Abstract
The 3′ end of histone mRNA is formed by an endonucleolytic cleavage of the primary transcript after a conserved stem-loop sequence. The cleavage reaction requires at least two trans-acting factors: the stem-loop binding protein (SLBP), which binds the stem-loop sequence, and the U7 snRNP that interacts with a sequence downstream from the cleavage site. Removal of SLBP from a nuclear extract abolishes 3′-end processing, and the addition of recombinant SLBP restores processing activity of the depleted extract. To determine the regions of human SLBP necessary for 3′ processing, various deletion mutants of the protein were tested for their ability to complement the SLBP-depleted extract. The entire N-terminal domain and the majority of the C-terminal domain of human SLBP are dispensable for processing. The minimal protein that efficiently supports cleavage of histone pre-mRNA consists of 93 amino acids containing the 73-amino-acid RNA-binding domain and 20 amino acids located immediately next to its C terminus. Replacement of these 20 residues with an unrelated sequence in the context of the full-length SLBP reduces processing >90%. Coimmunoprecipitation experiments with the anti-SLBP antibody demonstrated that SLBP and U7 snRNP form a stable complex only in the presence of pre-mRNA substrates containing a properly positioned U7 snRNP binding site. One role of SLBP is to stabilize the interaction of the histone pre-mRNA with U7 snRNP.
Unlike all other metazoan mRNAs, the mature 3′ ends of the replication-dependent histone mRNAs do not have poly(A) tails but are formed instead by an endonucleolytic cleavage of primary transcripts (pre-mRNAs) downstream from a highly conserved stem-loop sequence (11). This single-step processing reaction depends on two cis elements present in histone pre-mRNAs: the stem-loop sequence itself and a purine-rich sequence located 11 to 12 nucleotides downstream of the cleavage site and referred to as the histone downstream element (HDE) (26). The mature 3′ end of histone mRNA consists of a 26-nucleotide sequence including a 6-nucleotide stem and a 4-nucleotide loop (21) and is recognized by a protein termed the stem-loop binding protein (SLBP) (29, 45) or the hairpin binding protein (HBP) (20). Following the cleavage reaction, SLBP remains associated with the 3′ end of mature histone mRNAs (6, 15). SLBP probably participates in all critical events in histone mRNA metabolism, including nucleo-cytoplasmic transport (7, 46), translation (9, 40), and mRNA degradation (28).
The HDE interacts with the U7 small nuclear ribonucleoprotein (U7 snRNP), a low-abundance particle present in about 104 copies per mammalian nucleus, ∼1 to 3% of the amount of the major spliceosomal snRNPs (5, 14, 27). The U7 snRNP contains one molecule of the 63-nucleotide U7 snRNA and both the common Sm core and the U7 snRNP-specific proteins (34, 37). Genetic suppression experiments have revealed that binding of the U7 snRNP to the histone pre-mRNA is mediated at least in part through base pairing between the 5′ end of the U7 snRNA and the HDE (2, 39). The stem-loop sequence and the SLBP are absolutely required for 3′-end processing in vivo (30), but neither is essential for processing in vitro if the HDE has sufficient complementarity to U7 snRNA (38). Cleavage of the histone pre-mRNA occurs between the two cis elements in a favorable sequence context containing predominantly adenosine and cytidine residues (8) and requires a third factor, termed the heat-labile factor (HLF; 12). The heat labile factor, like U7 snRNP, is indispensable for the 3′-end processing and has been primarily characterized by its sensitivity to a mild heat treatment. Recent studies have shown that SLBP and the U7 snRNP, and possibly other components of the histone pre-mRNA 3′ processing machinery, are found colocalized in spheres, the equivalent of coiled bodies in Xenopus oocytes (1, 48).
Efficient 3′-end processing of the histone pre-mRNA depends on proper juxtaposition of the stem-loop sequence and the HDE. If the distance between the two cis-acting elements is too small (3) or too large (10, 31, 32), processing is abolished. Moving the HDE only several nucleotides 3′ of its normal position, resulting in a relatively small increase in the spacing between the stem-loop sequence and the HDE, leads to both a moderate inhibition of processing and a shift of the cleavage site by a comparable number of nucleotides (31). Scharl and Steitz have proposed that U7 snRNP acts as a molecular ruler, a measuring device which places the cleavage site at a fixed distance from the site of U7 snRNP binding to the histone pre-mRNA (31, 32).
SLBP from several species has recently been cloned by using the three-hybrid system (20, 45) designed for screening RNA binding proteins in the yeast Saccharomyces cerevisiae (33). This 31-kDa protein contains a centrally located RNA binding domain with no similarity to any known motifs that bind RNA (45). Thus far, SLBP is the only protein component of the histone pre-mRNA 3′ processing machinery available in large quantities for biochemical analysis. Here we describe experiments aimed at understanding the role of SLBP in the processing reaction and determining the minimal regions of the protein required for cleavage of the histone pre-mRNA. We show that a 93-amino-acid fragment retains most of the processing activity of SLBP. One role of SLBP is to stabilize the interaction of the U7 snRNP with the histone pre-mRNA, thereby increasing the efficiency of the cleavage reaction.
MATERIALS AND METHODS
Construction of the pre-mRNA substrates.
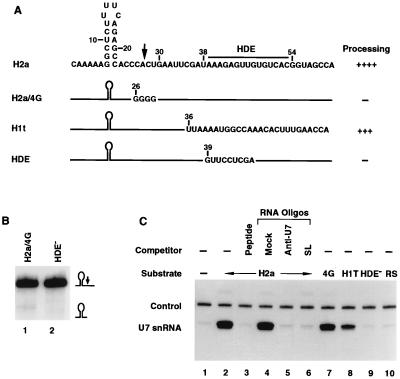
The H2a-614 pre-mRNA was generated by subcloning a 59-nucleotide fragment of the mouse histone H2a-614 gene encompassing the stem-loop structure, the cleavage site, and the HDE between the KpnI and HindIII sites of the pGEM3 vector (Promega). To facilitate construction of H2a derivatives, an EcoRI site was inserted between the cleavage site and the HDE by alteration of two nonconserved nucleotides. The H1t and HDE− clones were generated by replacement of the HDE in the H2a clone with appropriate sequences (see Fig. 5A) with double-stranded oligonucleotides which were inserted between the EcoRI and HindIII sites. The 4G clone and the clones with mutant stem-loops were constructed by subcloning the appropriate double-stranded oligonucleotides between the EcoRI and KpnI sites. The U7 snRNA clone was constructed by inserting a double-stranded oligonucleotide encoding the 63-nucleotide mouse U7 snRNA (35) between the EcoRI and HindIII sites of the pGEM3 vector. Clones encoding various HDEs were generated by inserting the EcoRI-HindIII restriction fragment of H2a, H1t, or HDE− constructs into EcoRI and HindIII sites of the pGEM3 vector. Clones encoding truncated versions of the SLBP were generated by deleting appropriate regions from the human SLBP gene by using restriction sites previously utilized to map the RNA binding domain (see Fig. 4A) (45). To construct the SLBP/20aa clone, the BamHI-PstI fragment in human SLBP cDNA was replaced by the PCR-amplified fragment encoding amino acids 174 to 193 of Xenopus SLBP2 (43).
FIG. 5.
