Abstract
A cDNA encoding a novel Krüppel-type zinc finger protein, FKLF, was cloned from fetal globin-expressing human fetal erythroid cells. The deduced polypeptide sequence composed of 512 amino acids revealed that, like Sp1 and EKLF, FKLF has three contiguous zinc fingers at the near carboxyl-terminal end. A long amino-terminal domain is characterized by the presence of two acidic and two proline-rich regions. Reverse transcription (RT)-PCR assays using various cell lines demonstrated that the FKLF mRNA is expressed predominantly in erythroid cells. FKLF message is detectable by RT-PCR in fetal liver but not in adult bone marrow cells. As predicted from its structural features, FKLF is a transcriptional activator. In luciferase assays FKLF activated the γ- and ɛ-globin gene promoters, and, to a much lower degree, the β-globin promoter. Studies of HS2-γ gene reporter constructs carrying CACCC box deletions revealed that the CACCC box sequence of the γ gene promoter mediates the activation of the γ gene by FKLF. Other erythroid promoters (GATA-1, glycophorin B, ferrochelatase, porphobilinogen deaminase, and 5-aminolevulinate synthase) containing CACCC elements or GC-rich potential Sp1-binding sites were activated minimally, if at all, by FKLF, indicating that FKLF is not a general activator of genes carrying the CACCC motifs. Transfection of K562 cells with FKLF cDNA enhanced the expression of the endogenous ɛ- and γ-globin genes, suggesting an in vivo role of FKLF in fetal and embryonic globin gene expression. Our results indicate that the protein potentially encoded by the FKLF cDNA acts as a transcriptional activator of embryonic and fetal β-like globin genes.
The programmed expression of globin genes is tissue and developmental stage specific. In humans, five β-like globin genes (ɛ, Aγ, Gγ, δ, and β) form a cluster on the short arm of chromosome 11, and their expression is characterized by two major switches initially from embryonic (ɛ) to fetal (Aγ and Gγ) and subsequently to adult (δ and β) globin gene expression (39). Although a number of cis-acting elements of globin genes and corresponding trans-acting factors have been identified (9, 27), the precise molecular mechanisms of globin gene regulation are still unclear. Especially limited is the information on the trans-acting factors involved in the developmental control of fetal and embryonic globin genes.
The CACCC (or GT) box is a cis-acting element found in a variety of genes expressed in erythroid and nonerythroid tissues. Each β-like globin gene (except the δ gene) has one or two CACCC boxes among the conserved promoter sequences. The importance of the CACCC box for β-globin gene transcription has been demonstrated by naturally occurring mutations which cause β thalassemia (20, 28, 29). A role of CACCC box in γ gene expression is supported by several pieces of evidence. First, CACCC box-deleted γ gene promoters display reduced activation of linked genes in K562 cells by transient (21, 45) and stable (6) transfection assays. Second, transgenic mice carrying a truncated Aγ promoter construct which lacks a functional CACCC box show decreased Aγ gene expression in all developmental stages (38). Third, in vivo footprinting studies have shown that the γ CACCC sequence shows significant protein binding in γ gene-expressing cells but not in β-gene-expressing cells (16). Collectively, these results suggest that the CACCC box is one of the key cis-acting elements for γ gene expression.
Among the proteins binding to the CACCC boxes of the globin genes, Sp1, a ubiquitous (18) Krüppel-like zinc finger protein, and EKLF, an erythroid tissue-specific (24), Krüppel-like zinc finger protein, are well characterized. Sp1 is known to interact with the ɛ (47)-, γ (13)-, and β (15)-globin gene CACCC boxes. However, Sp1−/− mice express embryonic globin genes at a reduced but still significant level (22), suggesting that this ubiquitous protein is not sufficient, at least in the embryonic stage, for globin gene transcription. In contrast, EKLF binds to the proximal β gene CACCC box (5, 24) and plays a critical role in β-globin gene expression (26, 32). Studies of EKLF-deficient mice carrying human β-globin loci, however, revealed that the γ gene expression is not dependent on EKLF (31, 46).
The specificity of interaction of EKLF with the β gene CACCC box is achieved by a 9-bp sequence (CCA CAC CCT), which can be recognized by the three zinc fingers of EKLF (24). The analogous sequence of the CACCC box of the γ-globin gene promoter is CTC CAC CCA, and it is also found in the mouse ɛy gene promoters. We assumed that a factor having Krüppel-type zinc finger structures such as Sp1 and EKLF exists and functions as an activator of the fetal globin gene promoter. Human fetal liver erythroid cells were screened for Sp1- or EKLF-type cDNAs by PCR (30), and a cDNA which encodes a novel Krüppel-type zinc finger protein, FKLF (embryonic/fetal β-like globin gene-activating Krüppel-like factor), was isolated. We show that the FKLF gene is preferentially expressed in erythroid cells and that it strongly activates ɛ- and γ-globin gene promoters.
MATERIALS AND METHODS
Cell culture and RNA extractions.
K562 cells were cultured in RPMI 1640 supplemented with 10% fetal calf serum (FCS). Total RNA was extracted from erythroid cells of day 67 human fetal liver BFU-E cultured for 11 days in RPMI 1640 supplemented with 10% FCS, stem cell factor, interleukin-6, and erythropoietin by standard methods (35). Poly(A)+ fraction of the total RNA was separated by an oligo(dT)-cellulose spun column (Pharmacia Biotech).
PCR screening for zinc finger motifs.
Poly(A)+ RNA (1.8 μg) was subjected to reverse transcription, with 50 pmol of a degenerate primer, 5′-AG(AG)TG(AG)TC(AG)(CG)(AT)IC(TG)I(AGC)(AT)(AG)AA-3′, in 20 μl of a reaction mixture (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 200 μM [each] deoxynucleotide triphosphate [dNTP]) using 200 U of reverse transcriptase (SuperScript; BRL). Following denaturation at 65°C for 5 min, the reaction mixture was incubated at 42°C for 2 h. Then, the mixture was heated at 94°C for 10 min to inactivate the reverse transcriptase. All of the reverse-transcribed products were used as template DNA and amplified in a reaction volume of 50 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% (wt/vol) gelatin, 200 μM (each) dNTP, 0.02 U of Taq DNA polymerase (AmpliTaq, Perkin-Elmer)/μl, 1 μM (each) degenerate primer described above, and 5′-CA(CT)AC(AGCT)GG(AGCT)GA(AG)(AC)(AG)(AG)CC-3′. The PCR conditions were as follows: cycle 1, 4 min at 94°C, 2 min at 40°C, heating for 2 min, and 3 min at 72°C; cycles 2 to 5, 1 min at 94°C, 2 min at 40°C, heating for 2 min, and 3 min at 72°C; cycles 6 to 45, 1 min at 94°C, 2 min at 50°C, and 3 min at 72°C. All of the PCR products were run on 2% low-melting-point agarose gel and stained with ethidium bromide. The expected 150-bp band was cut out, and the extracted DNA was used for ligation into a plasmid vector (T vector; Promega). Plasmid DNA was analyzed by restriction enzyme digestion with PstI and NcoI, and clones containing the insert were sequenced with a kit (Cyclist; Stratagene).
cDNA cloning.
