Abstract
Lung epithelial cell death is a prominent feature of acute lung injury and acute respiratory distress syndrome (ALI/ARDS), which results from severe pulmonary infection leading to respiratory failure. Multiple mechanisms are believed to contribute to the death of epithelia; however, limited data propose a role for epigenetic modifiers. In this study, we report that a chromatin modulator protein arginine N-methyltransferase 4/coactivator-associated arginine methyltransferase 1 (PRMT4/CARM1) is elevated in human lung tissues with pneumonia and in experimental lung injury models. Here PRMT4 is normally targeted for its degradation by an E3 ubiquitin ligase, SCFFBXO9, that interacts with PRMT4 via a phosphodegron to ubiquitinate the chromatin modulator at K228 leading to its proteasomal degradation. Bacterial-derived endotoxin reduced levels of SCFFBXO9 thus increasing PRMT4 cellular concentrations linked to epithelial cell death. Elevated PRMT4 protein caused substantial epithelial cell death via caspase 3-mediated cell death signaling, and depletion of PRMT4 abolished LPS-mediated epithelial cell death both in cellular and murine injury models. These findings implicate a unique molecular interaction between SCFFBXO9 and PRMT4 and its regulation by endotoxin that impacts the life span of lung epithelia, which may play a key role in the pathobiology of tissue injury observed during critical respiratory illness.
Subject terms: Ligases, Epigenetics, Ubiquitin ligases, Infection
Introduction
Acute lung injury and acute respiratory distress syndrome (ALI/ARDS) is a major public health concern with high mortality of approximately 30–40% [1–5]. Pathologically, ALI/ARDS is characterized by lung inflammatory cell infiltration, and alveolar-capillary leak of edema fluid into the airways leading to hypoxia and respiratory failure. Viral and bacterial lower-airway infection is a leading cause of ALI/ARDS, which contributes to approximately 40% of the total ALI/ARDS patients [6–9]. ALI/ARDS is also observed during pandemic coronavirus infection, caused by severe acute respiratory syndrome coronavirus 2 [10–13]. Among the mechanisms of the ALI/ARDS pathogenesis, lung epithelial cell death is a crucial signature event linked to loss of integrity of the lung lining leading to an alveolar–capillary leak with edema. Although many pathways have been described causing epithelial cell death [5, 14], there is limited investigation of unique molecular pathways involving epigenetic mechanisms. Molecular mechanistic studies are of particular importance in uncovering potential effective targets for therapeutic intervention in this illness.
Protein arginine methyltransferase 4 (PRMT4/CARM1) governs a range of life processes including gene transcription, proliferation, RNA splicing, development, nuclear factor (NF)-κB mediated inflammation, and p53-related signal transduction via its epigenetic and non-epigenetic functions [15–21]. PRMT4 is crucial in development, as knockout of PRMT4 in mice leads to neonatal death and developmental defects in respiratory system. In these mice, alveolar type II epithelial cells that are crucial for surfactant production and ion transport are smaller than normal but with increased numbers. Notably, depletion of PRMT4 blocks type II alveolar cell differentiation into type I cells, and no type I alveolar epithelial cells are generated in PRMT4 knockout mice lung [22]. Aberrant expression of PRMT4 proteins is identified in a number of neoplastic tissues that regulates many aspects of cancer, including carcinogenesis, metastasis, recurrence, and drug resistance [23, 24]. PRMT family members also regulate apoptosis as PRMT2 inhibits NF-κB signaling in mouse embryo fibroblasts [25]. PRMT4 expression induced by high-glucose loading triggers apoptosis of human retinal pigment epithelial cells via H3R17 di-methylation [26]. Recent studies reported that specific expression of PRMT4 in adipose tissue promotes lipolysis and increases serum triglycerides in type 1 diabetic mice [27]. Cardiac-specific expression of PRMT4 promotes apoptosis of cardiomyocytes in mouse models [28]. However, how bacterial pathogens regulate PRMT4 at the posttranslational level is not fully understood.
The ubiquitin proteasome system degrades most cellular proteins in living cells and at times in coordination with the gene transcriptional machinery. Ubiquitination proceeds in an enzymatic cascade, in which the final step involves an E3 ubiquitin ligase that recognizes a protein substrate to catalyze ligation of ubiquitin to the substrate. This process often results in substrate disposal within the proteasome. Among E3 ubiquitin ligases, a family of SCF (Skp1-Cullin1-Fbox) E3 proteins have been linked to bacterial infection, antiviral immunity, and host defense [29–36]. Interestingly, the F-box protein FBXO9, a component of SCFFBXO9 E3 ligase complex, was identified in a pooled RNAi screen as a lipopolysaccharide (LPS)-responsive protein in the context of interleukin (IL)-6 production to regulate innate immunity [37]. FBXO9 may be associated with patient survival in multiple myeloma, is linked to adipocyte development, and targets peroxisome proliferator-activated receptor gamma (PPARγ) for degradation [38–41]. Aside from these studies, our understanding of this E3 ligase subunit is limited, particularly as it relates to sepsis.
In this study, we identified that microbial factors (i.e., endotoxin) increase PRMT4 expression by extending its cellular protein life span thereby inducing lung epithelial cell death in experimental lung injury models. Our data show that LPS co-ordinately downregulates the expression of an E3 ubiquitin ligase SCFFBXO9 that normally targets PRMT4 for its elimination in cells. Hence, the molecular balance between SCFFBXO9 and PRMT4 is modulated during infection that alters viability of lung epithelia.
Results
Gram-negative bacterial-derived endotoxin increases PRMT4 protein expression
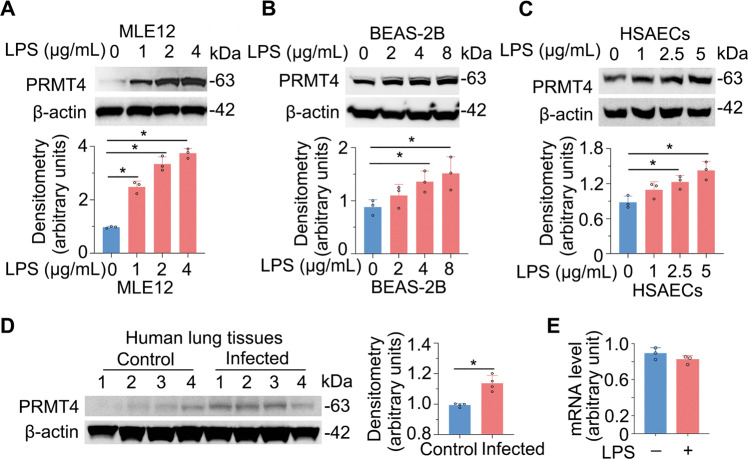
We identified that Escherichia coli-derived LPS increases PRMT4 protein levels in lung alveolar epithelial type II-like MLE12 cells (Fig. 1A). LPS-mediated PRMT4 accumulation was observed in a concentration-dependent manner in the cells. We also observed that LPS increased PRMT4 cellular concentrations in human lung epithelial BEAS-2B cells (Fig. 1B) and in primary human small airway epithelial cells (HSAECs; Fig. 1C). To assess clinical relevance, we investigated PRMT4 protein levels in de-identified human lung tissue samples [42]. Immunoblotting results showed that protein levels were higher in infected lung tissue samples (Fig. 1D). We then assayed PRMT4 mRNA levels using quantitative reverse transcription–polymerase chain reaction (qRT-PCR), with results showing no remarkable differences in LPS-treated and untreated BEAS-2B cells (Fig. 1E). These results suggest that bacteria elevate PRMT4 protein levels possibly by impairing its protein degradation.
Fig. 1. LPS increases PRMT4 protein in epithelia.