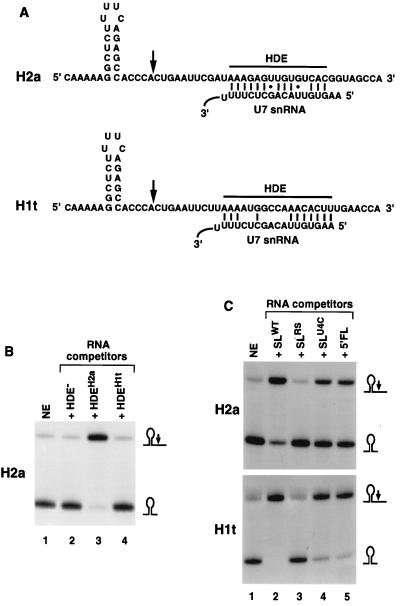
Formation of a stable complex containing the SLBP and the U7 snRNP on the pre-mRNA substrate. (A) Sequences of the substrates used for assaying U7 snRNP binding. The sequence of the parental H2a pre-mRNA comprising the stem-loop structure, the cleavage site represented by an arrow, and the U7 binding site are shown at the top. The nucleotides at the 5′ and 3′ ends of the RNA encoded in the pGEM3 vector are not included. The nucleotide substitutions introduced into the H2a pre-mRNA in order to generate the other pre-mRNA substrates are shown below the H2a sequence. The unchanged sequences are represented by solid lines. The RS mutant of the H2a-614 pre-mRNA contains reversed sequence of the stem-loop structure, as shown in Fig. 1B. (B) The H2a/4G (lane 1) and HDE− (lane 2) substrates shown in panel A were incubated in a nuclear extract under standard processing conditions. The RNA was analyzed as described in the legend to Fig. 2. (C) The H2a-614 substrate (lanes 2 to 6) and the indicated mutant substrates (lanes 7 to 10) were briefly incubated in nuclear extract to allow the formation of processing complexes. The complexes were immunoprecipitated with anti-SLBP antibody, RNA prepared, resolved by electrophoresis on 8.5% polyacrylamide gel containing 7 M urea, and assayed for the presence of U7 snRNA by Northern blotting. Immunoprecipitation was carried out in the presence of the H2a-614 pre-mRNA (lanes 2 to 6) or mutant pre-mRNAs, as indicated (lanes 7 to 10). Lane 1, no substrate added; lane 3, 10 μg of antigenic peptide was added to the reaction mixture prior to addition of the antibody; lanes 4 and 5, immunoprecipitation in the presence of 0.5 μg of a nonspecific 2′-O-methyl oligoribonucleotide or 2′-O-methyl oligoribonucleotide complementary to the 5′ end of U7 snRNA, respectively. Lane 6, immunoprecipitation in the presence of a 100-fold excess of the 30-nucleotide RNA containing the stem-loop sequence. Control, 0.1 ng of a synthetic 77-nucleotide RNA containing the complete sequence of the U7 snRNA added to each sample as an internal standard for RNA recovery and hybridization efficiency.
FIG. 4.
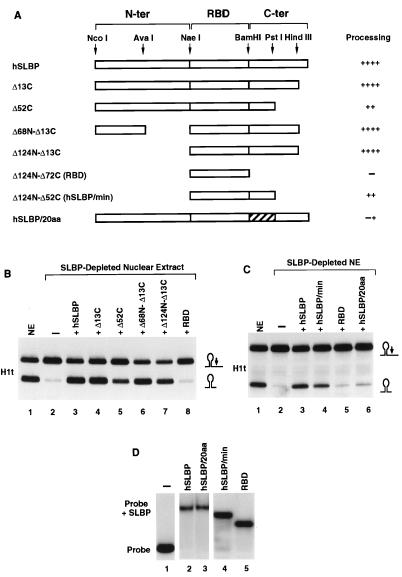
Regions of human SLBP required for histone pre-mRNA processing. (A) The restriction map of the human SLBP cDNA with the boundaries of the three regions of the protein corresponding to the N-terminal domain (N-ter), the RNA binding domain (RBD), and the C-terminal domain (C-ter) is outlined at the top. The constructs are named according to the number of amino acids deleted from the N or C terminus. The hSLBP/20aa has the 20 amino acids immediately after the RNA binding domain (196 to 215) changed to the 20 amino acids present at this position in frog XSLBP2, an SLBP which does not function in pre-mRNA processing (43). The ability of the mutant SLBP to restore processing in the SLBP-depleted extract is summarized to the right of each construct. (B) The nuclear extract (lane 1) was depleted of SLBP with anti-SLBP antibody and was assayed for the ability to cleave the H1t pre-mRNA when complemented with 50 ng of the indicated mutant human SLBP. Depletion of the extract resulted in an almost complete loss of processing activity (lane 2) which was fully restored when the depleted extract was supplemented with the full-length human SLBP (lane 3). Complementation of the depleted extract with the mutant proteins is shown in lanes 4 to 8. (C) Processing activity of the nuclear extract (NE; differing from that in Fig. 1B) before and after immunodepletion is shown in lanes 1 and 2, respectively. Fifty nanograms of human SLBP (lane 3) or mutant proteins was added to the depleted extract, as indicated (lanes 4 to 6). (D) The recombinant proteins were tested for their ability to bind to the stem-loop RNA oligonucleotide. Fifty nanograms of the recombinant protein indicated at the top of each lane was incubated with the radiolabeled stem-loop RNA, and the complexes were resolved by native gel electrophoresis.
Preparation of RNA.
The RNA was synthesized by either T7 or SP6 RNA polymerase with DNA templates linearized with either HindIII or EcoRI restriction enzymes, respectively. The transcription reaction was carried out in the absence of radioisotopes in a final volume of 150 μl according to a standard protocol. The RNA was subsequently gel purified to remove pretermination products. The 5′ phosphates of the RNA were removed by treatment with the calf intestinal phosphatase (Boehringer Mannheim), and 50 ng of dephosphorylated RNA was subsequently radiolabeled with 30 μCi of [γ-32P]ATP and 5 U of T4 polynucleotide kinase (New England Biolabs). The RNA was separated from the unincorporated ATP by G-50 spin columns (Pharmacia) and used without additional purification. The in vitro processing reactions were carried out in the presence of 86-nucleotide pre-mRNA substrates. Each substrate contained 59 nucleotides encompassing all cis elements required for processing and 22 and 5 nucleotides of pGEM3 vector (Promega) on 5′ and 3′ RNA flanks, respectively. The U7 snRNA was detected on Northern blots by hybridization with radiolabeled RNA containing a 63-nucleotide region complementary to the U7 snRNA and 13 and 5 nucleotides of vector sequences on the 5′ and 3′ ends, respectively. This RNA was synthesized in the antisense orientation from the U7 snRNA construct in the presence of 10 μM unlabeled CTP and 50 μCi of [α-32P]CTP. The remaining nucleoside triphosphates were used at a 500 μM concentration. The unlabeled RNA containing the U7 snRNA sequence used as an internal control for the Northern blots was synthesized from the U7 snRNA construct in the sense orientation. The reaction was carried out in the absence of radioisotopes in a final volume of 150 μl, and the RNA was purified as described above. The stem-loop RNA oligonucleotides used in band shift assays and competition experiments were synthesized by T7 RNA polymerase with the appropriate oligonucleotide templates (24).
Nuclear extract preparation and in vitro processing.
Nuclear extracts were prepared from mouse myeloma cells as described previously (6, 22). Each processing reaction was carried out in a total volume of 10 μl containing 5 μl of the nuclear extract, 5 ng of the pre-mRNA substrate labeled at the 5′ end with [γ-32P]ATP, and 20 mM EDTA. Samples were incubated at 32°C for 60 min, and the RNA was isolated and analyzed as previously described (6, 22). Since the efficiency of in vitro processing reactions varied significantly, depending on the preparation of the nuclear extract and the batch of the pre-mRNA substrate, each experiment was accompanied by appropriate controls. Whenever possible, a complete set of experiments was carried out with the same preparation of the nuclear extract.
Expression and purification of the SLBP.
Wild-type and mutant forms of the SLBP were expressed in Sf9 insect cells with the baculovirus expression system (Gibco BRL), as recommended by the manufacturer. For preparative purification, 200 ml of the infected cell culture was grown for 72 h, and the SLBP was purified by chromatography on Ni-agarose, as recommended. The typical yield was 200 to 500 μg of the pure protein.
Preparation of the SLBP-depleted extract and complementation of the in vitro processing reaction.
The endogenous SLBP was removed from the nuclear extract with either the biotinylated RNA containing the stem-loop structure, as previously described (6), or anti-SLBP antibody, essentially as previously described (22, 45). Each preparation of the depleted extract was tested for processing activity both alone and in the presence of the full-length human recombinant SLBP. To complement the in vitro processing reaction, approximately 20 to 50 ng of baculovirus-expressed SLBP was added to 5 μl of the depleted extract.
Mobility shift assay.
The nuclear extract (2.5 μl; 12.5 μg of protein) or pure SLBP expressed in Sf9 insect cells (50 ng) was mixed on ice with 1 ng of the 5′ labeled 30-nucleotide stem-loop RNA and immediately applied to 6 to 8% native polyacrylamide gels containing 1× Tris-borate–EDTA buffer. The complexes were resolved by electrophoresis and detected by autoradiography.
Western blots.
Proteins from nuclear extracts were separated on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels and transferred to nitrocellulose filters. The SLBP was detected with antibody against the C-terminal 13 amino acids of the protein (45) by using the ECL system (Amersham).
Immunoprecipitation of processing complexes and detection of the U7 snRNA.