The 3′ unknown sequence was obtained by rapid amplification of cDNA ends (10). First-strand cDNA of human fetal liver cells was synthesized from 100 ng of poly(A)+RNA by using an adapter-oligo(dT) primer (5′-GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT-3′) as described above. The 3′ unknown region was amplified by using an adapter primer (5′-GACTCGAGTCGACATCG-3′) and an outer gene-specific primer. A portion of 1/50 of the products was used as template DNA for the second-step PCR by using the adapter primer and an inner gene-specific primer.
For the 5′ unknown region, ligation-anchored (LA) PCR (43) was performed. Following ligation of 5′-phosphorylated, 3′-end-blocked anchor oligonucleotide (5′-pTTTAGTGAGGGTTAATAAGCGGCCGCGTCGTGACTGGGAGCGCddA-3′) to the first-strand cDNA, PCR was carried out under the same conditions as in the 3′-amplification, except that the primers were different. In this case, two—outer and inner—primers were prepared in the anchor sequence as follows: outer, 5′-CGCTCCCAGTCACGACGC-3′; inner, 5′-GCCGCTTATTAACCCTCACTAA-3′. The 5′ end of the cDNA was confirmed by repeated LA-PCR by using different gene-specific primers and cDNAs of different RNA preparation.
Based on the cDNA sequence determined from the above PCR, a 1.7-kb open reading frame (ORF) predicted to encode FKLF protein was amplified from random hexamer-primed cDNA by using the Expand Long PCR System (Boehringer Mannheim). The PCR product was digested with SacII and AvrII and cloned into the pGEM-5Zf(+) vector (Promega) cut with SacII-SpeI (pGEM/FKLF). The nucleotide sequence was checked for three clones, and one clone without mutation was used for further plasmid construction.
Plasmid constructions.
Transactivator plasmid of FKLF was prepared as follows: FKLF cDNA was cut as a SacII (blunted)-NotI fragment from the pGEM/FKLF and subcloned into pSPORT 1 vector (Life Technologies) digested with SmaI and NotI (pSPORT/FKLF). Subsequently, an EcoRI-HindIII fragment of the FKLF cDNA from the pSPORT/FKLF was inserted into an eukaryotic expression vector pSG5DD (a generous gift from T. Townes) digested with EcoRI and HindIII (pSG5/FKLF).
Various reporter plasmids were constructed from pHS2βCAT and pHS2γLuc (2, 8). A BglII site was created between the β promoter (−265 to +48) and the CAT gene of the pHS2βCAT by in vitro mutagenesis (Altered Sites II in vitro mutagenesis systems; Promega) according to the manufacturer’s instructions. Briefly, a PstI-BamHI fragment of pHS2βCAT was subcloned into PstI- and BamHI-digested pALTER-1 vector (Promega). The BglII site was generated by using the 5′ phosphorylated oligonucleotide 5′-pCGCCAAGCTCAGATCTAGGTGTCTGTT-3′. The BglII site-introduced PstI-BamHI fragment was reinserted into PstI-BamHI sites of the pHS2βCAT. Subsequently, the 1.5-kb HS2 fragment was cut out as a PstI (blunted)-BglII fragment, and the 0.3-kb β promoter fragment was cut out as a BglII fragment. The HS2 fragment was inserted into KpnI (blunted)-BglII sites of the pGL2-Basic vector (pHS2Luc), and then the β promoter was cloned into the BglII site of the pHS2Luc (pHS2βLuc). Similarly, CACCC box deletion mutants of pHS2γLuc were constructed by in vitro mutagenesis. Briefly, a KpnI-BamHI fragment of pHS2γLuc was subcloned into KpnI- and BamHI-digested pALTER-1 vector. Each CACCC box was deleted by using oligonucleotides 5′-pCATGCTGAGGCTTGCCCAGATGTTCTC-3′ for pHS2ΔCAC2γLuc, 5′-pGTCGGGGTCAGTGCGCCTTCTGGTTC-3′ for pHS2ΔCAC3γLuc, and 5′-pGTCCCTGGCTAAATGGGTTGGCCAG-3′ for pHS2γΔCACLuc. The CACCC-deleted KpnI-BamHI fragments were reinserted into the KpnI-BamHI sites of the pHS2γLuc. A luciferase reporter construct just driven by the γ gene promoter (pγLuc) was generated by deleting the HS2 fragment from the pHS2γLuc with KpnI and BglII digestion, followed by self-ligation. A BamHI-PvuII fragment of the ɛ gene promoter (−178 to +22) was subcloned into BamHI-EcoRV sites of pSP72 vector (Promega). Subsequently, the ɛ promoter was cut out as a BamHI-BglII fragment and inserted into the BglII site of pHS2Luc (pHS2ɛLuc). Constructs generated by in vitro mutagenesis were verified by sequencing.
DNA fragments of promoter of GATA-1 (−499 to −21) (48), porphobilinogen deaminase (PBGD; −244 to +59) (23), glycophorin B (GPB; −158 to +42) (34), ferrochelatase (FC; −181 to +49) (44), and 5-aminolevulinate synthase (ALAS; −298 to +100) (42) were obtained from HEL cell genomic DNA by PCR. Numbers represent base pair distances from the transcription start site except for in GATA-1 promoters, in which numbers represent the distance from the end of exon 1. The DNA fragments obtained by PCR were cloned into the BglII site of the pHS2Luc, generating pHS2-GATA-Luc, pHS2-PBGD-Luc, pHS2-GPB-Luc, pHS2-FC-Luc, and pHS2-ALAS-Luc. Each construct was verified by sequencing. Information on primers and PCR conditions that were used for amplification will be provided upon request.
Transactivation analysis.
Transient transfections of K562 cells were performed as previously described (2) with minor modifications. Briefly, reporter, expression (in molar concentration 10 times higher than that of the reporter plasmid), and pSG5 vectors (to total 50 μg) were electroporated at 960 μF and 320 V by Gene Pulser (Bio-Rad) into 1.5 × 107 to 2 × 107 log-phase K562 cells, in RPMI 1640 medium without serum. After being placed at room temperature for 10 min, the cells were plated in 10 ml of the complete medium and incubated at 37°C for 24 h. The cells were harvested, and cell extracts were prepared by using 400 μl of reporter lysis buffer (Promega). One hundred microliters, some of which was diluted to 1:10, 1:50, or 1:100, was analyzed by using the luciferase assay system (Promega). All transfection assays were performed multiple times and with different preparations of the same plasmid. Luciferase activities obtained were corrected for transfection efficiencies by protein concentration of the cell extract measured by A570 (protein assay; Bio-Rad). A series of experiments have shown that under the conditions of our studies, FKLF and EKLF are overexpressed relative to the promoters, resulting in maximal activation of the globin gene promoters, thus allowing rational comparisons of promoter activation by FKLF and EKLF.
To examine effects of FKLF on endogenous globin gene expression, K562 cells were transiently transfected with 40 μg of either pSG5/FKLF or pSG5DD and 10 μg of pSVβ-Gal. Each transfection was done in duplicate. On day 1 an aliquot of cells was harvested for β-galactosidase (β-Gal) assay (2), and cells which gave a higher β-Gal activity by the duplicate assays were further incubated. On day 3 the cells were harvested, and total RNA was extracted.
