Abstract
Exportins as the key mediators of nucleocytoplasmic transport have been identified as the controllers of the passage of numerous types of crucial cancer-related proteins. Targeting exportins in cancer cells might represent an emerging strategy in cancer intervention with the potential to affect clinical outcomes. Here, we focused on the prognostic and therapeutic values of Exportin-T (XPOT) in neuroblastoma. The correlation between the expression and prognostic values of XPOT in patients with neuroblastoma was investigated based on both published transcriptome data and our clinical data. Then, decision curve analysis (DCA) was implemented to identify a XPOT risk prediction model. In addition, RNA inference was performed to silence the expression of XPOT to further investigate the specific roles of XPOT in the progression of neuroblastoma in vitro. Overexpression of XPOT mRNA was associated with poor clinical characteristics, such as age at diagnosis more than 18 months, amplification of MYCN, and advanced International Neuroblastoma Staging System (INSS) stage, and XPOT expression was identified as an independent poor prognosis factor for neuroblastoma using Cox proportional hazards model (P < .001). DCA suggested that neuroblastoma patients could benefit from XPOT risk prediction model-guided interventions (status of MYCN + INSS stage + XPOT). Experimentally, knockdown of XPOT by small interfering RNA inhibited the proliferation and migration in neuroblastoma cells. XPOT is identified as a novel prognostic predictor and potential therapeutic target for neuroblastoma patients. Further investigation should focus on the profound molecular mechanism underlying the tumor inhibition activity of XPOT inhibitors.
Keywords: Exportin-T, neuroblastoma, prognostic marker, therapeutic target, prognosis
Introduction
Neuroblastomas are the most common extracranial solid malignancy in children originating from the adrenal glands or sympathetic ganglia.1,2 About 1.1 per 100 000 children are diagnosed with neuroblastoma annually, accounting for ∼6% of all childhood cancers.3 Neuroblastoma is a highly heterogeneous disease with a broad spectrum of clinical behavior.4 Tumors may spontaneously regress or mature, or they can progress and metastasize despite multimodality treatment.4 Therefore, pretreatment risk stratification is essential for determining the risk group and therapeutic strategy of patients.
Over the past decade, International Neuroblastoma Risk Group (INRG) classification system has provided invaluable information for determining the prognosis and therapy stratification for patients diagnosed with neuroblastoma.5 The INRG classification system is a strict schema, considering the criteria International Neuroblastoma Staging System (INSS) stage, age at diagnosis, histologic category, tumor differentiation, MYCN status, chromosomal aberrations, and tumor cell ploidy.5 However, this system has not yet been adopted by all regions,6 and its clinical application is limited internationally because of the difficulty of acquisition of some factors, such as tumor cell ploidy and chromosomal aberrations. Meanwhile, patients with low- and intermediate-risk neuroblastoma have achieved favorable prognosis, while the prognosis of treatment remains unfavorable for the high-risk ones, with the 5-year survival rate under 50%.7 This stagnation in therapeutic advances for high-risk neuroblastoma motivates efforts in excavating the underlying molecular biology of neuroblastoma and exploring novel therapeutic targets for further survival improvement.
Exportins are a group of karyopherins that regulate the transferring of proteins from nucleus to cytoplasm, which have been identified as the controllers of the passage of numerous types of crucial cancer-related proteins.8 Emerging evidence has considered exportin inhibitors as therapeutic targets against cancer and they have shown preclinical anticancer activity.9 Exportin-T (XPOT) is a member of the karyopherin-β family and acts as a specific mediator of the export of transfer RNA (tRNA).10,11 It shuttles from the nucleus to the cytoplasm bidirectionally through nuclear pore complexes by interacting with GTP-bound Ran (Ran-GTP).12 Previous studies have reported the critical roles of XPOT in the development and progression of various tumors, such as breast cancer,13 hepatocellular carcinoma,14 and promyelocytic leukemia.15 In addition, Exportin 1 (XPO1, another exportin family member, has been reported to play important roles in neuroblastoma. Inhibition of XPO1 suppresses neuroblastoma cell proliferation via reduction of the phosphorylation level of Forkhead box O3 (FOXO3a).16 The neuroblastoma animal models proved that the second-generation XPO1 inhibitor selinexor (KPT-330) showed anti-tumor effect in vivo.17 Despite these advances, XPOT remains 1 mysterious player in tumorigenesis, and only a few studies investigated the biological function and prognosis value of XPOT in the pathogenesis of neuroblastoma.
Here, we investigated the expression and prognostic value of XPOT in patients with neuroblastoma using 2 published datasets, and presented an XPOT risk prediction model by decision curve analysis (DCA). Then, its expression and the prognostic value of XPOT were verified clinically based on 64 neuroblastoma patients. In addition, the specific role of XPOT in the progression of neuroblastoma was analyzed using RNA inference by in vitro. Together, this study attempted to explore a promising predictor for predicting prognosis of neuroblastoma and reveal the significance of XPOT in neuroblastoma oncogenesis. Targeting XPOT in neuroblastoma cells might represent a novel strategy in treatment intervention of neuroblastoma.
Materials and Methods
Transcriptome Data
The mRNA expression of XPOT and clinical annotation were evaluated using R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl) on the datasets from the Sequencing Quality Control Consortium (SEQC, GEO accessing number GSE4971018) and the European Neuroblastoma Research Consortium (NRC, GEO accessing number GSE8504719)
Cell Lines and Cell Culture
The human neuroblastoma cell lines SK-N-BE(2), SH-SY5Y, and SK-N-SH were obtained from Guangzhou Jennio Biotech Co., Ltd and were cultured in Dulbecco’s modified Eagle medium (DMEM; HyClone) supplemented with 10% fetal bovine serum (FBS; Gibco) at 37 °C in a humidified incubator with 5% CO2.
Specimens and Tissue Microarray
Tissue samples were obtained from 64 neuroblastoma patients, who received radical resection at the Department of Pediatric Surgery, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine from 2012 to 2015. The diagnosis was confirmed by pathological biopsy. The study protocol was approved by the Ethics Committee of Xinhua Hospital (Approval number: XHEC-D-2018-034). Written informed consent was obtained from all donors and their relatives. The age at diagnosis ranged from 2 to 156 m, and the male:female ratio was 9:7. The 5-year follow-up overall survival was 26.7 ± 16.1 m. Tissue microarrays were constructed by Servicebio Technology Co., Ltd and scanned by Pannoramic MIDI20 (3D HISTECH).
