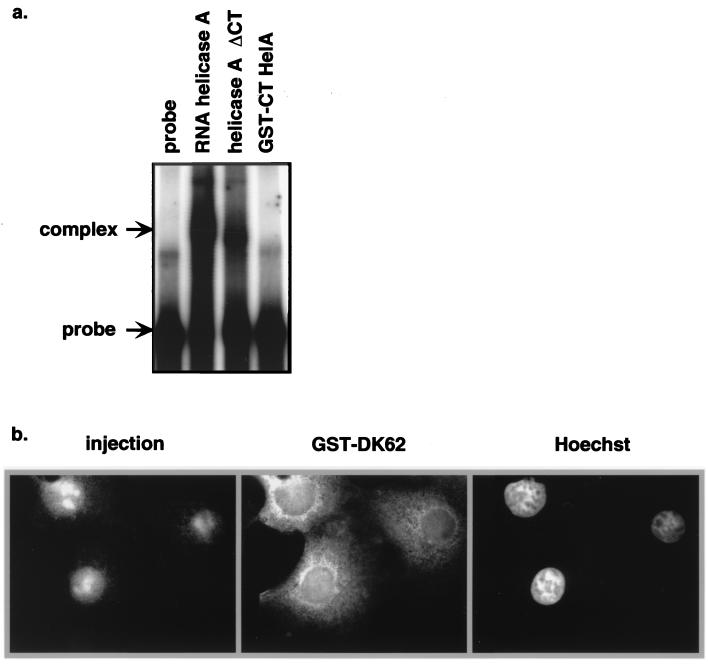
Since the NTD contains part of the RGG box that is implicated in RNA binding, it was possible that the export of NTD was bridged by RNA (e.g., CTE) which was being exported by other NES-containing proteins. To test this possibility, the ability of GST-CTD to bind to the CTE was analyzed in a gel mobility shift assay. As shown in Fig. 6a, although helicase A with CTD deleted had a weaker binding to CTE than did native helicase A, CTD itself was not sufficient for CTE binding in vitro. To further address the importance of cellular RNA in NTD export, RNase A was coinjected into the nucleus of Cos-1 cells with GST-ΔK62. The presence of RNase in the injected cells disrupted the integrity of the nucleus, as evidenced by the accumulation of the injection marker, rhodamine-dextran, in the nucleoli of these cells. However, GST-ΔK62 was exported efficiently in the same cells (Fig. 6b). Taken together, these results suggest that the export function of NTD is not mediated by binding to RNA.
FIG. 6.
RNA binding does not play a significant role in NTD export. (a) GST-CTD, helicase A protein, and helicase A with the C terminus deleted were tested for CTE binding in RNA gel shift assays. Full-length helicase A and the C-terminally deleted helicase A formed complexes with CTE probe, while GST-CTD was not able to interact with CTE in vitro. (b) RNase (5 mg/ml) was coinjected into Cos-1 nuclei with GST-ΔK62 and rhodamine-dextran. Cells were incubated for 30 min after injection before being fixed and stained for GST and DNA (Hoechst).
The nuclear import of NTD can be competed with NLS peptides.
