Abstract
The oral microbiome is a community of microorganisms, comprised of bacteria, fungi, viruses, archaea, and protozoa, that form a complex ecosystem within the oral cavity. Although minor perturbations in the environment are frequent and compensable, major shifts in the oral microbiome can promote an unbalanced state, known as dysbiosis. Dysbiosis can promote oral diseases, including periodontitis. In addition, oral dysbiosis has been associated with other systemic diseases, including cancer. The objective of this review is to evaluate the epidemiologic evidence linking periodontitis to oral, gastrointestinal, lung, breast, prostate, and uterine cancers, as well as describe new evidence and insights into the role of oral dysbiosis in the etiology and pathogenesis of the cancer types discussed. Finally, we discuss how antimicrobials, antimicrobial peptides, and probiotics may be promising tools to prevent and treat these cancers, targeting both the microbes and associated carcinogenesis processes. These findings represent a novel paradigm in the pathogenesis and treatment of cancer focused on the oral microbiome and antimicrobial‐based therapies.
Keywords: breast cancer, dysbiosis, gastrointestinal cancer, head and neck cancer, oral microbiome, probiotics, prostate cancer, uterine cancer
1. INTRODUCTION
1.1. Oral microbiome and its interactions with the host (oralome)
The oral cavity contains up to 1000 microbial species in total, comprised of bacteria, fungi, viral, archaea, and protozoan species that thrive in a very dynamic microenviroment.1, 2, 3, 4, 5 All of these microorganisms form a complex relationship among themselves, establishing a unique microbiome, known as the oral microbiome. Interestingly, the oral microbiome forms a close symbiotic relationship with human host cells in the oral cavity. Thus, the term oralome was coined to encompass not only the oral microbiome but also the host‐microbial interactions that take place in the human oral cavity.5 In this sense, healthy symbiotic host‐microbiome interactions between humans and these microorganisms are known as eubiosis.5, 6, 7
The microbial composition can be dramatically affected by interspecies and host‐microbial interactions. These microbial changes can impact the health and disease status of the host, since eubiosis plays an essential role both in the development of natural oral physiology and host defense mechanisms.5, 8, 9 Although the oral microbiome can compensate for most overall perturbations,5, 10 some changes can profoundly affect its composition, impacting the oral commensal populations and causing an unbalanced state known as dysbiosis.5, 11
1.2. Periodontitis and oral microbiome dysbiosis
Dysbiosis is an unbalanced microbiome state that is caused by internal and/or external microbial‐ecologic changes to the oral microbiome.5 This specific state has been described as capable of promoting diseases in the host.12, 13 Since periodontitis is considered an inflammatory disease that is initiated by pathogenic bacteria, the most accepted hypothesis for periodontitis initiation and progression is that there is a dysbiotic shift in the oral microbiome.5, 14 This shift is driven by an enrichment of Prevotella intermedia, Fusobacterium nucleatum, Porphyromonas gingivalis, Tannarella forsythia, and Treponema denticola species in the microbiome.14, 15, 16 Specifically, a dysbiotic oral biofilm infiltrates the gingival pocket, which then triggers the host immune response. This reaction leads to gingival tissue inflammation (gingivitis) and, ultimately, tissue degradation and periodontitis.14
Oral dysbiosis has been associated with a variety of systemic diseases and conditions, including Alzheimer's disease, diabetes, adverse pregnancy complications, and several types of cancer, including oral, gastrointestinal, lung, breast, prostate, and uterine cancer.5, 17, 18, 19, 20 Thus, the objective of this research is to (1) evaluate the epidemiologic evidence linking periodontitis to these types of cancer, (2) provide insights into the mechanisms by which oral microbial dysbiosis can cause these cancers, and (3) summarize the evolving evidence supporting the use of probiotics and related molecules (bacteriocins) for prevention and treatment of cancer. For more details on oral host‐microbial interactions and the role of oral dysbiosis on different systemic diseases, please refer to Radaic and Kapila.5
2. EVIDENCE LINKING PERIODONTITIS AND SEVERAL TYPES OF CANCER
Cancer, in a broad sense, refers to more than 277 different types of cancer diseases, each one caused by a series of genetic mutations that lead to abnormal cell proliferation and invasion.21, 22 In the United States, cancer is the second leading cause of death. It is estimated that there will be 1.8 million new cases of cancer and more than 600 000 deaths in the United States in 2020.23
Although genetic mutations are considered the main etiology agents of cancer overall, periodontitis has been recently associated with head and neck, gastrointestinal, lung, breast, prostate, and uterine cancers.18, 33 In the following sections, we will further discuss the association between periodontitis and these cancers.
2.1. Head and neck cancer
Head and neck cancer is a devastating disease, often disfiguring and debilitating affected patients. It is the sixth most common cancer worldwide, and it comprises cancers of the oral cavity, larynx, hypopharynx, and oropharynx.34, 35 In 2018, head and neck cancer accounted for approximately 706 000 new cases and 358 000 deaths worldwide.34 In the United States, head and neck cancer accounts for 3% of all cancers and approximately 65 000 Americans are diagnosed with head and neck cancer annually.23, 34, 36 Table 1 shows the estimated US incidence and the estimated new cases and deaths in the United States (for 2020)23 and worldwide (for 2018) for each head and neck cancer subtype.34 In this chapter, we will focus on oral cancer in particular.
TABLE 1.
Head and neck cancer estimated incidence and estimated new cases and deaths in the United States and worldwide
| Head and neck cancer classification | Estimated annual incidence in United States (per 100 000) | Estimated new cases | Estimated deaths | ||
|---|---|---|---|---|---|
| 2020 United States23 | 2018 worldwide34 | 2020 United States23 | 2018 worldwide34 | ||
| Oral cavity | 11.7 37 | 35 310 | 354 864 | 7110 | 177 098 |
| Larynx | 3.3 37 | 12 370 | 177 422 | 3750 | 94 771 |
| Hypopharynx | <1.0 38 | 17 950 | 80 608 | 3640 | 34 984 |
| Oropharynx | — | 92 887 | 51 005 | ||
Oral cancers have a complex etiology that includes lifestyle factors such as alcohol and tobacco usage, which are strongly associated with most head and neck cancers progression and aggressiveness.35, 39 It was recently demonstrated that alcohol consumption plus smoking have a synergetic effect in increasing head and neck cancer risk, particularly for oral and pharyngeal cancer.40 It has been demonstrated that that alcohol and tobacco‐induced head and neck cancer exhibit mutations in tumor suppressor protein p53 and inactivation of the tumor suppressor p16 gene via deletion of 9p21‐22.41, 42, 43
Besides tobacco and alcohol consumption, two recent cohort studies showed that poor oral hygiene decreased the survival rates of patients with oral cancers,44 whereas good oral health behaviors, such as daily tooth brushing and an annual dental visit, reduced the risk of head and neck cancer.45 These studies suggest that oral microbial dysbiosis may be an important contributor to oral cancer pathogenesis.
Recent studies have shed light on the possibility of periodontal disease–associated pathogenic bacteria having an important role in oral cancer tumorigenesis and aggressiveness. Anaerobic and facultative bacteria can colonize and grow in tumors.46, 47, 48 The possibility of pathogenic bacterial growth in tumors is currently attributed to unique pathophysiologic features known to many cancers that benefit the growth of these particular bacteria, such as impaired and abnormal vascular architecture, enhanced permeability and retention effect, low oxygen pressure/hypoxia and extensive necrosis.47 Interestingly, the main periodontal disease pathogens (ie, T. denticola, P. gingivalis, F. nucleatum, and T. forsythia) are considered facultative anaerobes and oxygen‐tolerant species.19, 49, 50, 51, 52
Particularly for oral cancer, increased salivary bacterial counts of Lactobacillus spp, Capnocytophaga gingivalis, Prevotella melaninogenica, and Streptococcus mitis and loss of Haemophilus, Neisseria, Gemella, and Aggregatibacter genera have been reported in oral cancer patients compared with normal controls.24, 25, 26, 53 Our group identified different bacterial species colonizing oral tumors compared with healthy sites and found a high fusobacterial and low streptococcal phenotype as part of the transition from primary to metastatic oral cancer.18 Interestingly, P. gingivalis and F. nucleatum (two periodontal pathogens) were detected up to 600 times more frequently in oral squamous cell carcinoma than in paracancerous and normal tissues.54, 55 Mechanistically, F. nucleatum and P. gingivalis downregulate p53 pathway56, 57, 58 and promote increased cell proliferation of tongue and oral squamous cell carcinoma up to 125 times compared with control conditions.56, 59
Dysregulation of toll‐like receptor expression may also influence the host response to periodontal pathogens, which then leads to an increase in inflammation and susceptibility to periodontitis.60, 61, 62 Periodontal pathogens predominately stimulate toll‐like receptor 2 and 4. This receptor activation then leads to the production of proinflammatory cytokines via regulation by transcription of nuclear factor kappa‐light‐chain‐enhancer of activated B cells and subsequent alveolar bone resorption through the production of matrix metalloproteinases and osteoclastogenesis.61, 63, 64, 65, 66, 67 Nuclear factor kappa‐light‐chain‐enhancer of activated B cells has also been identified as an integral factor in regulating various processes associated with cancer progression, such as cell survival,68 proliferation,69 and resistance to both targeted therapy and chemotherapy.70 Recently, Kamarajan et al71 demonstrated that T. denticola, P. gingivalis, and F. nucleatum enhance oral squamous cell carcinoma migration, invasion, and tumorsphere formation via integrin alpha V/focal adhesion kinase signaling; commensal bacteria were not able to trigger the same response. The authors also demonstrated that T. denticola triggers oral squamous cell carcinoma migration via crosstalk between toll‐like receptor 2 and 4/myeloid differentiation primary response 88 protein and integrin alpha V/focal adhesion kinase signaling, thereby contributing to the aggressive nature of the pathogen‐enhanced oral squamous cell carcinoma phenotype (Figure 1). These pathogen‐mediated cancer properties were abrogated by treatment with an antimicrobial bacteriocin. Huang et al72 demonstrated that Listeria monocytogenes had a direct tumor‐stimulating effect associated with its ability to activate toll‐like receptor 2–dependent signaling pathways in ovary cancer cells. Moreover, the toll‐like receptor 2–dependent activation of nuclear factor kappa‐light‐chain‐enhancer of activated B cells caused by L. monocytogenes resulted in an enhanced resistance of tumor cells to chemotherapeutics. Additionally, metastasis and progression of oral tumors were essentially retarded in toll‐like receptor 2 knockout mice, compared with wild‐type mice.73 Thus, toll‐like receptors have a tumor‐stimulating effect on a variety of cancer cell types, and this mechanism may play a direct role in driving periodontal inflammation–induced carcinogenesis.
