Abstract
The Rho3 protein plays a critical role in the budding yeast Saccharomyces cerevisiae by directing proper cell growth. Rho3 appears to influence cell growth by regulating polarized secretion and the actin cytoskeleton, since rho3 mutants exhibit large rounded cells with an aberrant actin cytoskeleton. To gain insights into how Rho3 influences these events, we have carried out a yeast two-hybrid screen using an S. cerevisiae cDNA library to identify proteins interacting with Rho3. Two proteins, Exo70 and Myo2, were identified in this screen. Interactions with these two proteins are greatly reduced or abolished when mutations are introduced into the Rho3 effector domain. In addition, a type of mutation known to produce dominant negative mutants of Rho proteins abolished the interaction with both of these proteins. In contrast, Rho3 did not interact with protein kinase C (Pkc1), an effector of another Rho family protein, Rho1, nor did Rho1 interact with Exo70 or Myo2. Rho3 did interact with Bni1, another effector of Rho1, but less efficiently than with Rho1. The interaction between Rho3 and Exo70 and between Rho3 and Myo2 was also demonstrated with purified proteins. The interaction between Exo70 and Rho3 in vitro was dependent on the presence of GTP, since Rho3 complexed with guanosine 5′-O-(3-thiotriphosphate) interacted more efficiently with Exo70 than Rho3 complexed with guanosine 5′-O-(3-thiodiphosphate). Overlapping subcellular localization of the Rho3 and Exo70 proteins was demonstrated by indirect immunofluorescence. In addition, patterns of localization of both Exo70 and Rho3 were altered when a dominant active allele of RHO3, RHO3E129,A131, which causes a morphological abnormality, was expressed. These results provide a direct molecular basis for the action of Rho3 on exocytosis and the actin cytoskeleton.
Polarized cell growth is a fundamental necessity for many cell types. Yeast cells must grow in a polarized fashion for proper bud formation, and likewise, nerve cells must also exhibit polarized growth during axon formation (6, 7, 12, 20, 21, 35). In yeast, this asymmetric pattern of growth is accomplished by the vectorial transport of secretory vesicles to the cell surface specifically at the bud site. Multiple genes are known to be responsible for ensuring the proper bud positioning and subsequent directed exocytosis of newly synthesized materials to the bud; however, the exact mechanism by which this is accomplished is poorly understood (6, 7, 12, 20, 21, 35).
One protein, of many, that seems to play a critical role in this process is Rho3 (36), a member of the Rho family of proteins which include Rho1 (33), Rho2 (33), Rho4 (36), and Cdc42 (1, 25). Rho3 and Rho4 are important for directing the deposition of newly synthesized materials to the bud site. Disruption of RHO3 results in slow growth, which is exacerbated by the additional disruption of RHO4, the combination of which causes lethality above 30°C (36). Morphologically, temperature-sensitive rho3-1 mutants appear as enlarged rounded cells with an aberrant actin cytoskeleton where actin patches are delocalized at nonpermissive temperatures (23). Furthermore, cells depleted in both Rho3 and Rho4 lyse at the small-budded stage (37). The addition of osmotic stabilizing agents partially suppresses this lethality (37), which supports the idea that the cell lysis is caused by mislocalization of materials necessary for bud growth. A weakened cell wall at the site of active growth could eventually cause the cell wall to rupture. Expression of an activated RHO3 allele, RHO3E129,A131, causes cold-sensitive growth and the appearance of elongated cells which are often bent at the sites where actin patches are observed (23). Genetic interactions of RHO3 with bud-site assembly genes such as CDC42 and BEM1 have been detected (37). In addition, RHO3 has been shown to interact genetically with SEC4 (23), a GTPase involved in the fusion of secretory vesicles with the plasma membrane (17, 48). From these studies, it was suggested that Rho3 plays a critical role in directing newly synthesized proteins to the bud site. This process involves organization of the actin cytoskeleton; when this function is disrupted, newly synthesized proteins are deposited in an isotropic fashion, leading to the appearance of uniformly enlarged cells or, once bud formation has begun, lysis of cells at the small-budded stage.
To gain further insights into the function of Rho3, we performed a yeast two-hybrid assay which led to identification of Exo70 and Myo2. Exo70 is a component of the exocyst, a multiprotein complex which is involved in exocytosis (53). Other components of the exocyst identified are Sec3, Sec5, Sec6, Sec8, Sec10, and Sec15 (53). This complex is believed to reside at the tip of the bud, enabling the fusion of secretory vesicles with the plasma membrane, a process which requires Sec4 (15). Myo2 is an unconventional myosin that is proposed to be important in the movement of secretory vesicles to the bud site (26). The myo2-66 mutant accumulates secretory vesicles in the mother cell at nonpermissive temperatures and, like rho3-1 temperature-sensitive mutants, exhibits an aberrant actin cytoskeleton including delocalized cortical actin patches (26). In this report, we also show that the interaction of Rho3 with these proteins requires the effector domain of Rho3 and that the interaction of Rho3 with Exo70 is dependent on the presence of the GTP-bound form of Rho3. In addition, we show that the localization of Exo70 largely follows that of Rho3, raising the possibility that Rho3 is required to direct the exocyst to areas of active cell growth.
MATERIALS AND METHODS
Construction of two-hybrid vectors and PCR mutagenesis.
rho3E129, an activating mutation of RHO3, was inserted into a modified version of the two-hybrid construct pGBT9 (13) which contains a KpnI site (44). A RHO3 fragment flanked at its 5′ end with a KpnI site and at its 3′ end with a SalI site was created by PCR amplifying the template pYO324-Rho3 (23), using primers 5′-ACTGTAGGTACCATGTCATTTCTATGTGGGTCAG-3′ and 5′-ATCTCCGTCGACTTACATAATGGTACAGCTGG-3′. The resulting fragment was then inserted into the corresponding sites of pGBT9-KpnI to create pGBRHO3E129. This construct was then sequenced to ensure that no additional mutations occurred during PCR amplification. The C-terminal CAAX sequence of Rho3 was deleted to prevent mislocalization of Rho3 in the two-hybrid system. This was accomplished by digesting pGBRHO3E129 with KpnI and BamHI and inserting the KpnI/BamHI fragment into the corresponding sites of pGBT9-KpnI. This created a plasmid, pGBRHO3E129Δ5, containing the RHO3 sequence missing the last five amino acids. Another CAAX-less construct, pGBRHO3E129Δ, missing the last four amino acids was also constructed by adding the linker 5′-GATCCAGCTAAAGATCTTG-3′, which is flanked by BamHI and SalI, into the BamHI- and SalI-digested pGBRHO3E129 vector. This resulted in the deletion of the last four amino acids corresponding to the CAAX motif followed by a stop codon.