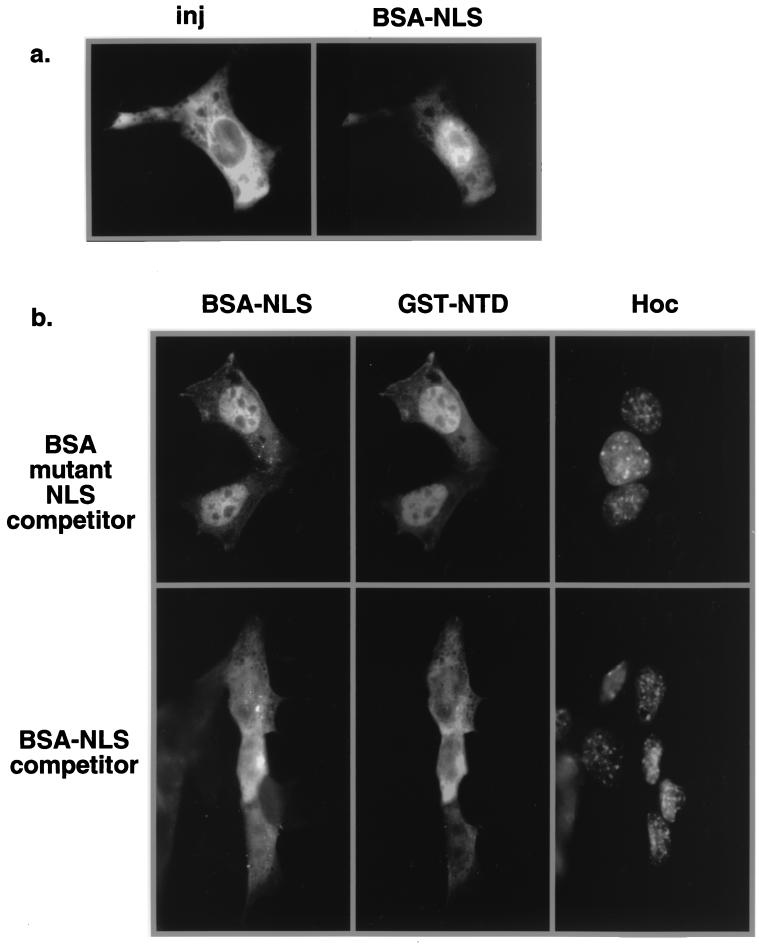
Although the NTD sequence bears no resemblance to a classical or bipartite NLS, the fact that closely spaced basic residues are critical for import raised the possibility that NTD uses the importin α/β pathway. To address this question, excess unlabeled TNLS peptides conjugated to BSA were injected into the cytoplasm of NIH 3T3 cells along with both labeled BSA-NLS and GST-NTD. BSA conjugates containing mutant NLS peptides were used as a control for the specificity of the competition. Injection of fluorescein isothiocyanate-labeled BSA-NLS into the cytoplasm resulted in the nuclear import of the conjugates after 30 min of incubation at 37°C (Fig. 7a), indicating that the BSA conjugates are functional as nuclear import substrates. In the presence of the BSA-mutant NLS conjugates, both the BSA-TNLS conjugates and GST-NTD were efficiently imported into the nuclei after 30 min (Fig. 7b, top panel). However, the nuclear import of both BSA-NLS and GST-NTD was competed by excess unlabeled BSA-NLS peptides (Fig. 7b, bottom panel). These results indicate that the nuclear import of NTD has the same sensitivity to excess NLS peptides as to conjugates containing TNLS. Microinjection experiments with T24N Ran also showed that the nuclear import of NTD is Ran dependent (data not shown), as expected for the classical NLS pathway. The glycine-rich feature of NTD also prompted us to determine if it interacts with transportin. NTD did not bind to transportin in the yeast two-hybrid assay, whereas the hnRNP A1 M9 region readily bound to transportin (data not shown).
FIG. 7.
Classical NLS peptides compete for NTD import. Peptides representing the wild-type or mutant NLS of SV40 T antigen were synthesized, purified, and conjugated to BSA. (a) Labeled BSA-NLS was imported into the nucleus after being injected (inj) into the cytoplasm of NIH 3T3 cells. The injection site was identified by the coinjected rhodamine-dextran. (b) Unlabeled BSA-mutant NLS (top) or BSA-NLS (bottom) was coinjected into the cytoplasm of NIH 3T3 cells with GST-NTD or fluorescein isothiocyanate-labeled BSA-NLS. The GST-NTD was stained with a monoclonal antibody against GST and detected by indirect fluorescence microscopy. The nuclei were identified by Hoechst staining.
The nuclear export of NTD is not sensitive to LMB treatment.