An equivalent of 10 individual processing reaction mixtures, containing 50 ng of unlabeled RNA substrate, 50 μl of the nuclear extract, and 20 mM EDTA, was set up on ice in a total volume of 100 μl and incubated for 5 min at 22°C to allow complex formation. Ten microliters of anti-SLBP antibody purified on protein A agarose was added, and the samples were rotated at 4°C for 1.5 h and then transferred to a new tube containing 20 μl of protein A agarose beads. The protein A agarose beads were preincubated in a nuclear extract from sea urchin blastula nuclei (17) and washed with buffer to reduce nonspecific binding. The samples were rotated for 1.5 h, and the beads were collected, rinsed twice with buffer D (20 mM HEPES-KOH [pH 7.9], 100 mM KCl, 0.5 mM dithiothreitol, 0.2 mM EDTA [pH 8.0], 20% glycerol) containing 0.1% Nonidet P-40 detergent, suspended in 1 ml of the same buffer, and rotated at 4°C for an additional 1.5 h. The protein A agarose beads were suspended in 100 μl of 0.3 M sodium acetate containing 10 μg of glycogen and 0.1 ng of the in vitro-synthesized U7 snRNA, and the mixture was extracted with phenol. The RNA was recovered by precipitation with ethanol, resolved on an 8.5% polyacrylamide–7 M urea denaturing gel, and transferred to a HyBond-N+ membrane (Amersham) by using the Genie electrophoretic blotter (Idea Scientific). The blot was irradiated with UV light in a Stratalinker apparatus (Stratagene), preincubated in Quikhyb solution (Stratagene) for 1 h at 62°C, and then hybridized overnight at the same temperature with antisense U7 snRNA probe (105 cpm). Following several washes in 0.015 M NaCl, 0.0015 M Na3 citrate, and 0.1% SDS at 62°C, the U7 snRNA was detected by autoradiography.
RESULTS
Importance of high-affinity binding of SLBP in histone pre-mRNA processing.
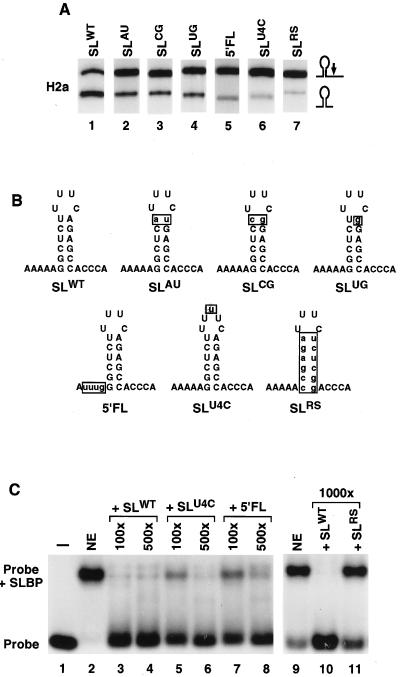
SLBP binds with high affinity to a conserved 26-nucleotide sequence immediately preceding the 3′ cleavage site in nonpolyadenylated histone pre-mRNAs. This sequence consists of a 16-nucleotide stem-loop structure flanked on either side by five conserved nucleotides (21). As shown by mobility shift assays and competition experiments, mutations in any region of this sequence affect its binding to SLBP (47). In vitro processing of commonly used histone pre-mRNA substrates, including the H2a-614 pre-mRNA, is not absolutely dependent on SLBP and the stem-loop sequence (6, 25, 38). When binding of SLBP to the H2a-614 substrate is prevented by removing the protein from the extract or by using a high molar excess of a 30-nucleotide competitor RNA containing the wild-type stem-loop sequence, the processing reaction still proceeds with 5 to 10% efficiency (6, 45). A similar reduction in processing efficiency is achieved by reversing the sequence of the stem in the H2a-614 pre-mRNA (RS; Fig. 1A, lane 7). The RS mutation virtually abolishes binding of SLBP to the stem-loop sequence as demonstrated by the inability of the mutant RNA oligonucleotide to form any detectable complex in a mobility shift assay and to effectively compete with the wild-type sequence for SLBP binding (47; Fig. 1C). In order to determine the importance of high-affinity binding of SLBP to the pre-mRNA in 3′-end processing, the stem-loop sequence of H2a pre-mRNA was altered by mutations having a less drastic effect on SLBP binding than the RS mutation. The mutant pre-mRNA substrates were subsequently tested in the in vitro processing assay with a nuclear extract from mouse myeloma cells. Base substitutions altering the conserved base pair at the top of the stem from UA to AU, CG, or UG result in 5- to 10-fold-lower affinity of SLBP for the stem-loop sequence (47). These mutations decreased processing efficiency from approximately 60% for the wild-type substrate (Fig. 1A, lane 1) to less than 20% (Fig. 1A, lanes 2 to 4). A much stronger effect on the in vitro processing was exerted by a 1-nucleotide expansion of the loop (U4C; Fig. 1A, lane 6) or alteration of the region 5′ to the stem-loop (5′FL; Fig. 1A, lane 5). These two mutations reduced cleavage efficiency to a level comparable with the RS mutation (5 to 10%). Interestingly, while the RNA oligonucleotide containing the RS mutation had very low affinity for SLBP (Fig. 1C, lanes 9 and 10), the U4C and 5′FL RNAs at high concentrations were able to compete binding of SLBP to the wild-type RNA (Fig. 1C, lanes 5 to 8). Binding of the wild-type sequence to SLBP was reduced at least 90% with a 100-fold excess of these RNAs and abolished with a 500-fold molar excess of these RNAs (Fig. 1C, lanes 5 and 8, respectively). The RS mutant RNA was inactive even at a 1,000-fold excess (Fig. 1C, lane 11) (47), while the wild-type competitor completely abolished binding of radiolabeled probe to SLBP at an excess of 20-fold (data not shown) and greater (Fig. 1C, lanes 3 and 4) (6, 47). These results indicate that high affinity of SLBP for the histone pre-mRNA substrate is required to effectively support 3′ processing in vitro as it does in vivo (30).
FIG. 1.
Effect of altering the affinity of the pre-mRNA for SLBP. (A) Each of the six mutant stem-loops shown was introduced into the mouse histone H2a-614 pre-mRNA. The radiolabeled synthetic pre-mRNAs were incubated for 30 min in a nuclear extract prepared from mouse myeloma cells, as described in Materials and Methods. The RNA was purified, resolved by gel electrophoresis, and detected by autoradiography. The input pre-mRNA (top band) and the shorter cleavage product (bottom band) are indicated. (B) Sequence of 26 nucleotides encompassing the stem-loop structure in the various mutants. (C) Thirty-nucleotide RNAs were synthesized by T7 RNA polymerase with the appropriate oligonucleotide templates. Each RNA consists of 26 nucleotides encompassing the stem-loop structure shown and the GCCC sequence at the 5′ end facilitating synthesis of RNA by T7 polymerase. The 30-nucleotide RNA containing the wild-type stem-loop structure was labeled at the 5′ end with [γ-32P]ATP and used to detect SLBP in the nuclear extract (NE) by using a mobility shift assay. The samples were analyzed on a 7% polyacrylamide gel under nondenaturing conditions. Unlabeled 30-nucleotide RNA competitors containing the wild-type or mutant stem-loop sequences were added to the reaction at a molar excess, as indicated above each lane. Lanes 2 and 9, no competitor added; lane 1, probe.
Construction of a processing substrate absolutely dependent on SLBP.
The requirement for SLBP in the processing reaction varies among different substrates and depends on the extent of complementarity between the U7 snRNA and the HDE (38). Substrates containing very strong HDEs, e.g., the mouse H4-12 pre-mRNA, are actually cleaved with normal efficiency in some nuclear extracts in which SLBP is sequestered by the wild-type stem-loop RNA oligonucleotide (38). Under the same conditions, the cleavage efficiency of the H2a-614 pre-mRNA containing a weaker HDE declines to 5 to 10% of the control level (6) (Fig. 2). Based on these observations it has been proposed that SLBP functions in processing by stabilizing the interaction of the U7 snRNP with the HDE in the histone pre-mRNA (38).
FIG. 2.
Comparison of H2a-614 and H1t pre-mRNAs as processing substrates. (A) The sequences of the H2a and H1t pre-mRNAs encompassing the stem-loop and HDE are shown. The sequence of the 5′ end of the U7 snRNA and its potential to form base pairs with the HDE are depicted below each pre-mRNA substrate. Watson-Crick base pairs are indicated by vertical lines, and GU base pairs are indicated by dots. (B) The 86-nucleotide H2a-614 pre-mRNA substrate was incubated in a nuclear extract (NE) for 60 min under processing conditions (lane 1). RNA oligonucleotides containing the sequence of either the mutant HDE (HDE−) (see Fig. 5A), the H2a-614 HDE, or the H1t HDE were added to the reaction samples at a 100-fold molar excess relative to the substrate (lanes 2, 3, and 4, respectively). The RNA was purified, resolved by gel electrophoresis, and detected by autoradiography. (C) The in vitro processing reaction was carried out with either the H2a-614 (top panel) or H1t (bottom panel) pre-mRNAs under standard conditions (lane 1) or in the presence of a 100-fold molar excess of the competing 30-nucleotide RNAs shown in Fig. 1B, as indicated above each lane (lanes 2 to 5).