The rho3E129,V25, rho3E129,A48, rho3E129,S47, and rho3E129,N30 mutations were created by site-directed mutagenesis with overlap extension using PCR (22). The mutagenic primers for the V25 mutation were B (5′-TGGGCGACGTTGCCTGTGGTAAAACTTCG-3′) and C (5′-CACAGGCAACGTCGCCCAAAATAACGATC-3′), those for the A48 mutation were B (5′-TTATGAGCCTGCTGTTTTTGAAAACTATATCC-3′) and C (5′-TCAAAAACAGCAGGCTCATAAACTTCGG-3′), those for the S47 mutation were B (5′-TTATGAGTCTACTGTTTTTGAAAACTATATCC-3′) and C (5′-TTCAAAAACAGTAGACTCATAAAC-3′), and those for the N30 mutation were B (5′-GTGGTAAAAATTCGTTGCTG-3′) and C (5′-GCAACGAATTTTTACCACAG-3′). The outside primers were A (5′-ACTGTAGGTACCATGTCATTTCTATGTGGGTCAG-3′) and D (5′-ATCTCCGTCGACTTACATAATGGTACAGCTGG-3′), and the template used was pGBRHO3E129Δ. The resulting PCR products were cloned into the corresponding sites of pGBT9-KpnI. These plasmids were sequenced to ensure that no additional mutations were introduced. Plasmids containing single mutations of V25, A48, S47, and N30 were produced by replacing the E129-containing fragment of RHO3 with a wild-type 3′ region with a BclI-to-SalI fragment.
pGBRHO1V19Δ was constructed using PCR mutagenesis as described above. The internal primers used were B (5′-GTTGGTGATGTTGCCTGTGGTAAGACATG-3′) and C (5′-CACAGGCAACATCACCAACGATTACCAGC-3′). The outside primers were A (5′-TATCGTTTCGACCATCG-3′) and D (5′-GGGATTGAAAAAGGGCAG-3′). The template used was pRS316-HA2-RHO1-SalI, which is a modified version of pRS316-HA2-RHO1 in which a SalI linker was inserted after the RHO1 stop codon (61). pGBT9-KpnI was digested with Acc65I and SalI, and the PCR product was inserted after digestion with Acc65I and SalI. The resulting vector, pGBRHO1V19, was sequenced to ensure that no additional mutations were introduced. The vector was then converted to a CAAL-less version by replacing the 3′ NheI-to-SalI fragment with a fragment missing the last four amino acids. The resulting plasmid, pGBRHO1V19Δ, was used in the two-hybrid analysis.
RBP4 was moved into the pGAD424 vector (4) to create pGAD-RBP4. The pGAD424 vector was digested with BamHI, and a BglII fragment of RBP4 obtained from the pACTI library vector was inserted. (This fragment encodes amino acids 275 to the stop codon of Exo70.) The full-length EXO70 plasmid was constructed by PCR amplification of the 5′ region of the EXO70 open reading frame from a Saccharomyces cerevisiae genomic library and cloning into pGAD424 vector. The primers used for this amplification were A (5′-TGTACCCGGGGATGCCAGCTGAAATTGAC-3′) and B (5′-CTTCTTGCGTACAATCTTGC-3′), which added a SmaI site at the 5′ region. This site was used to clone the 5′ SmaI-to-NdeI fragment of EXO70 into pGAD-RBP4 to make the full-length construct pGAD-EXO70. The PCR-amplified region was sequenced to confirm that no mutations were introduced. The MYO2 fragment isolated from the two-hybrid screen contained coding sequence for 11 residues of lysine after amino acid 1211. These lysine residues were removed by inserting a stop codon after amino acid 1211 via PCR, which resulted in the construction of pGAD-RBP26. The primers used for this amplification were A (5′-CCACTACAATGGATGATG-3′) and B (5′-GTACTAGGATCCGTCGACTTATTCACTTAAAACTATAATCAG-3′). This added a SalI site at the 3′ region of the PCR fragment, which was then digested with BamHI and SalI and cloned into corresponding sites of pGAD-GH (58). The PCR-amplified region was sequenced to ensure that no mutations were introduced. A truncated construct of MYO2, pACTRBP26ΔB, containing amino acids 871 to 1130 was also created. A BglII fragment was removed from the MYO2 clone isolated from the pACT1 library vector and cloned into the BamHI site of pACTII (31) to create pACTRBP26ΔB. pGAD424-PKC1 and pACTII-HK-BNI1 were provided by K. Tanaka (Osaka University).
Two-hybrid screening procedure.
The pGBRHO3E129Δ5 construct was used to screen a yeast cDNA library inserted into the two-hybrid construct pACT1 (13). The screen was carried out as follows. pGBRHO3E129Δ5 was transformed along with the yeast two-hybrid cDNA library into the yeast strain Y190 by the lithium acetate transformation method (16). The transformants were plated on synthetic complete medium (51) lacking tryptophan, leucine, and histidine but containing 25 mM 3-aminotriazole to minimize background. A total of approximately 2 × 106 colonies were screened. The pACT plasmids contained in colonies positive for both the histidine auxotrophy assay and the filter lift β-galactosidase assay were isolated and purified by transformation into Escherichia coli. The plasmid DNA was subsequently retransformed into Y190 with either pGBRHO3E129Δ5 or pGBT9-KpnI. Plasmids which were positive only in the presence of pGBRHO3E129Δ5 were further analyzed. The DNA from these candidates was then sequenced and analyzed with the BLAST program (2) against the S. cerevisiae genome database to determine the identity of the DNA. Filter lift assays and liquid β-galactosidase assays were performed as described previously (44).
Complementation studies.
A centromeric plasmid expressing full-length RHO3 was constructed by replacing RHO1 on plasmid pRS316-HA2-RHO1-SalI. pRS316-HA2-RHO1-SalI was digested with KpnI and SalI, and the KpnI-SalI RHO3 fragments of both the full-length wild type and various mutants were inserted to create pNRHO3, pNRHO3V25, pNRHO3A48, pNRHO3S47, and pNRHO3N30. This resulted in the addition of a double hemagglutinin (HA) epitope at the N terminus of RHO3. These constructs were transformed into yeast cells by the lithium acetate method (16), and the resulting transformants were examined for the ability to complement the rho3 null phenotype. The yeast strain YMR504 (MATα rho3::LEU2 GAL7p [GAL7 promoter]:RHO4 ura3::HIS3 leu2 his3 trp1 lys2 ade2) was used for this experiment. Colony size was assessed on both galactose- and dextrose-containing synthetic complete medium lacking uracil. Western analysis was carried out as described by Sambrook et al. (49).