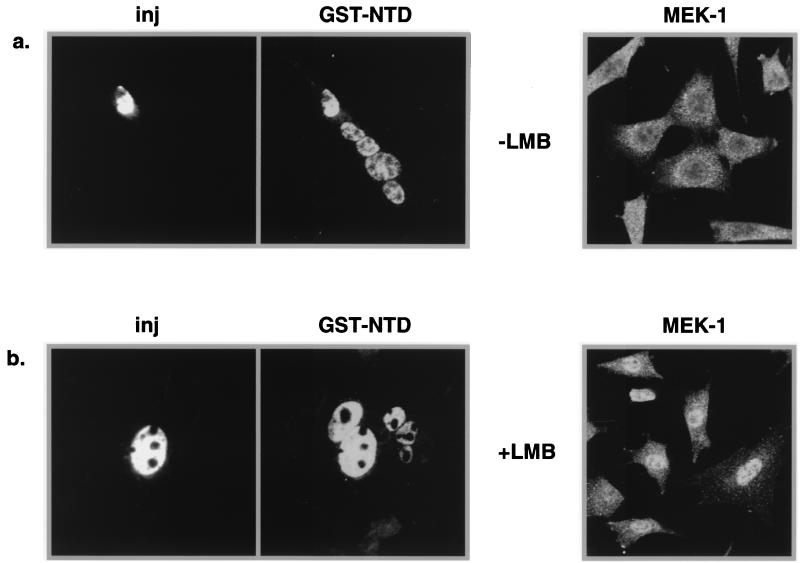
We tested if the nuclear export of helicase A NTD is sensitive to LMB, which disrupts the function of the leucine-rich NES. GST-NTD was injected into selected nuclei of NIH 3T3 cell polykaryons in the presence of 20 nM LMB, a concentration which is 10 times that used to inhibit Rev export (45). Normal shuttling of GST-NTD would be evidenced by its appearance in the uninjected nuclei within the polykaryon (Fig. 8a). The efficacy of LMB treatment on nuclear export in this study was controlled by analysis of the mitogen-activated protein kinase kinase (MEK-1) in parallel. MEK-1 is a shuttle protein with a steady-state cytoplasmic localization and contains a leucine-rich NES at the amino terminus (11). While LMB treatment relocalized MEK-1 to the nucleus by blocking the NES-mediated export as previously reported, it had no effect on GST-NTD export (Fig. 8b). This result indicates that the export mediated by NTD functions in a CRM1-independent manner.
FIG. 8.
NTD-mediated export is insensitive to LMB. (a) NIH 3T3 cells were grown on coverslips and treated with polyethylene glycol to form polykaryons. GST-NTD was then injected (inj) into selected nuclei of polykaryons along with the injection marker rhodamine-dextran. The cells were incubated at 37°C for 30 min before being fixed for immunofluorescence assay. (b) LMB (20 nM) was included to inhibit NES export. Cells stained for MEK-1 were similarly grown and treated with LMB, but no injections were performed.
DISCUSSION
RNA helicase A contains a sequence that specifies bidirectional nuclear-cytoplasmic trafficking.
ATP-dependent RNA helicase A was first purified from HeLa nuclear extracts as an in vitro RNA helicase activity (22). A subsequently described nuclear DNA helicase II (46) is the same protein. Helicase A contains two RNA-binding domains, a helicase core and an RGG box. Recent studies suggest that this protein may be involved in gene regulation at both the transcriptional and posttranscriptional levels (23, 29, 42). In spite of its predominantly nuclear localization (47), helicase A apparently shuttles between the nucleus and the cytoplasm (42). Here, we showed that a domain of 110 amino acids at the carboxyl terminus of helicase A is a bidirectional NTD. Helicase A with a deletion of this domain accumulated in the cytoplasm, in contrast to endogenous helicase A or GFP-helicase A fusion proteins that contain the NTD. NTD also targeted a completely cytoplasmic protein, pyruvate kinase, to the nucleus upon fusion to its carboxyl terminus. These observations suggest that NTD is both necessary and sufficient to mediate the nuclear localization of RNA helicase A. In addition, the NTD contains a nuclear export signal, since it was able to direct the rapid export of GST, GFP, a nuclear form of GFP (GFP-TNLS), and NPc-TNLS in a variety of assay systems. The import and export activities of NTD are separable by point mutations in two basic residues (K62 and R65), which abolished the ability of the NTD to direct nuclear import while sparing the export activity.
Nuclear import activity of the NTD.
Although no appreciable sequence homology was found between NTD and other NLSs, a short stretch of amino acids relatively rich in basic residues is found in NTD, the U1A signal, KNS, and the NLS of Sam68. An arginine-to-alanine substitution was shown to abolish the NLS function of Sam68 (16). While both K1162 and R1165 are crucial for the nuclear import function of NTD, R1158 is dispensable for this function. Interestingly, PK-ΔK62 and PK-ΔR65 mutants had a lower gel mobility than the nuclear counterparts (PK-NTD, PK-ΔR58), which had the expected mobility. It is possible that NTD is modified at the posttranslational level in the cytoplasm and that this putative modification is related to its NLS activity. We showed that this modification was not at the level of phosphorylation. Despite the lack of sequence resemblance to classical NLS, NTD apparently uses the same import pathway, since its nuclear import can be efficiently competed with an excess of BSA-conjugated NLS peptides.