Immunohistochemistry Staining
Immunohistochemistry staining was performed according to the procedure described by Meseure et al.21 XPOT antibodies for immunohistochemistry staining was obtained from Lifespan Biosciences (LS-C160669, rabbit, 1:1000). A semiquantitative histologic score (H-score) was applied for the evaluation of the immunohistochemical staining results. The H-score considers both the degrees of staining intensity and the %age of positive neoplastic cells. The intensity was scored on a scale of 0 to 3 (0, negative; 1, weak; 2, medium; 3, strong). The formula for the H-score is as follows: H-score = ∑(Pi × I) = (%age of cells with weak intensity × 1) + (%age of cells with moderate intensity × 2) + %age of cells with strong intensity × 3),22 where Pi = %age of positive tumor cells and I = staining intensity. A total of 2 pathologists assessed the scoring independently without prior knowledge of the clinical outcomes.
Small Interfering RNA Transfection
Small interfering RNAs (siRNAs) targeting XPOT were designed and synthesized by Huagene Biotech Co. Ltd. The sequences of XPOT-siRNAs were as follows: XPOT-siRNA1, 5′- GCUAGUGCUUUGCAGGAUATT-3′; XPOT-siRNA2, 5′- GCACAUUCCAUGUGUACUATT-3′; and XPOT-siRNA3, 5′- GCUGGAGUGCUGAUUGUUATT-3′. Negative control-siRNA (NC-siRNA) or XPOT-siRNA were transfected to SK-N-BE(2), SH-SY5Y, and SK-N-SH cells using the Lipofectamine™ RNAiMAX Transfection Reagent (Invitrogen).
Real-Time-Quantitative Polymerase Chain Reaction
Total RNA of SK-N-BE(2), SH-SY5Y, and SK-N-SH cells was extracted using Trizol (Takara) following the manufacturer’s instructions. Then, 1 μg of total RNA was extracted to synthesize cDNA as the polymerase chain reaction (PCR) template using PrimeScript Reverse Transcriptase (Takara). The primers used for amplification were as follows: XPOT 5′- AGGGAGACGCTCATATCATGG-3′ (forward), 5′-TTGGGCGGCTTTATTTCGTAT-3′ (reverse); glyceraldehyde 3-phosphate dehydrogenase 5′-CAACAGCCTCAAGATCATCAGC-3′ (forward), 5′-TTCTAGACGGCAGGTCAGGTC-3′ (reverse). Real-time (RT)-qPCR was conducted using the StepOnePlus™ RT-PCR system (Applied Biosystems) and XPOT expression levels were detected using the SYBR-Green kit (Takara). XPOT expression levels were calculated using the 2-ΔΔCq methods.23
Western Blot
Total protein was extracted from SK-N-BE(2), SH-SY5Y and SK-N-SH cell lines using radio immunoprecipitation assay (RIPA) buffer (Cell Signaling Technology) with 1 × phenylmethanesulfonyl fluoride (PMSF; Beyotime). Proteins (15 µg/lane) were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under constant voltage and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore) under constant current for 90 min. Then, all blots were blocked with fresh 5% skimmed milk for 2 h at room temperature. Next, PVDF membranes were incubated with polyclonal antibody against XPOT (LS-C160669, rabbit, 1:1000) purchased from Lifespan Biosciences and polyclonal antibody against β-actin (#4970, rabbit, 1:1000) acquired from Cell Signaling Technology overnight at 4 °C. Finally, all blots were incubated with a horseradish peroxidase-conjugated secondary antibody (A0208, rabbit, 1:1000) for 2 h at room temperature, and analyzed using BIO-RAD ChemiDoc XRS+(Bio-Rad).
Cell Proliferation Assays
Cell proliferation ability was tested by a Cell Counting Kit-8 reagent (Dojindo). In brief, cells were seeded in 96-well plates at a density of 1 × 103 cells per well and cultured with DMEM containing 10% FBS at day 0. Different rows are marked as day1 to day5 to be measured at different time points. For next every 24 h (total 5 days), after ensuring the cells were not contaminated by observation via a microscope, cells were cultured in DMEM with 10% Cell Counting Kit-8 (CCK-8) reagent for 2 h at 37 °C. Then, cell proliferation ability was assessed by the absorbance value at 450 nm using an Automated Microplate Reader (Bio-Rad).
Colony Formation Assay
Colony formation assay was used to assess single cell proliferation ability. Firstly, cells were cultured at a density of 1 × 104 cells per well in 6-well plates for 1 week. Next, cells were fixed by 4% paraformaldehyde (Sigma) for 15 min at room temperature. Then, cells were stained by 0.5% crystal violet (Beyotime) for 15 min at room temperature. Finally, after washing with water, colonies were observed under the optical microscope (Leica), and the number of colonies was counted.
Cell Migration Assays
Cell migration ability was measured in 24-well Transwell chambers (Corning). Cells were plated at a density of 2 × 104 per well onto the upper chamber with 200 μL serum-free DMEM. The lower chamber was filled with 500 μL DMEM supplemented with 10% FBS. After incubation for 48 h at 37 °C in a humidified incubator with 5% CO2, cells attached to the low surface of the membrane were fixed by 4% paraformaldehyde (Sigma) for 15 min at room temperature and stained with 0.5% crystal violet (Beyotime) for another 15 min. Finally, the migrated cells were visualized under an optical microscope (Leica), and counted from 5 randomly selected fields.
Statistical Analysis
Statistical analysis was conducted by SAS v8.0 (SAS Institute Inc.), Prism 6 software (GraphPad Software, Inc.) and R v3.5.0 (R Core Team, 2018). Pearson’s χ test or Cochran-Mantel-Haenszel (CMH) test was used to analyze the association between XPOT expression and clinical characteristics in neuroblastoma. Univariate and multivariate Cox regression models were performed for prognostic analysis. Log-rank (Mantel–Cox) test was used to determine the significant difference. DCA was performed by R package.24 X-tile software was used to determine the best cut-off value of XPOT expression in stratification analysis.25 All experiments were conducted independently 3 times. The results were considered statistically significant when P < .05.