Since the presence of SLBP-independent processing will obscure understanding of the role played by SLBP in the cleavage reaction, we constructed a substrate fully dependent on SLBP. Histone pre-mRNA encoded by the histone H1 gene specifically expressed in testis (H1t) is unique among natural processing substrates in that its HDE substantially departs from the consensus sequence and displays only limited complementarity to the U7 snRNA (44) (Fig. 2A). The HDE in the histone H2a-614 pre-mRNA can form a nearly perfect duplex with U7 snRNA, consisting of 14 bp (including 2 GU pairs), interrupted by only one mismatch. The HDE in the H1t pre-mRNA can form only 11 bp separated by several noncomplementary nucleotides (Fig. 2A). Moreover, the H1t HDE has relatively weak complementarity to the CUCUUU sequence (nucleotides 12 to 17 of the U7 snRNA) which base pairs with the core of the HDE in the somatic histone pre-mRNAs (2) and instead has the potential to base pair with the extreme 5′ end of the U7 snRNA. Additional base pairs with the pre-mRNA substrate and this region of the U7 snRNP have been reported previously for other histone pre-mRNAs (36). We reasoned that both the decreased number and more sparse distribution of base pairs formed between the H1t pre-mRNA and the U7 snRNA should result in a relatively weak interaction between the two RNAs. To demonstrate that the H1t HDE has a lower affinity for the U7 snRNP than the H2a-614 HDE, we synthesized RNA competitors containing either the H2a-614 or H1t HDE. Each RNA competitor was added to the in vitro processing reaction at a 100-fold molar excess over the H2a-614 pre-mRNA substrate and tested for its ability to affect efficiency of the cleavage. About 90% of the H2a-614 pre-mRNA was processed in the absence of any RNA competitor (Fig. 2B, lane 1). The cleavage reaction proceeded with similar efficiency in the presence of a 100-fold molar excess of a control RNA competitor with no complementarity to the U7 snRNA (Fig. 2B, lane 2). Competitor RNA containing the H2a-614 HDE virtually abolished processing (>95% reduction of the initial efficiency; Fig. 2B, lane 3), presumably by binding to the 5′ end of the U7 snRNA and inactivating the U7 snRNP in the nuclear extract. The RNA oligonucleotide containing the H1t HDE at the same concentration had no effect (Fig. 2B, lane 4). These results show that the extent of base pairing between the H1t HDE and the 5′ end of the U7 snRNA is not sufficient to allow stable binding of the U7 snRNP to this competitor RNA.
The 86-nucleotide histone H1t pre-mRNA substrate was constructed by substituting the HDE in the histone H2a-614 pre-mRNA with the corresponding element from the H1t pre-mRNA. The H1t pre-mRNA was processed in vitro at least 50% as efficiently as the H2a-614 substrate. To compare the dependence of processing of the H1t pre-mRNA on SLBP, we carried out the reaction in the presence of competitor RNA capable of titrating out SLBP from the nuclear extract. A 100-fold molar excess of 30-nucleotide RNA containing the wild-type stem-loop structure prevents binding of SLBP to the pre-mRNA substrate and causes substantial reduction, but not complete inhibition, of the processing of the H2a-614 pre-mRNA (6) (Fig. 2C, top panel, lane 2). Processing of the histone H1t pre-mRNA substrate is abolished under the same conditions (Fig. 2C, bottom panel, lane 2). Complete dependence of H1t pre-mRNA processing on SLBP was further confirmed by using RNA competitors that have lower affinity for SLBP due to either a uridine insertion in the loop (SLU4C, Fig. 1B) or placing a UUUG sequence 5′ to the stem-loop (5′ FL; Fig. 1B). Each competitor at a 100-fold molar excess almost completely inhibited cleavage of the H1t pre-mRNA (Fig. 2C, bottom panel, lanes 4 and 5) but had only a moderate effect on processing of the H2a substrate (Fig. 2C, top panel, lanes 4 and 5). At the same concentrations, these competitors reduced binding of SLBP to the wild-type stem-loop probe more than 95% as determined by the mobility shift assay (Fig. 1C). Addition of the RS RNA competitor, which has the stem sequence reversed (Fig. 1B) and at least a 100-fold-lower affinity for SLBP (47), did not affect processing of either substrate (Fig. 2C, lane 3, both panels). The differential effect of the U4C and 5′FL oligonucleotides on processing of the two substrates is discussed below (see Discussion). We conclude that SLBP is an essential and indispensable trans-acting factor in the in vitro 3′-end processing of the H1t pre-mRNA. This substrate was used in the majority of the subsequent experiments.
Complementation of the SLBP-depleted extract with recombinant baculovirus SLBP.
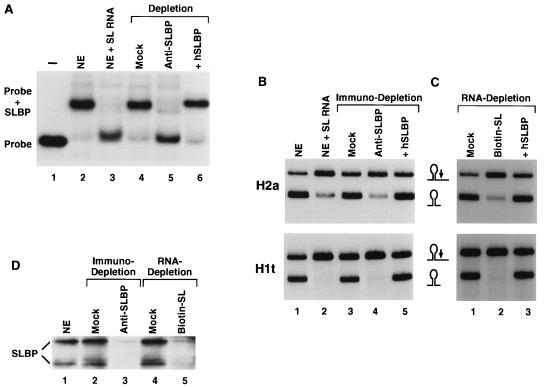
We have previously shown that depleting the SLBP from the nuclear extract with anti-SLBP antibody has an effect similar to that caused by the addition of an excess of the stem-loop RNA to the reaction and results in a substantial decrease in the processing of the histone H2a-614 pre-mRNA (45). To definitively show that removal of SLBP is solely responsible for the reduced efficiency of the reaction, we developed a complementation assay in which the depleted extract was supplemented with recombinant SLBP. Both the H2a-614 and the H1t pre-mRNAs were used as substrates to assess processing activity of the reconstituted nuclear extract. With an antibody to the 13 C-terminal amino acids of SLBP (45), more than 95% of SLBP was removed from the extract, as determined both by mobility shift assay (Fig. 3A, lane 5) and by Western blotting (Fig. 3D, lane 3). Depletion of SLBP resulted in a decrease in processing efficiency of the histone H2a pre-mRNA substrate from approximately 90 to about 10% (Fig. 3B, top panel, lane 4) and completely abolished processing of the H1t substrate (Fig. 3B, bottom panel, lane 4). An identical reduction in processing efficiency was caused by sequestering SLBP by addition of an excess of wild-type SL RNA competitor (Fig. 3B, lane 2). Mock depletion carried out with preimmune serum caused irreversible loss of some (about 25%) of the processing activity (Fig. 3B, lane 3) but did not significantly affect the amount of SLBP, as demonstrated by the mobility shift assay (Fig. 3A, lane 4). This loss of activity is most likely a result of prolonged incubation at 4°C or of nonspecific adsorption of processing factors to the protein A beads.
FIG. 3.
Depletion of SLBP from the nuclear extract and complementation with recombinant SLBP. (A) The 30-nucleotide radiolabeled wild-type stem-loop RNA (lane 1) was incubated in a nuclear extract (NE), and the complex was resolved by gel electrophoresis (lane 2). Lane 3, nuclear extract plus a 100-fold excess of unlabeled competitor 30-nucleotide stem-loop RNA; lane 4, extract treated with preimmune serum (mock depletion); lane 5, extract depleted with anti-SLBP antibody; lane 6, anti-SLBP-depleted extract supplemented with 50 ng of recombinant human SLBP. (B) The same extracts as those in panel A were used in processing reactions with the histone H2a-614 pre-mRNA (top panel) or the H1t pre-mRNA substrates (bottom panel). The RNAs were purified, resolved by electrophoresis in a denaturing polyacrylamide gel (8%; 7 M urea), and detected by autoradiography. Processing in the nuclear extract (NE) under standard conditions and in the presence of a 100-fold excess of 30-nucleotide competitor RNA with wild-type stem-loop sequence is shown in lanes 1 and 2, respectively. Lane 3, mock-depleted extract (preimmune serum); lane 4, extract depleted with anti-SLBP antibody; lane 5, anti-SLBP-depleted extract plus recombinant human SLBP. (C) Processing of the histone H2a-614 pre-mRNA (top panel) or the H1t pre-mRNA (bottom panel) in the nuclear extract depleted of SLBP with biotinylated RNA oligonucleotide containing the wild-type stem-loop. Lane 1, mock-depleted extract (nonspecific biotinylated oligonucleotide); lane 2, extract depleted with biotinylated stem-loop RNA; lane 3, depleted extract complemented with 50 ng of recombinant human SLBP. (D) Equal amounts (50 μg of protein) of the nuclear extracts (NE) used in panels B and C were resolved by electrophoresis on a 12% polyacrylamide–SDS gel, and SLBP was detected by Western blotting. The top band represents the intact 45-kDa SLBP, and the bottom band is a proteolytic cleavage product lacking part of the N terminus of the protein.