Northern blotting and mRNA detection by PCR.
Northern blotting was performed with RNA probes by the standard method (3). The RNA probes were prepared by using Riboprobe in vitro transcription systems (Promega) from a PCR fragment which was cloned into the T vector. Studies of gene expression were done by semiquantitative reverse transcription (RT)-PCR. First-strand cDNA was prepared from 2 μg of tRNA by using a random hexamer from various cell lines (see Results) and fetal liver and adult bone marrow cells. Each cDNA sample was appropriately diluted to give similar amplifications of 28S rRNA under the same PCR conditions. The primer sequences and cycling conditions were as follows: for 5′-ACGGTAACGCAGGTGTCCT-3′ and 5′-CCTCTCGTACTGAGCAGGA-3′, 95°C for 3 min followed by various cycles of 95°C for 40 s, 56°C for 30 s, and 72°C for 60 s for 28S rRNA; for 5′-TCTGACTCTGGGGATGTCAC-3′ and 5′-CGGCAATCTGGAGTCTGGA-3′, 95°C for 3 min followed by various cycles of 95°C for 40 s, 58°C for 40 s, and 72°C for 60 s for FKLF; for 5′-TGCTGAGGAGAAGGCTGCCG-3′ and 5′-GGCTTGAGGTTGTCCATGTTT-3′, 95°C for 3 min followed by various cycles of 95°C for 40 s, 56°C for 40 s, and 72°C for 40 s for ɛ globin; and for 5′-AGAGGAGGACAAGGCTACTA-3′ and 5′-CCCTTGAGATCATCCAGGTGC-3′, 95°C for 3 min followed by various cycles of 95°C 40 s, 58°C for 40 s, and 72°C for 40 s for γ globin.
Statistical analysis.
Data were analyzed by two sample t tests with STATISTICA (StatSoft) computer software.
RESULTS
Cloning of FKLF cDNA.
To clone cDNAs encoding EKLF- or Sp1-type zinc finger proteins by PCR, a set of degenerate primers was designed on the basis of the amino acid homology of zinc finger region of Sp1 family genes and EKLF. The upstream and the downstream primers were prepared from the conserved amino acid sequences HTGE(KR)P and F(SM)RSDEL, respectively (Fig. 1A). The PCR fragment amplified from human fetal liver cell cDNA was cloned into plasmid, and 51 individual clones were sequenced. Among them, 20 clones had deduced amino acid sequences which were compatible to Cys2-His2-type zinc finger motifs. Neither EKLF nor Sp1 was found.
FIG. 1.
(A) Deduced polypeptide sequence of FKLF. Proline residues are shown in boldface. The sequence of the three zinc fingers is underlined, and the conserved polypeptide regions used for the preparation of degenerate primers are shaded. (B) Schematic representation of structure of deduced FKLF protein. Acidic and proline-rich regions are indicated by shaded and hatched boxes, respectively. Three zinc fingers are represented by striped boxes. Numbers above the boxes represent amino acid positions from the first methionine.
Three types of zinc finger structures similar to those of EKLF or Sp1 were found in the 20 clones. One was the human homologue of BKLF (7), and the other two were novel proteins (data not shown). On the basis of its properties (see below), one of the two novel genes was designated FKLF. The 5′ and 3′ unknown regions of the cDNA were obtained by PCR, and the reconstituted 2,012-bp FKLF cDNA included an ORF which potentially encodes 512 amino acids (Fig. 1A).
FKLF is a new member of the Sp1- or EKLF-type zinc finger proteins.
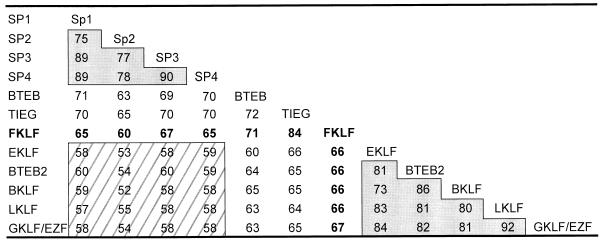
Inspection of the deduced polypeptide sequence of the FKLF reveals three contiguous zinc fingers present near the carboxyl-terminal end and two domains, a long amino-terminal domain and a short carboxyl-terminal domain, separated by the zinc finger domain (Fig. 1A and B). The structure of the zinc finger is C-X4-C-X12-H-X3-H-X7-C-X4-C-X12-H-X3-H-X7-C-X2-C-X12-H-X3-H (where X represents any amino acid residue), which is the same as those of Sp1 (18), EKLF (24), and other proteins of their family (1, 19). Figure 2 shows amino acid identities of zinc fingers of proteins which have the same finger structure. Notably, Sp1, Sp2, Sp3, and Sp4 form a high-homology group with 70 to 90% amino acid identities (Sp1 family). Similarly, EKLF, BTEB2, BKLF, LKLF, and GKLF form another high-homology group (the EKLF family). The amino acid identity across the two groups is relatively low, i.e., 50 to 60%. FKLF, TIEG, and BTEB show intermediate homologies (60 to 70%) to both the Sp1 and the EKLF protein families.
FIG. 2.
Percent amino acid identities of zinc fingers among Sp1- or EKLF-related proteins. Identities with FKLF zinc fingers are shown in boldface. Two shaded areas indicate high homology groups, i.e., the Sp1 family, including Sp1 (18), Sp2 (19), Sp3 (14, 19), and Sp4 (14), and the EKLF family, including EKLF (24), BTEB2 (37), LKLF (1), BKLF (7), and GKLF/EZF (11, 36). The identity between these two groups, indicated by striped area, is relatively low. Note that BTEB (17), TIEG (41), and FKLF show identities similar to both the Sp1 and the EKLF family proteins.
In contrast to the zinc finger region, the amino- and carboxyl-terminal domains of the FKLF protein show no homology to known proteins by GenBank homology search. The amino-terminal domain is characterized by high contents of proline and acidic amino acids (aspartic and glutamic acids) (Fig. 1A). The acidic residues are accumulated at amino acids 5 to 49 and 194 to 221 containing net charges of −8 and −6, respectively, and in these regions the proline residue is almost nonexistent. Therefore, the amino-terminal domain is composed of four subdomains, two acidic and two proline rich (Fig. 1B). Computer analyses of the polypeptide sequence with Wisconsin package version 9.1 (Genetics Computer Group) reveal that the acidic subdomains fold into α helices. Acidic amino acids occasionally appear to be accumulated on one face in a short stretch of the α helices, and in such a case hydrophobic amino acid residues appear on another face in the stretch. For example, an alignment around the α helix of 13 amino acid residues (no. 17 to 29), which comprises 3.6 α helical turns, shows that hydrophilic and hydrophobic residues are well separated and that acidic residues are accumulated on one face, suggesting that these amino acid residues form an amphipathic α helix.
The presence of acidic and proline-rich domains as well as that of the zinc finger domain suggests that the FKLF protein may function as a DNA-binding transcriptional activator (25).
FKLF is expressed predominantly in erythroid cells.