Results
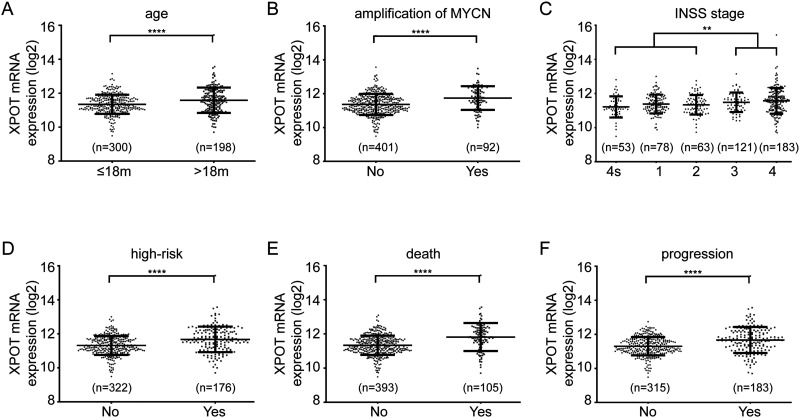
Patients with high-risk clinical characteristic have higher XPOT expression in neuroblastoma tissues than patients with low-risk clinical characteristics. To investigate the correlation between mRNA expression level of XPOT and clinical characteristics, 2 independent datasets, SEQC and NRC, were analyzed, respectively. According to the SEQC dataset, higher XPOT mRNA expression level was associated with several high-risk parameters, such as age at diagnosis >18 months, amplification of MYCN, high-risk group, and advanced INSS stage (Figure 1A to D, P < .05). Moreover, patients with death or tumor progression showed higher XPOT mRNA expression level (Figure 1E and F, P < .05). Further analysis of the NRC dataset identified consistent results (Figure S1).
Figure 1.
Patients with high-risk clinical characteristic have higher XPOT expression in neuroblastoma tissues than patients with low-risk clinical characteristics. Higher XPOT mRNA is expressed in patients with age at diagnosis > 18 months (A), amplification of MYCN (B), advanced INSS stage (C), high-risk group (D), death from disease (E), and tumor progression (F). *P < .05.
Abbreviations: INSS, International Neuroblastoma Staging System; XPOT, Exportin-T.
To further clarify the relevance between XPOT mRNA expression and these clinical characteristics, a stratification analysis was performed. The best cut-off value of XPOT mRNA expression level was determined by X-tile software (best P value). According to the SEQC dataset, higher XPOT mRNA expression was associated with age diagnosed >18 months, amplification of MYCN, advanced INSS stage, high-risk group, tumor progression, and death from disease (Table 1, P < .05). NRC analysis results also supported the above results (Table S1).
Table 1.
Association Between XPOT mRNA Expression and Clinical Characteristics in Neuroblastoma Patients (SEQC).
| XPOT expression | ||||
|---|---|---|---|---|
| Factors | Cases No. | low, No. (%) | high, No. (%) | P value |
| Gender | .544 | |||
| female | 211 | 188 (89.1) | 23 (10.9) | |
| male | 287 | 261 (90.9) | 26 (9.1) | |
| Age (month) | <.001 | |||
| ≤18 | 300 | 287 (95.7) | 13 (4.3) | |
| >18 | 198 | 162 (81.8) | 36 (18.2) | |
| MYCN amplification | <.001 | |||
| No | 401 | 388 (96.8) | 13 (3.2) | |
| Yes | 92 | 57 (62.0) | 35 (38.0) | |
| Tumor stage (INSS) | <.001 | |||
| 4s + 1 + 2 | 252 | 247 (98.0) | 5 (2.0) | |
| 3 + 4 | 246 | 202 (82.1) | 44 (17.9) | |
| High risk | <.001 | |||
| No | 322 | 317 (98.5) | 5 (1.5) | |
| Yes | 176 | 132 (75.0) | 44 (25.0) | |
| Progression | <.001 | |||
| No | 315 | 307 (97.5) | 8 (2.5) | |
| Yes | 183 | 142 (77.6) | 41 (22.4) | |
| Death | <.001 | |||
| No | 393 | 377 (95.9) | 16 (4.1) | |
| Yes | 105 | 72 (68.6) | 33 (31.4) | |
Pearson's χ2 test was used for statistical analysis.
Abbreviations: INSS, International Neuroblastoma Staging System; XPOT, Exportin-T; SEQC, Sequencing Quality Control Consortium.
Higher XPOT Level is Correlated to Poor Prognosis
Univariate and multivariate analysis suggested XPOT expression as an independent poor prognosis factor for neuroblastoma according to the SEQC dataset (Table 2, P < .001). NRC analysis results also supported the above results (Table S2). These results indicated that high XPOT expression level might be associated with poor clinical outcomes, and their correlation should be confirmed by further analysis.
Table 2.
Cox Proportional Hazards Model for Prognostic Factor Analysis in Neuroblastoma Patients (SEQC).
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Variables | Favorable/Unfavorable | HR (95% CI) | P value | HR (95% CI) | P value |
| Gender | female/male | 0.799 (0.545-1.173) | .253 | ||
| Age (month) | ≤18/>18 | 8.111 (4.978-13.214) | <.001 | 1.895(1.542-0.832) | .169 |
| MYCN amplification | No/Yes | 7.793 (5.262-11.541) | <.001 | 1.502(0.934-2.415) | .093 |
| Tumor stage (INSS) | 4s + 1 + 2/3 + 4 | 14.515 (7.323-28.768) | <.001 | 2.693(1.124-6.455) | .026 |
| High risk | No/Yes | 21.422 (11.931-38.462) | <.001 | 5.744(2.371-13.915) | <.001 |
| XPOT expression | low/high | 8.613 (5.644-13.142) | <.001 | 2.645(1.637-4.274) | <.001 |
Cox regression was used for statistical analysis.
Abbreviations: 95%CI, 95% confidence interval; HR, hazard ratio; INSS, International Neuroblastoma Staging System; XPOT, Exportin-T; SEQC, Sequencing Quality Control Consortium.