Recombinant human SLBP containing a histidine tag at the N terminus was expressed in Sf9 insect cells by using the baculovirus system and purified by Ni affinity chromatography. Complementation of the SLBP-depleted extract with the exogenous protein restored both SLBP binding activity (Fig. 3A, lane 6) and processing of the H2a-614 and H1t pre-mRNA substrates (Fig. 3B, lane 5, both panels). Addition of the SLBP to a complete extract resulted only in a slight stimulation of processing, suggesting that SLBP is not the limiting component of the processing extract (data not shown).
SLBP was also removed from the nuclear extract via binding to a biotinylated 30-nucleotide RNA containing the wild-type stem-loop sequence followed by adsorption of RNA-protein complexes to streptavidin beads (6, 20). Unlike immunodepletion, RNA-mediated depletion may result in removal of factors other than SLBP that bind to the stem-loop sequence. On the other hand, this approach would not lead to removal of inactive SLBP molecules unable to bind RNA. This procedure was equally effective in depleting SLBP from the nuclear extract, as assayed by mobility shift (6) and Western blotting (Fig. 3D, lane 5) and resulted in the same reduction of processing efficiency as immunodepletion (Fig. 3C, lane 2, both panels). Processing of both the H2a and the H1t pre-mRNA substrates was restored to the initial level by addition of the recombinant human SLBP to the depleted extract (Fig. 3C, lane 5). This result demonstrates that no other essential processing factors are quantitatively bound to the SLBP-RNA complex and removed from the extract during RNA-mediated SLBP depletion.
Mapping of the regions of SLBP involved in histone pre-mRNA processing.
In order to determine which regions of human SLBP participate in processing of the histone pre-mRNA, truncated versions of SLBP were expressed in the baculovirus system and tested for their ability to complement immunodepleted nuclear extract. All deletions were made outside the central region of SLBP encompassing the RNA binding domain (Fig. 4A). As determined by mobility shift assays, all the mutant proteins bound the stem-loop RNA (data not shown) (45). Complementation experiments were carried out in the immunodepleted nuclear extract by using the H1t pre-mRNA as a processing substrate. Due to a small amount of SLBP left in this depleted extract, processing of the H1t substrate was not completely abolished but proceeded with about 5% efficiency (Fig. 4B, lane 2).
The baculovirus-expressed deletion proteins Δ13C, Δ68N-Δ13C, and Δ124N-Δ13C restored activity of the depleted extract to control levels (Fig. 4B, lanes 4, 6, and 7, respectively), indicating that the last 13 amino acids of the protein and the entire 124 amino acids of the N-terminal domain are not necessary for processing. The Δ52C mutant protein, which contains only 20 amino acids after the RNA binding domain, restored processing activity to 40% of the control level (Fig. 4B, lane 5). Since deletion of the 13 C-terminal amino acids did not affect the activity of SLBP, we conclude that the remaining 39 amino acids (amino acids 218 to 256) play a role in enhancing the processing efficiency. A recombinant fragment of SLBP containing only the RNA binding domain did not have any activity in processing (Fig. 4B, lane 8; Fig. 4C, lane 5), although it bound the stem-loop RNA with high affinity (Fig. 4D, lane 5).
Deletion mutagenesis suggested that a 93-amino-acid fragment of SLBP, containing only the RNA binding domain and subsequent 20 amino acids, has processing activity. This protein (hSLBP/min) was expressed in baculovirus and tested for its ability to complement the immunodepleted extract. The hSLBP/min restored processing to 40% of the control level (Fig. 4C, lane 4), similar to the activity of the Δ52C deletion protein (Fig. 4B, lane 5). To definitively confirm the importance of the 20 amino acids after the RNA binding domain, we replaced this region in the intact SLBP with an unrelated sequence. We used the sequence found adjacent to the RNA binding domain of the Xenopus SLBP2, a protein that does not function in histone pre-mRNA processing (43). This protein, hSLBP/20aa, bound strongly to the stem-loop (Fig. 4D, lane 3) but retained only slight activity in stimulating the cleavage reaction (<5%; Fig. 4C, lane 6). Thus, in vitro processing requires only high-affinity binding of the SLBP to the RNA target via its RNA binding domain and the 20 amino acids adjacent to the C terminus of this domain.
Coprecipitation of the U7 snRNP with SLBP and histone pre-mRNA.
To initiate studies on the function of SLBP in the processing reaction, we used the anti-SLBP antibody to isolate any complexes that might form on the histone pre-mRNA. Since time course experiments indicate that histone pre-mRNA processing proceeds without a significant time lag (6, 11, 27), reactions containing an unlabeled pre-mRNA substrate and nuclear extract were briefly (5 min) incubated at 22°C (below the optimum 32°C) to allow complex formation and immediately cooled on ice. As determined by using radiolabeled substrate, incubation under these conditions resulted in binding of more than 95% of the pre-mRNA to SLBP and yielded less than 2% of the cleavage product (data not shown). Anti-SLBP antibody was then added to the reactions, and complexes were isolated by binding to protein A-agarose. RNA was recovered from the immunoprecipitates, and the U7 snRNA was detected by Northern analysis. More than 95% of the substrate RNA was recovered in the immunoprecipitates (data not shown), since SLBP is present in excess over the substrate in these reactions. An internal standard RNA containing the U7 snRNA sequence was added to each sample prior to phenol extraction to control for the efficiency of RNA isolation and hybridization.
Only a small amount of the U7 snRNA bound nonspecifically to the protein A-agarose in the absence of the pre-mRNA substrate (Fig. 5C, lane 1). After addition of unlabeled H2a pre-mRNA large amounts of U7 snRNP coimmunoprecipitated with SLBP (Fig. 5C, lane 2). Precipitation of the U7 snRNP was specific, since it was completely abolished by sequestering the antibody with the antigenic peptide (Fig. 5C, lane 3). Coimmunoprecipitation of the U7 snRNP in the presence of H2a pre-mRNA was also abolished by addition of a 2′-O-methyl oligoribonucleotide complementary to the first 19 nucleotides of the U7 snRNA (Fig. 5C, lane 5) and was not affected by a large excess of a nonspecific 2′-O-methyl oligoribonucleotide (Fig. 5C, lane 4). The anti-U7 2′-O-methyl oligoribonucleotide has been previously shown to completely inhibit the cleavage reaction by masking the 5′ end of the U7 snRNA and preventing the U7 snRNP from base pairing with the HDE (4, 6, 31). This result demonstrates that stable binding of the U7 snRNP to the substrate requires base pairing between the 5′ end of the U7 snRNA and the pre-mRNA substrate. Consistently, the HDE mutant substrate, which cannot base pair with the U7 snRNA (Fig. 5A), is inactive in processing (Fig. 5B, lane 2) and is not able to form a stable complex with the U7 snRNP (Fig. 5C, lane 9). No U7 snRNA was recovered when the H2a/RS substrate containing the reversed stem sequence was used (Fig. 5C, lane 10). This mutation virtually abolishes binding of SLBP to the histone pre-mRNA and prevents immunoprecipitation of the substrate by anti-SLBP antibody. Finally, coimmunoprecipitation of the U7 snRNP was completely blocked by the addition of excess stem-loop RNA oligonucleotide which sequestered all free SLBP and prevented binding of SLBP to the pre-mRNA substrate (Fig. 5C, lane 6). This result demonstrates that the SLBP–stem-loop RNA complex formed under these conditions does not stably associate with the U7 snRNP and confirms that stable interaction between the two factors requires that both cis-acting elements be present on the same pre-mRNA molecule.