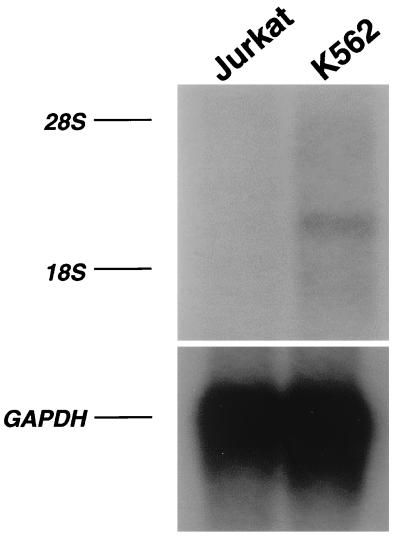
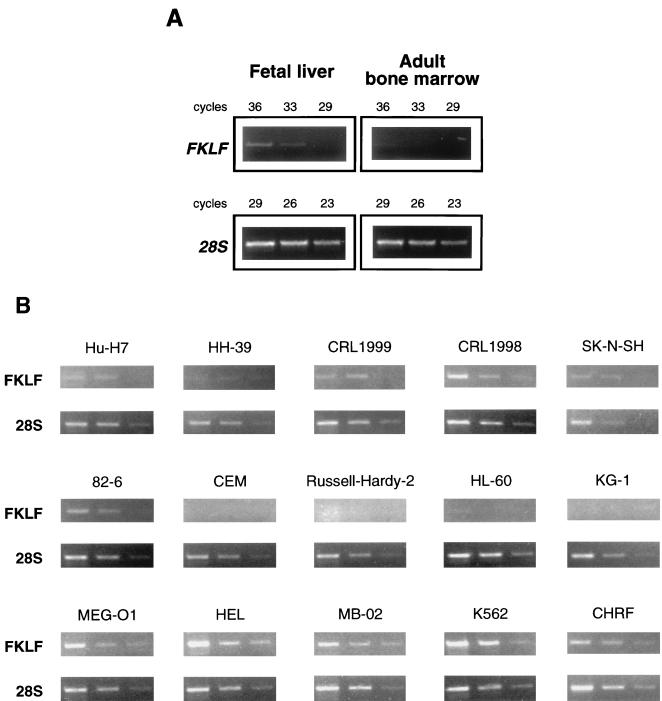
To examine the tissue specificity of FKLF mRNA expression, Northern blotting analyses were performed. Firstly, poly(A)+RNA of K562 (erythroid phenotype) and Jurkat (T-cell phenotype) lines was analyzed. As shown in Fig. 3, a 2.3-kb band (estimated from the positions of 18S and 28S ribosomal RNA) was detected in the RNA of K562 cells but not in that of Jurkat cells. Subsequently, we blotted a commercially available membrane (MTN Blots Human III), which contains poly(A)+RNA extracted from human stomach, thyroid gland, spinal cord, lymph node, trachea, adrenal gland, and bone marrow. We failed to detect a significant band in any of these tissues (data not shown), suggesting that FKLF has a restricted pattern of expression or that it is expressed at a very low level, if at all, in adult human tissues. Subsequently, the expression of FKLF mRNA in human fetal liver and adult bone marrow was compared by RT-PCR. As shown in Fig. 4A, the amplification of FKLF gene from the bone marrow cDNA was inefficient compared with that from the fetal liver cDNA, whereas these cDNAs gave similar band patterns in the amplification of 28S rRNA. These results confirm the very low level of expression, if any, of FKLF mRNA in adult human bone marrow and its expression in the cells of the erythroid human fetal liver.
FIG. 3.
FKLF mRNA expression in K562 and Jurkat cells by Northern blotting. Four micrograms of poly(A)+RNA was run on each lane. After standard capillary transfer to a nylon membrane, the RNA was blotted with a specific FKLF probe (upper panel). Subsequently, the FKLF probe was stripped off, and the membrane was reblotted with a murine GAPDH probe (lower panel). 28S and 18S rRNA positions are indicated.
FIG. 4.
FKLF mRNA expression in primary cells and established cell lines. First-strand cDNA was transcribed from total RNA by using random hexamers. The cDNA solution was diluted to give similar band intensities in the same cycles of amplification of 28S rRNA. (A) FKLF expression in adult human bone marrow and human fetal liver. Note that the amplification of FKLF message is less efficient in the cDNA of the adult bone marrow than in that of the fetal liver, while these cDNAs gave similar amplification of 28S rRNA. (B) FKLF expression in various cell lines. Total RNA used for the RT-PCR assays was prepared from erythroid lines (K562, CHRF, MB-02, HEL, and MEG-O1), a T-cell line (CEM), an Epstein-Barr virus-transformed B-cell line (Russell-Hardy-2), myeloid lines (KG-1 and HL-60), a fibroblastic line (82-6), a neuroblastoma line (SK-N-SH), an endothelial line (CRL1998), a smooth muscle line (CRL1999), a kidney epithelial line (HH-39), and a hepatoma line (Hu-H7). Three bands in each picture show the results of amplification in different cycles, i.e., from left to right, 35, 33, and 31 cycles for FKLF, and 22, 20, and 18 cycles for 28S rRNA. Note that the FKLF gene was amplified from RNA of all erythroid lines but not from RNA of lymphoid or myeloid lines. FKLF was also amplified in the endothelial line, which is noteworthy in view of the fact that endothelial cells express other hemopoietic lineage characters such as the erythropoietin receptor, kit and kit ligand, CD34, etc. Amplification of FKLF cDNA in the other nonhemopoietic cell lines was less efficient.
To further test the expression pattern of FKLF, RNA from a series of human cell lines was analyzed by semiquantitative RT-PCR. In this assay, template cDNAs were appropriately diluted to give similar band patterns of 28S rRNA (Fig. 4B). FKLF cDNA was efficiently amplified from cell lines with erythroid features (K562, HEL, CHRF, MB-02, and MEG-O1), a finding which was consistent in multiple experiments. In contrast, we failed to amplify the cDNA from myeloid lines (KG-1 and HL-60) or lymphoid lines (CEM and Russell-Hardy-2; T- and B-cell phenotypes, respectively). In cell lines originating from neuroblastoma, kidney epithelium, smooth muscle, fibroblasts, and hepatoma, the amplification of FKLF cDNA was less efficient. The only exception was the endothelial line, which gave a band pattern similar to that of the erythroid lines.
In conclusion, our expression data based on Northern blotting analyses of primary human tissues, semiquantitative RT-PCR of primary human hematopoietic tissues, Northern blotting analyses of established cell lines, and semiquantitative RT-PCR of established cell lines show (i) that FKLF is predominantly expressed in cell lines of the erythroid lineage and (ii) that in humans, in vivo expression of FKLF is limited to the cells of fetal liver erythropoiesis.
FKLF is a transcriptional activator.