A MLE12 cells were treated with LPS as indicated; cell lysates were immunoblotted with PRMT4 and β-actin antibodies. B BEAS-2B cells were treated with LPS as indicated; cell lysates were immunoblotted with PRMT4 and β-actin antibodies. C Primary human small airway epithelial cells (HSAECs) were treated with LPS as indicated; cell lysate was collected and analyzed with PRMT4 and β-actin antibodies. For A–C, relative expression of PRMT4 was plotted in the below panels. D Lung tissue lysates from deidentified human lung samples from normal and infected patients were analyzed for PRMT4 and β-actin by immunoblotting. Densitometry was presented in the right panel. E PRMT4 mRNA levels were determined by qRT-PCR in LPS-treated bronchial epithelial cells. Experiments n = 3. Statistical significance was indicated with asterisk “*” between groups where P < 0.05 vs. control (0).
PRMT4 is a labile protein degraded via the ubiquitin proteasomal machinery
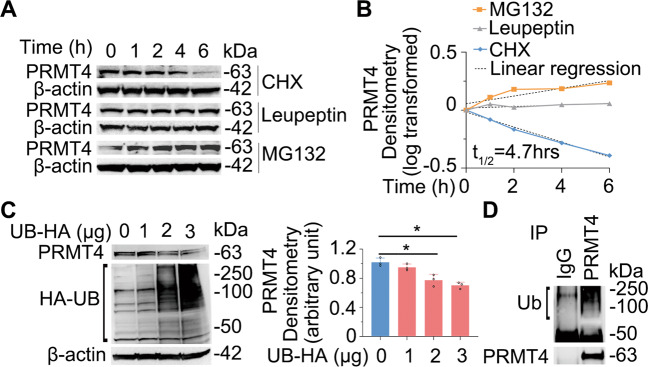
The availability of a specific proteins in cells is the result of dynamic homeostasis between its gene transcription, translational efficiency, and protein decay. However, the molecular mechanism(s) of PRMT4 protein degradation is not fully elucidated [43]. Immunoblotting results confirmed that the half-life of PRMT4 protein is approximately 4.7 h in human lung epithelial BEAS-2B cells (Fig. 2A, B). PRMT4 degradation was via the proteasome but not the lysosome, as the proteasome inhibitor MG132 but not the lysosome inhibitor leupeptin accumulated PRMT4. Proteins degraded via the proteasome are generally poly-ubiquitinated and ubiquitin could be a limiting factor for protein degradation in a specified cellular compartment as the distribution of ubiquitin can be spatially uneven. Ectopic expression of ubiquitin accelerated PRMT4 turnover in a ubiquitin-dependent manner (Fig. 2C). Results from immunoprecipitation (IP) studies showed that PRMT4 was also poly-ubiquitinated in BEAS-2B cells (Fig. 2D). In all, these data indicate that PRMT4 is a labile protein with a half-life of about 4.7 h degraded via a ubiquitin proteasomal machinery.
Fig. 2. PRMT4 is a labile protein degraded via the ubiquitin proteasomal machinery.
A BEAS-2B cells were treated with cycloheximide (CHX), leupeptin, or MG132 as indicated. Cell lysates were immunoblotted with PRMT4 and β-actin antibodies. B Densitometry results in A were analyzed in semi-log format with GraphPad prism 5 and the results were plotted. C Ectopic expression of ubiquitin reduces PRMT4 protein in a concentration-dependent manner. Densitometry was plotted in the right panel. D PRMT4 is polyubiquitinated as shown by co-immunoprecipitation (Co-IP) of precipitates analyzed with ubiquitin and PRMT4 antibodies.
SCFFBXO9 E3 ubiquitin ligase targets PRMT4 in lung epithelial cells
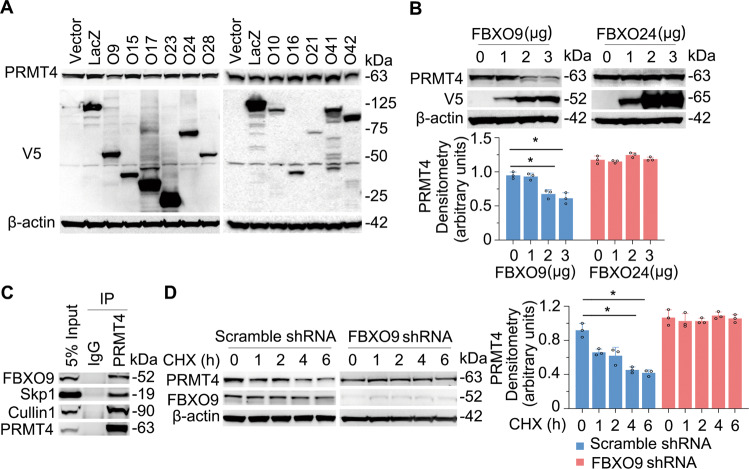
Protein ubiquitination is a cascade involving E1, E2, and E3 enzymes, in which an E3 ubiquitin ligase is substrate specific. By screening a library of SCF-E3 ubiquitin ligases, we identified that SCFFBXO9 specifically degrades PRMT4 (Fig. 3A). Further, PRMT4 protein was degraded in an FBXO9 plasmid concentration-dependent manner (Fig. 3B, left panels). As a control, FBXO24, another F-box protein, did not degrade PRMT4 (Fig. 3B, right panels). IP studies showed that FBXO9 protein associates with PRMT4, as do the SCF complex components Skp1 and Cullin1 (Fig. 3C). Moreover, knockdown of FBXO9 with small hairpin RNA (shRNA) stabilized PRMT4 as compared with that of a scrambled shRNA control (Fig. 3D). Overall, these data indicate that PRMT4 is specifically targeted for its cellular disposal by the SCFFBXO9 ubiquitin proteasomal machinery.
Fig. 3. SCFFBXO9 E3 ubiquitin ligase specifically degrades PRMT4 in lung epithelia.
A Screen of F-box (FBXO) family members revealed that overexpression of FBXO9 selectively degrades PRMT4 in BEAS-2B cells. B Ectopically expressed FBXO9 plasmid degrades PRMT4 in a concentration-dependent manner not observed with FBXO24. Densitometric results are presented in the bottom panels. C Co-IP studies showing that PRMT4 interacts with Skp1 and Cullin 1, components of SCF E3 ligase. D Knockdown of FBXO9 stabilizes PRMT4 using protein half-life studies with cellular exposure to cycloheximide (CHX) for various times to inhibit protein synthesis. The relative expression of PRMT4 was plotted in the right panel. *P < 0.05 vs. time 0. Experiments n = 3.