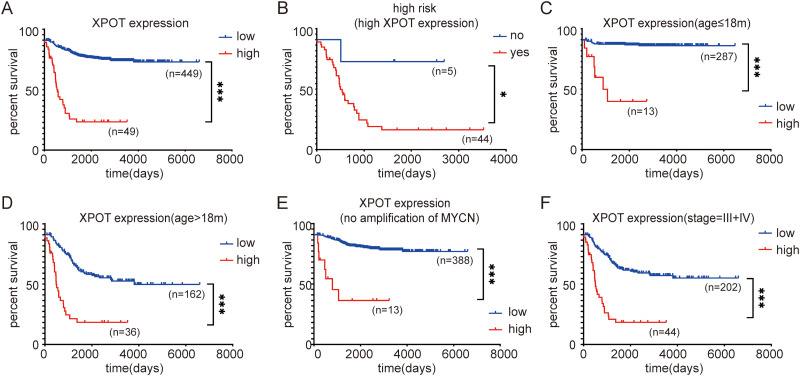
Overall survival analysis was performed to evaluate the prognostic impact of XPOT mRNA expression in neuroblastoma patients. According to the SEQC dataset, patients with high XPOT mRNA expression presented shorter overall survival time relative to those with low XPOT expression (Figure 2A, P < .05). For the subgroup with high XPOT expression, patients with high risk presented shorter overall survival time (Figure 2B, P < .05). In the stratification analysis for clinical parameters, higher XPOT level was related to poor prognosis in patients with age diagnosed <18 months, with age diagnosed >18 months, without amplification of MYCN, and with advanced INSS stage (Figure 2C to F, P < .05). NRC analysis showed consistent results (Figure S2). These results demonstrated that neuroblastoma patients with higher XPOT levels presented poor prognosis.
Figure 2.
Higher XPOT level is correlated to poor prognosis. (A) Patients with high XPOT mRNA expression present shorter overall survival time relative to those with low XPOT expression. (B) For the subgroup with high XPOT expression, patients with high risk have a poor prognosis. (C) For patients with age diagnosed <18 months, higher XPOT expression shows lower overall survival. (D) For patients with age diagnosed >18 months, higher XPOT expression shows lower overall survival. (E) For patients without amplification of MYCN, higher XPOT expression achieves lower overall survival. (F) For patients with advanced INSS stage, higher XPOT expression achieves lower overall survival. INSS, International Neuroblastoma Staging System.
* P < .05.
Abbreviations: INSS, International Neuroblastoma Staging System; XPOT, Exportin-T.
Neuroblastoma Patients Could Benefit From XPOT Risk Prediction Model-Guided Interventions
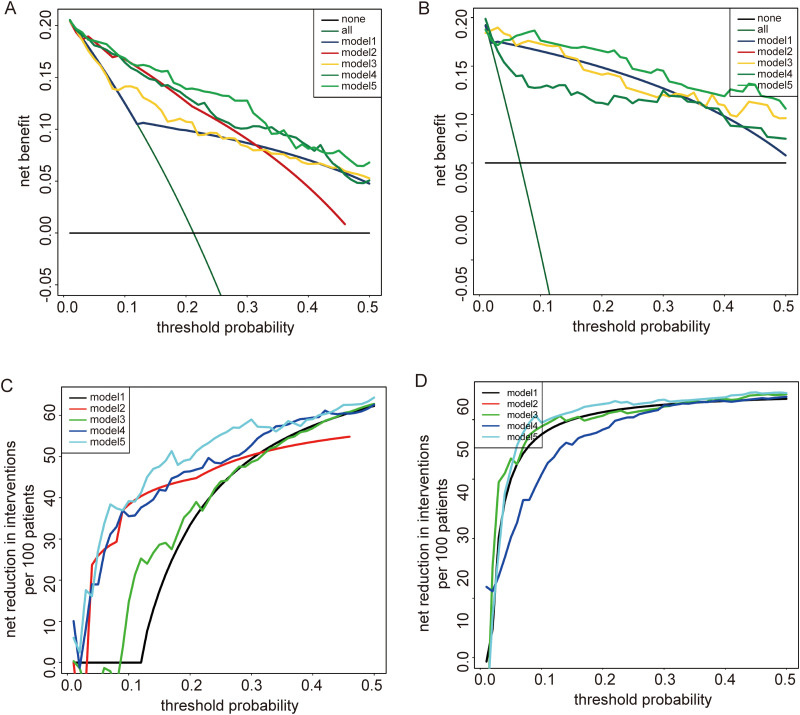
DCA analysis was used to detect whether patients could benefit from a risk prediction model that included XPOT. According to the SEQC dataset, DCA analysis identified that model 5 (status of MYCN + stage + XPOT) had a higher net benefit compared with other models in the threshold probability of 0–0.5 (Figure 3A). Given that age at diagnosis <18 months is a favorable prognostic factor, we performed a stratification analysis. For patients with diagnosed age <18 months, model 5 also had a higher net benefit (Figure 3B). Besides, net reduction curves, which show the potential to avoid unnecessary intervention, implied that model 5 had the best reduction rates compared with other models (Figure 3C). For patients with diagnosed age <18 months, model 5 also had excellent reduction rates (Figure 3D). In the NRC dataset, models 2, 4, and 5 showed good net benefit and reduction rates in the threshold probability of 0–0.5, with similar net benefit among 3 models (Figure S3A and S3B) for all patients. Although, for patients with diagnosed age <18 months, model 5 (threshold probability 0–0.25) had a higher net benefit and good reduction rates (Figure S3C and S3D). These results suggested that neuroblastoma patients, especially those with diagnosed age <18 months, could benefit from XPOT risk prediction model-guided interventions.
Figure 3.
Decision curve analysis and net reduction curves show that neuroblastoma patients can benefit from XPOT risk prediction model-guided interventions. For all patients, model 5 achieves the highest net benefit (A) and the greatest ability to avoid unnecessary intervention (C) in the threshold probability of 0 to 0.5. For patients with diagnosed age <18 months, model 5 has a higher net benefit (B) and greater ability to avoid unnecessary intervention (D) than the other models in the threshold probability of 0 to 0.5.