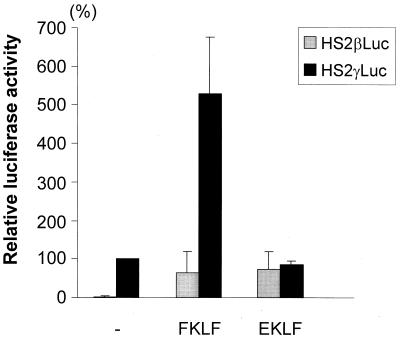
The structural features of the potential FKLF protein (Fig. 1 and 2) and the preferential expression pattern of the FKLF gene indicated that FKLF functions as a transcriptional activator of a gene(s) expressed in erythroid cells. Therefore, we tested the potential role of FKLF in globin gene regulation by transient-transfection assays. The FKLF cDNA in a mammalian expression vector pSG5DD (pSG5/FKLF) was cotransfected into K562 cells with a reporter construct containing a luciferase gene which was driven by the β-globin-hypersensitive site 2 (HS2) and either the β- or the γ-globin gene promoter. The HS2 was chosen as an enhancer because it has been used extensively in studies of other Krüppel-like factors (1, 2, 8). The luciferase activities are depicted in Fig. 5 along with those obtained without transactivator plasmid or with plasmid pSG5/EKLF, which activates the β gene promoter (2, 8).
FIG. 5.
trans activation of β- and γ-globin gene promoters by FKLF compared with EKLF. Reporter constructs containing a luciferase gene driven by HS2 and either the β or the γ gene promoter were transfected into K562 cells with or without the activator plasmid, pSG5/FKLF or pSG5/EKLF. Luciferase activities were corrected by protein concentrations and expressed as relative percentages of luciferase activity of pHS2γLuc which were not cotransfected by transactivator plasmid (100%). Data are expressed as means (columns) ± standard deviations (error bars) derived from multiple transfections with two different plasmid sets. Note that FKLF activates γ- and β (at a lower level)-globin gene promoters, while EKLF activates only the β gene promoter.
FKLF markedly activated the γ gene promoter (Fig. 5). In the presence of exogenous FKLF, the mean luciferase activity obtained from the γ promoter construct (pHS2γLuc) was 528% of that without FKLF, considered as 100% (P = 0.0001). With respect to the β-globin gene promoter construct, the average luciferase activity without FKLF was 1% of that of the γ gene construct lacking stimulation by FKLF (100%), demonstrating that the β promoter activity is considerably lower in K562 cells than γ promoter activity. In the presence of FKLF, the mean luciferase activity driven by the β promoter was 65%, while in the presence of EKLF the mean luciferase activity of the β promoter was 74% and that of the γ promoter was 86% (the luciferase activity of a γ construct lacking FKLF was considered to be 100%). The same pattern of activation of the promoters was observed in transfections using half amounts of EKLF and EKLF plasmids (data not shown). Thus, FKLF activated both β- and γ-globin promoters in K562 cells, showing that it functions as a transcriptional activator.
FKLF activates the γ-globin gene promoter through its CACCC box.
The fact that FKLF has zinc finger motifs as the proteins listed in Fig. 2 strongly suggests that FKLF achieves its function as a transcriptional activator by binding to a target DNA sequence with its zinc fingers. By analogy to Sp1 and EKLF, it is reasonable to infer that the CACCC element is the target DNA sequence of FKLF.
There are four CACCC sequences in the reporter plasmid pHS2γLuc described in the previous section, three are located in the HS2 fragment (hereafter referred to as HS-CAC1, HS-CAC2, and HS-CAC3 [300, 830, and 920 bp downstream of the KpnI site, respectively]), and the other is located in the γ gene promoter. To clarify whether FKLF activates the γ-globin gene through interaction with the HS2 or γ gene promoter CACCC box and, if so, which CACCC box FKLF uses for globin promoter activation, we generated deletion mutants of the reporter construct (Fig. 6) and examined the activation of the γ promoter by FKLF, using transfections of K562 cells.
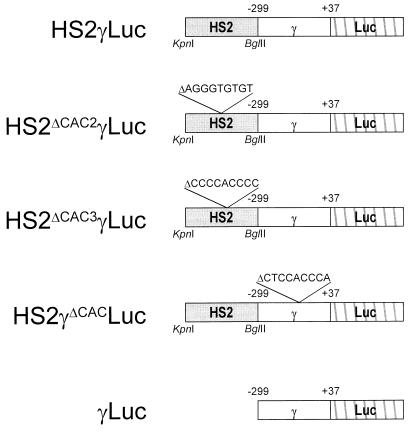
FIG. 6.
Structures of γ gene reporter constructs with deletion of a CACCC box or a whole HS2 sequence. Deletions of 9-bp CACCC sequences of HS2 (HS-CAC2 and HS-CAC3) and the γ gene promoter are indicated.
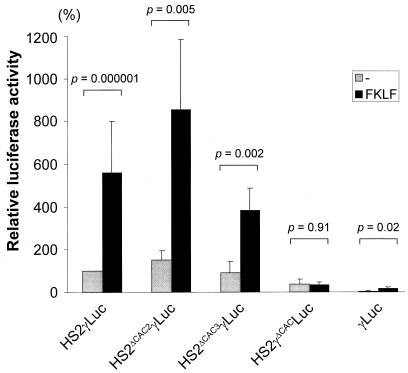
As shown in Fig. 7, the deletion of HS-CAC2 (pHS2ΔCAC2γLuc) and HS-CAC3 (pHS2ΔCAC3γLuc) only slightly affected the basal luciferase activities obtained in the absence of exogenous FKLF. By cotransfection with FKLF the mean luciferase activity from pHS2ΔCAC2γLuc was 853% of the activity without FKLF, and that from pHS2ΔCAC3γLuc was 382%. Both these values are significantly higher than those obtained without FKLF (P = 0.005 and P = 0.002, respectively). FKLF still raised the luciferase activity from the construct pγLuc, containing just γ gene promoter without HS2 (16 versus 5% without FKLF stimulation; P = 0.02 [Fig. 7]). This last result excludes the possibility that HS-CAC1 mediated the γ gene activation by FKLF, because all HS-CAC sites are absent from the pγLuc construct. By contrast, the reporter construct composed of the HS2 and a CACCC sequence-deleted γ gene promoter (pHS2γΔCACLuc) gave no increased luciferase activity as a result of the addition of FKLF (P = 0.91) (Fig. 7).
FIG. 7.
γ gene activation by FKLF with reporter constructs with deletions of a CACCC sequence or an HS2. Reporter constructs depicted in Fig. 7 were transiently transfected into K562 cells with or without pSG5/FKLF. Luciferase activities were corrected by protein concentrations and expressed as relative percentages of luciferase activity of pHS2γLuc which were not cotransfected by transactivator plasmid (100%). P values given by statistical comparison of data of two groups, i.e., the presence and the absence of FKLF, are shown above the columns. Notice that FKLF activates the γ gene promoter of the constructs pHS2ΔCAC2γLuc, pHS2ΔCAC3γLuc, and pγLuc lacking the HS2 element but failed to activate the γ gene promoter in the construct pHS2γΔCACLuc.
These data show that the CACCC element of the γ gene promoter is the mediator of γ gene activation by FKLF.
FKLF is not a general activator of promoters containing the CACCC element.