FIGURE 1.
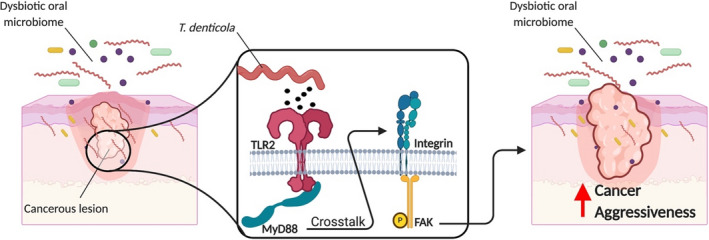
Treponema denticola drives cancer aggressiveness through toll‐like receptor 2 and 4/myeloid differentiation primary response 88 protein and Integrin/focal adhesion kinase crosstalk.71 TRL2, toll‐like receptor 2; MyD88, myeloid differentiation primary response 88 protein; FAK, focal adhesion kinase
Among periodontal pathogens, T. denticola has also been implicated in oropharyngeal squamous cell carcinoma, since dentilisin, a major virulence factor of T. denticola,74, 75 was found inside of the cellular cytoplasm of the majority (87%) of oropharyngeal squamous cell carcinoma tissues.76
The focus has traditionally been on bacteria when discussing microbiological aspects of oral diseases.77 However, a causal link between various microbes in human immunodeficiency virus–infected individuals has been documented.78 Specifically, inflammation is known to stimulate human immunodeficiency virus type‐1 gene expression and replication, and infection by bacterial pathogens usually involves production of proinflammatory cytokines that are associated with nuclear factor kappa‐light‐chain‐enhancer of activated B cells activation.79 Additionally, human immunodeficiency virus–infected patients show a higher incidence of squamous cell carcinoma of the oral cavity and anus.80, 81 As a means of establishing a link between the two diseases, Imai et al78 examined the effects of P. gingivalis on human immunodeficiency virus type‐1 replication. The group readily demonstrated that butyric acid produced by P. gingivalis promoted increased expression of latent human immunodeficiency virus type‐1–associated genes by inhibiting histone deacetylases and enriching for acetylation at histone 3, highlighting the role of bacteria as a risk factor for promoting acquired immune‐deficiency syndrome progression. Similar findings in the intestine with other butyric acid–producing bacteria, such as Clostridium, Fusobacterium, and Eubacterium, suggest that these bacteria might also be involved in the accelerated replication of human immunodeficiency virus type‐1.82 Latent human immunodeficiency virus type‐1 proviruses also carry methylated histone H3, which has been either trimethylated on lysine 9 or lysine 2783, 84 or dimethylated on lysine 9.85 Each of these modified histones is considered to be a repressive mark for cellular genes.86 Immunohistochemical staining of diseased periodontal epithelium revealed an increased abundance of the histone lysine‐specific demethylase 4B that correlates with inflammation in murine sections exposed to Aggregatibacter actinomycetemcomitans lipopolysaccharide.85, 87 Taken together, these data suggest that bacteria‐virus interactions play an integral role in promoting carcinogenesis in the oral cavity.
Alcohol88 and tobacco89 exposure influence the oral microbiome, increasing the prevalence of periodontal pathogens from Fusobacterium, Cardiobacterium, Synergistes, Atopobium, Bifidobacterium, Lactobacillus, and Selenomonas genera and decreasing the levels of Capnocytophaga, Neisseria, Haemophilus, and Aggregatibacter compared with nonsmokers.89, 90 Among the enriched bacteria, F. nucleatum is also able to damage host cell deoxyribonucleic acid (DNA).56 Thus, it is possible that F. nucleatum may play a more prominent role in alcohol and tobacco‐induced oral cancer, although these associations need further testing. On the other hand, a nested case‐control study with 129 head and neck cancer patients demonstrated that Corynebacterium and Kingella species were associated with a strongly reduced risk of oral cancer in those with a history of tobacco use.91 Interestingly, these genera are functionally associated with xenobiotic biodegradative metabolic pathways, including the capacity to metabolize toxicants found in cigarette smoke.90
Despite numerous studies focusing on dysbiosis in oral cancer, most of these studies did not test for the human papillomavirus status of the samples.92 Among those that did, a distinct oral microbiome composition for human papillomavirus–positive oral cancer compared with human papillomavirus–negative oral cancer was revealed, namely an enrichment in Lactobacillus, Gemella, and Leuconostoc and Weeksellaceae genera in human papillomavirus–positive tumors compared with human papillomavirus–negative tumors.25, 55, 92, 93, 94 Interestingly, species from these four bacterial genera have been recently associated with the establishment and progression of oral cancer,95, 96, 97, 98 implicating human papillomavirus as a driver of oropharyngeal and oral cancer tumorigenesis by influencing the composition of the oral microbiome.25, 92, 93
2.1.1. Mechanisms of oral microbiome dysbiosis–induced head and neck cancer
At least four main mechanisms have been proposed to explain how oral microbial dysbiosis can induce head and neck cancer carcinogenesis (Figure 2). All these mechanisms are not mutually exclusive, and they might even occur concurrently in mediating carcinogenesis.
FIGURE 2.
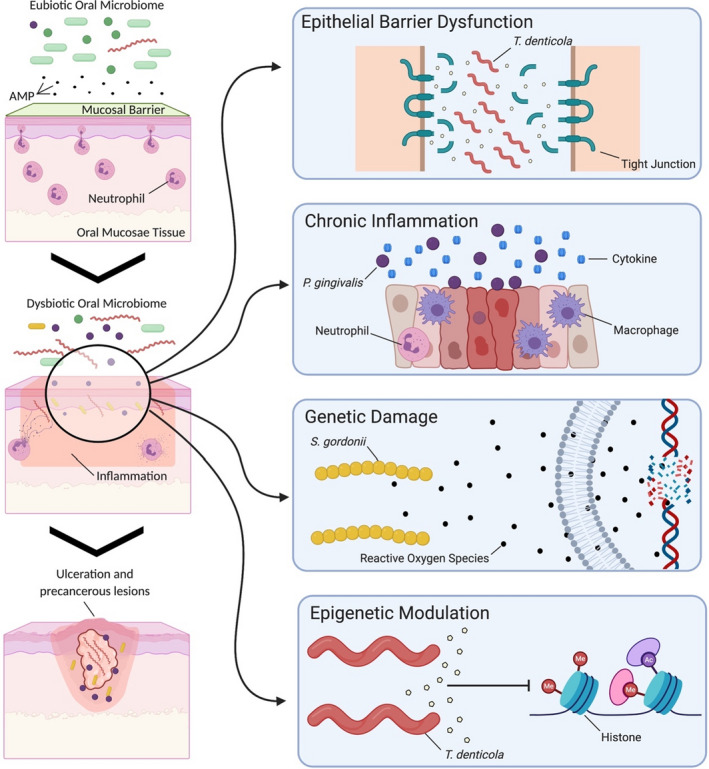
Epithelial barrier disruption, bacterial invasion, chronic inflammation, and genetic and epigenetic modulation are mechanisms by which an oral microbiome dysbiosis can promote carcinogenesis. AMP, antimicrobial peptides
Epithelial barrier disruption
To safeguard tissue homeostasis, the oral cavity relies on an anatomic separation between the host and the microbes, known as the oral and gingival epithelial barrier, comprised of a complex immunological network (eg, continuous neutrophil recruitment and extravasation into healthy sites) and antimicrobial peptides (eg, histatins and LL‐37 produced by salivary glands).99, 100, 101 Maintaining the integrity of this barrier is key to promoting healthy host‐microbial interactions.98
Several studies have demonstrated that microbes can affect this epithelial barrier function.101, 102 A proposed mechanism highlights that oral microbial dysbiosis induces epithelial barrier dysfunction, leading to head and neck cancer. A key example of this mechanism has been demonstrated with mucin‐2 knockout mice, wherein gastrointestinal mucosa lacking mucin spontaneously develop colorectal cancer, but antibiotic treatment or a germ‐free environment significantly reduced tumorigenesis.103 From a mechanistic standpoint, several oral microorganisms—such as Bifidobacterium, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, Peptostreptococcus, and Streptococcus spp—produce compounds, such as acids (eg, lactic, acetic, butyric, and isobutyric acids), that can alter the homeostasis of epithelial barriers, thereby disrupting the mucous barrier and leading to mucosal dysfunction.104, 105, 106 Although this mechanism has not yet been confirmed in the oral cavity, this seems relevant to head and neck cancer, since oral pathogens can degrade tight junction–associated proteins that regulate epithelial barrier function. For example, P. gingivalis can degrade gingival epithelial junctional adhesion molecule 1 in gingival cells, and T. denticola can degrade zonula occludens‐1, claudin‐1, and occludin.107, 108 The degradation of epithelial tight junctions enables increased permeability of the gingival epithelium, allowing bacterial virulence factors to penetrate further into the tissue and leading to bacterial invasion of the tissue,107 as T. denticola, P. gingivalis, and F. nucleatum are also able to intracellularly invade epithelial and gingival tissues.109, 110, 111, 112, 113
Chronic inflammation
A second main mechanism is chronic inflammation. Interestingly, periodontal disease is characterized by chronic inflammation of the supporting tissues of the teeth, which can lead to loss of periodontal ligament and alveolar bone.114, 115 The disease is driven by a dysbiotic oral microbiome that interacts with the human host and leads to inflammation of the surrounding tissues.114, 116, 117 In this context, several pathogenic bacteria, such as P. gingivalis and F. nucleatum, are enriched. These pathogens are able to upregulate several cytokines and inflammatory mediators (eg, interleukins, matrix‐metalloproteinases, and tumor necrosis factor alpha) in the surrounding tissues, but also facilitate invading, persisting, and spreading to adjacent cells, promoting chronic inflammation. This chronic inflammation can lead to alterations in cell metabolism, proliferation, and tumorigenesis.109, 110, 114, 118, 119, 120, 121, 122
Genetic damage
A third main mechanism is genetic damage. Several lactobacilli (eg, Lactobacillus acidophilus, Lactobacillus fermentum, Lactobacillus jensenii, and Lactobacillus minutus) and streptococci species (eg, Streptococcus gordonii, S. mitis, Streptococcus oligofermentans, and Streptococcus oralis) produce reactive oxygen species, nitrogen reactive species, sulfides, nitrosamines, and acetaldehyde98, 132 that can lead to DNA damage in epithelial cells and, thus, promote tumorigenesis.125, 126, 127, 131, 133 For example, in alcohol‐induced oral cancer, some oral microbial species, such as S. gordonii, S. mitis, S. oralis, Streptococcus salivarius, and Candida albicans, can metabolize ethanol to acetaldehyde. Acetaldehyde is an electrophilic molecule that reacts with nucleosides, forming DNA adducts.134, 135, 136 DNA adducts are sections of the DNA strand that are covalently bound to chemical compounds, which cause abnormalities during DNA replication that can lead to genetic mutations.137
Epigenetic modulation
Epigenetic modifications are defined as heritable alterations, not coincident with alterations in the underlying DNA sequence, that allow biologic systems to interfere with transcription in response to a variety of environmental stimuli.138 Altered expression of DNA methylation138, 139, 140, 141 and histone modifications142, 143, 144, 145, 146 have been reported to play critical roles in the onset and progression of both chronic periodontitis and oral squamous cell carcinoma.147 Thus, a fourth mechanism is related to epigenetic modulation of the hosts’ gene expression.