Protein purification and in vitro binding.
The plasmid for expression of the glutathione S-transferase (GST)–Rho3 fusion protein was constructed by inserting a full-length RHO3 fragment into the KpnI and SalI sites of a modified pGEX-5X-3 vector (Amersham Pharmacia, Piscataway, N.J.) containing a KpnI linker. GST-Rho3 was purified as described previously (52, 60). Maltose binding protein (MBP)-RBP4 was constructed by inserting a BglII (blunt-ended)-to-SalI fragment of RBP4 into the BamHI (blunt-ended) and SalI sites of pMAL-c2, an MBP vector (New England Biolabs). MBP-Exo70 was constructed by inserting a SmaI-to-PstI full-length fragment of EXO70 into the BamHI (blunt-ended) and PstI sites of pMAL-c2. MBP-RBP26 was constructed by inserting an XhoI (blunt-ended)-to-XhoI (blunt-ended) fragment into the BamHI (blunt-ended) site of pMAL-c2. MBP-RBP4, MBP-Exo70, and MBP-RBP26 were purified as described previously (57). The MBP-RBP26 protein was purified as above except that the cells were induced at an optical density (600 nm) of 0.1 and harvested at an optical density of 0.5. In vitro binding was carried out by using a modified version of a previously described method (62). First, GST-Rho3 was preloaded for 15 min at 37°C with the nonhydrolyzable guanine nucleotide guanosine 5′-O-(3-thiotriphosphate) (GTPγS) or guanosine 5′-O-(3-thiodiphosphate) (GDPβS) to exchange any bound nucleotides. The loading buffer contained 20 mM Tris-HCl (pH 7.4), 50 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, and 1 mM GTPγS. After the addition of 12.5 mM MgCl2, GST-Rho3 was incubated with either MBP, MBP-RBP4, MBP-Exo70, or MBP-RBP26 bound to amylose resin for 2 h at 4°C. The binding buffer contained 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM dithiothreitol, 1% Triton X-100, 10 mM MgCl2, and 2 mg of bovine serum albumin (BSA) per ml. The protein-bound resin was then collected, washed with binding buffer, and resuspended in Laemmli buffer. The samples were boiled, separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and then subjected to Western analysis performed as described elsewhere (49). The presence of GST-Rho3 was detected by using an anti-GST antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.). The same protocol was used to test binding with GST, GST-Kir, GST-Rad, GST–H-Ras, and GST-Rho4.
Immunofluorescence.
A yeast strain containing an HA-tagged version of EXO70 was constructed as follows. A 1.9-kb DNA fragment of the EXO70 coding region was amplified by PCR and cloned into pRS303. The sequence for three copies of tandemly repeated HA epitope was inserted in frame just in front of the stop codon of EXO70. The resulting plasmid was then integrated into the XhoI site of the coding region of the EXO70 allele in the wild-type yeast strain YPH499 (23) to produce HA-tagged Exo70 from the EXO70 promoter. These cells were then transformed with plasmids containing either GAL7p:RHO3E129 or GAL7p:RHO3E129,A131 and prepared for indirect immunofluorescence.
Cells were grown to mid-log phase in synthetic complete medium without uracil containing 2% galactose and 0.3% sucrose as carbon sources (SCGal-U) and stained with both rabbit anti-Rho3 antibody (23) and monoclonal mouse anti-HA antibody (Berkeley Antibody Company, Berkeley, Calif.) as previously described (45). Cells were fixed with 3.6% formaldehyde for 120 min, treated to form spheroplasts, and fixed onto polylysine-coated slides. The cells were then treated with 0.2% SDS in 100 mM potassium phosphate buffer (pH 7.5) containing 1 M sorbitol for 5 min, washed five times with G1T (1% gelatin, 0.5 mg of BSA per ml, 150 mM NaCl, 50 mM HEPES [pH 7.5], 0.1% Tween 20, 1 mM NaN3), submerged in methanol (−20°C) for 6 min and then in acetone (−20°C) for 30 s, and subsequently incubated in G3T (3% gelatin, 0.5 mg of BSA per ml, 150 mM NaCl, 50 mM HEPES [pH 7.5], 0.1% Tween 20, 1 mM NaN3) for 30 min. Cells were then incubated with anti-Rho3 antibody (1:1,000 dilution in G1T) at 25°C for 1 h followed by fluorescein isothiocyanate-labeled sheep anti-rabbit immunoglobulin G (1:500 dilution in G1T) for 1 h. Next, cells were incubated with G3T for 1 h, mouse anti-HA (1:400 dilution in G1T) for 1 h, and Cy3-labeled sheep anti-mouse immunoglobulin G (1:500 dilution in G1T; Chemicon International Inc., Temecula, Calif.) for 1 h. After each incubation, cells were washed five times with G1T buffer. The samples were then mounted in p-phenylenediamine (1 mg/ml in 90% glycerol) and observed with a model BH-2 epifluorescence microscope.
RESULTS
Rho3 interacts with Exo70 and Myo2.