SCFFBXO9 ubiquitinates PRMT4 at K228
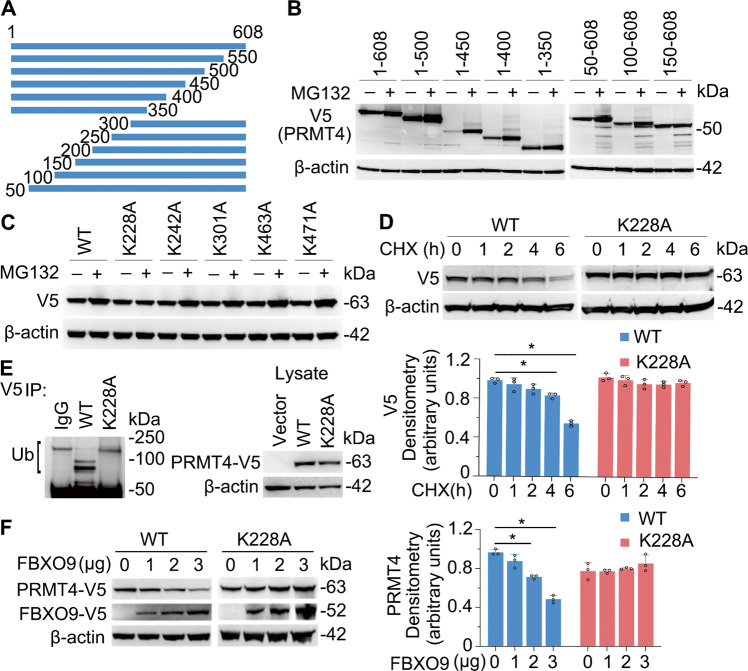
We next fine mapped the ubiquitination site(s) within PRMT4. We generated and expressed truncated PRMT4 constructs in cells to examine protein degradation of the truncations (Fig. 4A). Cellular expression of constructs harboring carboxyl terminal deletions resulted in PRMT4 accumulation suggesting that molecular signatures that mediate protein disposal are not localized within these fragments (Fig. 4B, left panels). We next examined NH2-terminal deletions and observed that fragments 50–608 and 100–608 also resulted in PRMT4 accumulation after MG132 treatment. However, fragment 150–608 prevented PRMT4 accumulation (Fig. 4B, right panels), suggesting that aa100–150 contains a degradation element. We analyzed a series of lysine residues resided in the truncation mutant containing aa100–608. As compared with that of wild-type (WT) PRMT4 and other mutants, a K228A mutant was stable from proteasomal degradation (Fig. 4C, D). The K228A mutant was less ubiquitinated as compared with that of WT PRMT4 (Fig. 4E, left panels), which indicated that K228 is a ubiquitin acceptor site. In addition, mutation of K228 abrogated FBXO9-mediated PRMT4 degradation (Fig. 4F). These data suggest that K228 may be one authentic SCFFBXO9-catalyzed ubiquitin acceptor site.
Fig. 4. SCFFBXO9 ubiquitinates PRMT4 at K228.
A Schematic presentation of PRMT4 truncation constructs that were expressed in BEAS-2B cells. B Fragment 150–608 does not accumulate after MG132 treatment. C Cellular expression of a K228A point mutant does not accumulate after MG132 treatment. D Cellular expression of a K228 mutant of PRMT4 is stable after CHX treatment. E Cellular expression of a V5-PRMT4 K228A mutant vs. a V5 wild-type (WT) PRMT4 is not ubiquitinated. Constructs were expressed in cells and immunoprecipitated using V5 antibodies and probed with ubiquitin antibodies. The right blot shows the levels of WT and mutant construct protein expression in cells. F Ectopic expression of FBXO9 does not degrade the PRMT4 K228A mutant. *P < 0.05 vs. “0.” Experiments n = 3.
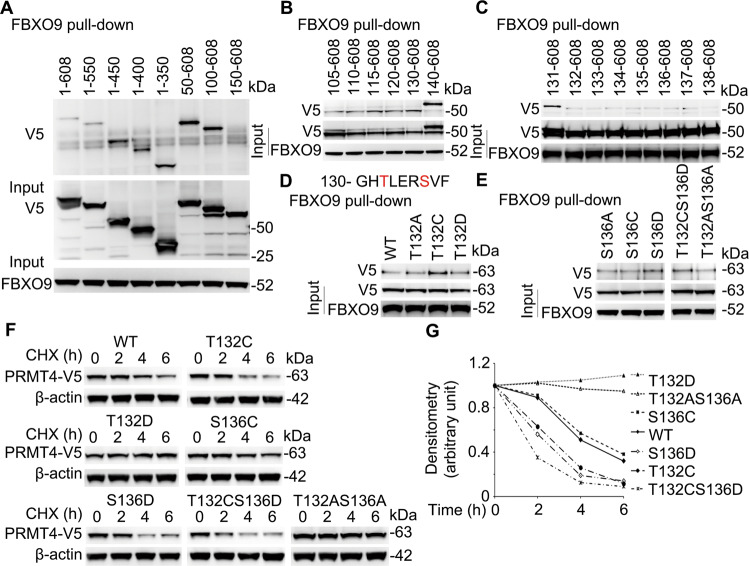
SCFFBXO9 recognizes a PRMT4 phosphodegron
As SCF E3 ubiquitin ligases specifically interact with their substrates, we next studied the FBXO9 docking site within PRMT4. We focused on the NH2-terminus because the above studies suggested that aa100–150 contains a degradation element, possibly an E3 ligase docking site. Results from in vitro pull-down assays showed that deletion of NH2-terminal 150 aa impairs PRMT4 and FBXO9 binding (Fig. 5A). We further fine mapped the docking site for FBXO9 within PRMT4 by pull-down assays (Fig. 5B, C). Pull-down results showed that amino acids after aa131 were critical for FBXO9 binding. We analyzed the sequence and identified a phosphorylation motif in this region: 132-TxxxS. To test whether this motif is a potential phosphodegron, we substituted T132 and S136 with a cysteine to mimic dephosphorylation or introduced an aspartic acid at T132 to mimic a phosphorylation site. Pull-down results suggest that a dephosphorylated T132 and phosphorylated S136 sites constituted an optimal FBXO9-binding motif constituting a phosphodegron for E3 SCF ligase interaction (Fig. 5D, E). Consistent with these observations, our recent published study showed that glycogen synthase kinase-3 beta (GSK-3β)-mediated T132 phosphorylation stabilized PRMT4 [43]. We further tested the protein stability with these mutations (Fig. 5F, G). T132C and S136D mutants degraded faster than that of WT PRMT4. A S136C mutant displayed greater protein stability. T132D and T132AS136A double mutants completely stabilized the protein from degradation. However, a T132CS136D PRMT4 variant when expressed in cells degraded most rapidly (Fig. 5F, G). In all, these data indicate that SCFFBXO9 engages a phosphodegron 132-TdxxxSp to ubiquitinate PRMT4 at K228.
Fig. 5. FBXO9 docks on a phosphodegron 132TxxxS within PRMT4.
A BEAS-2B cells were ectopically expressed with various deletion V5-PRMT4 plasmid constructs and lysates subjected to FBXO9 immunoprecipitation from MLE12 cell lysates and subsequent V5 immunoblotting to assess FBXO9:V5 interaction. Pull-down assays suggest that aa100–150 of PRMT4 is important for FBXO9 binding. B, C Fine mapping the FBXO9-binding sites with pull-down assays indicate that a phosphodegron 132TxxxS is critical for FBXO9 binding. D, E Pull-down assays with phosphorylation/dephosphorylation mimics indicate that T132 dephosphorylation and S136 phosphorylation promote the degron binding to FBXO9. F T132 dephosphorylation and S136 phosphorylation accelerate PRMT4 degradation using cycloheximide (CHX). G Densitometric results of F were plotted. Experiments n = 3.