Multiple studies have suggested that the hypomethylation status of the interleukin‐6 and interleukin‐8 gene promoters may be related to an overexpression of these cytokines in inflamed periodontal disease tissues compared with controls.148, 149, 150 It has been found that human gingival epithelial cells may be triggered by the release of these proinflammatory mediators, which may promote the recruitment and activation of inflammatory cells to facilitate oral malignant transformation,151, 152 further supporting a link between periodontal infection and induction of cancer‐related cellular processes. Although the carcinogenesis‐associated epigenetic components governing interleukin‐6 and interleukin‐8 have yet to be fully determined, one might consider the connections between bacterial/microbial infection, cytokines, and cancer development as intriguing and intersecting points for systems biology that may reveal novel insights for the development of better diagnostics and gene and drug therapies.153
DNA methylation and histone modifications are not separate events; they are linked and result in a unique tissue and cell‐specific gene expression.154 A recent study showed that the activation of toll‐like receptors by periodontal pathogens not only induced activation of nuclear factor kappa‐light‐chain‐enhancer of activated B cells but also led to an enrichment in histone acetylation in oral epithelial cells.144 Histones are subjected to a myriad of post‐translational modifications, which can open and closed regions of DNA that regulate the accessibility of transcription factors to bind to their targets.155 Interestingly, P. gingivalis and F. nucleatum can induce histone modifications, such as induction of histone acetylation, in oral epithelial cells.144 On the other hand, T. denticola seems to regulate cellular division/chromatid segregation (potentially inducing genetic instability) and histone methylation and acetylation, as aurora kinase, histone methyltransferase, and histone acetyltransferases inhibition seem to modulate matrix metalloproteinase‐2 activation and expression in periodontal ligament cells in the context of T. denticola infection.156 Yet, few studies are available on histone modifications in periodontitis.
Although genetic and epigenetic similarities have been reported between chronic periodontitis and head and neck cancer,152, 157 knowledge in this domain lacks deeper mechanistic insight and is largely comprised of correlative findings.
2.2. Gastrointestinal cancer
Gastrointestinal cancer, one of the most common causes of cancer worldwide, is often subdivided by anatomic location: the esophagus, stomach, liver, gallbladder, pancreas, colon, rectum, anus cancer. Combined, gastrointestinal cancers accounted for almost 5 million new cases and more than 3.5 million deaths worldwide in 2018.34 In the United States in 2020, more than 330 000 Americans are expected to be diagnosed with gastrointestinal cancer and more than 155 000 deaths from gastrointestinal cancer are expected.23, 34 Table 2 shows the estimated new cases and deaths in the United States (for 2020)23 and worldwide (for 2018) for each gastrointestinal cancer subtype.34 In this particular section, we will focus on gastric, pancreatic, and colon cancers.
TABLE 2.
Gastrointestinal cancer estimated new cases and deaths in the United States and worldwide
| Gastrointestinal cancer classification | Estimated new cases | Estimated deaths | ||
|---|---|---|---|---|
| 2020 United States23 | 2018 worldwide34 | 2020 United States23 | 2018 worldwide34 | |
| Esophagus | 18 440 | 572 034 | 16 170 | 508 585 |
| Stomach | 27 600 | 1 033 701 | 11 010 | 782 685 |
| Colon | 104 610 | 1 096 601 | 53 200 | 551 269 |
| Other intestines | 11 110 | — | 1 700 | — |
| Rectum | 43 340 | 704 376 | — | 310 394 |
| Anus | 8 590 | 48 541 | 1 350 | 19 129 |
| Liver | 42 810 | 841 080 | 30 160 | 781 631 |
| Gallbladder and other biliary ducts | 11 980 | 219 420 | 4090 | 165 087 |
| Pancreas | 57 600 | 458 918 | 47 050 | 432 242 |
| Other organs | 7 600 | — | 3 060 | — |
Recently, Zhang et al158 meta‐analyzed 10 cohort studies and demonstrated a 23% increased risk for overall gastrointestinal cancer in periodontitis patients, compared with normal patients (hazard ratio, 1.23; 95% confidence interval, 1.10‐1.37). Also, the authors found 59% increased mortality from gastrointestinal cancer in patients with periodontitis compared with healthy patients (hazard ratio, 1.59; 95% confidence interval, 1.16‐2.16).
Regarding gastric cancer, in a meta‐analysis, Zhang et al158, found a 12% increased risk of gastric cancer (hazard ratio, 1.12; 95% confidence interval, 0.88‐1.42) and a 28% increased mortality rate for gastric cancer in patients with periodontitis compared with normal patients (hazard ratio, 1.28; 95% confidence interval, 0.71‐3.37). Another meta‐analysis159 showed that patients with lost teeth, a marker for severe periodontitis, have increased risk for gastric cancer. Unfortunately, lost teeth are not a unique marker for periodontitis; thus, these specific data must be taken carefully.
Interestingly, Salazar et al27 demonstrated that plaque colonization by periodontal pathogens (ie, P. gingivalis, T. forsythia, T. denticola, and A. actinomycetemcomitans) was associated with increased risk of precancerous lesions of gastric cancer, thereby suggesting that periodontal pathogens may have a role in gastric cancer. However, most patients included in this study were Hispanic (45.4%), female (63.0%), and had a mean age of 57 years, with high prevalence of smoking, and so the results may be more relevant to this demographic.
Regarding pancreatic cancer, a prospective study with more than 700 patients reported that the presence of P. gingivalis and A. actinomycetemcomitans in the oral cavity was associated with a 60% increased risk of pancreatic cancer (adjusted odds ratio, 1.60; 95% confidence interval, 1.15‐2.22).160 The study reported that patients positive for the presence of P. gingivalis in the oral cavity have a 59% greater risk of developing pancreatic cancer than those who were negative for the pathogen. Even after excluding pancreatic cancer cases that occurred less than 2 years after oral samples were obtained, no significant changes were found in the risk factor, demonstrating that it is very unlikely that the oral dysbiosis occurred after or concurrently with pancreatic cancer.160, 161 Similarly, a prospective cohort study found that patients with higher levels of P. gingivalis antibodies in the blood (>200 ng/mL) had a twofold greater risk of developing pancreatic cancer than those with lower levels of the same antibody.162 Zhang et al158 found a 25% increased risk on having pancreatic cancer (hazard ratio, 1.25; 95% confidence interval, 0.84‐1.85) and a remarkable 120% increased mortality rate for pancreatic cancer in patients with periodontitis compared with those without periodontitis (hazard ratio, 2.20; 95% confidence interval, 1.44‐3.37).
Regarding colon cancer, a prospective study with 17 904 women and 24 582 men found increased risks for colon precursor lesions—that is, serrated polyps and conventional adenomas, with hazard ratios of 1.17 (95% confidence interval, 1.06‐1.29) and 1.11 (95% confidence interval, 1.02‐1.19), respectively—in patients with chronic periodontitis compared with healthy patients. Losing teeth (four or more teeth) increased the risk for colon cancer by 20% for those with serrated polyps (odds ratio, 1.20; 95% confidence interval, 1.03‐1.39).163 In contrast, in a meta‐analysis, Zhang et al158 found no significant difference for risk of colon cancer but found a 66% increased mortality rate for colon cancer in patients with periodontitis compared with those without periodontitis (hazard ratio, 1.66; 95% confidence interval, 0.44‐6.27).
Accumulating evidence demonstrates that microbes have a more potent role in gastrointestinal carcinogenesis than previously thought. In particular, various members of the oral microbiota have been linked to cancers of the gastrointestinal tract.