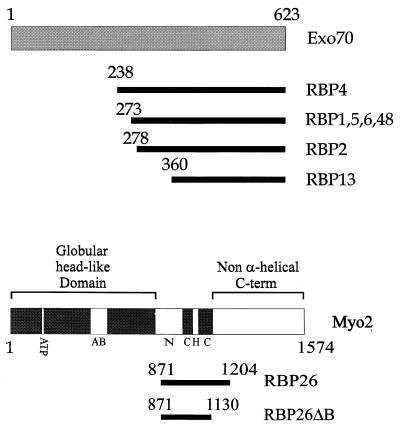
To identify proteins interacting with Rho3, we performed a yeast two-hybrid screen (8). An S. cerevisiae cDNA library was screened by using an activated mutant of RHO3 fused with the DNA binding domain of GAL4 as bait. Using an activated mutant of RHO3 is important, since we found that only the activated Rho3 protein interacts with its downstream effectors in the two-hybrid system. Similar results were obtained with RHO1, which also requires an activated form for detectable levels of effector binding (30, 43). The mutations rho3E129 (the original clone of RHO3 that was isolated carries the 129th codon as Glu [36], but RHO3 in the standard wild-type strain, W303 [38], carries the 129th codon as Lys) and rho3V25 were used as activating mutations in this study. The 129th residue of Rho3 is located in the G-4 region and the 25th residue is located in the G-1 region, both of which are necessary for GTP and GDP binding of small G proteins (5). Such mutations in Ras superfamily G proteins result in constitutive activation (10, 50). Finally, the CAAX box at the C terminus of RHO3 was deleted to prevent possible interference with the nuclear localization of the fusion protein. A total of 2 × 106 colonies were screened with His+ selection. Of the 190 candidate clones obtained from this screen, 24 have been scored as β-galactosidase positive by filter lift assay (44) upon isolation and retransformation. Eight clones contained fragments of the EXO2 and Myo2 genes and were further characterized. The interactions between activated Rho3 and two of these clones, RBP4 and RBP26, are shown in Fig. 1 and 2 and Table 1. Fragments of Exo70 were identified seven times in our screen. Exo70 is a component of a multiprotein complex called the exocyst which functions at the late stage of protein secretion from the Golgi complex to the plasma membrane (53). Another clone, RBP26, encodes a fragment of the Myo2 protein, an unconventional myosin believed to be involved in the regulation of the actin cytoskeleton and translocation of secretory vesicles along the actin cytoskeleton to the bud (26). Figure 1 shows the regions of Exo70 and Myo2 which interact with Rho3. The fragments of Exo70 that we identified map to the C-terminal half of the protein, the smallest of which contains amino acids 360 through 623. This finding indicates that the C-terminal region of Exo70 is sufficient for Rho3 binding. Full-length Exo70 also interacts with Rho3 (Table 1). In the case of Myo2, the RBP26 fragment encompasses a region including the coiled coil and a part of the non-α-helical C-terminal region spanning amino acids 871 to 1204. In addition, the smaller construct RBP26ΔB, which contains the region between amino acids 871 and 1130, also interacts with activated Rho3. This region is outside the regions of actin and ATP binding which are located in the N-terminal half of the protein (26). The interacting region consists of the C-terminal part of the neck region, which contains two of the six IQ motifs found in the protein, as well as the hinge and N-terminal region of the non-α-helical C terminus. Neither RBP4, full-length Exo70, nor RBP26 interacts with wild-type Rho3 in the two-hybrid system, presumably because the GTP-bound form of Rho3 is necessary for sustained interaction (data not shown). Similar observations have been made for other small G proteins, including Rho1 and RhoA, which need to be activated in order to produce measurable two-hybrid signals with Bni1 or Pkc1 or with myosin phosphatase, respectively (29, 30, 43). RBP4 interaction was also detected with an activated version of Rho3 containing a CAAX box. On the other hand, RBP26 interaction was seen only with the CAAX-less construct (data not shown).
FIG. 1.
Fragments of Exo70 and Myo2 which interact with Rho3. Regions of Exo70 and Myo2 contained in the two-hybrid clones identified to interact with Rho3 are shown as bars below the intact molecules. ATP, ATP binding region; AB, actin binding region; N, neck region; C, coiled-coil region; H, hinge region.
FIG. 2.
The yeast two-hybrid interaction between both Rho3 and Rho1 with their downstream effectors. Rho3 and Rho1 were fused to the DNA binding domain of GAL4. Their interactions with Pkc1, RBP4, RBP26, and Bni1, all of which are fused to the activation domain of GAL4, are demonstrated by the plate β-galactosidase assay.
TABLE 1.
Two-hybrid interaction of Rho3 with Exo70 and Myo2a
| Construct | β-Galactosidase activity (Miller units; mean ± SD)
|
||
|---|---|---|---|
| Exo70 | RBP4 | RBP26 | |
| Rho3E129 | 3.28 ± 0.10 | 48.09 ± 12.7 | 3.48 ± 0.83 |
| Rho3E129/A48 | 0.20 ± 0.02 | 0.17 ± 0.04 | 0.11 ± 0.02 |
| Rho3E129/S47 | 0.26 ± 0.02 | 6.79 ± 1.13 | 0.35 ± 0.18 |
| Rho3E129/N30 | 0.20 ± 0.03 | 0.23 ± 0.01 | 0.19 ± 0.03 |
The liquid β-galactosidase assay was carried out as described by Poullet and Tamanoi (44). RHO3 was fused to the DNA binding domain vector pGBT9. EXO70, RBP4, and RBP26 were fused to the activation domain vector pGAD-GH or pGAD424. The interaction between pGBT9 and pGAD-GH (0.19 ± 0.04 U) was used as a negative control, and the interaction between pGBT9-NF1 and pGAD-HrasV12 (activated Hras) (4.35 ± 2.11) was used as a positive control as previously described (44). These experiments were repeated three times with similar results.
Effector domain and dominant negative mutants of Rho3.
Mutants of rho3 can be used to examine whether Exo70 and Myo2 are downstream effectors of Rho3. A region called the effector domain which encompasses residues 43 to 53 of Rho3 is known to be critical for interaction with downstream effector molecules (23). Mutations in this domain result in the loss of function of Rho3, Rho1, or RhoA (3, 23, 30). We introduced two effector loop mutations into rho3E129 and examined their effects on the interaction with Exo70 as well as with Myo2 in the two-hybrid system. To provide a quantitative comparison of the interaction, a two-hybrid liquid assay which measures β-galactosidase activity (41, 44) was carried out. As shown in Table 1, no interaction was seen between the Rho3E129,A48 mutant and full-length Exo70, the Exo70 fragment (RBP4), or the Myo2 fragment (RBP26). Similarly, the Rho3E129,S47 mutant did not interact with full-length Exo70. We did detect an interaction between the Rho3E129,S47 mutant and the Exo70 fragment, but the interaction was significantly less than that of Rho3E129. A weak interaction was also detected with Rho3E129,S47 and the Myo2 fragment (RBP26). Interactions between Rho3E129,S47 and full-length Exo70 as well as the Myo2 fragment (RBP26) were detected by using the Lex A two-hybrid system (19), which is more sensitive than the Gal4 version (data not shown). We also examined a form of Rho3, Rho3N30, which is analogous to dominant negative forms of other Rho proteins. Analogous mutants of Rho1, Rho4, and RhoA do not interact with downstream effectors (24, 30, 59). Furthermore, the dominant negative form of RhoA was shown to remain preferentially in the GDP-bound form (63). As shown in Table 1, Rho3E129,N30 does not interact with full-length Exo70, the Exo70 fragment, or the Myo2 fragment. This mutant was also tested in the LexA two-hybrid system, and no interaction was observed (data not shown). Western analyses of the Rho3 proteins suggest that the expression levels of the Rho3 mutants are comparable to that of the wild type in these experiments (data not shown). In addition, all aforementioned two-hybrid constructs were tested for self-activation and gave negative results (data not shown). These results suggest that Exo70 and Myo2 are downstream effectors of Rho3.