Abbreviations: INSS, International Neuroblastoma Staging System; none = no patients received interventions; all = all patients received interventions, model 1 = status of MYCN, model 2 = INSS stage, model 3 = status of MYCN + XPOT, model 4 = INSS stage + XPOT, model 5 = status of MYCN + INSS stage + XPOT; XPOT, Exportin-T.
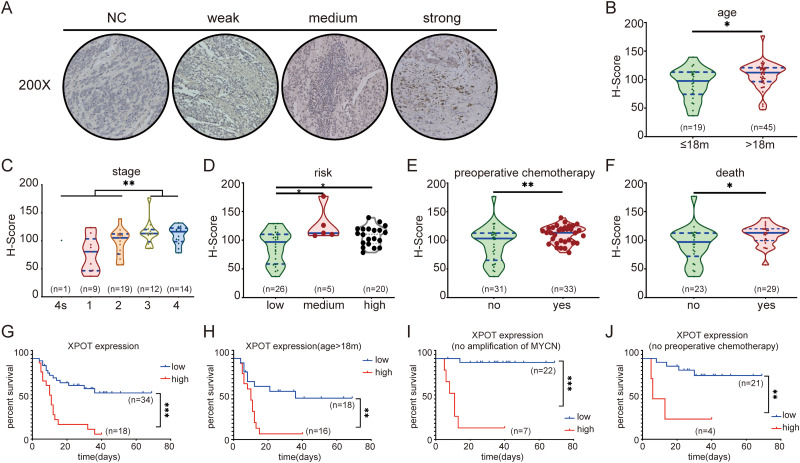
Higher XPOT Protein Expression is Associated with Poor Prognosis
Staining intensity was divided into weak, medium, and strong under an optical microscope (Figure 4A). XPOT protein was significantly overexpressed in patients with diagnosed age >18 months, advanced INSS stage, advanced risk group, preoperative chemotherapy, death due to disease (Figure 4B to F, P < .05), suggesting that higher XPOT protein expression was correlated with poor clinical characteristics. To further investigate association between XPOT protein expression and clinical characteristics, the best cut-off value of XPOT protein expression level was determined by X-tile software (best P value), and 64 neuroblastoma patients were divided into high XPOT protein expression group and low XPOT protein expression group. Overall survival analysis showed that patients with high XPOT protein expression presented lower overall survival (Figure 4G, P < .05). In the stratification analysis for those with age diagnosed >18 months, without amplification of MYCN and without preoperative chemotherapy, high XPOT expression presented shorter survival time (Figure 4H to J, P < .05).
Figure 4.
Higher XPOT protein expression is associated with poor prognosis. (A) Representative photographs of different XPOT staining intensity of neuroblastoma tissue immunohistochemistry. Higher XPOT protein is expressed in patients with age at diagnosis >18 months (B), advanced INSS stage (C), advanced risk group (D), preoperative chemotherapy (E), and death from disease (F). Overall survival analysis shows that patients with high XPOT protein expression present lower overall survival (G). For patients with age diagnosed >18 months (H), without amplification of MYCN (I), and without preoperative chemotherapy (J), high XPOT expression present shorter survival time.
Abbreviation: INSS, International Neuroblastoma Staging System; XPOT, Exportin-T.
* P < .05.
Tissue microarray showed that higher XPOT protein expression was associated with diagnosed age >18 months (P = .013), advanced INSS stage (P = .008), preoperative chemotherapy (P = .006), and death from disease (P < .001) (Table S3). As showed in Table S4, univariate analysis suggested XPOT as a risk factor with a hazard ratio (HR) (95% CI) of 4.008 (1.892-8.491). However, multivariate analysis did not support XPOT as an independent prognostic factor. This might be due to too few samples.
Knockdown of XPOT Inhibits Neuroblastoma Cell Proliferation and Migration
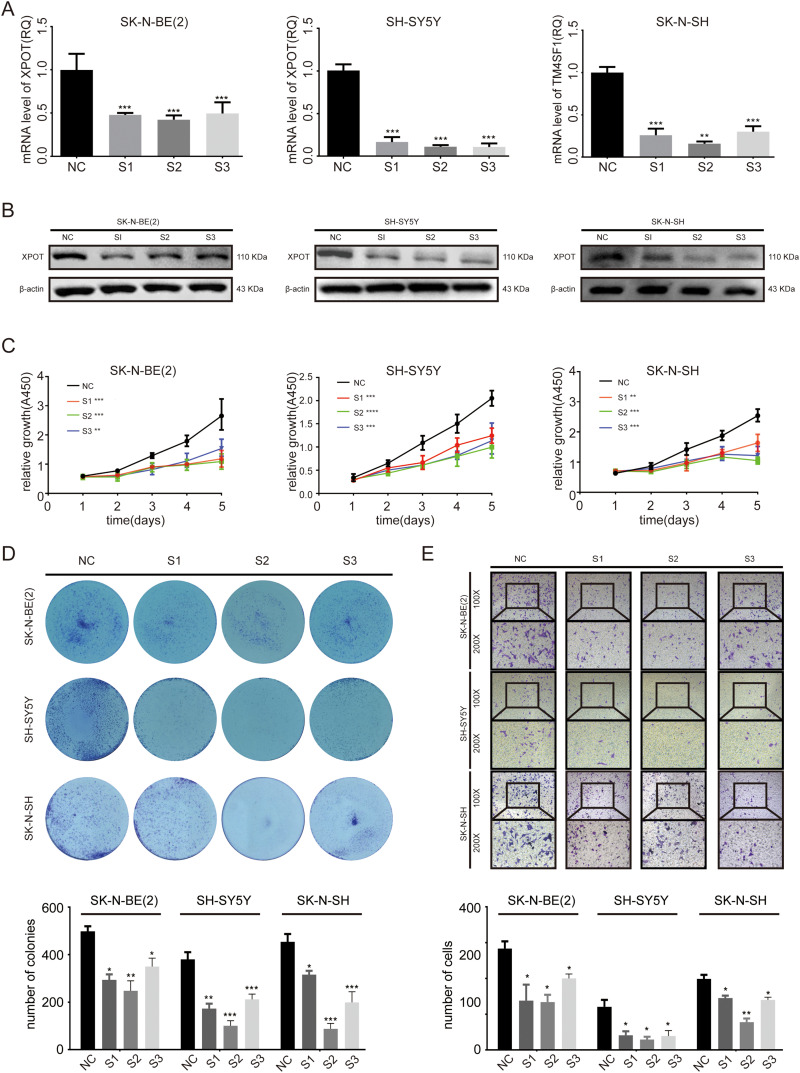
To evaluate the biological function of XPOT in the development of neuroblastomas, SK-N-BE(2), SH-SY5Y, and SK-N-SH were transfected with 3 siRNAs targeting XPOT. XPOT mRNA and protein expression were significantly downregulated after transfected with siRNAs in SK-N-BE(2), SH-SY5Y, and SK-N-SH cells (Figure 5A to B, P < .05). The CCK-8 assay indicated that knockdown of XPOT significantly decreased cell proliferation ability in SK-N-BE(2), SH-SY5Y, and SK-N-SH cells after transfection (Figure 5C, P < .05). The colony formation assay identified that the number and the size of colonies formed by neuroblastoma cells transfected with XPOT-siRNA were significantly reduced compared to those transfected with control siRNA (Figure 5D, P < .05). A Transwell assay was conducted to investigate the effect of XPOT knockdown on cell migration ability. As shown in Figure 5E, the migratory ability from the upper chamber to the lower chamber was significantly suppressed in SK-N-BE(2), SH-SY5Y, and SK-N-SH cells transfected with XPOT–siRNA (P < .05). These results suggested that knockdown of XPOT inhibited neuroblastoma cell proliferation and migration.