2.2.1. Gastrointestinal microbiome
Gastric microbiome
Despite being considered sterile for many years due to the low pH of the gastric lumen, it is now known that the stomach possesses a very specific microbiome.164, 165 This gastric microbiome is mainly composed of Streptococcus, Veillonella, Prevotella, Fusobacterium, and Rothia genera,165, 166 and more than 65% of these species have been previously described as oral commensal bacteria.164, 165 Colonization by Helicobacter pylori leads to dramatic changes in the gastric microbiome, including a higher abundance of Proteobacteria, Spirochaetes, and Acidobacteria and a decreased abundance of Actinobacteria, Bacteroidetes, and Firmicutes. H. pylori infection can cause chronic gastritis, peptic ulcer, atrophic gastritis, gastric adenocarcinoma, and mucosa‐associated lymphoid tissue lymphoma. Still, the interaction between the bacterium and gastric microbiota in the pathogenesis of these conditions has to be fully elucidated.165, 166
Regarding the contribution of oral pathogens to gastric cancer, very little has been studied so far. Yuan et al167 reported very low levels of P. gingivalis in stomach samples; the authors tested the survival rates of the bacteria in acidic buffers and concluded that the bacteria cannot survive in highly acidic environments, such as the stomach. On the other hand, F. nucleatum was recently detected in higher abundance in gastric cancer tissues than in normal tissue.168, 169 Recently, Boehm et al169 demonstrated that F. nucleatum is associated with lower long interspersed nuclear element‐1 DNA methylation in gastric cancer tissues. Thus, more studies are needed to stablish whether oral bacteria have tumorigenic effects in the context of gastric cancer. Recently, Nieminen et al170 found the presence of the T. denticola dentilisin protease in all gastric cancer tissues tested, indicating that the bacterium may have an important role in gastric cancer. However, more studies are still needed to determine the contribution of all these oral bacteria to gastrointestinal carcinogenesis.
Pancreatic microbiome
Once thought to be a sterile organ, a number of studies have now established that microbes are present within this organ in normal, nonpathologic states. However, the composition of the microbiota is still debatable.171
Recently, Pushalkar et al172 demonstrated that pancreatic tumors harbor a specific microbiome, which is distinct from the gut and normal pancreatic microbiome. Specifically, there is an increase in the genus Brevibacterium and order Chlamydiales in pancreatic cancer compared with normal controls. This tumor‐associated microbiome is markedly more abundant than in normal pancreas in both mice and humans and is capable of promoting oncogenesis via significant immune suppression in the tissue. Despite not directly finding P. gingivalis in the tissue, Gnanasekaran et al173 recently demonstrated that the bacterium is capable of invading and surviving inside pancreatic cancer cells. Also, the authors showed that P. gingivalis enhances pancreatic tumor growth in vivo. Interestingly, previous studies reported high rates of mutations in tumor suppressor protein p53 and Kirsten rat sarcoma viral oncogene homologue genes for pancreatic cancer.174, 175, 176, 177 This suggests that P. gingivalis may cause genetic damage to pancreatic cells, thereby inducing aggressive pancreatic cancer; however, further studies are necessary to stablish this hypothesis.
Recently, Nieminen et al170 found the presence of T. denticola dentilisin in 65% of the pancreatic cancer tissues tested, indicating that the bacterium may have a role in pancreatic cancer as well. However, more tests are warranted to determine the contribution of T. denticola to pancreatic cancer.
Intestinal microbiome
The human intestinal microbiome comprises about 1013 microbial species, including bacteria, viruses, fungi, and other members, which play a significant role in normal human physiology, by contributing to metabolism and digestion, gut homeostasis, and tissue development.178 In this chapter, we will focus on the large intestine microbiome and its association with colon cancer.
Among all species found in the large intestine, F. nucleatum seems to be increasingly reported in gut infections, such as intestinal abscesses and acute appendicitis.178 Castellarin et al179 was one of the first groups to point out that F. nucleatum infection is prevalent in colorectal cancer. Subsequently, metagenomic and transcriptomic studies have demonstrated an enrichment in Fusobacteria species, specifically F. nucleatum in colorectal cancer, compared with adjacent normal tissues. Further, F. nucleatum accelerates colorectal cancer cell growth and progression in vitro and in vivo.180, 181 The bacterium also seems to suppress anti–tumor immunity by inhibiting immune natural killer cells.182 All these findings support that F. nucleatum can colonize and is enriched in colorectal cancer, and it may also play a major role in tumor growth and survival. Similar to the mechanism proposed by Kamarajan et al71 for oral squamous cell carcinoma, Sun et al178 demonstrated that F. nucleatum induces colorectal cancer proliferation through toll‐like receptor 4 and myeloid differentiation primary response 88 protein activation and upregulation of micro–ribonucleic acids 18a and 4802.
On the other hand, Abed et al183 questioned the ability of F. nucleatum to reach the colon from the oral cavity; thus, the authors hypothesized that a bacteremia may be responsible for the presence of the bacterium in the colon and rectum. In brief, transient bacteremia is a common finding during periodontal disease, in which periodontal pathogens are able to reach the circulatory system via the periodontitis‐infected sites. It has been reported that bacterial loads reach up to 104 bacteria per milliliter of blood.184 Within the circulatory system, these bacteria would have access to other tissues, wherein they could colonize and infect. To test this hypothesis, Abed and colleagues intravenously injected mice with F. nucleatum and found significantly higher counts of the bacteria on colorectal tumors than in adjacent normal sites. The authors found that a fusobacterial fatty acid–binding protein 2 mediates F. nucleatum binding to tumor cells by binding d‐galactose‐beta(1‐3)‐N‐acetyl‐d‐galactosamine, a polysaccharide overexpressed in colorectal cancer, whereas fatty acid–binding protein 2–deficient bacteria show reduced d‐galactose‐beta(1‐3)‐N‐acetyl‐d‐galactosamine binding. Although this finding is very interesting from the standpoint of demonstrating bacteremia in gastrointestinal cancer, one must also consider the possibility of cancer‐enhanced permeability and retention effects. The enhanced permeability and retention effect, first reported by Matsumura and Maeda in 1986,185 indicates that there is a greater accumulation and prolonged circulation of macromolecules in cancer tissues than in normal tissues due to the enhanced permeability of blood vessels inside the cancer tissues. Although this effect has been demonstrated for macromolecules and nanoparticles, Fang et al47 demonstrated that bacteria can be taken up by tumors through the enhanced permeability and retention effect, and the effect can be enhanced with the use of nitroglycerin, a known vasodilator. Thus, F. nucleatum accumulation within cancer tissues can be mediated by active processes of bacterial internalization and/or by an enhanced permeability and retention effect. This hypothesis could be tested by performing the same procedure proposed by Abed et al but comparing the amount of heat‐treated vs live bacteria found in tumor tissues. The enhanced permeability and retention effect hypothesis would be rejected if significantly fewer heat‐treated bacteria are found in tumors sites compared with live bacteria.
Recently, Nieminen et al170 found the presence of T. denticola dentilisin in half of the colon cancer tissues tested, indicating that the bacterium may play a role in colon cancer. However, more tests are necessary to determine its contribution.
2.3. Lung cancer
Lung cancer is the leading cause of new cancer cases annually and annual cancer deaths worldwide, leading to more than 2.09 million new cases and more than 1.7 million deaths in 2018.34 Lung cancer is expected to cause almost 230 000 new cancer cases and more than 135 000 deaths in 2020 in the United States.23
Associations between periodontal disease and lung cancer have been suggested in the literature. In 2003, Hujoel et al20 demonstrated that general lung cancer presented the strongest correlation with periodontal disease, among the most fatal cancers; the study found a 1.97‐fold increase risk for lung cancer in patients with periodontitis. In 2016, a meta‐analysis of five different cohort studies, evaluating more than 320 000 patients, demonstrated that individuals with periodontal disease had a 1.24‐fold increased rate of general lung cancer. Also, the study pointed out that women with periodontitis have higher risks of developing lung cancer than men do.186
Interestingly, Yan et al187 were the first study to demonstrate that salivary bacteria (ie, Capnocytophaga, Selenomonas, Veillonella, and Neisseria genera) were significantly altered in patients with both squamous cell and adenocarcinoma lung cancer compared with control patients. The authors even validated this Capnocytophaga and Veillonella genera alteration as a potential biomarker for lung cancer. Similarly, Zhang et al28 also found an increased abundance in Veillonella and Streptococcus genera in non–small cell lung cancer patients, compared with healthy patients. Additionally, the authors also found Fusobacterium, Prevotella, Bacteroides, and Faecalibacterium genera decreased in non–small cell lung cancer patients, compared with healthy patients.
2.3.1. Lung microbiome
Although the lungs were also regarded as sterile for a long time, several recent studies demonstrated the presence of a lung microbiome.29, 188 This microbiome seems to be comprised of Acinetobacter, Pseudomonas, Ralstonia, Prevotella, Veillonella, and Streptococcus genera.29, 188, 189 Microbial species from the oral cavity are thought to migrate from the oral cavities to the lungs via microaspiration, as they are more similar to oropharynx microbiota than nasopharynx microbiota. There is also a fine balance between microaspirated species and microbial elimination via mucociliary clearance, cough, or host immune defense processes.29, 188, 190 Despite the similarities and a direct impact by the oral microbiome, the lung microbiome has been characterized as a distinct microbiome, with a higher abundance of Thermi and Cyanobacteria phyla than in other human microbiomes.189 Nevertheless, a precise description of the lung microbiome and the influence of the oral microbiome on the lung microbiome is still lacking as the field is just emerging.191
Similar to the oral microbiome, lung microbiome dysbiosis seems to be corelated with the emergence of respiratory diseases, such as severe asthma and lung cancer.191 There is emerging evidence that the lung microbiome seems to play an important role in lung cancer progression. For example, patients with unspecified lung cancer have lower microbial diversity than nonmalignant controls do, with Thermus and Legionella genera being more abundant in advanced and metastatic lung tissues.189 Also, a cohort study with 64 patients evaluated differences in the lung microbiome of smokers vs nonsmokers.192 They found a relatively lower abundance of Bacteroidetes and Proteobacteria phyla in smokers than in nonsmokers, with no significant changes, suggesting that smoking may have a more pronounced effect on the microbiota of the head and neck area than on that of the lower respiratory tract. Nevertheless, data on the impact and mechanisms of the lung microbiome on lung cancer pathogenesis remain limited.