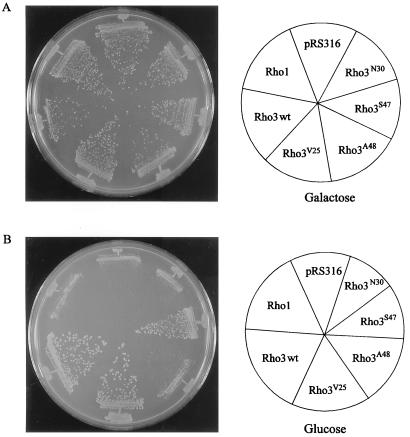
The ability of various Rho3 mutants to interact with downstream effectors correlates well with their ability to complement the loss of Rho3 function in vivo (Fig. 3). A rho3 disruption strain containing a copy of RHO4 under the control of the GAL7 promoter was used, and all mutants were expressed on a centromeric vector under the control of the RHO1 promoter. On galactose-containing medium where overproduction of Rho4 complements the growth defect exhibited by the loss of Rho3, there is no detectable difference in colony size in transformants expressing either vector (pRS316) alone, wild-type Rho3, Rho3V25, Rho3A48, Rho3S47, Rho3N30, or Rho1 (Fig. 3A). However, when Rho4 overproduction is shut down by a shift to dextrose-containing medium, the differences become apparent. Both wild-type Rho3 and Rho3V25 can complement the growth defect, while neither Rho3A48 nor Rho3N30 shows any difference in colony size from Rho1-expressing cells or those transformed with vector alone (Fig. 3B). Cells expressing Rho3S47 show almost wild-type levels of complementation, with a barely discernible decrease in colony size compared with wild-type Rho3. Similar results were observed with these mutations in the E129 background (data not shown). Western analysis showed that all Rho3 proteins were expressed at similar levels (data not shown). These observations suggest that the differences in binding ability among the different mutants correlates with their ability to properly function in vivo.
FIG. 3.
Complementation of a Δrho3 yeast with various RHO3 mutants. Yeast strain YMR504 was transformed with RHO3, rho3V25, rho3A48, rho3S47, and rho3N30, as well as RHO1, all of which were inserted into the centromeric vector pRS316. Transformants were grown on medium containing either galactose (A) or glucose (B) and assessed for the ability to complement the growth defect exhibited by Δrho3 cells. wt, wild type.
Exo70 and Myo2 interact specifically with Rho3, while Bni1 interacts with both Rho3 and Rho1.
While Rho3 plays critical roles in directing exocytosis to the bud, another Rho protein, Rho1, is important for bud site maintenance as well as for cell wall synthesis (11, 46). Two downstream effectors, Pkc1 and Bni1, are known to interact with activated Rho1 in the two-hybrid assay (30, 43). Bni1 contains formin homology domains and interacts with profilin, an actin binding protein, while Pkc1 participates in a protein kinase cascade regulating cell wall integrity (27, 43). To investigate the specificity of Rho3 for Myo2 and Exo70, Rho1 effectors were tested with Rho3 and vice versa. The activating mutation Rho3V25 was used, as was the corresponding mutation in Rho1, Rho1V19. As can be seen from Fig. 2, Rho3 does not interact with Pkc1 whereas Rho1 shows strong interaction with Pkc1. In addition, Rho1 does not bind to either Myo2 or Exo70. Thus, Rho3 interacts with a separate and distinct set of proteins compared with Rho1. Rho3 does interact with Bni1, but the interaction is weaker than that seen between Rho1 and Bni1. We also examined the interaction of Bni1 with the various Rho3 mutants described above and obtained results similar to those for Exo70: Bni1 interacted with Rho3E129 and Rho3E129,S47 but not with Rho3E129,A48 or Rho3E129,N30 (data not shown). The yeast Rho family members include Cdc42 (1, 25), Rho2 (33), and Rho4 (37), which we also tested for binding with Myo2 and Exo70 in the two-hybrid system. No interaction was observed for Cdc42 or Rho2 with either Myo2 or Exo70. Rho4, on the other hand, interacted with Exo70 but not with Myo2 (data not shown). The interaction between Rho4 and Exo70 was also confirmed by in vitro analysis (see below).
Rho3 interacts with Myo2 and Exo70 in vitro.
The interactions between Rho3 and Myo2 and Exo70 were confirmed by in vitro binding experiments (Fig. 4). Rho3 was purified as a fusion protein with GST. RBP26 (fragment of Myo2) and RBP4 (fragment of Exo70) were purified as fusion proteins with MBP. GST-Rho3 was complexed with a nonhydrolyzable guanine nucleotide analogue, GTPγS, and then mixed with MBP-RBP26 or MBP-RBP4 bound to amylose resin. After incubation, the resin was collected and washed. GST-Rho3 bound to the resin was recovered by boiling with Laemmli buffer and detected by SDS-PAGE and Western blotting using an anti-GST antibody. As shown in Fig. 4D, GST-Rho3 complexed with GTPγS binds to both MBP-RBP26 and MBP-RBP4. No GST-Rho3 binding is observed with MBP alone. Furthermore, no binding of GST with MBP-RBP26 or MBP-RBP4 is observed (data not shown).
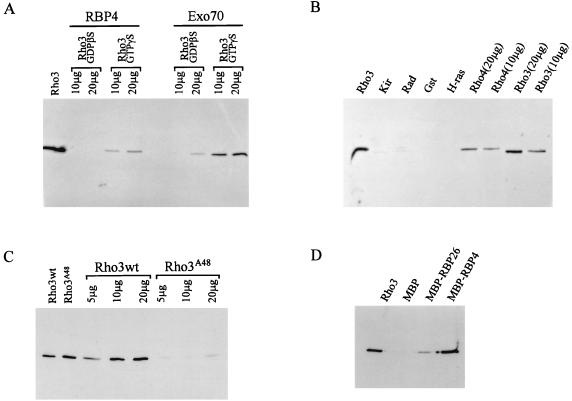
FIG. 4.