Nuclear export activity of NTD.
Nuclear export activity of NTD was demonstrated in several different systems. First, NTD was able to direct the export of three different heterologous proteins in interspecies heterokaryon assays. GFP-NTD, GFP-TNLS-NTD, and NPc-TNLS-NTD all shuttled when tested, indicating the NTD can direct nuclear export even in the presence of a strong NLS. Second, export and reimport of GST-NTD were detected within 30 min after being injected into selected nuclei of NIH 3T3 polykaryons, demonstrating the rapid shuttling ability of NTD. Finally, a mutant form of NTD that lost its NLS activity directed nuclear export upon injection into the nucleus. This export function of NTD is not mediated by RNA binding. The NTD showed no ability to bind to the CTE in vitro, and coinjection of RNase had no effect on export function.
Helicase A and Rev use distinct nuclear export pathways.
Leucine-rich NES have been found in a variety of cellular and viral proteins (4, 7, 8, 15, 28, 37, 44). The human immunodeficiency virus Rev activation domain is a prototypic NES that is involved in the nuclear export of intron-containing human immunodeficiency virus mRNAs. It has been shown recently that CRM1, a cellular protein distantly related to importin β, serves as the functional export receptor for Rev NES. Competition experiments with Xenopus oocytes suggest that CTE and Rev-RRE utilize different cellular factors for RNA export (34, 38). Therefore, if helicase A plays a role in CTE-mediated export, it should function in a CRM1-independent manner. LMB disrupts the complex formation of NES, CRM1, and RanGTP by binding to CRM1, thus blocking Rev export. In functional studies, LMB blocked Rev-RRE-mediated but not CTE-mediated gene expression (33). We found that NTD-mediated export was resistant to 20 nM LMB, a concentration 10-fold higher than that previously observed to block Rev-dependent mRNA export (45), indicating that NTD does not use CRM1 as its export receptor. This observation is consistent with a possible role for helicase A in CTE-mediated export. Our recent observation that the injection of antisera specific for helicase A perturbs CTE function in human somatic cells further supports an important role of this protein in CTE function. The human protein TAP was recently also identified as a cellular CTE-binding protein that is involved in CTE-mediated export in Xenopus oocytes (13). The importance of TAP and that of helicase A in CTE function are not mutually exclusive. Rather, both proteins may be required to facilitate efficient RNA export. Alternatively, the two proteins may function at different steps in the CTE-mediated processing and export of unspliced RNA expressed by type D retroviruses. Further functional studies will help address the biological significance of helicase A in CTE-mediated RNA export.
Why simple and complex retroviruses have evolved different pathways to achieve the same function is not clear. By requiring the accumulation of an early viral protein, Rev, to direct the nuclear export of partially spliced and unspliced RNA, complex retroviruses are temporally regulated and may thus have an advantage in delaying immune system surveillance by the host and building up a larger burst size for viral production. Simple retroviruses are often less cytopathic and express lower levels of virus. For this type of chronic infection, a constitutive cellular export pathway seems to be sufficient. Although simple and complex retrovirus mRNAs apparently use distinct cellular receptors for nuclear export of unspliced mRNA, they may still have some common step(s) in their posttranscriptional regulation, e.g., in their interfacing with the splicing machinery to make unspliced RNA available for export. Our recent data (23) that helicase A may be involved in the Rev transactivation pathway at a step prior to nuclear export are consistent with such a hypothesis.
ACKNOWLEDGMENTS
We thank C. G. Lee and J. Hurwitz for the helicase A cDNA, Gideon Dreyfuss for Myc-PK and Myc-NPc-TNLS plasmids, B. Wolff for LMB, M. Vodika and M. Emerman for NLS peptides, T. R. Reddy for help with the constructions of pGST-NTD, W. D. Xu for help with site-directed mutagenesis, K. Kuhen for critical reading of the manuscript, and F. Gage and the James B. Pendleton Trust for the use of their microscopic facilities. We also thank C. Goodwin for his excellent technical assistance throughout the study.
This study was supported in part by NIH grant AI35477 to T. Hope and NIH grant GM56089 to F. Wong-Staal.