Despite advances in the field, some studies used either sputum or bronchoalveolar fluid collection during bronchoscopy, risking a potential contamination of the samples with oral bacteria, since collection instruments pass through the naso or oropharynx first before reaching the lungs.188, 189, 193 Thus, avoiding the use of these collection methods is essential to effectively evaluate the lung microbiome.
2.4. Other cancer types
Periodontal disease has been recently associated with other cancer types, such as prostate, breast, and uterine cancer. However, more studies are still needed to determine whether this association is true, and, if so, to determine the possible underlying mechanisms.
2.4.1. Prostate cancer
Periodontal disease has been associated with prostate cancer.20, 194, 195, 196 For instance, Hujoel et al20 reported a 1.81‐fold increased risk for prostate cancer in patients with periodontal disease, compared with patients without it. This was the highest fold risk from all the cancers studied. Recently, the presence of oral bacterial DNA (ie, P. intermedia, P. gingivalis, and T. denticola) was found in expressed prostatic secretion of patients with periodontitis.197 Interestingly, patients with periodontitis and prostate inflammation had greater levels of prostate‐specific antigen than those with either disease alone did; and periodontal treatment decreased prostate‐specific antigen levels in those patients with both conditions, improving prostate inflammation symptoms.198, 199 These data support the concept of a bacteremia mechanism whereby oral pathogens would colonize and infect the prostate. This infection would, then, lead to a chronic inflammatory response in the prostate, which, in turn, would induce neoplastic transformations; growing evidence implicates chronic prostate inflammation as one of the main contributors for prostate cancer.30, 196, 200, 201 Specifically, Simons et al30 provided direct evidence that bacterial‐induced prostate inflammation accelerates prostate cancer progression and sheds light on how changes in the prostate microenvironment caused by prostate inflammation may accelerate tumor progression.
2.4.2. Breast cancer
Literature is still inconsistent with regard to associations between periodontitis and breast cancer. For instance, in 2011, a longitudinal prospective study in Sweden with more than 3000 subjects between 30 and 40 years of age reported that chronic periodontitis patients were 2.3 times more likely to be diagnosed with general breast cancer than those with healthy gums were.33 However, the conclusions were limited by the fact that the authors did not clinically examine some of the patients for periodontitis, but instead used missing molars as a proxy to distinguish patients with or without periodontitis. Since caries can also lead to tooth loss, periodontitis may have been overestimated in these patients. Similarly, Sfreddo et al202 reported that women diagnosed with periodontitis had two to threefold more chances of having invasive ductal breast carcinoma than healthy patients did, even after adjusting for confounding variables. In this study, patients were subjected to full‐mouth periodontal examinations to diagnose periodontitis. Recently, a meta‐analysis study with more than 1.5 million participants investigated the association between periodontitis and general breast cancer and found a 1.2‐fold increased risk for general breast cancer in periodontitis cases compared with controls.203 Interestingly, no significant association was found among patients with periodontitis and history of periodontal therapy and general breast cancer. If this association is accurate, this report suggests that periodontitis therapy can decrease the higher risk of having general breast cancer. In contrast, Mai and coworkers204, 205 evaluated 1337 postmenopausal women and found no significant association between the incidence of invasive general breast cancer and either alveolar bone loss or presence of periodontal pathogens. Hujoel et al20 also reported no significant association between periodontitis and higher risk of general breast cancer (hazard ratio, 1.32; 95% confidence interval, 0.74‐2.28). Thus, more studies are needed to determine whether the association between periodontitis and breast cancer is true and, if so, to determine the possible underlying mechanism.
Although the association between periodontitis and breast cancer is unclear, Parhi et al206 recently tested whether an F. nucleatum bacteremia could lead to tumorigenesis and increase the aggressiveness of triple‐negative breast cancer in mice. The authors observed a 100‐fold increase in abundance in bacteria 24 h after the intravenous injection of the bacterium within triple‐negative breast cell line (AT3) tissues compared with normal tissues. They reported that F. nucleatum colonizes the triple‐negative breast tumor tissue, accelerates tumor growth and progression, and inhibits tumor immune‐modulator cells, such as T, B, and natural killer cells, thus indicating that F. nucleatum, via a bacteremia, can reach the breast tumor tissue and induce tumor growth and progression. However, an enhanced permeability and retention effect is also possible, and it is not known whether this bacterial accumulation is driven by the bacterium itself or by an enhanced permeability and retention effect.
2.4.3. Uterine cancers
Few studies have examined the correlation between periodontitis and uterine cancer. In a study by Arora et al207 that examined twins (to control for confounding factors), periodontitis was positively associated with increased risk of uterine corpus cancer (hazard ratio, 2.20). Interestingly, Han et al208, demonstrated that intravenous tail vein injection with F. nucleatum led to uterine bacterial colonization, proliferation, and intrauterine infection. Nevertheless, this result may be due to the tail vain draining more to the uterus than other tissues, thus resulting in higher uterine colonization of the bacterium than in other tissues. In his context, gingival inoculation may be more suitable to replicate periodontal bacteremia and evaluate whether uterine colonization by periodontal pathogens is, indeed, possible. Interestingly, though, recent reports have detected periodontal pathogens on placental tissue associated with multiple adverse pregnancy outcomes,208, 209, 210, 211, 212 and preliminary data from our group using an in vivo polymicrobial periodontal disease model described by Gao et al213 show higher counts of periodontal pathogens in the mouse uterus than in control mice. These data indicate that uterine colonization by periodontal pathogens is, indeed, possible. However, further studies are still necessary to better understand how periodontitis may be associated with uterine cancer and whether F. nucleatum intrauterine infection can lead to uterine cancer.
3. POTENTIAL THERAPEUTICS FOR ORAL DYSBIOSIS–RELATED CANCERS
3.1. Antimicrobial peptides
Antimicrobial peptides are peptides composed of 12‐100 amino acids with a cationic net charge and amphiphilic characteristics. These peptides are produced by most organisms (from bacteria to humans) as primary defense molecules that target a broad spectrum of pathogens.214, 215 In this sense, antimicrobial peptides s can be used as antimicrobials to treat dysbiotic microbiomes, thus possibly preventing the formation of dysbiosis‐related cancers. For instance, Shin et al216 demonstrated that nisin, an antimicrobial peptide produced by the bacteria Lactococcus lactis, can effectively inhibit and kill periodontal disease pathogens, including T. denticola, P. gingivalis, F. nucleatum, A. actinomycetemcomitans, and P. intermedia, without affecting normal human oral cells, such as primary periodontal ligament, gingival fibroblasts, and oral keratinocyte cells. Similarly, Radaic et al19 recently demonstrated that nisin is able to revert the diversity levels of in vitro human saliva–based dysbiotic oral biofilms back to control levels, promoting healthier oral biofilms.
Although their main role is in defending against pathogens, several antimicrobial peptides have been reported to selectively target human tumor cells.215 As far as we know, our group was the first to highlight the potential of antimicrobial peptides in cancer treatment in vivo.217 To date, we have demonstrated that nisin inhibits pathogen‐induced oral squamous cell carcinoma migration, invasion, tumorsphere formation, and angiogenic sprouting, and it promotes oral squamous cell carcinoma apoptosis in vitro while reducing overall tumor size and prolonging survival in vivo.71, 217, 218, 219 Importantly, nisin had no cytotoxic effects on normal human oral keratinocytes, periodontal ligament cells, gingival fibroblasts, and osteoblasts.216, 218 Magainin 2, an antimicrobial peptide isolated from the African frog Xenopus laevis, has antitumor activity against a human lung cancer cell line,220 cytotoxic and antiproliferative effects on bladder cancer cells, and has no effect on normal human fibroblasts.221 LL‐37, an antimicrobial peptide produced in humans, promotes caspase‐independent calpain‐mediated apoptosis in an acute T cell leukemia cell line and mediates mitochondrial depolarization and caspase‐independent apoptosis in an oral squamous cell carcinoma cell line.222 However, LL‐37 had no effect on a human keratinocyte cancer cell line, indicating possible tissue‐specific effects.223 Interestingly, LL‐37 is a highly expressed antimicrobial peptide in normal mucosa, especially in lung, breast, and colon tissues.215 However, LL‐37 was downregulated in colon cancer tissues due to DNA methylation of the cathelicidin antimicrobial peptide gene promoter, which facilitates colon cancer growth.224 This suggests a possible role for LL‐37 in colon tumorigenesis and cancer progression.
These studies illustrate the ability of antimicrobial peptide to specifically bind to and affect tumors, but not normal tissue. This is thought to be related to antimicrobial peptide binding to phosphatidylserine lipids on the outer membranes of tumor cells, which are often not present on normal cells. Briefly, normal cell membranes exhibit an asymmetry in the lipids forming their outer and inner layers; neutral lipids, such as phosphatidylcholines and sphingomyelins, are primarily located in the outer leaflet of the membrane bilayer, whereas most phosphatidylserine molecules are located in the inner leaflet of the membrane.215, 225 Then, due to the effect of inflammatory cytokines, oxidative stress, and acidity, tumors lose this membrane asymmetry, thus displaying most of their phosphatidylserine molecules on the outer layer of the cell membrane.215, 226, 227 Other proposed mechanisms include decreased levels of cholesterol in cancer cells that facilitate antimicrobial peptide binding and cancer cell apoptosis228; electrostatic attraction between other negatively charged components of cancer cells and antimicrobial peptides, facilitating binding and selective disruption of cell membrane229, 230; and the ability of antimicrobial peptide to selectively interact with ion channels.217, 231 However, all these mechanisms may occur concurrently, contributing more to a set of mechanisms constituting the action of antimicrobial peptides on tumors.