In vitro interaction of Rho3 with RBP26, Exo70, and RBP4. (A) GST-Rho3 preincubated with a nonhydrolyzable analogue of GDP or GTP was mixed with either MBP-RBP4 or MBP-Exo70 bound to amylose resin. After rinsing, the resin was collected, loading buffer was added, and the samples were boiled. The proteins were then separated on an SDS-polyacrylamide gel, and the presence of GST-Rho3 was examined by using an anti-GST antibody. The Rho3 lane contains purified GST-Rho3 and is intended to show the position of GST-Rho3. (B) GST-Kir (20 μg), GST-Rad (20 μg), GST (20 μg), GST–H-Ras (20 μg), GST-Rho4 (10 and 20 μg), and GST-Rho3 (10 and 20 μg) were loaded with GTPγS and assessed for the ability to bind MBP-Exo70 as described above. The Rho3 lane contains purified GST-Rho3 as in panel A. (C) GST-Rho3 and GST-Rho3A48 were loaded with GTPγS and assessed for the ability to bind MBP-RBP4 as described above. The Rho3wt (wild-type Rho3) and Rho3A48 lanes contain purified Rho3 proteins. (D) GST-Rho3 was loaded with GTPγS and assessed for its ability to bind to either MBP, MBP-RBP26, or MBP-RBP4 as described above. The Rho3 lane contains purified GST-Rho3 as in panel A.
Because RBP4 shows stronger interaction with Rho3, we decided to further characterize this interaction. As shown in Fig. 4A, MBP-RBP4 binds specifically to GST-Rho3 complexed with GTPγS but does not bind Rho3 complexed with GDPβS. Furthermore, the full-length MBP-Exo70 protein also prefers the GTPγS-bound form of GST-Rho3, although low levels of binding with GDPβS are still observed. To test the specificity of Exo70 binding, other small G proteins were also evaluated (Fig. 4B). GST, GST-Kir (9, 34), GST-Rad (47), GST–H-Ras (5), GST-Rho3, and GST-Rho4 (37) were loaded with GTPγS and tested for binding with MBP-Exo70 as described above. Neither GST, GST-Kir, GST-Rad, nor GST-H-Ras bound MBP-Exo70. GST-Rho4, on the other hand, did bind MBP-Exo70, albeit less efficiently than Rho3.
To further confirm the specificity of the binding, the effector loop mutant GST-Rho3A48 was tested for interaction with MBP-RBP4. GST-Rho3 and GST-Rho3A48 were loaded with GTPγS and tested for binding with MBP-RBP4 as described above. As shown in Fig. 4C, the wild-type GST-Rho3 protein interacts with MBP-RBP4 whereas minimal interaction is observed with the effector loop mutant GST-Rho3A48.
These results establish that Rho3 interacts with Exo70 directly in a GTP-dependent manner. Furthermore, the lack of binding observed with other small G proteins as well as Rho3A48 enforces the specificity of this interaction. The binding observed with Rho4 correlates well with the two-hybrid data (see above) and the complementation data showing that overexpression of Rho4 suppresses the growth defect exhibited by Rho3-deficient cells (36).
Rho3 localization overlaps with the localization of Exo70.
The above results suggest that Exo70 is a downstream effector of Rho3; thus, Rho3 may be responsible for directing the localization of Exo70. Subcellular localization studies were carried out to examine this point. As shown in Fig. 5A, when Rho3E129 and HA-Exo70 were expressed, they exhibited similar patterns of localization. The staining pattern of Rho3E129 as detected by indirect immunofluorescence showed a patch-like appearance throughout the cell, with fluorescence in the bud. When the patterns of localization of Rho3E129 and Exo70 were compared, Exo70 staining also exhibited a patch-like appearance, with strong fluorescence detected in the bud. Thus, the two proteins overlap in cellular localization. We also examined the localization of both HA-Exo70 and endogenous wild-type Rho3. In these cells, Exo70 fluorescence is concentrated more at the bud tip, while Rho3 is dispersed uniformly throughout the cell surface and the bud (data not shown). Thus, localization of wild-type Rho3 overlaps with that of Exo70, but the two patterns also show differences. Activated Rho3 shows more pronounced colocalization with Exo70, presumably because Rho3 interacts with Exo70 only when it is in the GTP-bound form.
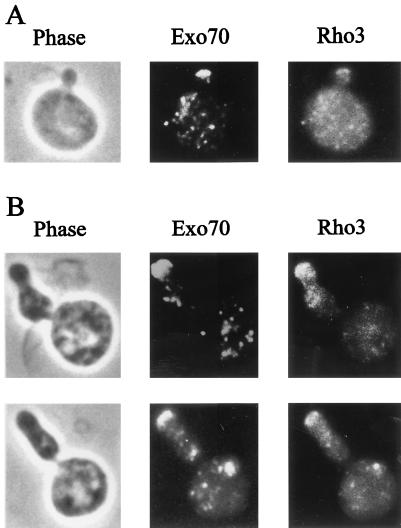
FIG. 5.
Localization of Exo70 and Rho3. (A) HA-Exo70-producing cells carrying GAL7p:RHO3E129 were cultured in SCGal-U at 30°C. (B) HA-Exo70-producing cells carrying GAL7p:RHO3E129,A131 growing exponentially in SCGal-U at 30°C were shifted to 15°C and harvested 48 h after the shift. Cells were fixed and stained with anti-HA (middle) and anti-Rho3 (right) antibodies. Phase photographs are also shown (left).
To further investigate the overlapping localization of Rho3 and Exo70, we took advantage of a Rho3 double mutant, Rho3E129,A131. Expression of this mutant results in the generation of cold-sensitive cells with an aberrant morphology; they are elongated and bent (23) (Fig. 5B). Also, these cells exhibit a staining pattern in which accumulation of Rho3 is observed at several points on the cell surface (Fig. 5B), in contrast to the pattern observed for Rho3E129, which exhibits a relatively uniform distribution of staining (Fig. 5A). Comparison of the localization of Rho3 and Exo70 in yeast cells expressing the mutant Rho3E129,A131 shows that Rho3E129,A131 accumulates at several sites as well as at the bud tip and that this distribution pattern is similar to that observed with Exo70. These results further confirm that the localization of activated Rho3 overlaps with that of Exo70.
DISCUSSION
In this report, we have presented evidence that Exo70 and Myo2 are downstream effectors of the Rho3 GTPase, which plays a critical role in the regulation of bud growth. Together with the effector Bni1, it appears that this GTPase has at least three different effector molecules which have all been implicated in aspects of actin cytoskeletal organization and exocytosis, as shown in Fig. 6.
FIG. 6.
Schematic showing the various effectors of Rho3 and the cellular processes that they regulate. See Discussion for an explanation of each interaction.