Figure 5.
Knockdown of XPOT inhibits neuroblastoma cell proliferation and migration. (A) RT-qPCR identifies the efficiency of siRNA against XPOT in SK-N-BE(2), SH-SY5Y, and SK-N-SH cells. (B) Western blot identifies the knockdown efficiency of siRNA against XPOT. (C) A CCK-8 array indicates the relative growth of SK-N-BE(2), SH-SY5Y, and SK-N-SH cells after knockdown by siRNA against XPOT. (D) Colony images and histogram for SK-N-BE(2), SH-SY5Y, and SK-N-SH cell lines after knockdown of XPOT. (E) Cell migration images and histogram for SK-N-BE(2), SH-SY5Y, and SK-N-SH cell lines after knockdown of XPOT. All experiments were performed 3 times independently. Error bars represent the mean ± SD. * P < .05.
Abbreviations: XPOT, Exportin-T; RT-qPCR, quantitative real-time- polymerase chain reaction.
Discussion
In eukaryotic cells, RNAs needed to be transported to the cytoplasm to function.26 The regulation of genes in biological processes depends in part on the controlled exchange of those RNAs between the nucleus and the cytoplasm.27 Therefore, understanding the molecular pathways of nuclear export receptor in tumor development could contribute to the development of novel clinical intervention.28 In the present study, XPOT, as one of the key nuclear export receptors, was investigated to explore its biological functions and prognostic value in neuroblastomas. Our study indicated that XPOT overexpression was associated with poor clinical characteristics, such as age at diagnosis >18 months, amplification of MYCN, and advanced INSS stage, and poor prognosis in both bioinformatics and clinical experiment analyses. Moreover, we built a risk prediction model based on the status of MYCN, clinical stage (INSS), and XPOT mRNA expression using DCA analysis. In addition, RNA interference studied in vitro indicated that knockdown of XPOT inhibited the proliferation and migration of neuroblastoma cells.
Inhibition of nuclear export results in subsequent change of gene expression by disordering RNAs bidirectionally transportation. It has become a promising therapeutic strategy for several tumors.29,30 Selinexor, also called KPT-330, is a selective inhibitor of nuclear export (SINE) against nuclear export protein exportin 1, and has been proven to exhibit broad antitumor activity.31 In myelofibrosis CD34 + cells, inhibition of nuclear export causes nuclear accumulation of p53 and enhanced ruxolitinib-mediated proliferation suppression and apoptosis.32 Inhibition of XPO1 has also been indicated to decrease myeloid cell leukemia sequence 1 (Mcl-1) levels and enhances cell death induced by Venetoclax (ABT-199), a B-cell lymphoma 2 (Bcl-2)-selective inhibitor, in acute myeloid leukemia.33 XPO1 inhibitor impedes Mcl-1 and the B-cell lymphoma-extra large (Bcl-xL) complex, causes increased mitochondrial membrane permeability, and therefore triggers cell apoptosis.34 The combination of XPO1 and fms like tyrosine kinase 3 (FLT3) exerts synergistic pro-apoptotic effects through elevated nuclear levels of extracellular regulated protein kinases (ERK), AKT, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), and FOXO3a in FLT3-mutated acute myeloid leukemias.35 These studies demonstrate the enormous clinical potential of SINE combination therapy. However, few studies focus on the molecular action of XPOT in tumorigenesis currently, and the underlying mechanism of XPOT in the progression of tumor remains unknown. Given that XPOT is also a key member of nuclear export receptors, it might play an important role in cancer by transporting key mediators of oncogenesis across the nuclear membrane in cancer cells.
In our study, we built a risk prediction model based on status of MYCN, clinical stage (INSS), and XPOT mRNA expression using DCA analysis. Compared with receiver operating characteristic curve analysis, DCA analysis focuses on clinical net benefit. According to DCA analysis, neuroblastoma patients could benefit from the XPOT risk prediction model-guided interventions. Compared with other models, the accession of XPOT significantly increased the clinical net benefit of neuroblastoma patients. At the translation level, XPOT protein overexpression was also significantly associated with poor prognosis through tissue microarray analysis. These results implied that XPOT might be a novel predictor of neuroblastoma prognosis. Considering the convenience and feasibility of XPOT testing, this could be more practical in clinic.