Use of nano‐sized drug delivery systems is a potential strategy to improve the delivery of drugs, peptides, and genes into host cells.214, 232, 233, 234 These systems are able to increase their load potency, area under the curve, and peak serum concentration, decrease the time to reach the maximum serum concentration, and reduce load toxicity and food interactions.214, 232 Recently, our group delineated the state of the art for nano‐sized drug delivery systems of antimicrobial peptide delivery, focusing specifically on bacteriocins, and we found throughout the literature that associations of antimicrobial peptides with nano‐sized drug delivery systems seem to be beneficial, since the delivery systems were able to significantly increase microbial inhibition compared with free bacteriocin.214 Similar effects were found by Goudarzi et al235 for nisin and Niemirowicz et al236 for LL‐37. The previous studies demonstrate the increased efficacy of nisin on gastrointestinal, hepatic, and leukemia cancer cell lines after loading the bacteriocin in polylactic acid‐polyethylene glycol‐polylactic acid nanoparticles; the latter demonstrate that immobilization of LL‐37 on the surface of magnetic nanoparticles significantly increases antitumor activity of LL‐37 against colorectal adenocarcinoma cell lines (DLD‐1 and HT‐29) compared with free antimicrobial peptide. These examples illustrate the potential use for antimicrobial peptide–loaded nano‐sized drug delivery systems to enhance antimicrobial peptide capabilities for cancer treatment. However, very little has been tested in the field.
Although activity is found for antimicrobial peptides in cancer, the particular mechanisms by which these molecules mediate their actions are still poorly understood.215 For more details on antimicrobial peptides and cancer, please refer to Tornesello et al.215
3.2. Probiotics
The term “probiotic” was first coined by Lilly and Stillwell237 in 1965 to distinguish it from the term “antibiotics” and thus define substances produced by protozoa that were able to support the growth of other microorganisms. In 1998, the term was refined by Guarner and Schaafsma238 as live cultures of microorganisms, which confer a health benefit to the host when administered in adequate amounts. Since then, these definitions have been adopted by the World Health Organization to define probiotics.239, 240
Probiotics have gained attention owing to their ability to modulate cancer proliferation and apoptosis in vitro and in vivo, turning them into potential chemotherapeutic alternatives.240, 241 For instance, Thirabunyanon and Hongwittayakorn242 showed that Pediococcus pentosaceus FP3, Lactobacillus salivarius FP25, Lactobacillus salivarius FP35, and Enterococcus faecium FP51 promote antiproliferative effects and induction of apoptosis in a colon cancer cell line (Caco‐2) in vitro. The mechanism by which these bacteria promote their anticancer effect may be due to short‐chain fatty acid production, mainly butyric and propionic acids. Appleyard et al243 demonstrated that pretreating rats for 1 week with VSL#3, a commercial highly concentrated probiotic preparation containing eight live freeze‐dried bacterial species normally found in the human gastrointestinal microbiome, including strains of Lactobacillus, Bifidobacterium, and S. salivarius subsp thermophilus,244 prevented the development of colitis‐associated colon dysplasia.
Recently, our group reported that a nisin‐producing L. lactis probiotic can modulate in vitro human‐based dysbiotic oral biofilms toward health.19 Specifically, we demonstrated that L. lactis can modulate cancer‐related bacteria (ie, F. nucleatum and T. forsythia) levels back to control levels, thus promoting a healthier oral biofilm. In this context, a nisin‐producing L. lactis probiotic has the potential to prevent several types of cancer, as described in this review, by specifically inhibiting these cancer‐related pathogens.
Interestingly, nano‐sized drug delivery systems were used to successfully deliver probiotics into cancer cells. Zheng et al245 encapsulated Clostridium butyricum spores into chemically modified dextran and administrated nano‐sized drug delivery system to mice. Interestingly, the spore‐loaded nano‐sized drug delivery system was specifically found enriched in colon tumor sites after oral administration. Inside the tumors, C. butyricum cells were reconstituted and they fermented the dextran encapsulation, producing short‐chain fatty acid in the process and reducing tumors up to 89%. The nano‐sized drug delivery system was also able to increase gut microbiome richness, by increasing the abundance of short‐chain fatty acid–producing bacteria.
Although probiotics show promise in preventing and treating cancer, their mechanism of action is still largely unknown. Part of their mechanism of action is thought to be an ability to modulate pathogenic bacteria, the local pH, harmful enzymes, proinflammatory cytokine levels, production of short‐chain fatty acid, and degradation of potential carcinogens.241 For more details on how probiotics are a promising tool for the prevention and treatment of cancer and other diseases, please refer to Nguyen et al,240 Shin et al,219 and Górska et al241; for more details specifically about probiotics in gastrointestinal cancer, please refer to Javanmard et al.246
3.3. Epigenetic drugs
The fact that epigenetic mechanisms are reversible makes them attractive targets for new treatment models in both cancer and inflammatory diseases, garnering significant research interest.247, 248, 249 The term “epidrugs” was coined by Ivanov et al250 and used to describe drugs that inhibit or modulate disease‐associated epigenetic proteins, thereby ameliorating, curing, or preventing the disease. The discovery of specific bromodomain and extraterminal motif protein inhibitors acting as acetylated histone mimetics (ie, I‐BET151, and (+)‐JQ‐1)251 has not only allowed for therapeutic targeting of bromodomain and extraterminal motif proteins in cancer, but also provided insight into contributions of bromodomain‐containing proteins in the pathogenesis of inflammatory disorders that are associated with an altered epigenetic landscape.252 Bromodomain and extraterminal motif protein inhibitors suppress lipopolysaccharide and cytokine‐induced expression of inflammatory cytokines and chemokines in monocytes and macrophages in vitro and in vivo, and they protect mice from lethal endotoxic shock and sepsis.253, 254 Inhibition of bromodomain and extraterminal motif proteins also ameliorates inflammation and resulting pathology in animal models of several inflammatory diseases, including rheumatoid arthritis, graft‐vs‐host disease, and multiple sclerosis.254, 255, 256 There are two studies to date that demonstrate that the bromodomain and extraterminal motif protein inhibitor (+)‐JQ‐1 ameliorates gingival inflammation and alveolar bone destruction in P. gingivalis–induced experimental periodontitis in mice.247, 249 In this model, the therapeutic effects of bromodomain and extraterminal motif protein inhibition were attributed to diminished inflammatory cytokine production by macrophages and reduced osteoclast formation. However, despite extensive efforts toward understanding bromodomain protein function in health and disease, little is still known about the role of bromodomain and extraterminal motif proteins in the pathogenesis of periodontitis. Thus, there is a need to investigate to what extent xenobiotic exposure can influence epigenetic signatures in the human body, and how epigenetic variability in disease‐related genes can be translated into individual differences in drug pharmacokinetics, toxicity, and responsivity.
Epigenetic modifications within periodontal tissues that are influenced by dysbiotic biofilms and the effects of environmental exposures lead to an infectious‐inflammatory condition and transgenerational changes in the genome stability that drive disease progression and a favorable landscape for cancer development.152 Therefore, epigenomic approaches will enable a better understanding of the epigenetic mechanisms regulating the dynamics of complex diseases. The next challenge will be to perform integrated basic and clinical studies in order to comprehensively explore epigenetic regulatory pathways as markers of disease and potential therapeutic targets.152
Lastly, targeting super‐enhancer regions could be a useful strategy to return whole sets of disease‐related genes back to normal physiologic levels. Epigenetic modifications associated with the regulation of human immunodeficiency virus‐1 latency may also be interesting targets that hold promise for the discovery of novel drug targets.
4. SUMMARY
Oral microbial dysbiosis emerges as a result of an unbalanced oral microbial state that is capable of promoting diseases in the host. This unbalanced state is the most accepted paradigm for explaining the initiation and progression of periodontal disease, and it is thought to be driven by an enrichment of pathogens, including P. intermedia, F. nucleatum, P. gingivalis, T. forsythia, and T. denticola.
Although findings are still emerging for breast and uterine cancer, periodontitis has been positively associated with an increased risk for specific subtypes of head and neck, gastrointestinal, lung, and prostate cancers. In all these cancers, oral bacteria are present and positively associated with tumor growth and progression.
Finally, antimicrobial peptides, probiotics, and epidrugs show significant promise for cancer treatment, since they have a dual therapeutic potential in treating the tumors and tumor microenvironment directly (antimicrobial peptides, probiotics, epidrugs) and also modulating the oral dysbiosis state back to health (antimicrobial peptides and probiotics).
ACKNOWLEDGMENTS
This work was supported by funding from the AAP Sunstar Innovation Grant and NIH R01 DE025225 grant to YLK and from the 2019 IADR/Philips Oral Healthcare Young Investigator Research Grant to AR. All original figures in this article were created with BioRender.com.