PRMT4 promotes caspase 3 activation and cell death in lung epithelial cells
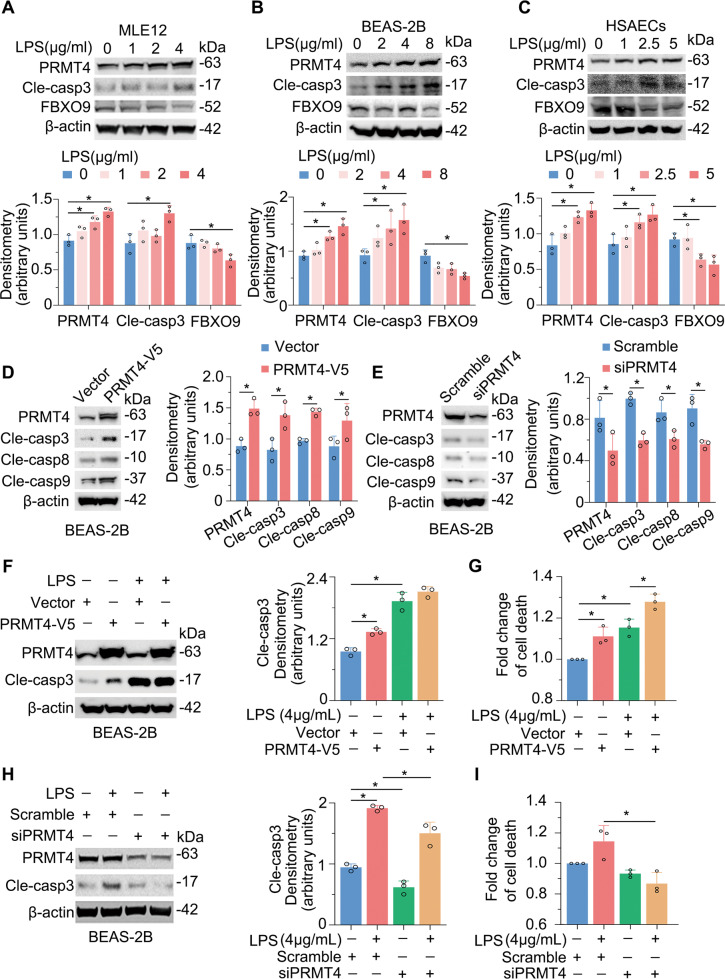
We next assessed a potential pathophysiological role for elevated PRMT4 after bacterial infection. We tested whether actions of LPS on lung epithelial cell death are mediated by PRMT4 through its E3 ligase, SCFFBXO9. We studied cell death using activated caspase 3 as a marker in a variety of cell types including mouse type II MLE12 cells, human airway epithelial BEAS-2B cells, and human primary small airway epithelial cells after 8 h of LPS stimulation. As predicted, caspase 3 was activated in all of these cells, and its activation occurred in an LPS concentration-dependent manner (Fig. 6A-C, second upper panels). Notably, LPS downregulated FBXO9 protein variably in all the tested cells, suggesting a mechanism whereby LPS stabilizes PRMT4 in cells via depletion of SCFFBXO9 E3 ubiquitin ligase (Fig. 6A–C, second to lowest panels). To understand the potential role(s) of PRMT4 in caspase cleavage in lung epithelial cells, we ectopically expressed PRMT4 in BEAS-2B cells. Immunoblotting results showed that PRMT4 ectopic expression was sufficient to activate caspase 3, 8, and 9 in BEAS-2B cells (Fig. 6D). We then depleted PRMT4 expression using small interfering RNA (siRNA) to further examine our findings. Immunoblotting results showed that depletion of PRMT4 by siRNA reduced caspase 3, 8, and 9 activation to lower levels compared to the scrambled control (Fig. 6E). Ectopic expression of PRMT4 promoted LPS-induced caspase 3 activation (Fig. 6F) as well as epithelial cell death (Fig. 6G). In contrast, depletion of PRMT4 blocked LPS-mediated caspase 3 activation to baseline levels (Fig. 6H) and cell death of epithelia (Fig. 6I). Thus, these data suggest that LPS increases PRMT4 protein level possibly via downregulation of FBXO9, thereby facilitating the methyltransferase to trigger caspase 3 signaling in lung epithelial cells.
Fig. 6. Endotoxin reduces FBXO9 levels leading to PRMT4 accumulation and caspase 3 activation in lung epithelia.
A–C MLE12 cells, BEAS-2B cells, and human primary small airway epithelial cells (HSAECs) were treated with LPS as indicated. Cell lysates were analyzed by PRMT4, cleaved caspase 3, FBXO9, and β-actin immunoblotting. Shown below is densitometric analysis of immunoblots. D Overexpression of PRMT4 increases cleaved caspase 3, 8, and 9 baseline levels in BEAS-2B lung epithelial cells. E Knockdown of PRMT4 in BEAS-2B epithelial cells with shRNA reduces cleaved caspase 3, 8, and 9 expression. F, G Overexpression of PRMT4 enhances LPS-induced caspase 3 activation (F) and causes BEAS-2B lung epithelial cell death. Cell death is determined using LDH assay and the data are normalized with that from untreated control cells. H, I Silencing of PRMT4 inhibits LPS-induced caspase 3 activation (H) and cell death (I) in BEAS-2B cells. *P < 0.05. Experiments n = 3.
PRMT4 mediates lung epithelial cell death in experimental ALI
To test our hypothesis biologically, we assessed PRMT4 in an LPS lung injury mouse model. We overexpressed PRMT4 or knocked down PRMT4 using lentiviral constructs (1 × 107 colony-forming units/mouse, intratracheal [i.t.] administration) in mice (C57BL/6J) for 14 days. LPS (5 mg/kg, i.t. in phosphate-buffered saline (PBS) buffer) was administrated (i.t.) into mice for 24 h. Results from histological studies showed that administration of LPS caused lung inflammation with inflammatory cell infiltration. Overexpression of PRMT4 enhanced lung inflammation and knockdown of PRMT4 or application of PRMT4 inhibitor TP064 (10 μg/kg) attenuated lung inflammation (Fig. 7A, upper panels). Results from terminal deoxynucleotidyl transferase-mediated dUTP-fluorescein nick end labeling (TUNEL) staining showed that LPS caused cell death in lung tissues, in which overexpression of PRMT4 enhanced cell death and knockdown of PRMT4 or application of PRMT4 inhibitor attenuated cell death in the lung (Fig. 7A, lower panels, and Fig. 7B). Immunoblotting results from lung indicated that i.t. administration of LPS or lentiviral particles did modulate PRMT4 protein levels in lung tissues (Fig. 7C, D). A PRMT4-specific inhibitor TP064 partially protected lung epithelial cells from death. Overall, these data suggest that LPS-enhanced PRMT4 protein expression promotes lung epithelial cell death and targeting PRMT4 attenuates severity of cellular injury in an experimental mouse lung inflammation model.
Fig. 7. Inhibition of PRMT4 suppresses cell death in a LPS lung injury model.
A–D PRMT4 was silenced or overexpressed by i.t. administered lentiviral constructs for 14 days. Mice were then given (i.t.) LPS with or without a PRMT4 inhibitor as indicated for 24 h (n = 8). Lung tissues were stained with hematoxylin–eosin (H&E) (A, upper panels) and TUNEL staining (A, lower panels). B TUNEL-positive cells in lung tissues were quantitated. C PRMT4 overexpression and knockdown in lung tissues from A were analyzed by immunoblotting. The data from C were quantitated and plotted in D (n = 3). *P < 0.05 vs. scrambled control. Scale bar = 100 μm.
Discussion
Lung epithelial cell death is one of the mechanistic centerpieces of ALI/ARDS, a disorder resulting from severe pulmonary infection underscoring an unmet need to identify new molecular targets for therapeutic intervention. The fundamentally new findings in this study are that (i) PRMT4 protein is elevated in infected lung tissue of human subjects and lung epithelial cells exposed to bacterial endotoxin; (ii) SCFFBXO9, via unique molecular signatures, normally targets the labile protein PRMT4 for its cellular elimination, but the E3 ligase is decreased in cells by endotoxin resulting in stabilized PRMT4 that mediates lung epithelial cell death; and (iii) genetic depletion or chemical inhibition of PRMT4 attenuates lung epithelial cell death and protects mice after endotoxin-induced lung injury (Fig. 8). Dysregulation of PRMT4 has been reported in many pathophysiological settings. PRMT4 is aberrantly expressed in breast, prostate, and colorectal cancers that is associated with a poor prognosis by promoting tumor progress and cancer metastasis [24, 44–49]. Consistent with these observations, we identified that PRMT4 expression is increased in ALI. Here the proinflammatory PRMT4 protein was upregulated in cellular and mouse injury models and in human infected lung tissue samples. Increased PRMT4 protein was both required and sufficient to cause lung epithelial cell death after endotoxin exposure and this finding may be one mechanism underlying the pathogenesis of ALI [50].