Rho3 directly interacts with Exo70, a component of the exocyst (a complex of Exo70, Sec3, Sec5, Sec6, Sec8, Sec10, and Sec15), which appears to reside in the acceptor membrane and is thought to facilitate docking of secretory vesicles, since a high concentration of Sec6, Sec8, and Sec15 was found at the tips of small buds, which represent active sites of exocytosis (53, 54). We have shown that the interaction of Rho3 with Exo70 requires the intact effector domain of Rho3 and is dependent on the GTP-bound form of Rho3 and that the patterns of localization of the two proteins overlap. It is possible that the interaction between Rho3 and Exo70 serves to properly localize Exo70. This hypothesis is supported by our demonstration that the localization patterns of these proteins overlap and that the localization of Exo70 is altered by the expression of an activated allele of RHO3, rho3E129,A131. This activated allele of RHO3 causes cells to become cold sensitive and elongated as well as bent at the sites where actin patches are localized (23). Exo70 appears to accumulate at various regions, including the bud tips in Rho3E129,A131-expressing cells. This localization pattern is also observed for Rho3E129,A131. Thus, activated Rho3E129,A131 appears to affect the localization of Exo70, which may result in misdirection of the exocytosis machinery. Rho3 may exert additional effects on exocyst function by influencing the process of vesicle fusion; however, additional work is needed to clarify these points.
Rho3 also interacts with Myo2, an essential myosin in yeast that appears to play critical roles in the organization of the actin cytoskeleton. During polarized growth of yeast cells, actin patches, which represent the cytoskeleton-plasma membrane interface (42), accumulate in the bud where cell growth is taking place; in addition, actin cables orient toward the bud (28, 32). These specialized structures do not form in the absence of Myo2 function, as the temperature-sensitive myo2-66 mutant displays delocalized actin patches at the restrictive temperature (26). In addition, the mutants arrest as large unbudded cells. These phenotypes are similar to the rho3-1 mutant in which the cells become enlarged and rounded at the restrictive temperature, and actin patches lie scattered throughout the cell surface (23). Furthermore, both the myo2-66 and rho3 rho4 mutants display delocalized deposition of chitin as well as a multinucleate phenotype (37). This overlap of myo2-66 and rho3-1 phenotypes is consistent with our assignment of Myo2 as an effector of Rho3 and raises the possibility that Rho3 affects the activity of Myo2. Finally, Rho3 may also influence changes in the actin cytoskeleton through its interaction with the Bni1 protein. The Bni1 protein contains two formin homology domains which are responsible for the binding of profilin, an actin binding protein (24).
In addition to its role in actin cytoskeletal organization, Myo2 appears to be critical for the transport of a class of secretory vesicles to the bud, as secretory vesicles accumulate predominantly in the mother cell in the myo2-66 mutant (18, 26). Myo2 belongs to the class V unconventional myosin family which includes mouse dilute (39) and chicken brain myosin V (14), proteins also implicated in the transport of membrane-bound organelles (55, 56). Genetic interactions between MYO2 and seven late-acting sec genes including four components of the exocyst have been detected (18). Furthermore, both rho3-1 and myo2-66 cells display synthetic lethality with sec4 mutants (18, 23). While the precise mechanism remains unclear, these observations raise the possibility that Myo2 and the exocyst work in concert to ensure polarized cell growth by properly depositing newly synthesized proteins at the bud site. Rho3 may be the G protein which regulates this process. It is interesting that the region of Myo2 where Rho3 interacts is located in the coiled-coil and non-α-helical C-terminal domain, a region that is thought to specify cargo loading in the class V myosins (40).
How might Myo2, Rho3, and Exo70 work in concert to bring about proper bud growth? As discussed above, Myo2 may be responsible for transporting vesicles to the bud. Once in the bud, the vesicles then interact with proteins necessary to promote vesicle fusion, a process requiring both Sec4 and likely the exocyst (15, 53). Rho3 could function in this process early on by assisting Myo2 in transport of vesicles to the bud. This hypothesis is supported by the fact that Rho3-depleted cells undergo lysis with small buds, which could be indicative of the cargo never reaching its destination. One possibility is that Rho3 is actually inserted into the vesicular membrane and functions as the anchor to which both the vesicle and Myo2 bind. Upon arriving at the bud tip, Rho3 could then interact with Exo70 to facilitate the fusion event. Another possibility is that Rho3 located at the bud tip membrane can interact with Exo70. Upon arrival of the vesicle, Rho3, through its interaction with Myo2, could then function to facilitate vesicle fusion. The validity of these ideas awaits the results of genetic analyses which will give further insight into the exact role that these proteins play in the process of polarized cell growth.
ACKNOWLEDGMENTS
We thank Greg Payne for critical reading of the manuscript and for valuable suggestions. We also thank T. Ito for his photographic contributions. We thank Kazuma Tanaka for constructs containing Rho proteins and their effectors. We also thank Trisha Davis for advice on Myo2 protein purification.
This work was supported by NIH grant CA41996. N.G.G.R. was supported in part by Institutional National Research Service Award GM08375 from USHHS and by a Warsaw Fellowship. This work was also supported in part by a grant for scientific work from Monbusho. J.I. is a recipient of the Fellowship of the Japan Society for the Promotion of Science for Japanese Junior Scientists.