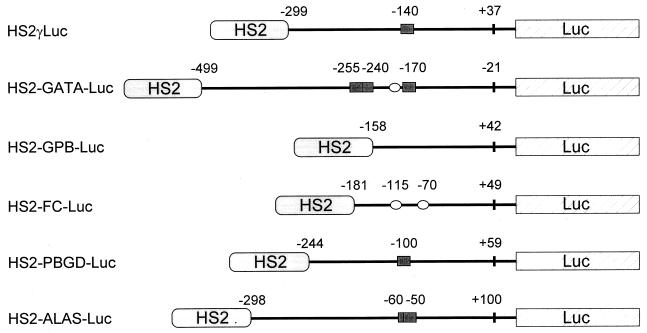
CACCC elements are found in the cis-regulatory sequences of a variety of genes expressed in both erythroid and nonerythroid cells. Hence, our finding that FKLF activates the γ-globin gene promoter through its CACCC element may be interpreted as evidence that (i) FKLF functions as a general activator of genes carrying a CACCC sequence or that (ii) FKLF activates limited repertoire of genes carrying a CACCC element. To address this question, we tested the activation by FKLF of five different promoters of genes expressed in erythroid tissues, i.e., GATA-1, GPB, FC, PBGD, and ALAS. The γ gene promoter of pHS2γLuc was replaced by these promoters, generating pHS2-GATA-Luc, pHS2-GPB-Luc, pHS2-FC-Luc, pHS2-PBGD-Luc, and pHS2-ALAS-Luc (Fig. 8). One to three CACCC elements are found in the GATA-1, PBGD, and ALAS gene promoters (Fig. 8). GC-rich sequences (possible binding sites for Sp1 and FKLF) are found in the GATA-1 and FC promoters (Fig. 8).
FIG. 8.
Reporter constructs containing a luciferase gene driven by the HS2 and a promoter of a gene expressed in erythroid cells. CACCC sequences and GC-rich potential Sp1-binding sites in the promoters are illustrated with solid rectangles and open ellipses, respectively. Numbers above the promoters are base pair distances from the cap site (γ, GPB, FC, PBGD, and ALAS) or from the end of exon 1 (GATA-1 [GATA]) and indicate the upstream and the downstream ends of the promoter sequences cloned and the positions of the CACCC or the GC-rich sequences.
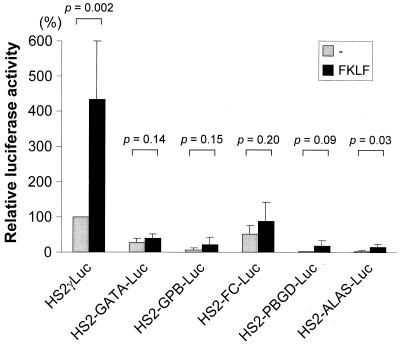
These reporter constructs were transiently transfected into K562 cells with or without pSG5/FKLF. Luciferase activities driven by the HS2 and six different promoters are shown in Fig. 9. The expected γ gene promoter activation by FKLF was observed; in contrast, FKLF failed to activate the GATA-1 promoter (P = 0.14) in spite of the fact that it carries three CACCC elements and one potential Sp1-binding site (Fig. 8). FKLF could not significantly activate the FC promoter (P = 0.20), although it has two Sp1-binding sites. Similarly, there was no significant activation of GPB or PBGD promoters by FKLF (P = 0.15 or 0.09, respectively). The ALAS promoter carrying two CACCC elements was slightly activated by FKLF (P = 0.03; Fig. 9). Thus, the mere presence of a CACCC motif or a GC-rich potentially Sp1-binding site is not sufficient for activation of a promoter by FKLF. Previously we have shown that the substitution of the β CACCC box for the γ CACCC box does not result in activation of the γ gene by EKLF, indicating that the activation of the γ-globin gene promoter depends on promoter context (2). A similar conclusion can be reached on the basis of the results described here. Our results allow us to conclude that FKLF is not a general activator of promoters containing CACCC motifs.
FIG. 9.
trans activation of promoters of various genes expressed in erythroid cells by FKLF. Reporter constructs depicted in Fig. 9 were transiently transfected into K562 cells with or without pSG5/FKLF. Luciferase activities were corrected by protein concentrations and expressed as relative percentages of luciferase activity of pHS2γLuc which was not cotransfected by transactivator plasmid (100%). P values given by statistical comparison of data of two groups, i.e., the presence and the absence of FKLF, are shown above the columns. Notice that FKLF activates the γ gene promoter as expected, but minimal (if any) activation of other erythroid promoters by FKLF was detected in spite of the fact that those promoters contained a CACCC or a GC-rich sequence.
FKLF activates the ɛ-globin gene promoter.
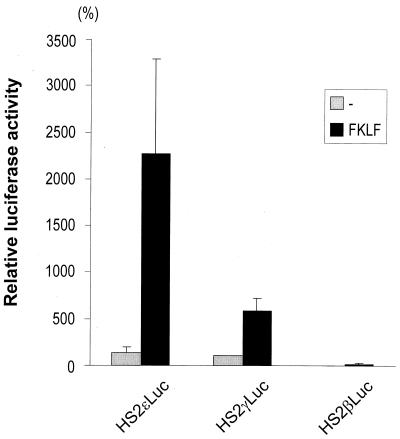
To determine whether FKLF is capable of activating the ɛ gene promoter, we performed transient-transfection experiments by using K562 cells and reporter constructs with either the ɛ, γ, or β promoter. Luciferase activities obtained from each construct with or without FKLF are depicted in Fig. 10. The highest activation was observed in the ɛ-globin gene promoter, while the levels of activation of the γ and the β gene promoters were consistent with the data described in the previous sections. Original luciferase activities (transfections without FKLF) were essentially the same in the ɛ and the γ gene constructs (135 and 100% average, respectively). In the presence of exogenous FKLF the luciferase activity from the ɛ gene construct was 2,270% of the activity driven by the γ promoter without FKLF, i.e., approximately fourfold higher than that of the γ gene construct (579%).
FIG. 10.
Comparison of activities of FKLF on ɛ-, γ-, and β-globin gene promoters. Reporter constructs containing a luciferase gene driven by HS2 and either the ɛ, the γ, or the β gene promoter were transfected into K562 cells with or without the activator plasmid, pSG5/FKLF. Luciferase activities were corrected by protein concentrations and expressed as relative percentages of luciferase activity of pHS2γLuc which was not cotransfected by transactivator plasmid (100%). Note that FKLF activates the ɛ gene promoter more strongly than the γ gene promoter.
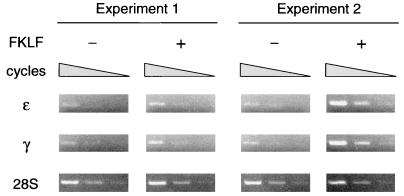
FKLF enhances endogenous ɛ- and γ-globin gene expression in K562 cells.
To analyze the potential in vivo role of FKLF in the regulation of ɛ- and γ-globin genes, we transiently transfected FKLF cDNA into K562 cells and assayed the resulting endogenous ɛ- and γ-globin gene expression by semiquantitative RT-PCR. As shown in Fig. 11, in two independent experiments both the ɛ- and the γ-globin gene were amplified more efficiently with exogenous FKLF than without it. The band intensities of ɛ gene amplicon relative to those of 28S rRNA (considered as 1) increased from 0.07 and 0.02 without FKLF to 0.29 and 1.94 with FKLF, and those of the γ gene increased from 0.07 and 0.1 without FKLF to 0.36 and 2.15 with FKLF. Thus, expression of exogenous FKLF enhanced endogenous expression of the ɛ- and γ-globin genes in K562 cells, providing further evidence that FKLF is a transcriptional activator of these globin genes in vivo.