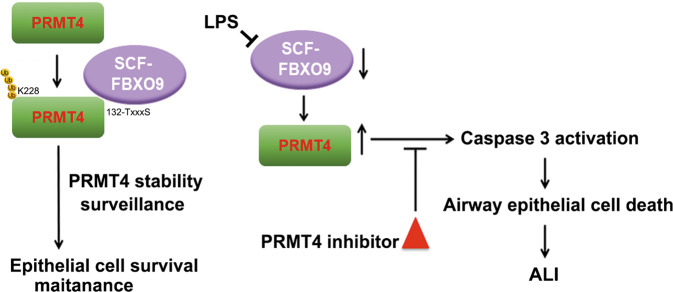
Fig. 8. Schematic presentation of the role of PRMT4 and SCFFBXO9 in airway cell death.
Left: PRMT4 is normally degraded via a SCFFBXO9 E3 ligase-mediated ubiquitin proteasomal pathway to maintain airway epithelial cell survival. SCFFBXO9 recognizes PRMT4 via a phosphodegron and poly-ubiquitinates PRMT4 at K228. Bacterial endotoxin reduces FBXO9 levels that results in PRMT4 protein accumulation. Increased PRMT4 activates caspase 3 to induce lung epithelial apoptosis during bacterial infection.
A number of SCF-E3 ubiquitin ligases or deubiquitinases are found to regulate protein stability and availability of key molecules in the pathogenesis of ALI/ARDS. SCFFBXL2 regulates the protein stability of TRAF proteins in viral infection; mutations in FBXL2 may impair TRAF protein stability that augments inflammatory response [25, 51]. SCFFBXL19 specifically interacts with ST2L receptor for IL-33 that abrogates the proapoptotic and inflammatory effects of IL-33 in ALI models [23]. In previous work, we identified that SCFFBXL18 turns over a chromatin modulator Mortality factor 4 like 1 (MORF4L1) to regulate the life span of lung epithelial cells. The Gram-negative bacterium Pseudomonas aeruginosa disrupts the interaction between FBXL18 and MORF4L1 that elevates cellular MORF4L1 protein levels to cause lung epithelial cell death [42]. FBXO9 is a less studied E3 ubiquitin ligase with functions in muscle and neuronal development [52, 53]. Aberrant expression of FBXO9 to regulate mammalian target of rapamycin signaling by targeting Tel2 and Tti1 degradation promotes survival of multiple myeloma cells [38]. FBXO9 has been shown to be related to innate immunity by responding to LPS stimulation in mouse models [37]. FBXO9 interacts with PPARγ to ubiquitinate this receptor for proteasome degradation [41]. PPARγ is an important player in the immune system for maturation and function of various immune system-related cell types, such as monocytes/microphages, dendritic cells, and lymphocytes [54].
Here we identified a new molecular target of FBXO9 in its control of a protein arginine methyltransferase, PRMT4, that participates in ALI pathogenesis through control of lung epithelial cell viability. Endotoxin LPS decreases FBXO9 that impairs degradation of PRMT4 protein. Elevated PRMT4 activates both intrinsic and external apoptotic pathways to cause epithelia death. Before PRMT4 is targeted for ubiquitination and subsequent degradation, PRMT4 is recognized by the E3 ubiquitin ligase SCFFBXO9 through a phosphodegron. In this motif (132-TdxxxSp), the phosphorylation status of serine and threonine residues appear to be differentially modified for optimal FBXO9 binding and subsequent ubiquitination. Phosphodegrons have been widely cited in governing the stability of crucial molecules in the pathogenesis of diseases. In this regard, we previously identified that oxidative stress reduces PRMT4 levels via downregulation of a protein kinase GSK-3β. GSK-3β phosphorylates T132 that is crucial in regulating the affinity between the phosphodegron and the E3 ubiquitin ligase [43]. Interestingly, in the present study, we observed that LPS downregulates the E3 ubiquitin ligase component FBXO9 at the protein level to stabilize PRMT4 protein. Depletion of PRMT4 by shRNA or with a PRMT4 small-molecule inhibitor improved mouse lung cell viability in an LPS lung injury mouse model, suggesting that lung epithelial cell death mediated by this protein arginine methyltransferase may be a critical factor underlying the pathogenesis of endotoxin-mediated injury. Thus, it is reasonable to postulate that there exists a delicate balance between SCFFBXO9 and PRMT4 that, in part, governs the life span of epithelia during endotoxin stress. The results suggest a potential for pharmaceutical targeting of PRMT4 to reduce the severity of tissue injury in ALI/ARDS.
Materials and methods
Cell lines and reagents
Human lung epithelial BEAS-2B cells and mouse alveolar epithelial MLE12 cells were from ATCC, maintained with HITES medium (Gibco) containing 10% fetal bovine serum. Cells were maintained in a 37 °C incubator in the presence of 5% CO2. Human primary small airway epithelial cells were from ATCC and cultured with airway cell basal medium (Cat#: ATCC PCS-300-040) accompanied with bronchial epithelial growth kit (Cat#: ATCC PCS-300-040). V5 antibody (Cat#: 37–7500), pcDNA3.1D-His-V5-TOPO cloning kit (Cat#: K490001) and E. coli Top10 competent cells (Cat#: C404006) were from Invitrogen (St. Louis, MO). PRMT4 (Cat#:12495), cleaved caspase 3 (Cat#: 9661), cleaved caspase 8 (Cat#: 9748), and cleaved caspase 9 (Cat#: 9505) antibodies were from Cell Signaling (Danvers, MA). Ubiquitin (Cat#: sc-166553, lot: A2710) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). FBXO9 (Cat#: PA5-23474) antibody was from Thermo Fisher (Rockford, IL). The PRMT4 shRNA was from Origene (Rockville, MD). Cycloheximide (Cat#: ALX-380-269-G001, lot: 01061518) and Ubiquitin aldehyde (Cat#: BML-UW8450-0050, lot: 07021447) were from Enzo Life Sciences (Farmingdale, NY). β-Actin (Cat#: A3853) antibody and bacterial lipopolysaccharide (LPS) from E. coli O111:B4 (Cat#: L4391, lot: 115M4090V) were from Sigma (Carlsbad, CA). MG132 (Cat#: F1101, lot: F11052079) was from UBPBio (Aurora, CO). TP064 (Cat#: 6008) was from Tocris Bioscience (Ellisville, MO). QuikChange II XL site-directed mutagenesis kits (Cat#: 200522) were from Agilent Technologies (Santa Clara, CA). TnT Quick Coupled Transcription/Translation Systems (Cat#: L1170) were from Promega (Madison, WI). Immobilized protein A/G agarose beads (Cat#: 20421) were from Pierce (Rockford, IL). All other reagents were of the highest grade available commercially.
Cloning and mutagenesis
V5-tagged PRMT4 truncations were cloned into pcDNA3.1D-His-V5-TOPO plasmid using PCR-based approaches as previously described [55]. Mutagenesis was introduced by using a QuickChange II XL site-directed mutagenesis kit according to the manufacturer’s instructions. The accuracy of the mutagenesis was confirmed by sequencing. The primers used in the construction of PRMT4 truncations and site-directed mutagenesis are listed in Table 1.
Table 1.