Guanosines are not found at the cleavage site of any natural histone pre-mRNAs, and substitution of guanosine for cytosine at this site greatly reduces processing (8). We constructed the H2a/4G mutant substrate derived from the H2a pre-mRNA by replacing the CACU sequence at the cleavage site with four guanosines (Fig. 5A). The H2a/4G substrate was inactive in processing (Fig. 5B, lane 1). In spite of that, the H2a/4G pre-mRNA supported formation of stable complexes with U7 snRNP with an efficiency similar to that of the parental H2a substrate (Fig. 5C, lane 7). Thus, stable association with the U7 snRNP is necessary but not sufficient for cleavage of histone pre-mRNAs. The H1t pre-mRNA, which contains a weaker HDE than the H2a and H2a/4G pre-mRNAs, was significantly less efficient in the formation of stable complexes with U7 snRNP. This substrate bound about 25% of the amount of U7 snRNA immunoprecipitated with H2a-614 pre-mRNA (Fig. 5C, lane 8).
We conclude from the above results that histone pre-mRNAs support formation of a stable processing complex containing SLBP and the U7 snRNP.
Function of SLBP in processing.
The above immunoprecipitation experiments confirmed previous results that the U7 snRNP and the histone pre-mRNA can form a stable complex dependent on base pairing between the U7 snRNA and the HDE but did not definitively show that SLBP is required for this interaction. Since in our experiments isolation of the processing complexes is dependent on the anti-SLBP antibody, mutating the stem-loop structure to prevent binding of the SLBP to the pre-mRNA or removing SLBP from the extract prevents subsequent isolation of the histone pre-mRNA substrate.
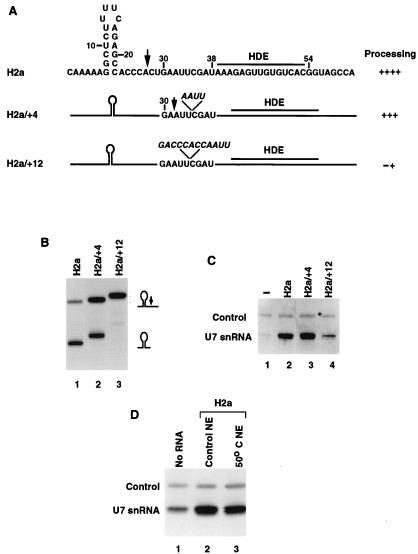
Increasing the distance between the stem-loop structure and the HDE results in a shift of the cleavage site by a comparable number of nucleotides (31, 32). In addition to movement of the cleavage site, insertions between the stem-loop and the HDE result in a progressive decline of processing efficiency as the distance between the two elements increases, leading eventually to a complete loss of activity (31, 32). To determine whether the spacing between the stem-loop and the HDE was critical for stable association of U7 snRNP with the pre-mRNA, we constructed the H2a/+4 and H2a/+12 pre-mRNA substrates by inserting 4 or 12 nucleotides between the cleavage site and the HDE, respectively (Fig. 6A). The inserted sequences were compatible with known cleavage specificity, containing highly preferred adenosine residues (8) or a duplicated cleavage site (H2a/+4 and H2a/+12 pre-mRNA, respectively; Fig. 6A). The H2a/+4 construct was processed relatively efficiently, and in agreement with the results of Scharl and Steitz (31), the cleavage site was shifted about four nucleotides downstream of the wild-type site (Fig. 6B, lane 2). The H2a/+12 substrate was essentially inactive in processing (Fig. 6B, lane 3), although longer exposures resulted in detection of minor products resulting from cleavage 10 to 15 nucleotides 3′ of the wild-type site (data not shown). The overall efficiency of processing at these sites approximately equaled the efficiency of SLBP-independent processing of the H2a-614 substrate, suggesting that large insertions between the stem-loop and the HDE are functionally equivalent to mutating the stem-loop or depleting SLBP from the nuclear extract.
FIG. 6.
The stimulation of binding of the U7 snRNP by SLBP is distance dependent. (A) The sequence of the H2a-614 mutant substrates containing either 4- or 12-nucleotide insertions between the normal cleavage site and the HDE. (B) The radiolabeled substrates shown in panel A and indicated above each lane were incubated in nuclear extract under standard processing conditions, and RNA was analyzed as described above. (C) The anti-SLBP antibody was used to isolate processing complexes assembled on the pre-mRNA substrates, as described in Fig. 5C. Immunoprecipitation with no pre-mRNA added (lane 1) or in the presence of the substrates shown in panel A (lanes 2 to 4). (D) The anti-SLBP antibody was used to isolate processing complexes assembled on the H2a-614 substrate (lanes 2 and 3) in the control nuclear extract (NE) or in extract which had been heat inactivated at 50° for 15 min. Lane 1, no RNA was added to the control extract.
In order to determine the ability of the H2a/+4 and H2a/+12 pre-mRNAs to form stable complexes with U7 snRNP, each substrate was incubated in the nuclear extract to form processing complexes that were subsequently immunoprecipitated by the anti-SLBP antibody and analyzed for the presence of the U7 snRNA. The H2a/+4 pre-mRNA supported formation of processing complexes containing the U7 snRNP at least as efficiently as the control H2a pre-mRNA (Fig. 6C, lanes 2 and 3, respectively), although it was less efficiently processed (Fig. 6B, lane 2). Strikingly, immunoprecipitation of processing complexes assembled on the H2a/+12 pre-mRNA containing the same HDE yielded only a small amount of the U7 snRNA, about 10% of the control levels but significantly higher than the background level (Fig. 6C, lane 4). The small amount of U7 snRNP stably associated with the H2a/+12 pre-mRNA probably represents a complex formed independently of SLBP. The basal level of the U7 snRNP binding to the HDE is most likely responsible for residual processing of the H2a/+12 pre-mRNA at heterogeneous sites and less than 10% efficient processing of the H2a pre-mRNA at the normal site in the absence of bound SLBP. We conclude that formation of a stable complex of U7 snRNP with the HDE is very inefficient without the assistance of SLBP bound close to the HDE. Thus, one role of the SLBP in histone pre-mRNA processing is to help recruit U7 snRNP into a stable complex on histone pre-mRNA. SLBP-mediated stabilization of the interaction between the U7 snRNP and the histone pre-mRNA is not dependent on the HLF. Brief treatment of a nuclear extract at 50°C results in inactivation of this essential processing factor (12), and in our hands resulted in only a 50% reduction in the amount of SLBP activity as judged by mobility shift and has no effect on the U7 snRNP (12, 23). Although the heat-treated extract was completely inactive in processing (data not shown), the U7 snRNP still formed a stable complex with the histone pre-mRNA with only a slightly reduced efficiency compared to the control extract (Fig. 6D, lanes 3 and 2, respectively). Thus, the heat-labile factor is not essential for formation of the complex containing the histone pre-mRNA, SLBP, and U7 snRNP.
DISCUSSION
We (45) and others (20) have previously reported the cloning and preliminary analysis of the SLBP, a trans-acting factor which plays an important role in 3′-end processing of the replication-dependent histone pre-mRNAs. SLBP is not similar to any other protein in the database and contains a novel RNA binding domain. Here we have determined the regions of SLBP required for histone pre-mRNA processing and a role of SLBP in the processing reaction.
A 93-amino-acid region of SLBP supports efficient in vitro processing.
The processing activity of extracts depleted by either a biotinylated stem-loop or anti-SLBP was efficiently restored by addition of the full-length human SLBP expressed in Sf9 insect cells by using the baculovirus system. The ability of purified SLBP to complement the depleted extract indicates that no other component of the processing machinery is removed from the extract during the depletion procedure. This suggests that the HBF, which has previously been functionally defined as the factor which interacts with the stem-loop (41), is comprised only of SLBP.
SLBP contains a 73-amino-acid region in the center of the protein responsible for binding to the stem-loop sequence in histone pre-mRNAs. The entire amino-terminal region of the SLBP can be removed without affecting 3′-end histone pre-mRNA processing in vitro. This 124-amino-acid region may play a role in some of the other processes in histone mRNA metabolism, such as nucleocytoplasmic transport of the histone mRNP, translation, or regulation of histone mRNA stability. The RNA binding domain alone was not sufficient to support processing, but the addition of only 20 amino acids to the C-terminal side of the RNA binding domain restored about 50% of the activity of the intact SLBP. Additional amino acids in the C-terminal region of SLBP must contribute to the maximal activity of the protein. Replacement of this entire 20-amino-acid region in the context of the full-length SLBP greatly reduced processing without affecting its ability to bind RNA.
Processing of the H1t pre-mRNA is fully dependent on SLBP.