Radaic A, Ganther S, Kamarajan P, Grandis J, Yom SS, Kapila YL. Paradigm shift in the pathogenesis and treatment of oral cancer and other cancers focused on the oralome and antimicrobial‐based therapeutics. Periodontol 2000. 2021;87:76–93. 10.1111/prd.12388
Funding information
Larry Berkelhammer funds to Yvonne L. Kapila
REFERENCES
- 1.Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. 2019;23:122‐128. 10.4103/jomfp.JOMFP_304_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandara HMHN, Panduwawala CP, Samaranayake LP. Biodiversity of the human oral mycobiome in health and disease. Oral Dis. 2019;25:363‐371. 10.1111/odi.12899 [DOI] [PubMed] [Google Scholar]
- 3.Berger D, Rakhamimova A, Pollack A, Loewy Z. Oral biofilms: development, control, and analysis. High‐Throughput. 2018;7(24):1–8. 10.3390/ht7030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morse DJ, Wilson MJ, Wei X, et al. Denture‐associated biofilm infection in three‐dimensional oral mucosal tissue models. J Med Microbiol. 2018;67:364‐375. 10.1099/jmm.0.000677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radaic A, Kapila Y. The oralome and its dysbiosis: new insights in oral biofilm host‐microbe interactions. Comput Struct Biotechnol J 2021;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dovrolis N, Kolios G, Spyrou GM, Maroulakou I. Computational profiling of the gut‐brain axis: microflora dysbiosis insights to neurological disorders. Brief Bioinformatics. 2019;20:825‐841. [DOI] [PubMed] [Google Scholar]
- 7.Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Do T, Devine D, Marsh PD. Oral biofilms: molecular analysis, challenges, and future prospects in dental diagnostics. Clin Cosmet Investig Dent. 2013;5:11‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28:397‐403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosier BT, Marsh PD, Mira A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J Dent Res. 2018;97:371‐380. [DOI] [PubMed] [Google Scholar]
- 11.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Méthot P‐O, Alizon S. What is a pathogen? Toward a process view of host‐parasite interactions. Virulence. 2014;5:775‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kilian M, Chapple ILC, Hannig M, et al. The oral microbiome—an update for oral healthcare professionals. Br Dent J. 2016;221:657‐666. [DOI] [PubMed] [Google Scholar]
- 14.Van Dyke TE, Bartold PM, Reynolds EC. The nexus between periodontal inflammation and dysbiosis. Front Immunol. 2020;11:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He J, Li Y, Cao Y, Xue J, Zhou X. The oral microbiome diversity and its relation to human diseases. Folia Microbiol (Praha). 2015;60:69‐80. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W‐L, Wang S‐S, Wang H‐F, Tang Y‐J, Tang Y‐L, Liang X‐H. Who is who in oral cancer? Exp Cell Res. 2019;384:111634. [DOI] [PubMed] [Google Scholar]
- 17.Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22‐38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin JM, Luo T, Kamarajan P, Fenno JC, Rickard AH, Kapila YL. Microbial communities associated with primary and metastatic head and neck squamous cell carcinoma—a high fusobacterial and low streptococcal signature. Sci Rep. 2017;7:9934. 10.1038/s41598-017-09786-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radaic A, Ye C, Parks B, et al. Modulation of pathogenic oral biofilms towards health with nisin probiotic. J Oral Microbiol. 2020;12:1809302. 10.1080/20002297.2020.1809302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hujoel PP, Drangsholt M, Spiekerman C, Weiss NS. An exploration of the periodontitis‐cancer association. Ann Epidemiol. 2003;13:312‐316. [DOI] [PubMed] [Google Scholar]
- 21.Vago R, Collico V, Zuppone S, Prosperi D, Colombo M. Nanoparticle‐mediated delivery of suicide genes in cancer therapy. Pharmacol Res. 2016;111:619‐641. [DOI] [PubMed] [Google Scholar]
- 22.Hassanpour SH, Dehghani M. Review of cancer from perspective of molecular. J Cancer Res Pract. 2017;4:127‐129. Published Online First: July 2017. doi:10.1016/j.jcrpr.2017.07.001. [Google Scholar]
- 23.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7‐30. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Liu Y, Zheng HJ, Zhang CP. The oral microbiota may have influence on oral cancer. Front Cell Infect Microbiol. 2019;9:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero‐Preston R, Godoy‐Vitorino F, Jedlicka A, et al. 16S rRNA amplicon sequencing identifies microbiota associated with oral cancer, human papilloma virus infection and surgical treatment. Oncotarget. 2016;7:51320‐51334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaonkar PP, Patankar SR, Tripathi N, Sridharan G. Oral bacterial flora and oral cancer: the possible link? J Oral Maxillofac Pathol. 2018;22:234‐238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar CR, Sun J, Li Y, et al. Association between selected oral pathogens and gastric precancerous lesions. PLoS One. 2013;8:e51604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Luo J, Dong X, et al. Salivary microbial dysbiosis is associated with systemic inflammatory markers and predicted oral metabolites in non–small cell lung cancer patients. J Cancer. 2019;10:1651‐1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu N, Wang L, Li C, et al. Microbiota dysbiosis in lung cancer: evidence of association and potential mechanisms. Transl Lung Cancer Res. 2020;9:1554‐1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons BW, Durham NM, Bruno TC, et al. A human prostatic bacterial isolate alters the prostatic microenvironment and accelerates prostate cancer progression. J Pathol. 2015;235:478‐489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker JM, Chase DM, Herbst‐Kralovetz MM. Uterine microbiota: residents, tourists, or invaders? Front Immunol. 2018;9:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xuan C, Shamonki JM, Chung A, et al. Microbial dysbiosis is associated with human breast cancer. PLoS One. 2014;9:e83744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söder B, Yakob M, Meurman JH, Andersson LC, Klinge B, Söder P‐Ö. Periodontal disease may associate with breast cancer. Breast Cancer Res Treat. 2011;127:497‐502. [DOI] [PubMed] [Google Scholar]
- 34.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 35.Jethwa AR, Khariwala SS. Tobacco‐related carcinogenesis in head and neck cancer. Cancer Metastasis Rev. 2017;36:411‐423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Global Burden of Disease Cancer Collaboration , Fitzmaurice C, Allen C, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability‐adjusted life‐years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524‐548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.American Cancer Society . 2020 estimated cancer statistics. https://cancerstatisticscenter.cancer.org/#!/. Accessed February 17, 2020. [Google Scholar]
- 38.American Speech‐Language‐Hearing Association . Head and Neck Cancer Incidence. https://www.asha.org/PRPSpecificTopic.aspx?folderid=8589943346§ion=Incidence_and_Prevalence. Accessed February 17, 2020.
- 39.Reichart PA, Nguyen XH. Betel quid chewing, oral cancer and other oral mucosal diseases in Vietnam: a review. J Oral Pathol Med. 2008;37:511‐514. [DOI] [PubMed] [Google Scholar]
- 40.Hashibe M, Brennan P, Chuang S‐C, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18:541‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed AL, Califano J, Cairns P, et al. High frequency of p16 (CDKN2/MTS‐1/INK4A) inactivation in head and neck squamous cell carcinoma. Cancer Res. 1996;56:3630‐3633. [PubMed] [Google Scholar]
- 42.Carlos de Vicente J, Junquera Gutiérrez LM, Zapatero AH, Fresno Forcelledo MF, Hernández‐Vallejo G, López Arranz JS. Prognostic significance of p53 expression in oral squamous cell carcinoma without neck node metastases. Head Neck. 2004;26:22‐30. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi K, Hisamatsu K, Suzui N, Hara A, Tomita H, Miyazaki T. A review of HPV‐related head and neck cancer. J Clin Med. 2018;7:241. 10.3390/jcm7090241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farquhar DR, Divaris K, Mazul AL, Weissler MC, Zevallos JP, Olshan AF. Poor oral health affects survival in head and neck cancer. Oral Oncol. 2017;73:111‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashim D, Sartori S, Brennan P, et al. The role of oral hygiene in head and neck cancer: results from International Head and Neck Cancer Epidemiology (INHANCE) Consortium. Ann Oncol. 2016;27:1619‐1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao M, Yang M, Li X‐M, et al. Tumor‐targeting bacterial therapy with amino acid auxotrophs of GFP‐expressing Salmonella typhimurium . Proc Natl Acad Sci USA. 2005;102:755‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang J, Long L, Maeda H. Enhancement of tumor‐targeted delivery of bacteria with nitroglycerin involving augmentation of the EPR effect. Methods Mol Biol. 2016;1409:9‐23. [DOI] [PubMed] [Google Scholar]
- 48.Malmgren RA, Flanigan CC. Localization of the vegetative form of Clostridium tetani in mouse tumors following intravenous spore administration. Cancer Res. 1955;15:473‐478. [PubMed] [Google Scholar]
- 49.Suzuki N, Yoneda M, Hirofuji T. Mixed red‐complex bacterial infection in periodontitis. Int J Dent. 2013;2013:587279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lai Y, Chu L. Novel mechanism for conditional aerobic growth of the anaerobic bacterium Treponema denticola . Appl Environ Microbiol. 2008;74:73‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGraw WT, Potempa J, Farley D, Travis J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect Immun. 1999;67:3248‐3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou X, Li Y.Subgingival microbes. In: Zhou X, Li Y, eds. Atlas of oral microbiology: from healthy microflora to disease. London, UK: Academic Press; 2015: 67‐93. [Google Scholar]
- 53.Mager DL. Bacteria and cancer: cause, coincidence or cure? A review. J Transl Med. 2006;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang C, Geng F, Shi X, et al. The prevalence rate of periodontal pathogens and its association with oral squamous cell carcinoma. Appl Microbiol Biotechnol. 2019;103:1393‐1404. [DOI] [PubMed] [Google Scholar]