REFERENCES
- 1.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 4.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 5.Fischer U, Meyer S, Teufel M, Heckel C, Luhrmann R, Rautmann G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 7.Fridell R A, Benson R E, Hua J, Bogerd H P, Cullen B R. A nuclear role for the Fragile X mental retardation protein. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- 8.Fridell R A, Fischer U, Luhrmann R, Meyer B E, Meinkoth J L, Malim M H, Cullen B R. Amphibian transcription factor IIIA proteins contain a sequence element functionally equivalent to the nuclear export signal of human immunodeficiency virus type 1 Rev. Proc Natl Acad Sci USA. 1996;93:2936–2940. doi: 10.1073/pnas.93.7.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci 110. 1997;11:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 10.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda M, Gotoh I, Gotoh Y, Nishida E. Cytoplasmic localization of mitogen-activated protein kinase kinase directed by its NH2-terminal, leucine-rich short amino acid sequence, which acts as a nuclear export signal. J Biol Chem. 1996;271:20024–20028. doi: 10.1074/jbc.271.33.20024. [DOI] [PubMed] [Google Scholar]
- 12.Gorlich D, Mattaj I W. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 13.Gruter P, et al. TAP, the human homologue of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 14.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors: the Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 15.Hope T J. Viral RNA export. Chem Biol. 1997;4:335–344. doi: 10.1016/s1074-5521(97)90124-1. [DOI] [PubMed] [Google Scholar]
- 16.Ishidate T, Yoshihara S, Kawasaki Y, Roy B C, Toyoshima K, Akiyama T. Identification of a novel nuclear localization signal in Sam68. FEBS Lett. 1997;409:237–241. doi: 10.1016/s0014-5793(97)00455-9. [DOI] [PubMed] [Google Scholar]
- 17.Izaurralde E, Jarmolowski A, Beisel C, Mattaj I W, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kambach C, Mattaj I W. Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J Cell Biol. 1992;118:11–21. doi: 10.1083/jcb.118.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim F J, Beeche A A, Hunter J J, Chin D J, Hope T J. Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol Cell Biol. 1996;16:5147–5155. doi: 10.1128/mcb.16.9.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartman E, Gorlich D. Identification of tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 22.Lee C G, Hurwitz J. Human RNA helicase A is homologous to the maleless protein of Drosophila. J Biol Chem. 1993;268:16822–16830. [PubMed] [Google Scholar]
- 23.Li J, Tang H, Mullen T M, Westberg C, Reddy T R, Rose D W, Wong-Staal F. A role for RNA helicase A in post-transcriptional regulation of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malim M H, Cullen B R. HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell. 1991;65:241–248. doi: 10.1016/0092-8674(91)90158-u. [DOI] [PubMed] [Google Scholar]
- 25.Meyer B E, Malim M H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 26.Michael W M, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 27.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy R, Wente S R. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T, Uchida C, Anderson S F, Lee C G, Hurwitz J, Parvin J D, Montminy M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 30.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 31.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 32.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 33.Otero G C, Harris M E, Donello J E, Hope T J. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593–7597. doi: 10.1128/jvi.72.9.7593-7597.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. The constitutive transport element CTE of Mason-Pfizer monkey virus MPMV accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 36.Pollard V W, Michael W M, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 37.Richards S A, Lounsbury K M, Carey K L, Macara I G. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 39.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stade K, Ford C S, Guthrie C, Weis K. Exportin 1 Crm1p is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 41.Tang H, Xu Y, Wong-Staal F. Identification and purification of cellular proteins that specifically interact with the RNA constitutive transport elements from retrovirus D. Virology. 1997;228:333–339. doi: 10.1006/viro.1996.8407. [DOI] [PubMed] [Google Scholar]
- 42.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]
- 43.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 44.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 45.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S, Grosse F. Domain structure of human nuclear DNA helicase II RNA helicase A. J Biol Chem. 1997;272:11487–11494. doi: 10.1074/jbc.272.17.11487. [DOI] [PubMed] [Google Scholar]
- 47.Zhang S, Maacke H, Grosse F. Molecular cloning of the gene encoding nuclear DNA helicase II. A bovine homologue of human RNA helicase A and Drosophila Mle protein. J Biol Chem. 1995;270:16422–16427. doi: 10.1074/jbc.270.27.16422. [DOI] [PubMed] [Google Scholar]
- 48.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]