Primers used in the study.
| Truncation | PRMT4F1 | CACCATGGCAGCGGCGGCAGCG |
| PRMT4F2 | CACCATGCACGCGGAGCAGCAG | |
| PRMT4F3 | CACCATGATCACCCTGGGCTGCAAC | |
| PRMT4F4 | CACCATGTTCCAGTTCTATGGC | |
| PRMT4F5 | CACCATGTTTTTTGCTGCTCAAGC | |
| PRMT4F6 | CACCATGCAAGTGGACATTATC | |
| PRMT4F7 | CACCATGGAACAGCTCTACATGG | |
| PRMT4R1 | ACTCCCATAGTGCATGGTGTTG | |
| PRMT4R2 | GGAGTGGGTGTGATTGACAATC | |
| PRMT4R3 | TGTTCCACATATTCTCCGAGG | |
| PRMT4R4 | GTCATAGCTCTGTCTTTTG | |
| PRMT4R5 | TATGGAGCCAATGAAAGCAAC | |
| PRMT4R6 | CATCAGGATCCGGATGTCAAATG | |
| DN131f | CACCATGACACTGGAGCGCTCTGTGTTC | |
| DN132f | CACCATGCTGGAGCGCTCTGTGTTCAG | |
| DN133f | CACCATGGAGCGCTCTGTGTTCAGTG | |
| DN134f | CACCATGCGCTCTGTGTTCAGTGAG | |
| DN135f | CACCATGTCTGTGTTCAGTGAGCGGAC | |
| DN136f | CACCATGGTGTTCAGTGAGCGGACAG | |
| DN137f | CACCATGTTCAGTGAGCGGACAGAGG | |
| DN138f | CACCATGAGTGAGCGGACAGAGGAATC | |
| DN105f | CACCATGAACAGCGTCCTCATCCAG | |
| DN110f | CACCATGCAGTTTGCCACACCCCACG | |
| DN115f | CACCATGCACGATTTCTGTTCTTTCTAC | |
| DN120f | CACCATGTTCTACAACATCCTGAAAAC | |
| DN130f | CACCATGCACACACTGGAGCGCTCTG | |
| DN140f | CACCATGCGGACAGAGGAATCCTCAG | |
| Mutagenesis | T132Cf | CCTGTCGGGGCCACTGCCTGGAGCGCTCTGTG |
| T132Cr | CACAGAGCGCTCCAGGCAGTGGCCCCGACAGG | |
| T132Df | CCTGTCGGGGCCACGACCTGGAGCGCTCTGTG | |
| T132Dr | CACAGAGCGCTCCAGGTCGTGGCCCCGACAGG | |
| S136Cf | CCACACACTGGAGCGCTGTGTGTTCAGTGAGCGG | |
| S136Cr | CCGCTCACTGAACACACAGCGCTCCAGTGTGTGG | |
| S136Df | CCACACACTGGAGCGCGATGTGTTCAGTGAGCGG | |
| S136Dr | CCGCTCACTGAACACATCGCGCTCCAGTGTGTGG | |
| T132AS136Af | CCACGCACTGGAGCGCGCTGTGTTCAGTGAGCGG | |
| T132AS136Ar | CCGCTCACTGAACACAGCGCGCTCCAGTGCGTGG | |
| T132CS136Df | CCACTGCCTGGAGCGCGATGTGTTCAGTGAGCGG | |
| T132CS136Dr | CCGCTCACTGAACACATCGCGCTCCAGGCAGTGG | |
| K463Af | GACCAGACAGGCTCCGCGTCCAGTAACCTGCTGG | |
| K463Ar | CCAGCAGGTTACTGGACGCGGAGCCTGTCTGGTC | |
| K471Af | GTAACCTGCTGGATCTAGCGAACCCCTTCTTCAGG | |
| K471Ar | CCTGAAGAAGGGGTTCGCTAGATCCAGCAGGTTAC | |
| K310Af | CATGGAGCAGTTCACCGCAGCCAACTTCTGGTACC | |
| K310Ar | GGTACCAGAAGTTGGCTGCGGTGAACTGCTCCATG | |
| K228Af | GCAGAGGTCCTGGTGGCGAGTAACAATCTGACAG | |
| K228Ar | CTGTCAGATTGTTACTCGCCACCAGGACCTCTGC | |
| K242Af | CGTGGTCATCCCTGGCGCAGTAGAGGAGGTCTC | |
| K242Ar | GAGACCTCCTCTACTGCGCCAGGGATGACCACG |
The orientation of all primers is in 5′ to 3′ direction.
Plasmid transfection
All plasmids were introduced into cells using electroporation executed with a nuclear transfection apparatus (Amaxa Biosystems, Gaithersburg, MD) with a preset program (T-013 for MLE12 and BEAS-2B cells), following the manufacturer’s instructions as previously described [55]. Briefly, one million cells in 100 μL of transfection buffer (20 mM Hepes in PBS buffer) were mixed with 3 μg of plasmids (including expression and shRNA constructs). After electroporation, the cells were cultured with 2 mL conditional medium in 6-well plates for 24 h for further analysis.
Immunoblotting and co-IP
Immunoblotting and co-IP were conducted as previously described [55]. Briefly, for immunoblotting, whole-cell extracts (normalized to total protein concentration) were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to membranes by electroblotting. The membranes were blocked with 5% (w/v) non-fat milk in Tris-buffered saline and probed with primary antibodies as indicated. Membranes were developed by an enhanced chemiluminescence (ECL) system. For IP, 1 mg of cell lysates (in PBS with 0.5% Tween 20 plus protease inhibitors) were incubated with specific primary antibodies for 2 h at room temperature. The mixture was added with 35 µL of protein A/G agarose beads for an additional 2 h at room temperature. The precipitated complex was washed for three times with 0.5% Tween 20 in PBS and analyzed by immunoblotting with ECL system.
Real‐time PCR
Total RNA from cells was isolated with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and reverse-transcribed by using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s manual instructions. Single-stranded cDNA was then amplified by RT‐PCR with specific primers of PRMT4 and GAPDH. RT‐PCR was performed on an ABI Prism 7000 thermocycler (Applied Biosystems, Thermo-Fisher Scientific, Waltham, MA, USA) with SYBR Green PCR Master Mix (Roche, Basel, Switzerland). For each experiment, samples (n = 3) were run in triplicate. Relative gene expression was calculated using the comparative CT method.
In vitro pull-down assay
We conducted in vitro binding assays to identify the FBXO9-binding domain within PRMT4. V5-tagged PRMT4 truncated or site-directed mutant proteins were in vitro expressed using a TnT coupled reticulocyte system. Endogenous FBXO9 protein was obtained by FBXO9 IP from MLE12 cell lysate. FBXO9-precipitated protein A/G agarose beads were incubated with a variety of PRMT4 truncations or mutants for 2 h. The beads were washed extensively with 0.5% Tween 20 in PBS and analyzed by V5 and FBXO9 immunoblotting.
Cell death detection
Lactate dehydrogenase (LDH) assay kit (Takara, Japan) was used to detect the epithelial cell death according to the manufacturer’s instructions. Absorbance of the samples at wavelength of 490 and 620 nm (background) were measured using a SpectraMax M2 microplate reader (Molecular Devices, CA, USA). The LDH activity was determined by subtracting the optical density (OD) at 620 nm absorbance value from the OD at 490 nm. The fold change of epithelial cell death was indicated with the relative LDH activity comparing to the control group and plotted in a bar graph.
Lentivirus production
Lentiviruses were generated by Lenti-X Packaging Single Shots VSV-G (Cat#: 631276, Clontech, CA) according to the manufacturer’s procedures. Lentivirus-containing supernatants were concentrated using Lenti-X Concentrator (Cat#: 631231). Concentrated lentiviruses were resuspended in PBS and titrated by Lenti-X GoStix (Cat#: 631280). The samples were aliquoted and stored at −80 °C.
Mouse LPS-induced lung injury procedures
All animal protocols and procedures were reviewed and approved by the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC protocol #: IS00010084). LPS-induced lung injury model was conducted as previously described. Briefly, C57BL/6J mice at the age of 10 weeks were anesthetized by injecting intraperitoneally a solution of 1:1 ketamine (100 mg/kg) and xylazine (20 mg/kg) throughout the experiment. LPS (7 mg/kg) were i.t. administrated. The mice were observed for 48 h and sacrificed. Lung tissues were collected and applied for hematoxylin–eosin staining, TUNEL assay, and western blotting.