FIG. 11.
Activation of the endogenous ɛ- and γ-globin genes by FKLF. FKLF cDNA was transiently transfected into K562 cells, and the expression of ɛ- and γ-globin genes was analyzed at day 3 by RT-PCR. Amplifications of the ɛ- and the γ-globin cDNAs with and without exogenous FKLF cDNA were compared under conditions which give rise to similar band patterns of 28S rRNA. Three bands in each picture show the results of amplification in different cycles; i.e., from left to right, 28, 26, and 24 cycles for the ɛ gene; 27, 25, and 23 cycles for the γ gene; and 21, 19, and 17 cycles for the 28S rRNA. The specificity of amplifications of the ɛ- and the γ-globin genes was confirmed by NcoI digestion of the PCR products, resulting in 177- and 60-bp bands for the ɛ gene and 140- and 97-bp bands for the γ gene (data not shown). Note that both the ɛ- and the γ-globin genes were more efficiently amplified in the presence of exogenous FKLF than in its absence, indicating that FKLF up-regulated the transcription of these genes.
DISCUSSION
The identification of transcription factors involved in the globin gene expression would advance our understanding of the mechanisms that guarantee the tissue and developmental stage specificity of globin genes. In view of the evidence that EKLF activates the β-globin gene through the CACCC element (5, 24), we postulated that γ gene expression is activated by a CACCC box binding trans-acting factor. Based on the expectation that such a factor should have an EKLF-like or Sp1-like zinc finger domain, we searched for such structures by screening cDNAs from 50- to 60-day-old human fetal liver. At this stage of development, the human fetal liver is predominantly hematopoietic, and over 90% of the cells are of the erythroid lineage. During this study, a cDNA which potentially encodes a novel Krüppel-type zinc finger protein, FKLF, was cloned.
A distinct feature of the family of Sp1- or EKLF-related zinc finger proteins is the presence of three contiguous Krüppel-type zinc fingers composed of two cysteine and two histidine residues near the carboxyl-terminal end (18). The deduced polypeptide sequence of FKLF cDNA showed the presence of such a zinc finger motif, indicating that FKLF is a member of the multigene EKLF-Sp1 family. Based on the amino acid homology of zinc fingers, the proteins of this family have been further classified into two categories, the Sp1 subfamily (19) and the EKLF subfamily (1), each characterized by a high degree of amino acid identity among its members. The zinc fingers of FKLF, BTEB, and TIEG are only 60 to 70% homologous to those of the Sp1 or the EKLF subfamilies, suggesting that FKLF, BTEB, and TIEG form a third group, with intermediate homologies to Sp1 and EKLF.
Aside from the zinc finger domain, FKLF shows no significant homologies to known proteins, a common characteristic of all the EKLF-type zinc finger proteins. Two features, the presence of acidic domains and proline-rich domains, which are found in a number of transcriptional activators (25), including EKLF (24), are characteristic of the amino-terminal domain of FKLF. A net negative charge and an amphipathic α-helical structure in a short stretch of amino acids are typical of the acidic transcriptional activation domain (12). Two acidic domains of FKLF possess these features, suggesting that FKLF functions as a transcriptional activator. Transient-transfection assays with β- and γ-globin promoter constructs confirmed this prediction. Experiments with deletion mutants of FKLF gene revealed that the main transcriptional activity of FKLF is present in the amino-terminal domain (data not shown) and that the two acidic domains constitute a key element for the trans-activation function of FKLF.
Our results suggest that the embryonic and fetal globin gene promoters are preferential target DNA sequences for FKLF among a broad repertoire of genes carrying CACCC motifs in their promoters. Using CACCC box deletion mutants of reporter constructs, we demonstrated that FKLF activates the γ-globin gene promoter through the CACCC element and not through the CACCC elements present in the HS2 enhancer. It remains to be clarified whether the interaction between the γ gene CACCC box and FKLF is direct or indirect. Considering that (i) Sp1 binds to the γ CACCC box (13), (ii) the zinc fingers of FKLF have the same amino acid residues, with Sp1 at the position most directly determining the site preference to the target DNA (data not shown [4]), and that (iii) generally, a DNA-binding trans-activating factor has separable DNA-binding and transactivation units (25), it is reasonable to assume that FKLF directly binds to the γ CACCC element.
Unlike EKLF, which is a pure β gene activator, FKLF activates not only the γ and the ɛ gene promoters, but also, to a much lesser degree, the β gene promoter in K562 cells. Previously (2) we attributed the specificity of the activation of β-globin gene promoter by EKLF to the recruitment of transcriptional activators (33, 40) and suggested that the transcriptional machinery recruited by EKLF is functional on the β gene promoter but not in the γ gene promoter. The transcriptional machinery recruited by FKLF may be different from that recruited by EKLF, which might be the reason for the preferential activation of the fetal and embryonic globin genes by this transcriptional factor.
The finding that FKLF can increase the transcription of endogenous ɛ- and γ-globin genes of K562 cells suggests that FKLF is one of the in vivo regulators of embryonic and fetal globin gene expression. Our data do not answer the question of whether the primary target of FKLF is the embryonic rather than the fetal globin gene, a possibility raised by the higher degree of activation of the ɛ-globin gene promoter as measured by the luciferase assays. The ontogenetic expression pattern of FKLF (very low expression in adult bone marrow compared to that in fetal liver) is compatible with a role of FKLF in the regulation of either the ɛ- or the γ-globin genes. Knockout experiments may provide an answer to this question.
ACKNOWLEDGMENTS
This study was supported by grants from the National Institute of Diabetes Digestive and Kidney Diseases and the National Heart, Lung, and Blood Institute, National Institutes of Health.