XPOT depletion by siRNA could significantly suppressed neuroblastoma cell proliferation and migration, highlighting XPOT as a therapeutic target in neuroblastoma. However, siRNA as a drug remains far to clinical application due to the off-target effects as well as the difficulties of siRNA delivery.36 Unfortunately, specific inhibitors directly against XPOT are not available yet. Recently, 1-α,25-dihydroxyvitamin D3 (1α,25[OH]2D3) has been reported to suppress the expression of XPOT in human promyelocytic leukemia HL-60 cells and consequently inhibited the proliferation of HL-60 cells.15 This implied that 1α,25(OH)2D3 can be administrated as an XPOT inhibitor. However, the therapeutic window of 1α,25(OH)2D3 is very narrow in clinical practice because of dose-limiting hypercalcemia. Fortunately, several low-calcemic analogs with vitamin D3-mediated anticancer have been reported.15,37 24R,25(OH)2D3, another metabolite of 25(OH)D3, potentiates a great ability of inducing cell apoptosis and suppressing metastasis in breast cancer,37 although it is still unclear if the anti-tumor activity of 24R,25(OH)2D3 is achieved by suppressing XPOT. For neuroblastoma, a novel 1α,25(OH)2D3 analog, QW1624F2 to 2 induced cell-cycle arrest in the G1 phase and induced cell differentiation by increasing neurite length.38 Considering its role in human promyelocytic leukemia, we highly believe that these analogs may provide an alternative approach to inhibit neuroblastoma by downregulating XPOT. Further research can focus on the interaction between these analogs and XPOT.
Conclusions
In this study, we systematically mined 2 transcriptome datasets of neuroblastoma and identified XPOT as a novel prognostic predictor of neuroblastoma. Tissue microarray analysis including 64 neuroblastoma patients confirmed the results. Moreover, DCA analysis suggested neuroblastoma patients benefited from XPOT risk prediction model-guided interventions. Knockdown of XPOT presented anti-tumor effect in neuroblastoma in vitro, highlighting the potential therapeutic target of XPOT in neuroblastoma. However, profound molecular mechanism underlying the tumor inhibition activity of XPOT needs more work for further investigation. Another logical extension of this work would be to investigate the molecular roles of XPOT in the anti-tumor activity of the 1α,25(OH)2D3 analogs.
Supplemental Material
Supplemental material, sj-tif-1-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-tif-2-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-tif-3-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-4-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-5-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-6-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-7-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-tiff-8-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Abbreviations
- DCA
Decision curve analysis
- DMEM
Dulbecco’s modified eagle medium
- FBS
Fetal bovine serum
- INRG
International Neuroblastoma Risk Group
- INSS
International Neuroblastoma Staging System
- PMSF
Phenylmethanesulfonyl fluoride
- RT-qPCR
Quantitative real-time PCR
- SDS-PAGE
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SINE
Inhibitors of nuclear export
- XPOT
Exportin-T
- 1α,25[OH]2D3
1alfa,25-dihydroxyvitamin D3
Footnotes
Ethics Approval: The study protocol was approved by the Ethics Committee of Xinhua Hospital (XHEC-D-2018-034). Written informed consent was obtained from all donors and their relatives.
Availability of Data and Materials: The datasets analyzed during the current study are available in the Gene Expression Omnibus repository, (GSE85047, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85047; GSE49710, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49710).
All data generated and analyzed during this study are included in this published article and its supplementary information files.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article. This work is supported by Natural Science Foundation of China (No. 81874234) to ZW, Suzhou Clinical Medicine Innovation Team Introduction Project (SZYJTD201706), Emerging Cutting-edge Technology Joint Research Project of Shanghai (No.SHDC12018123), Science, Technology Support Projects in Biomedicine Field of Shanghai Science and Technology Commission (No. 18401931800), and Translational Medicine Collaborative Innovation Project of Shanghai Jiao Tong University School of Medicine (TM201809) to YW.
ORCID iD: Li-Jia Pan https://orcid.org/0000-0001-5977-9555
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Maris JM, Hogarty MD, Bagatell R, et al. Neuroblastoma. The Lancet. 2007;369(9579):2106-2120. [DOI] [PubMed] [Google Scholar]
- 2.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3(3):203-216. [DOI] [PubMed] [Google Scholar]
- 3.Pastor ER, Mousa SA. Current management of neuroblastoma and future direction. Crit Rev Oncol Hematol. 2019;138:38-43. [DOI] [PubMed] [Google Scholar]
- 4.Whittle SB, Smith V, Doherty E, et al. Overview and recent advances in the treatment of neuroblastoma. Expert Rev Anticancer Ther. 2017;17(4):369-386. [DOI] [PubMed] [Google Scholar]
- 5.Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG task force report. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology. 2009;27(2):289-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sokol E, Desai AV. The evolution of risk classification for neuroblastoma. Children (Basel, Switzerland). 2019;6(2):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgenstern DA, Bagatell R, Cohn SL, et al. The challenge of defining “ultra-high-risk” neuroblastoma. Pediatr Blood Cancer. 2019;66(4):e27556. [DOI] [PubMed] [Google Scholar]
- 8.Turner JG, Dawson J, Sullivan DM. Nuclear export of proteins and drug resistance in cancer. Biochem Pharmacol. 2012;83(8):1021-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depping R, von Fallois M, Landesman Y, et al. The nuclear export inhibitor selinexor inhibits hypoxia signaling pathways And 3D spheroid growth Of cancer cells. Onco Targets Ther. 2019;12:8387-8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arts GJ, Fornerod M, Mattaj IW. Identification of a nuclear export receptor for tRNA. Current Biology : CB. 1998;8(6):305-314. [DOI] [PubMed] [Google Scholar]
- 11.Kutay U, Lipowsky G, Izaurralde E, et al. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1(3):359-369. [DOI] [PubMed] [Google Scholar]
- 12.Kuersten S, Arts GJ, Walther TC, et al. Steady-state nuclear localization of exportin-t involves RanGTP binding and two distinct nuclear pore complex interaction domains. Mol Cell Biol. 2002;22(16):5708-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaidyanathan S, Thangavelu PU, Duijf PH. Overexpression of Ran GTPase components regulating nuclear export, but not mitotic spindle assembly, marks chromosome instability and poor prognosis in breast cancer. Target Oncol. 2016;11(5):677-686. [DOI] [PubMed] [Google Scholar]