TUNEL assay
Mouse lung tissues were fixed in 10% formalin for 48 h and then embedded in paraffin and sectioned. Slides were deparaffinized and permeabilized with 0.1% Triton X-100 in citrate buffer (0.1%, pH 6.0). TUNEL staining was performed on these slides using an In Situ Cell Death Detection Kit (Sigma-Aldrich, Cat#: 11684795910) according to the manufacturer’s instructions. The slides were visualized by a confocal microscope (NIKON A1 Spectral Confocal, Japan) with excitation wavelength in the range of 450–500 nm and detection in the range of 515–565 nm.
Statistics
The data represent the mean ± standard deviation in the graphs depicting the error bars or as specifically indicated. Prism 7 (GraphPad software, San Diego, CA) was used to determine statistical significance. Comparisons between groups were made using unpaired, two-tailed Student’s t test (two groups) and one-way analysis of variance with post hoc Tukey honestly significant difference or Bonferroni and Holm multiple comparisons. Kaplan–Meier estimate was used for survival analysis in mouse septic models. P < 0.05 indicates statistical significance.
Acknowledgements
The US National Heart, Lung and Blood institute and the US Department of Veterans Affairs have been gratefully acknowledged. We thank Dr. Prabir Ray and Dr. Anuradha Ray for the in-depth discussion in science.
Author contributions
CZ conceived science and designed the experiments. YL, XL, and TL conducted immunoblotting analysis. YL and XL performed animal studies. BJM, YZ, SMN, and GDK provided human samples. JSL and RKM helped to develop the science and interpreted the results. The manuscript was written by YL and CZ and edited by KC, TN, YZ, BJM, GDK, SMN, JSL, and RKM. All authors read and approved the final manuscript.
Funding information
This work was in part, supported by a National Institutes of Health R01 grants HL125435 and HL142997 (to CZ), HL096376, HL097376, HL098174, HL081784, and P01 HL114453 (to RKM), HL136143, HL142084, HL143285, and P01 HL114453 (to JSL), and grants 5I01CX000105-06 and CX001048 from the US Department of Veterans Affairs (to TN/RKM). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or any other sponsoring agency.
Data availability
The data that supporting the finding of this study are available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This paper includes the use of deidentified human lung samples. All animal protocols and procedures were reviewed and approved by the University of Pittsburgh Institutional Animal Care and Use Committee (IACUC protocol #: IS00010084).
Footnotes
Edited by Professor Hans-Uwe Simon
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yandong Lai, Xiuying Li, Tiao Li, Rama K. Mallampalli, Chunbin Zou.
References
- 1.Han S, Mallampalli RK. The acute respiratory distress syndrome: from mechanism to translation. J Immunol. 2015;194:855–60. doi: 10.4049/jimmunol.1402513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ARDS Definition Task Force. Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, et al. Acute respiratory distress syndrome: the Berlin Definition. JAMA. 2012;307:2526–33. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 3.Matthay MA, Song Y, Bai C, Jones KD. The acute respiratory distress syndrome in 2013. Transl Respir Med. 2013;1:10. doi: 10.1186/2213-0802-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Confalonieri M, Salton F, Fabiano F. Acute respiratory distress syndrome. Eur Respir Rev. 2017;26:160116.. doi: 10.1183/16000617.0116-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthay MA, Zemans RL, Zimmerman GA, Arabi YM, Beitler JR, Mercat A, et al. Acute respiratory distress syndrome. Nat Rev Dis Prim. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iscimen R, Cartin-Ceba R, Yilmaz M, Khan H, Hubmayr RD, Afessa B, et al. Risk factors for the development of acute lung injury in patients with septic shock: an observational cohort study. Crit Care Med. 2008;36:1518–22. doi: 10.1097/CCM.0b013e31816fc2c0. [DOI] [PubMed] [Google Scholar]
- 7.Kojicic M, Li G, Hanson AC, Lee KM, Thakur L, Vedre J, et al. Risk factors for the development of acute lung injury in patients with infectious pneumonia. Crit Care. 2012;16:R46. doi: 10.1186/cc11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubenfeld GD, Caldwell E, Peabody E, Weaver J, Martin DP, Neff M, et al. Incidence and outcomes of acute lung injury. N. Engl J Med. 2005;353:1685–93. doi: 10.1056/NEJMoa050333. [DOI] [PubMed] [Google Scholar]
- 9.Li T, Zou C. The role of deubiquitinating enzymes in acute lung injury and acute respiratory distress syndrome. Int J Mol Sci. 2020;21:4842.. doi: 10.3390/ijms21144842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martines RB, Ritter JM, Matkovic E, Gary J, Bollweg BC, Bullock H, et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect Dis. 2020;26:2005–15. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–54. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calabrese F, Pezzuto F, Fortarezza F, Hofman P, Kern I, Panizo A, et al. Pulmonary pathology and COVID-19: lessons from autopsy. The experience of European Pulmonary Pathologists. Virchows Arch. 2020;477:359–72. doi: 10.1007/s00428-020-02886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acosta MAT, Singer BD. Pathogenesis of COVID-19-induced ARDS: implications for an ageing population. Eur Respir J. 2020;56:2002049.. doi: 10.1183/13993003.02049-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson ER, Matthay MA. Acute lung injury: epidemiology, pathogenesis, and treatment. J Aerosol Med Pulm Drug Deliv. 2010;23:243–52. doi: 10.1089/jamp.2009.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Covic M, Hassa PO, Saccani S, Buerki C, Meier NI, Lombardi C, et al. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 2005;24:85–96. doi: 10.1038/sj.emboj.7600500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc Natl Acad Sci USA. 2003;100:6464–8. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36:3202–13. doi: 10.1093/nar/gkn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 20.Wei Y, Horng JC, Vendel AC, Raleigh DP, Lumb KJ. Contribution to stability and folding of a buried polar residue at the CARM1 methylation site of the KIX domain of CBP. Biochemistry. 2003;42:7044–9. doi: 10.1021/bi0343976. [DOI] [PubMed] [Google Scholar]
- 21.Shishkova E, Zeng H, Liu F, Kwiecien NW, Hebert AS, Coon JJ, et al. Global mapping of CARM1 substrates defines enzyme specificity and substrate recognition. Nat Commun. 2017;8:15571. doi: 10.1038/ncomms15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'brien KB, Alberich-Jordà M, Yadav N, Kocher O, Diruscio A, Ebralidze A, et al. CARM1 is required for proper control of proliferation and differentiation of pulmonary epithelial cells. Development. 2010;137:2147–56. doi: 10.1242/dev.037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis MB, Liu X, Wang S, Reeves J, Khramtsov A, Huo D, et al. Expression and sub-cellular localization of an epigenetic regulator, co-activator arginine methyltransferase 1 (CARM1), is associated with specific breast cancer subtypes and ethnicity. Mol Cancer. 2013;12:40. doi: 10.1186/1476-4598-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Roberts CW. CARMA: CARM1 methylation of SWI/SNF in breast cancer. Cancer Cell. 2014;25:3–4. doi: 10.1016/j.ccr.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganesh L, Yoshimoto T, Moorthy NC, Akahata W, Boehm M, Nabel EG, et al. Protein methyltransferase 2 inhibits NF-κB function and promotes apoptosis. Mol Cell Biol. 2006;26:3864–74. doi: 10.1128/MCB.26.10.3864-3874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D-i, Park MJ, Lim SK, Choi JH, Kim JC, Han HJ, et al. High-glucose-induced CARM1 expression regulates apoptosis of human retinal pigment epithelial cells via histone 3 arginine 17 dimethylation: role in diabetic retinopathy. Arch Biochem Biophysics. 2014;560:36–43. doi: 10.1016/j.abb.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Peng M, Zeng T, Zheng J, Liao Y, Zhang H, et al. Protein arginine methyltransferase 4 regulates adipose tissue lipolysis in type 1 diabetic mice. Diabetes Metab Syndr Obes. 2020;13:535–44. doi: 10.2147/DMSO.S235869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Ju C, Hu J, Huang K, Yang L. PRMT4 overexpression aggravates cardiac remodeling following myocardial infarction by promoting cardiomyocyte apoptosis. Biochem Biophys Res Commun. 2019;520:645–50. doi: 10.1016/j.bbrc.2019.10.085. [DOI] [PubMed] [Google Scholar]