SLBP (HBF) is required for maximal processing efficiency of all commonly used histone pre-mRNA substrates, including H2a-614, H3-614, H3-53, H4-1, and H4-12 pre-mRNAs (38). In the absence of SLBP, processing occurs at a reduced rate. In contrast, the H1t pre-mRNA substrate is processed with efficiency similar to that of the H2a-614 histone pre-mRNA in the complete extract but is absolutely dependent on SLBP for processing. The strength of the HDE is also important for high efficiency of processing in vivo. For example, the H2a-614 pre-mRNA capable of forming with the U7 snRNA 14 bp (including 2 GU bp) is expressed very efficiently in vivo (13). Other histone pre-mRNAs that have much weaker HDEs (18, 45) are expressed at a lower level in vivo, partly due to a decreased efficiency of 3′-end formation (18).
The U7 snRNP forms a stable complex with the histone pre-mRNA and SLBP.
Processing of histone pre-mRNA involves assembly of a multifactor complex on the histone pre-mRNA, followed by cleavage of the histone pre-mRNA (23). Since SLBP alone can form a stable complex with histone pre-mRNA, the initial event in processing in vivo is likely binding of the SLBP. The immunoprecipitation experiments with the anti-SLBP antibody allowed us to detect formation of a stable complex containing at least three components: SLBP, the pre-mRNA substrate, and the U7 snRNP. This complex was formed rapidly and was resistant to extensive washing with isotonic solutions containing nonionic detergents. Stable binding of the U7 snRNP to the pre-mRNA was absolutely dependent on formation of a duplex RNA between the U7 snRNA and the HDE. While these results confirm that the HDE is a primary site of binding for the U7 snRNP, they do not preclude existence of an independent but weaker binding of the U7 snRNP to the stem-loop (or stem-loop–SLBP complex) in the initial phase of the processing reaction, as suggested previously by a combination of RNase protection and anti-Sm-mediated immunoprecipitation experiments (26). Such an interaction, which could involve protein-protein interactions between SLBP and a protein in U7 snRNP, might be disrupted by the stringent washing conditions used during our immunoprecipitation experiments.
In agreement with previous observations (23), the efficiency of complex formation was dependent on the stability of the RNA duplex formed between the 5′ end of the U7 snRNA and the HDE. In the immunoprecipitation experiments, about 20% of the total U7 snRNP in the extract and 3% of the H2a-614 pre-mRNA substrate are involved in formation of stable complexes. SLBP was present in the extract in excess of the substrate, and all of the substrate was precipitated by the anti-SLBP antibody, ruling out the possibility that there are substrate molecules that form a stable complex only with U7 snRNP. The histone H1t pre-mRNA also formed a stable complex with SLBP and U7 snRNP, although less efficiently, despite the fact that the H1t HDE alone could not stably interact with the U7 snRNP (Fig. 2B). These results suggest that stable binding of the U7 snRNP to the H1t pre-mRNA is dependent on additional interactions with SLBP, presumably due to protein-protein interactions with a protein(s) in the U7 snRNP.
Subsequent to formation of the complex containing the pre-mRNA, SLBP and the U7 snRNP, additional factors must be recruited for cleavage to occur. Since only a small fraction of the substrate and a large fraction of the U7 snRNP are present in the complex, the same U7 snRNP must function in cleavage of multiple substrates during the 1 h in vitro reaction incubation, which normally yields more than 50% of the mature product. It is not necessary for assembly of the complex that the substrate be cleavable.
Proper spacing between the stem-loop and the HDE is necessary for stable binding of U7 snRNP.
A role for SLBP in stabilizing the interaction between the U7 snRNP and pre-mRNA was first suggested by the observation that pre-mRNAs with weak HDEs are more dependent on SLBP (HBF) (38). This role of SLBP (HBF) was further supported by demonstrating that mutations within the HDE which allowed more extensive base pairing with the U7 snRNA resulted in a reduced requirement for SLBP (38). Additional experimental support for the role of SLBP in recruiting the U7 snRNP to pre-mRNA substrates with weak HDE is provided by the immunoprecipitation experiments with the H2a/+12 pre-mRNA. Insertion of 12 nucleotides between the stem-loop sequence and the HDE virtually abolished processing, most likely by precluding productive interaction between factors binding to both elements. The failure to process the H2a/+12 pre-mRNA was accompanied by significant reduction in the amount of the U7 snRNP coassembled on this substrate. These experiments provide direct biochemical evidence that stable association of the U7 snRNP with the histone pre-mRNA is not determined solely by the sequence of the HDE but is stimulated by SLBP bound to the stem-loop sequence at the appropriate distance from the HDE.
In a previous study with psoralen cross-linking and immunoprecipitation with antitrimethylguanosine antibodies, no difference was observed in the ability of the U7 snRNP to bind H2a-614 mutant pre-mRNAs containing variable insertions between the stem-loop sequence and the HDE (31). These experiments were performed under processing conditions, and indeed there was more U7 snRNP bound to the substrates that were not efficiently cleaved than to the wild-type H2a-614 substrate (31). Our results show a clear dependence on the proper positioning of the HDE for stable U7 snRNP binding to the H2a-614 pre-mRNA (Fig. 6). This discrepancy is most likely due to efficient processing of the wild-type substrate under the conditions used by Scharl and Steitz, which would result in rapid dissociation of complexes containing the U7 snRNP (31). During the short time and at the lower temperature (22°C) we used, there is complex formation but virtually no cleavage. We believe this complex is on the pathway to histone pre-mRNA processing, since subsequent warming of the reaction sample results in efficient processing. We observed a low level of binding of the U7 snRNP to the H2a/+12 substrate, which is probably due solely to base pairing of the U7 snRNA with the HDE and occurs independently of SLBP.
Efficient processing requires high-affinity binding of SLBP.
Stabilization of binding of the U7 snRNP to the HDE by SLBP requires that SLBP binds to the stem-loop sequence on the same pre-mRNA molecule. Substrates which lack strong SLBP binding sites are processed less efficiently, and a reduction of 5- to 10-fold in the affinity of SLBP for the stem-loop has a large effect on processing efficiency, even on the efficient H2a-614 substrate, in vitro (Fig. 1) and in vivo (30). An increase in the off rate of SLBP effected by a reduction in affinity suggests that the formation and assembly of the initial stable processing complex requires binding of SLBP to the pre-mRNA for a significant amount of time. The affinity of both SLBP and U7 snRNP for their respective sites on the histone pre-mRNA is critical for determining the rate of the in vitro processing reaction. If the U7 snRNP binds stably to the pre-mRNA due to a high degree of complementarity between the HDE and the 5′ end of U7 snRNA, then it can also recruit the additional factors necessary for processing, albeit with lower efficiency.
An initially puzzling result, that the U4C and 5′FL RNA competitors have very different effects on processing of the H2a-614 pre-mRNA and the H1t mRNA, is understandable in this scenario. In the case of the H2a-614 pre-mRNA, U7 snRNP associates with the HDE relatively stably. Binding of the small amount of free SLBP present in the reaction mixture containing competitors is sufficient to form significant amounts of a stable processing complex, which is resistant to the competitors. In contrast, the U7 snRNP does not associate for a significant amount of time with the H1t pre-mRNA; rather, the formation of the processing complex is absolutely dependent on bound SLBP, which is greatly reduced in the presence of the competitors. U7 snRNP bound stably to the histone pre-mRNA is also capable of ultimately recruiting the other factors necessary for processing in the absence of SLBP, although the formation of the stable complex is greatly enhanced by the presence of SLBP. These results are consistent with previous results that suggested cooperative binding of SLBP and U7 snRNP to the histone pre-mRNA (25, 26, 41) and provide additional evidence for interaction between these two trans-acting factors in 3′-end processing.
Other trans-acting factors involved in histone pre-mRNA processing.
The immunoprecipitation experiments with anti-SLBP antibody revealed that formation of stable processing complexes containing the histone pre-mRNA and the U7 snRNP is not dependent on the HLF, the third known trans-acting processing factor (12). How many other components are necessary for 3′-end processing of the histone pre-mRNAs? A large number of polypeptides are required for cleavage of the other pre-mRNAs prior to polyadenylation, although no snRNA is required in this reaction (16, 19, 42). The coimmunoprecipitation procedure with anti-SLBP and a nuclear extract labeled with [35S]methionine may allow detection of other factors coassembled with U7 snRNP on the histone pre-mRNA. These experiments are in progress.
ACKNOWLEDGMENTS
This work was supported by a grant from the NIH (GM29832) to W.F.M. and a faculty research award from the University of North Carolina to Z.D.