REFERENCES
- 1.Anderson K P, Kern C B, Crable S C, Lingrel J B. Isolation of a gene encoding a functional zinc finger protein homologous to erythroid Krüppel-like factor: identification of a new multigene family. Mol Cell Biol. 1995;15:5957–5965. doi: 10.1128/mcb.15.11.5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asano H, Stamatoyannopoulos G. Activation of β-globin promoter by erythroid Krüppel-like factor. Mol Cell Biol. 1998;18:102–109. doi: 10.1128/mcb.18.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 4.Berg J M. Sp1 and the subfamily of zinc finger proteins with guanine-rich binding sites. Proc Natl Acad Sci USA. 1992;89:11109–11110. doi: 10.1073/pnas.89.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieker J J. Role of erythroid Krüppel-like factor (EKLF) in erythroid-specific transcription. In: Stamatoyannopoulos G, editor. Molecular biology of hemoglobin switching. Andover, United Kingdom: Intercept; 1995. pp. 231–241. [Google Scholar]
- 6.Catala F, deBoer E, Habets G, Grosveld F. Nuclear protein factors and erythroid transcription of the human Aγ-globin gene. Nucleic Acids Res. 1989;17:3811–3827. doi: 10.1093/nar/17.10.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crossley M, Whitelaw E, Perkins A, Williams G, Fujiwara Y, Orkin S H. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol Cell Biol. 1996;16:1695–1705. doi: 10.1128/mcb.16.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donze D, Townes T M, Bieker J J. Role of erythroid Kruppel-like factor in human γ- to β-globin gene switching. J Biol Chem. 1995;270:1955–1959. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- 9.Evans T, Felsenfeld G, Reitman M. Control of globin gene transcription. Annu Rev Cell Biol. 1990;6:95–124. doi: 10.1146/annurev.cb.06.110190.000523. [DOI] [PubMed] [Google Scholar]
- 10.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett-Sinha L A, Eberspaecher H, Seldin M F, de Crombrugghe B. A gene for a novel zinc-finger protein expressed in differentiated epithelial cells and transiently in certain mesenchymal cells. J Biol Chem. 1996;271:31384–31390. doi: 10.1074/jbc.271.49.31384. [DOI] [PubMed] [Google Scholar]
- 12.Giniger E, Ptashne M. Transcription in yeast activated by a putative amphipathic α helix linked to a DNA binding unit. Nature (London) 1987;330:670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- 13.Gumucio D L, Rood K L, Blanchard-McQuate K L, Gray T A, Saulino A, Collins F S. Interaction of Sp1 with the human γ globin promoter: binding and transactivation of normal and mutant promoters. Blood. 1991;78:1853–1863. [PubMed] [Google Scholar]
- 14.Hagen G, Müller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;21:5519–5525. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hartzog G A, Myers R M. Discrimination among potential activators of the β-globin CACCC element by correlation of binding and transcriptional properties. Mol Cell Biol. 1993;13:44–56. doi: 10.1128/mcb.13.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikuta T, Papayannopoulou T, Stamatoyannopoulos G, Kan Y W. Globin gene switching. In vivo protein-DNA interactions of the human β-globin locus in erythroid cells expressing the fetal or the adult globin gene program. J Biol Chem. 1996;271:14082–14091. doi: 10.1074/jbc.271.24.14082. [DOI] [PubMed] [Google Scholar]
- 17.Imataka H, Sogawa K, Yasumoto K, Kikuchi Y, Sasano K, Kobayashi A, Hayami M, Fujii-Kuriyama Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11:3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadonaga J T, Carner K R, Masiarz F, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 19.Kingsley C, Winoto A. Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol Cell Biol. 1992;12:4251–4261. doi: 10.1128/mcb.12.10.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulozik A E, Bellan-Koch A, Bail S, Kohne E, Kleihauer E. Thalassemia intermedia: moderate reduction of β globin gene transcriptional activity by a novel mutation of the proximal CACCC promoter element. Blood. 1991;77:2054–2058. [PubMed] [Google Scholar]
- 21.Lin H J, Han C Y, Nienhuis A W. Functional profile of the human fetal γ-globin gene upstream promoter region. Am J Hum Genet. 1992;51:363–370. [PMC free article] [PubMed] [Google Scholar]
- 22.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 23.Mignotte V, Eleouet J F, Raich N, Romeo P-H. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci USA. 1989;86:6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller I J, Bieker J J. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 26.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature (London) 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 27.Orkin S H. Transcriptional factors and hematopoietic development. J Biol Chem. 1995;270:4955–4958. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- 28.Orkin S H, Antonarakis S E, Kazazian H H., Jr Base substitution at position −88 in a β-thalassemic globin gene. Further evidence for the role of distal promoter element ACACCC. J Biol Chem. 1984;259:8679–8681. [PubMed] [Google Scholar]
- 29.Orkin S H, Kazazian H H, Jr, Antonarakis S E, Goff S C, Boehm C D, Sexton J P, Waber P G, Giardina P J V. Linkage of β-thalassemia mutations and β-globin gene polymorphisms with DNA polymorphisms in human β-globin gene cluster. Nature (London) 1982;296:627–631. doi: 10.1038/296627a0. [DOI] [PubMed] [Google Scholar]
- 30.Pellegrino G R, Berg J M. Identification and characterization of “zinc-finger” domains by the polymerase chain reaction. Proc Natl Acad Sci USA. 1991;88:671–675. doi: 10.1073/pnas.88.2.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins A C, Gaensler K M L, Orkin S H. Silencing of human fetal globin expression is impaired in the absence of the adult β-globin gene activator protein EKLF. Proc Natl Acad Sci USA. 1996;93:12267–12271. doi: 10.1073/pnas.93.22.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perkins A C, Sharpe A H, Orkin S H. Lethal β-thalassemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature (London) 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 33.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 34.Rahuel C, Vinit M-A, Lemarchandel V, Cartron J-P, Roméo P-H. Erythroid-specific activity of the glycophorin B promoter requires GATA-1 mediated displacement of a repressor. EMBO J. 1992;11:4095–4102. doi: 10.1002/j.1460-2075.1992.tb05502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Shields J M, Christy R J, Yang V W. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sogawa K, Imataka H, Yamasaki Y, Kusume H, Abe H, Fujii-Kuriyama Y. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res. 1993;7:1527–1532. doi: 10.1093/nar/21.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stamatoyannopoulos G, Josephson B, Zhang J-W, Li Q. Developmental regulation of human γ-globin genes in transgenic mice. Mol Cell Biol. 1993;13:7636–7644. doi: 10.1128/mcb.13.12.7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatoyannopoulos G, Nienhuis A W. Hemoglobin switching. In: Stamatoyannopoulos G, Nienhuis A W, Majerus P, Varmus H, editors. The molecular basis of blood diseases. W. B. Philadelphia, Pa: Saunders; 1993. pp. 107–154. [Google Scholar]
- 40.Stargell L A, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- 41.Subramaniam M, Harris S A, Oursler M J, Rasmussen K, Riggs B L, Spelsberg T C. Identification of a novel TGF-β-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–4912. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Surinya K H, Cox T C, May B K. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. J Biol Chem. 1997;272:26585–26594. doi: 10.1074/jbc.272.42.26585. [DOI] [PubMed] [Google Scholar]
- 43.Troutt A B, McHeyzer-Williams M G, Pulendran B, Nossal G J V. Ligation-anchored PCR: a simple amplification technique with single-sided specificity. Proc Natl Acad Sci USA. 1992;89:9823–9825. doi: 10.1073/pnas.89.20.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tugores A, Magness S T, Brenner D A. A single promoter directs both housekeeping and erythroid preferential expression of the human ferrochelatase gene. J Biol Chem. 1994;269:30789–30797. [PubMed] [Google Scholar]
- 45.Ulrich M J, Ley T J. Function of normal and mutated γ-globin gene promoters in electroporated K562 erythroleukemia cells. Blood. 1990;75:990–999. [PubMed] [Google Scholar]
- 46.Wijgerde M, Gribnau J, Trimborn T, Nuez B, Philipsen S, Grosveld F, Fraser P. The role of EKLF in human β-globin gene competition. Genes Dev. 1996;10:2894–2902. doi: 10.1101/gad.10.22.2894. [DOI] [PubMed] [Google Scholar]
- 47.Yu C-Y, Motamed K, Chen J, Bailey A D, Shen C-K J. The CACC box upstream of human embryonic ɛ globin gene binds Sp1 and is a functional promoter element in vitro and in vivo. J Biol Chem. 1991;266:8907–8915. [PubMed] [Google Scholar]
- 48.Zon L I, Orkin S H. Sequence of the human GATA-1 promoter. Nucleic Acids Res. 1992;20:1812. doi: 10.1093/nar/20.7.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]