REFERENCES
- 1.Adams A E, Johnson D I, Longnecker R M, Sloat B F, Pringle J R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Amano M, Mukai H, Ono Y, Chihara K, Matsui T, Hamajima Y, Okawa K, Iwamatsu A, Kaibuchi K. Identification of a putative target for Rho as the serine-threonine kinase protein kinase N. Science. 1996;271:648–650. doi: 10.1126/science.271.5249.648. [DOI] [PubMed] [Google Scholar]
- 4.Bartel P L. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interactions in development: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 5.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 6.Chant J. Generation of cell polarity in yeast. Curr Opin Cell Biol. 1996;8:557–565. doi: 10.1016/s0955-0674(96)80035-4. [DOI] [PubMed] [Google Scholar]
- 7.Chant J, Pringle J R. Budding and cell polarity in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1991;1:342–350. doi: 10.1016/s0959-437x(05)80298-9. [DOI] [PubMed] [Google Scholar]
- 8.Chien C-T, Bartel P L, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen L, Mohr R, Chen Y-Y, Huang M, Kato R, Dorin D, Tamanoi F, Goga A, Afar D, Rosenberg N, Witte O. Transcriptional activation of a ras-like gene (kir) by oncogenic tyrosine kinases. Proc Natl Acad Sci USA. 1994;91:12448–12452. doi: 10.1073/pnas.91.26.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Der C J, Pan B-T, Cooper G M. rasH mutants deficient in GTP binding. Mol Cell Biol. 1986;6:3291–3294. doi: 10.1128/mcb.6.9.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drgonova J, Drgon T, Tanaka K, Kollar R, Chen G C, Ford R A, Chan C S M, Takai Y, Cabib E. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- 12.Drubin D G. Development of cell polarity in budding yeast. Cell. 1991;65:1093–1096. doi: 10.1016/0092-8674(91)90001-f. [DOI] [PubMed] [Google Scholar]
- 13.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 14.Espindola F S, Espreafico E M, Coelho M V, Martins A R, Costa F R, Mooseker M S, Larson R E. Biochemical and immunological characterization of p190-calmodulin complex from vertebrate brain: a novel calmodulin-binding myosin. J Cell Biol. 1992;118:359–368. doi: 10.1083/jcb.118.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- 16.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goud B, Salminen A, Walworth N C, Novick P J. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- 18.Govindan B, Bowser R, Novick P. The role of Myo2, a yeast class V myosin, in vesicular transport. J Cell Biol. 1995;128:1055–1068. doi: 10.1083/jcb.128.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 20.Herskowitz I. Building organs and organisms: elements of morphogenesis exhibited by budding yeast. Cold Spring Harbor Symp Quant Biol. 1997;62:57–63. [PubMed] [Google Scholar]
- 21.Herskowitz I, Park H O, Sanders S, Valtz N, Peter M. Programming of cell polarity in budding yeast by endogenous and exogenous signals. Cold Spring Harbor Symp Quant Biol. 1995;60:717–727. doi: 10.1101/sqb.1995.060.01.078. [DOI] [PubMed] [Google Scholar]
- 22.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Imai J, Toh-e A, Matsui Y. Genetic analysis of the Saccharomyces cerevisiae RHO3 gene, encoding a rho-type small GTPase, provides evidence for a role in bud formation. Genetics. 1996;142:359–369. doi: 10.1093/genetics/142.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson D I, Pringle J R. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston G C, Prendergast J A, Singer R A. The Saccharomyces cerevisiae MYO2 gene encodes an essential myosin for vectorial transport of vesicles. J Cell Biol. 1991;113:539–551. doi: 10.1083/jcb.113.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 28.Kilmartin J V, Adams A E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 30.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Elledge S J, Peterson C A, Bales E S, Legerski R J. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R, Zheng Y, Drubin D G. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madaule P, Axel R, Myers A M. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1987;84:779–783. doi: 10.1073/pnas.84.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maguire J, Santoro T, Jensen P, Siebenlist U, Yewdell J, Kelly K. Gem: an induced, immediate early protein belonging to the Ras family. Science. 1994;265:241–244. doi: 10.1126/science.7912851. [DOI] [PubMed] [Google Scholar]
- 35.Mata J, Nurse P. Discovering the poles in yeast. Trends Cell Biol. 1998;8:163–167. doi: 10.1016/s0962-8924(98)01224-0. [DOI] [PubMed] [Google Scholar]
- 36.Matsui Y, Toh-e A. Isolation and characterization of two novel ras superfamily genes in Saccharomyces cerevisiae. Gene. 1992;114:43–49. doi: 10.1016/0378-1119(92)90705-t. [DOI] [PubMed] [Google Scholar]
- 37.Matsui Y, Toh-e A. Yeast RHO3 and RHO4 ras superfamily genes are necessary for bud growth, and their defect is suppressed by a high dose of bud formation genes CDC42 and BEM1. Mol Cell Biol. 1992;12:5690–5699. doi: 10.1128/mcb.12.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsui Y, Matsui R, Akada R, Toh-e A. Yeast src homology region 3 domain-binding proteins involved in bud formation. J Cell Biol. 1996;133:865–878. doi: 10.1083/jcb.133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercer J A, Seperack P K, Strobel M C, Copeland N G, Jenkins N A. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 40.Mermall V, Post P L, Mooseker M S. Unconventional myosins in cell movement, membrane traffic and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- 41.Morcos P, Thapar N, Tusneem N, Stacey D, Tamanoi F. Identification of neurofibromin mutants that exhibit allele specificity or increased Ras affinity resulting in suppression of activated ras alleles. Mol Cell Biol. 1996;16:2496–2503. doi: 10.1128/mcb.16.5.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poullet P, Tamanoi F. Use of yeast two-hybrid system to evaluate Ras interactions with neurofibromin-GTPase-activating protein. Methods Enzymol. 1995;255:488–497. doi: 10.1016/s0076-6879(95)55051-8. [DOI] [PubMed] [Google Scholar]
- 45.Pringle J R, Preston R A, Adams A E, Stearns T, Drubin D G, Haarer B K, Jones E W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 46.Qadota H, Python C P, Inoue S, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin D E, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 47.Reynet C, Kahn C R. Rad: a member of the Ras family overexpressed in muscle type II diabetic humans. Science. 1993;262:1441–1444. doi: 10.1126/science.8248782. [DOI] [PubMed] [Google Scholar]
- 48.Salminen A, Novick P J. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell. 1987;49:527–538. doi: 10.1016/0092-8674(87)90455-7. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Seeburg P H, Colby W W, Capon D J, Goeddel D V, Levinson A D. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 51.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 52.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 53.TerBush D R, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- 54.TerBush D R, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Titus M A. From fat yeast and nervous mice to brain myosin-V. Cell. 1993;75:9–11. [PubMed] [Google Scholar]
- 56.Titus M A. Motor proteins: myosin V—the multi-purpose transport motor. Curr Biol. 1997;7:R301–R304. doi: 10.1016/s0960-9822(06)00143-6. [DOI] [PubMed] [Google Scholar]
- 57.Urano, J., and F. Tamanoi. Reconstitution of yeast farnesyltransferase from individually purified subunits. In M. Gelb (ed.), Protein lipidation protocols, in press. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 58.Van Aelst L, Barr M, Marcus S, Polverino A, Wigler M. Complex formation between RAS and RAF and other protein kinases. Proc Natl Acad Sci USA. 1993;90:6213–6217. doi: 10.1073/pnas.90.13.6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S. Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science. 1996;271:645–648. doi: 10.1126/science.271.5249.645. [DOI] [PubMed] [Google Scholar]
- 60.Xu G F, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, White R, Tamanoi F. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63:835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 61.Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X F, Marshall M S, Avruch J. Ras-Raf complexes in vitro. Methods Enzymol. 1995;255:323–331. doi: 10.1016/s0076-6879(95)55035-6. [DOI] [PubMed] [Google Scholar]
- 63.Zheng Y, Olson M F, Hall A, Cerione R A, Toksoz D. Direct involvement of the small GTP-binding protein Rho in lbc oncogene function. J Biol Chem. 1995;270:9031–9034. doi: 10.1074/jbc.270.16.9031. [DOI] [PubMed] [Google Scholar]