- 14.Lin J, Hou Y, Huang S, et al. Exportin-T promotes tumor proliferation and invasion in hepatocellular carcinoma. Mol Carcinog. 2019;58(2):293-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki T, Koyama Y, Hayakawa S, et al. 1,25-Dihydroxyvitamin D3 suppresses exportin expression in human promyelocytic leukemia HL-60 cells. Biomedical Research (Tokyo, Japan). 2006;27(2):89-92. [DOI] [PubMed] [Google Scholar]
- 16.Schwab TS, Madison BB, Grauman AR, et al. Insulin-like growth factor-I induces the phosphorylation and nuclear exclusion of forkhead transcription factors in human neuroblastoma cells. Apoptosis. 2005;10(4):831-840. [DOI] [PubMed] [Google Scholar]
- 17.Attiyeh EF, Maris JM, Lock R, et al. Pharmacodynamic and genomic markers associated with response to the XPO1/CRM1 inhibitor selinexor (KPT-330): a report from the pediatric preclinical testing program. Pediatr Blood Cancer. 2016;63(2):276-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium SM-I. A comprehensive assessment of RNA-seq accuracy, reproducibility and information content by the Sequencing Quality Control Consortium. Nat Biotechnol. 2014;32(9):903-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajbhandari P, Lopez G, Capdevila C, et al. Cross-Cohort analysis identifies a TEAD4-MYCN positive feedback loop as the core regulatory element of high-risk neuroblastoma. Cancer Discov. 2018;8(5):582-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunes C, Rocha R, Buzelin M, et al. High agreement between whole slide imaging and optical microscopy for assessment of HER2 expression in breast cancer: whole slide imaging for the assessment of HER2 expression. Pathol Res Pract. 2014;210(11):713-718. [DOI] [PubMed] [Google Scholar]
- 21.Meseure D, Vacher S, Alsibai KD, et al. Expression of ANRIL-polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a Two-gene (EZH2/CBX7) signature with independent prognostic value. Molecular Cancer Research : MCR. 2016;14(7):623-633. [DOI] [PubMed] [Google Scholar]
- 22.Yeo W, Chan SL, Mo FK, et al. Phase I/II study of temsirolimus for patients with unresectable hepatocellular carcinoma (HCC)- a correlative study to explore potential biomarkers for response. BMC cancer. 2015;15:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenneth J, Livak TD. Analysis of relative gene expression data using real—time quantitative PCR and the 2 一 ct method. Method. 2001;25(4):402-8. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald M, Saville BR, Lewis RJ. Decision curve analysis. Jama. 2015;313(4):409-410. [DOI] [PubMed] [Google Scholar]
- 25.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clinical Cancer Research : an Official Journal of the American Association for Cancer Research. 2004;10(21):7252-7259. [DOI] [PubMed] [Google Scholar]
- 26.Okamura M, Inose H, Masuda S. RNA export through the NPC in eukaryotes. Genes (Basel). 2015;6(1):124-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez MS, Dargemont C, Stutz F. Nuclear export of RNA. Biology of the Cell. 2004;96(8):639-655. [DOI] [PubMed] [Google Scholar]
- 28.Conforti F, Wang Y, Rodriguez JA, et al. Molecular pathways: anticancer activity by inhibition of nucleocytoplasmic shuttling. Clinical Cancer Research : an Official Journal of the American Association for Cancer Research. 2015;21(20):4508-4513. [DOI] [PubMed] [Google Scholar]
- 29.Muqbil I, Azmi AS, Mohammad RM. Nuclear export inhibition for pancreatic cancer therapy. Cancers (Basel). 2018;10(5):138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta A, Saltarski JM, White MA, et al. Therapeutic targeting of nuclear export inhibition in lung cancer. Journal of Thoracic Oncology : Official Publication of the International Association for the Study of Lung Cancer. 2017;12(9):1446-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdul Razak AR, Mau-Soerensen M, Gabrail NY, et al. First-in-Class, first-in-human phase I study of selinexor, a selective inhibitor of nuclear export, in patients With advanced solid tumors. Journal of Clinical Oncology : Official Journal of the American Society of Clinical Oncology. 2016;34(34):4142-4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan D, Pomicter AD, Tantravahi S, et al. Nuclear-Cytoplasmic transport Is a therapeutic target in myelofibrosis. Clinical Cancer Research : an Official Journal of the American Association for Cancer Research. 2019;25(7):2323-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luedtke DA, Su Y, Liu S, et al. Inhibition of XPO1 enhances cell death induced by ABT-199 in acute myeloid leukaemia via Mcl-1. J Cell Mol Med. 2018;22(12):6099-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu ZC, Liu JW, Yang C, et al. XPO1 Inhibitor KPT-330 synergizes with Bcl-xL inhibitor to induce cancer cell apoptosis by perturbing rRNA processing and Mcl-1 protein synthesis. Cell Death Dis. 2019;10(6):395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang W, Ly C, Ishizawa J, et al. Combinatorial targeting of XPO1 and FLT3 exerts synergistic anti-leukemia effects through induction of differentiation and apoptosis in FLT3-mutated acute myeloid leukemias: from concept to clinical trial. Haematologica. 2018;103(10):1642-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B, Weng Y, Xia XH, et al. Clinical advances of siRNA therapeutics. J Gene Med. 2019;21(7):e3097. [DOI] [PubMed] [Google Scholar]
- 37.Verma A, Cohen DJ, Schwartz N, et al. 24R,25-Dihydroxyvitamin D3 regulates breast cancer cells in vitro and in vivo. Biochimica et Biophysica Acta General Subjects. 2019;1863(10):1498–1512. [DOI] [PubMed] [Google Scholar]
- 38.Reddy CD, Patti R, Guttapalli A, et al. Anticancer effects of the novel 1alpha, 25-dihydroxyvitamin D3 hybrid analog QW1624F2-2 in human neuroblastoma. J Cell Biochem. 2006;97(1):198-206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-tif-1-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-tif-2-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-tif-3-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-4-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-5-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-6-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-7-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment
Supplemental material, sj-tiff-8-tct-10.1177_15330338211039132 for Exportin-T: A Novel Prognostic Predictor and Potential Therapeutic Target for Neuroblastoma by Li-Jia Pan, Jian-Lei Chen, Zhi-Xiang Wu and Ye-Ming Wu in Technology in Cancer Research & Treatment