We thank members of the Marzluff laboratory for comments on the manuscript.
REFERENCES
- 1.Abbott J, Marzluff W F, Gall J G. The stem loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell. 1999;10:487–499. doi: 10.1091/mbc.10.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bond U M, Yario T A, Steitz J A. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- 3.Cho D C, Scharl E C, Steitz J A. Decreasing the distance between the two conserved sequence elements of histone pre-messenger RNA interferes with 3′ processing in vitro. RNA. 1995;1:905–914. [PMC free article] [PubMed] [Google Scholar]
- 4.Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel M L. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro RNA processing reaction. EMBO J. 1988;7:801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotten M, Oberhauser B, Brunar H, Holzner A, Issakides G, Noe C R, Schaffner G, Wagner E, Birnstiel M L. 2′-O-Methyl, 2′-O-ethyl oligoribonucleotides and phosphorothioate oligodeoxyribonucleotides as inhibitors of the in vitro U7 snRNP-dependent mRNA processing event. Nucleic Acids Res. 1991;19:2629–2635. doi: 10.1093/nar/19.10.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominski Z, Sumerel J, Hanson R J, Marzluff W F. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. RNA. 1995;1:915–923. [PMC free article] [PubMed] [Google Scholar]
- 7.Eckner R, Ellmeier W, Birnstiel M L. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furger A, Schaller A, Schümperli D. Functional importance of conserved nucleotides at the histone RNA 3′ processing site. RNA. 1998;4:246–256. [PMC free article] [PubMed] [Google Scholar]
- 9.Gallie D R, Lewis N J, Marzluff W F. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgiev O, Birnstiel M L. The conserved CAAGAAAGA spacer sequence is an essential element for the formation of 3′ termini of the sea urchin H3 histone mRNA by RNA processing. EMBO J. 1985;4:481–489. doi: 10.1002/j.1460-2075.1985.tb03654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gick O, Krämer A, Keller W, Birnstiel M L. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gick O, Krämer A, Vasserot A, Birnstiel M L. Heat-labile regulatory factor is required for 3′ processing of histone precursor mRNAs. Proc Natl Acad Sci USA. 1987;84:8937–8940. doi: 10.1073/pnas.84.24.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graves R A, Wellman S E, Chiu I-M, Marzluff W F. Differential expression of two clusters of mouse histone genes. J Mol Biol. 1985;183:179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- 14.Grimm C, Stefanovic B, Schümperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanson R J, Sun J-H, Willis D G, Marzluff W F. Efficient extraction and partial purification of the polyribosomal-associated stem-loop binding protein bound to the 3′ end of histone mRNA. Biochemistry. 1996;35:2146–2156. doi: 10.1021/bi9521856. [DOI] [PubMed] [Google Scholar]
- 16.Keller W. No end yet to messenger RNA 3′ processing! Cell. 1995;81:829–832. doi: 10.1016/0092-8674(95)90001-2. [DOI] [PubMed] [Google Scholar]
- 17.Li J-M, Parsons R A, Marzluff W F. Transcription of the sea urchin U6 gene in vitro requires a TATA-like box, a proximal sequence element, and sea urchin USF, which binds an essential E box. Mol Cell Biol. 1994;14:2191–2200. doi: 10.1128/mcb.14.3.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu T-J, Levine B J, Skoultchi A I, Marzluff W F. The efficiency of 3′-end formation contributes to the relative levels of different histone mRNAs. Mol Cell Biol. 1989;9:3499–3508. doi: 10.1128/mcb.9.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manley J L. Messenger RNA polyadenylation: a universal modification. Proc Natl Acad Sci USA. 1995;92:1800–1801. doi: 10.1073/pnas.92.6.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin F, Schaller A, Eglite S, Schümperli D, Müller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marzluff W F. Histone 3′ ends: essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- 22.Marzluff W F, Whitfield M L, Dominski Z, Wang Z-F. Identification of the protein that interacts with the 3′ end of histone mRNA. In: Richter J D, editor. mRNA formation and function. San Diego, Calif: Academic Press; 1997. pp. 163–193. [Google Scholar]
- 23.Melin L, Soldati D, Mital R, Streit A, Schümperli D. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8794. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mowry K L, Oh R, Steitz J A. Each of the conserved sequence elements flanking the cleavage site of mammalian histone pre-mRNAs has a distinct role in the 3′-end processing reaction. Mol Cell Biol. 1989;9:3105–3108. doi: 10.1128/mcb.9.7.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mowry K L, Steitz J A. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3′ end formation in vitro. Mol Cell Biol. 1987;7:1663–1672. doi: 10.1128/mcb.7.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mowry K L, Steitz J A. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA’s. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- 28.Pandey N B, Marzluff W F. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandey N B, Sun J-H, Marzluff W F. Different complexes are formed on the 3′ end of histone mRNA in nuclear and polysomal extracts. Nucleic Acids Res. 1991;19:5653–5659. doi: 10.1093/nar/19.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandey N B, Williams A S, Sun J-H, Brown V D, Bond U, Marzluff W F. Point mutations in the stem-loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol Cell Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharl E C, Steitz J A. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 1994;13:2432–2440. doi: 10.1002/j.1460-2075.1994.tb06528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scharl E C, Steitz J A. Length suppression in histone messenger RNA 3′-end maturation: processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc Natl Acad Sci USA. 1996;93:14659–14664. doi: 10.1073/pnas.93.25.14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.SenGupta D J, Zhang B L, Kraemer B, Prochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc Natl Acad Sci USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith H O, Tabiti K, Schaffner G, Soldati D, Albrecht U, Birnstiel M L. Two-step affinity purification of U7 small nuclear ribonucleoprotein particles using complementary biotinylated 2′-O-methyl oligoribonucleotides. Proc Natl Acad Sci USA. 1991;88:9784–9788. doi: 10.1073/pnas.88.21.9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soldati D, Schümperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol Cell Biol. 1988;8:1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spycher C, Streit A, Stefanovic B, Albrecht D, Koning T H W, Schümperli D. 3′ end processing of mouse histone pre-mRNA: evidence for additional base-pairing between U7 snRNA and pre-mRNA. Nucleic Acids Res. 1994;22:4023–4030. doi: 10.1093/nar/22.20.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefanovic B, Hackl W, Lührmann R, Schümperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Streit A, Koning T W, Soldati D, Melin L, Schümperli D. Variable effects of the conserved RNA hairpin element upon 3′ end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993;21:1569–1575. doi: 10.1093/nar/21.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strub K, Galli G, Busslinger M, Birnstiel M L. The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J. 1984;3:2801–2807. doi: 10.1002/j.1460-2075.1984.tb02212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun J-H, Pilch D R, Marzluff W F. The histone mRNA 3′ end is required for localization of histone mRNA to polyribosomes. Nucleic Acids Res. 1992;20:6057–6066. doi: 10.1093/nar/20.22.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasserot A P, Schaufele F J, Birnstiel M L. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci USA. 1989;86:4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wahle E. 3′-end cleavage and polyadenylation of mRNA precursors. Biochim Biophys Acta Gene Struct Expr. 1995;1261:183–194. doi: 10.1016/0167-4781(94)00248-2. [DOI] [PubMed] [Google Scholar]
- 43.Wang Z-F, Ingledue T C, Dominski Z, Sanchez R, Marzluff W F. Two Xenopus proteins that bind the 3′ end of histone mRNA: implications for translational control of histone synthesis during oogenesis. Mol Cell Biol. 1999;19:835–845. doi: 10.1128/mcb.19.1.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z-F, Krasikov T, Frey M R, Wang J, Matera A G, Marzluff W F. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over one megabase. Genome Res. 1996;6:688–701. doi: 10.1101/gr.6.8.688. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z-F, Whitfield M L, Ingledue T I, Dominski Z, Marzluff W F. The protein which binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 46.Williams A S, Ingledue T C, Kay B K, Marzluff W F. Changes in the stem-loop at the 3′ terminus of histone mRNA affects its nucleocytoplasmic transport and cytoplasmic regulation. Nucleic Acids Res. 1994;22:4660–4666. doi: 10.1093/nar/22.22.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams A S, Marzluff W F. The sequence of the stem and flanking sequences at the 3′ end of histone mRNA are critical determinants for the binding of the stem-loop binding protein. Nucleic Acids Res. 1995;23:654–662. doi: 10.1093/nar/23.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C-H H, Gall J G. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci USA. 1993;90:6257–6259. doi: 10.1073/pnas.90.13.6257. [DOI] [PMC free article] [PubMed] [Google Scholar]