- 29.Zou C, Chen Y, Smith RM, Snavely C, Li J, Coon TA, et al. SCF(Fbxw15) mediates histone acetyltransferase binding to origin recognition complex (HBO1) ubiquitin-proteasomal degradation to regulate cell proliferation. J Biol Chem. 2013;288:6306–16. doi: 10.1074/jbc.M112.426882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou C, Butler PL, Coon TA, Smith RM, Hammen G, Zhao Y, et al. LPS impairs phospholipid synthesis by triggering beta-transducin repeat-containing protein (beta-TrCP)-mediated polyubiquitination and degradation of the surfactant enzyme acyl-CoA:lysophosphatidylcholine acyltransferase I (LPCAT1) J Biol Chem. 2011;286:2719–27. doi: 10.1074/jbc.M110.192377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Wei J, Mialki RK, Mallampalli DF, Chen BB, Coon T, et al. F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat Immunol. 2012;13:651–8. doi: 10.1038/ni.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen BB, Glasser JR, Coon TA, Zou C, Miller HL, Fenton M, et al. F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood. 2012;119:3132–41. doi: 10.1182/blood-2011-06-358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen BB, Coon TA, Glasser JR, McVerry BJ, Zhao J, Zhao Y, et al. A combinatorial F box protein directed pathway controls TRAF adaptor stability to regulate inflammation. Nat Immunol. 2013;14:470–9. doi: 10.1038/ni.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Li DC, Li ZF, Liu CX, Xiao YM, Zhang B, et al. Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene. 2011;30:3875–86. doi: 10.1038/onc.2011.103. [DOI] [PubMed] [Google Scholar]
- 35.Du X, Meng F, Peng D, Wang Z, Ouyang W, Han Y, et al. Noncanonical role of FBXO6 in regulating antiviral immunity. J Immunol. 2019;203:1012–20. doi: 10.4049/jimmunol.1801557. [DOI] [PubMed] [Google Scholar]
- 36.Lin M, Wang ZW, Zhu X. FBXO45 is a potential therapeutic target for cancer therapy. Cell Death Discov. 2020;6:55. doi: 10.1038/s41420-020-0291-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang IV, Wade CM, Kang HM, Alper S, Rutledge H, Lackford B, et al. Identification of novel genes that mediate innate immunity using inbred mice. Genetics. 2009;183:1535–44. doi: 10.1534/genetics.109.107540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernández-Sáiz V, Targosz BS, Lemeer S, Eichner R, Langer C, Bullinger L, et al. SCFFbxo9 and CK2 direct the cellular response to growth factor withdrawal via Tel2/Tti1 degradation and promote survival in multiple myeloma. Nat Cell Biol. 2013;15:72–81. doi: 10.1038/ncb2651. [DOI] [PubMed] [Google Scholar]
- 39.Cheng H, Ma Y, Ni X, Jiang M, Guo L, Jin W, et al. cDNA cloning and expression analysis of a novel human F-box only protein. Mol Cells. 2002;14:56–9. [PubMed] [Google Scholar]
- 40.Lee KW, Kwak SH, Ahn BY, Lee HM, Jung HS, Cho YM, et al. F-box only protein 9 is required for adipocyte differentiation. Biochem Biophys Res Commun. 2013;435:239–43. doi: 10.1016/j.bbrc.2013.04.072. [DOI] [PubMed] [Google Scholar]
- 41.Lee KW, Kwak SH, Koo YD, Cho YK, Lee HM, Jung HS, et al. F-box only protein 9 is an E3 ubiquitin ligase of PPARgamma. Exp Mol Med. 2016;48:e234. doi: 10.1038/emm.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zou C, Li J, Xiong S, Chen Y, Wu Q, Li X, et al. Mortality factor 4 like 1 protein mediates epithelial cell death in a mouse model of pneumonia. Sci Transl Med. 2015;7:311ra171. doi: 10.1126/scitranslmed.aac7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X, Lai Y, Li J, Zou M, Zou C. Oxidative stress destabilizes protein arginine methyltransferase 4 via glycogen synthase kinase 3beta to impede lung epithelial cell migration. Am J Physiol Cell Physiol. 2017;313:C285–94. doi: 10.1152/ajpcell.00073.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng H, Qin Y, Fan H, Su P, Zhang X, Zhang H, et al. Overexpression of CARM1 in breast cancer is correlated with poorly characterized clinicopathologic parameters and molecular subtypes. Diagn Pathol. 2013;8:129. doi: 10.1186/1746-1596-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong H, Kao C, Jeng MH, Eble JN, Koch MO, Gardner TA, et al. Aberrant expression of CARM1, a transcriptional coactivator of androgen receptor, in the development of prostate carcinoma and androgen-independent status. Cancer. 2004;101:83–9. doi: 10.1002/cncr.20327. [DOI] [PubMed] [Google Scholar]
- 46.Kim YR, Lee BK, Park RY, Nguyen NT, Bae JA, Kwon DD, et al. Differential CARM1 expression in prostate and colorectal cancers. BMC Cancer. 2010;10:197. doi: 10.1186/1471-2407-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habashy HO, Rakha EA, Ellis IO, Powe DG. The oestrogen receptor coactivator CARM1 has an oncogenic effect and is associated with poor prognosis in breast cancer. Breast Cancer Res Treat. 2013;140:307–16. doi: 10.1007/s10549-013-2614-y. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, Zhao Z, Meyer MB, Saha S, Yu M, Guo A, et al. CARM1 methylates chromatin remodeling factor BAF155 to enhance tumor progression and metastasis. Cancer Cell. 2014;25:21–36. doi: 10.1016/j.ccr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng L, Chen J, Zhou Z, He Z. miR-195 enhances the radiosensitivity of colorectal cancer cells by suppressing CARM1. Onco Targets Ther. 2017;10:1027–38. doi: 10.2147/OTT.S125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McDunn JE, Hotchkiss RS. Leukocyte phenotyping to stratify septic shock patients. Crit Care. 2009;13:127. doi: 10.1186/cc7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma J, Zou C, Guo L, Seneviratne DS, Tan X, Kwon YK, et al. Novel death defying domain in Met entraps the active site of caspase-3 and blocks apoptosis in hepatocytes. Hepatology. 2014;59:2010–21. doi: 10.1002/hep.26769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki T, Delgado-Escueta AV, Alonso ME, Morita R, Okamura N, Sugimoto Y, et al. Mutation analyses of genes on 6p12-p11 in patients with juvenile myoclonic epilepsy. Neurosci Lett. 2006;405:126–31. doi: 10.1016/j.neulet.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 53.Zhang DM, He T, Katusic ZS, Lee HC, Lu T. Muscle-specific f-box only proteins facilitate bk channel beta(1) subunit downregulation in vascular smooth muscle cells of diabetes mellitus. Circ Res. 2010;107:1454–9. doi: 10.1161/CIRCRESAHA.110.228361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hernandez-Quiles M, Broekema MF, Kalkhoven E. PPARgamma in metabolism, immunity, and cancer: unified and diverse mechanisms of action. Front Endocrinol. 2021;12:624112. doi: 10.3389/fendo.2021.624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou C, Synan MJ, Li J, Xiong S, Manni ML, Liu Y, et al. LPS impairs oxygen utilization in epithelia by triggering degradation of the mitochondrial enzyme Alcat1. J Cell Sci. 2016;129:51–64. doi: 10.1242/jcs.176701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that supporting the finding of this study are available from the corresponding author upon reasonable request.