- 55.Guerrero‐Preston R, White JR, Godoy‐Vitorino F, et al. High‐resolution microbiome profiling uncovers Fusobacterium nucleatum, Lactobacillus gasseri/johnsonii, and Lactobacillus vaginalis associated to oral and oropharyngeal cancer in saliva from HPV positive and HPV negative patients treated with surgery and chemo‐radiation. Oncotarget. 2017;8:110931‐110948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geng F, Zhang Y, Lu Z, Zhang S, Pan Y. Fusobacterium nucleatum caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA Cell Biol. 2020;39:144‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang W, Jia Z, Tang D, et al. Fusobacterium nucleatum facilitates apoptosis, ROS generation, and inflammatory cytokine production by activating AKT/MAPK and NF‐κB signaling pathways in human gingival fibroblasts. Oxid Med Cell Longev. 2019;2019:1681972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuboniwa M, Hasegawa Y, Mao S, et al. P. gingivalis accelerates gingival epithelial cell progression through the cell cycle. Microbes Infect. 2008;10:122‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hoppe T, Kraus D, Novak N, et al. Oral pathogens change proliferation properties of oral tumor cells by affecting gene expression of human defensins. Tumour Biol. 2016;37:13789‐13798. [DOI] [PubMed] [Google Scholar]
- 60.de Faria Amormino SA, Arão TC, Saraiva AM, et al. Hypermethylation and low transcription of TLR2 gene in chronic periodontitis. Hum Immunol. 2013;74:1231‐1236. [DOI] [PubMed] [Google Scholar]
- 61.De Oliveira NFP, Andia DC, Planello AC, et al. TLR2 and TLR4 gene promoter methylation status during chronic periodontitis. J Clin Periodontol. 2011;38:975‐983. [DOI] [PubMed] [Google Scholar]
- 62.Benakanakere M, Abdolhosseini M, Hosur K, Finoti LS, Kinane DF. TLR2 promoter hypermethylation creates innate immune dysbiosis. J Dent Res. 2015;94:183‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun Y, Shu R, Li C‐L, Zhang M‐Z. Gram‐negative periodontal bacteria induce the activation of toll‐like receptors 2 and 4, and cytokine production in human periodontal ligament cells. J Periodontol. 2010;81:1488‐1496. [DOI] [PubMed] [Google Scholar]
- 64.Nussbaum G, Ben‐Adi S, Genzler T, Sela M, Rosen G. Involvement of toll‐like receptors 2 and 4 in the innate immune response to Treponema denticola and its outer sheath components. Infect Immun. 2009;77:3939‐3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asai Y, Jinno T, Ogawa T. Oral treponemes and their outer membrane extracts activate human gingival epithelial cells through toll‐like receptor 2. Infect Immun. 2003;71:717‐725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Promsudthi A, Poomsawat S, Limsricharoen W. The role of toll‐like receptor 2 and 4 in gingival tissues of chronic periodontitis subjects with type 2 diabetes. J Periodontal Res. 2014;49:346‐354. [DOI] [PubMed] [Google Scholar]
- 67.Lappin DF, Sherrabeh S, Erridge C. Stimulants of toll‐like receptors 2 and 4 are elevated in saliva of periodontitis patients compared with healthy subjects. J Clin Periodontol. 2011;38:318‐325. [DOI] [PubMed] [Google Scholar]
- 68.Baldwin AS. Regulation of cell death and autophagy by IKK and NF‐κB: critical mechanisms in immune function and cancer. Immunol Rev. 2012;246:327‐345. [DOI] [PubMed] [Google Scholar]
- 69.Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NF‐κB addiction and its role in cancer: “one size does not fit all”. Oncogene. 2011;30:1615‐1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DiDonato JA, Mercurio F, Karin M. NF‐κB and the link between inflammation and cancer. Immunol Rev. 2012;246:379‐400. [DOI] [PubMed] [Google Scholar]
- 71.Kamarajan P, Ateia I, Shin JM, et al. Periodontal pathogens promote cancer aggressivity via TLR/MyD88 triggered activation of Integrin/FAK signaling that is therapeutically reversible by a probiotic bacteriocin. PLoS Pathog. 2020;16:e1008881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang B, Zhao J, Shen S, et al. Listeria monocytogenes promotes tumor growth via tumor cell toll‐like receptor 2 signaling. Cancer Res. 2007;67:4346‐4352. [DOI] [PubMed] [Google Scholar]
- 73.Karin M, Yamamoto Y, Wang QM. The IKK NF‐kappa B system: a treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17‐26. [DOI] [PubMed] [Google Scholar]
- 74.Dashper SG, Seers CA, Tan KH, Reynolds EC. Virulence factors of the oral spirochete Treponema denticola . J Dent Res. 2011;90:691‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chi B, Qi M, Kuramitsu HK. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol. 2003;154:637‐643. [DOI] [PubMed] [Google Scholar]
- 76.Kylmä AK, Jouhi L, Listyarifah D, et al. Treponema denticola chymotrypsin‐like protease as associated with HPV‐negative oropharyngeal squamous cell carcinoma. Br J Cancer. 2018;119:89‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grinde B, Olsen I. The role of viruses in oral disease. J Oral Microbiol. 2010;2:2127. 10.3402/jom.v2i0.2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Imai K, Ochiai K, Okamoto T. Reactivation of latent HIV‐1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J Immunol. 2009;182:3688‐3695. [DOI] [PubMed] [Google Scholar]
- 79.Doolittle JM, Webster‐Cyriaque J. Polymicrobial infection and bacterium‐mediated epigenetic modification of DNA tumor viruses contribute to pathogenesis. MBio. 2014;5:e01015‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ficarra G, Eversole LE. HIV‐related tumors of the oral cavity. Crit Rev Oral Biol Med. 1994;5:159‐185. [DOI] [PubMed] [Google Scholar]
- 81.Kao GD, Devine P, Mirza N. Oral cavity and oropharyngeal tumors in human immunodeficiency virus–positive patients: acute response to radiation therapy. Arch Otolaryngol Head Neck Surg. 1999;125:873‐876. [DOI] [PubMed] [Google Scholar]
- 82.Read NW. Physiological and clinical aspects of short chain fatty acids. Gut. 1996;38:156–157. [Google Scholar]
- 83.Pearson R, Kim YK, Hokello J, et al. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol. 2008;82:12291‐12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han Y, Lassen K, Monie D, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV‐1)–infected individuals carry integrated HIV‐1 genomes within actively transcribed host genes. J Virol. 2004;78:6122‐6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Imai K, Togami H, Okamoto T. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV‐1 latency and its reactivation by BIX01294. J Biol Chem. 2010;285:16538‐16545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198‐209. [DOI] [PubMed] [Google Scholar]
- 87.Kirkpatrick JE, Kirkwood KL, Woster PM. Inhibition of the histone demethylase KDM4B leads to activation of KDM1A, attenuates bacterial‐induced pro‐inflammatory cytokine release, and reduces osteoclastogenesis. Epigenetics. 2018;13:557‐572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fan X, Peters BA, Jacobs EJ, et al. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome. 2018;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar PS, Matthews CR, Joshi V, de Jager M, Aspiras M. Tobacco smoking affects bacterial acquisition and colonization in oral biofilms. Infect Immun. 2011;79:4730‐4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu J, Peters BA, Dominianni C, et al. Cigarette smoking and the oral microbiome in a large study of American adults. ISME J. 2016;10:2435‐2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hayes RB, Ahn J, Fan X, et al. Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 2018;4:358‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lin D, Kouzy R, Abi Jaoude J, et al. Microbiome factors in HPV‐driven carcinogenesis and cancers. PLoS Pathog. 2020;16:e1008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim Y, Fukuma N, Totsika M, Kenny L, Morrison M, Punyadeera C. The performance of an oral microbiome biomarker panel in predicting oral cavity and oropharyngeal cancers. Front Cell Infect Microbiol. 2018;8:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. 2019;18:1–18. 10.1177/1533033819867354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Furquim CP, Soares GMS, Ribeiro LL, et al. The salivary microbiome and oral cancer risk: a pilot study in Fanconi anemia. J Dent Res. 2017;96:292‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pushalkar S, Ji X, Li Y, et al. Comparison of oral microbiota in tumor and non‐tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012;12:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Y, Xiang Q, Zhang Q, Huang Y, Su Z. Overview on the recent study of antimicrobial peptides: origins, functions, relative mechanisms and application. Peptides. 2012;37:207‐215. [DOI] [PubMed] [Google Scholar]
- 98.La Rosa GRM, Gattuso G, Pedullà E, Rapisarda E, Nicolosi D, Salmeri M. Association of oral dysbiosis with oral cancer development. Oncol Lett. 2020;19:3045‐3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moutsopoulos NM, Konkel JE. Tissue‐specific immunity at the oral mucosal barrier. Trends Immunol. 2018;39:276‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khurshid Z, Naseem M, Sheikh Z, Najeeb S, Shahab S, Zafar MS. Oral antimicrobial peptides: types and role in the oral cavity. Saudi Pharm J. 2016;24:515‐524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Takahashi N, Sulijaya B, Yamada‐Hara M, Tsuzuno T, Tabeta K, Yamazaki K. Gingival epithelial barrier: regulation by beneficial and harmful microbes. Tissue Barriers. 2019;7:e1651158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799‐809. [DOI] [PubMed] [Google Scholar]
- 103.Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295:1726‐1729. [DOI] [PubMed] [Google Scholar]
- 104.Pang X, Tang Y‐J, Ren X‐H, Chen Q‐M, Tang Y‐L, Liang X‐H. Microbiota, epithelium, inflammation, and TGF‐β signaling: an intricate interaction in oncogenesis. Front Microbiol. 2018;9:1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Taur Y, Pamer EG. Microbiome mediation of infections in the cancer setting. Genome Med. 2016;8:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takeuchi H, Sasaki N, Yamaga S, Kuboniwa M, Matsusaki M, Amano A. Porphyromonas gingivalis induces penetration of lipopolysaccharide and peptidoglycan through the gingival epithelium via degradation of junctional adhesion molecule 1. PLoS Pathog. 2019;15:e1008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kikuchi Y, Kimizuka R, Kato T, Okuda K, Kokubu E, Ishihara K. Treponema denticola induces epithelial barrier dysfunction in polarized epithelial cells. Bull Tokyo Dent Coll. 2018;59:265‐275. [DOI] [PubMed] [Google Scholar]
- 109.Ji S, Choi YS, Choi Y. Bacterial invasion and persistence: critical events in the pathogenesis of periodontitis? J Periodontal Res. 2015;50:570‐585. [DOI] [PubMed] [Google Scholar]
- 110.Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol 2000 2010;52:68‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Colombo AV, da Silva CM, Haffajee A, Colombo APV. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodontal Res. 2007;42:236‐243. [DOI] [PubMed] [Google Scholar]
- 112.Vitkov L, Krautgartner WD, Hannig M. Bacterial internalization in periodontitis. Oral Microbiol Immunol. 2005;20:317‐321. [DOI] [PubMed] [Google Scholar]
- 113.Rautemaa R, Järvensivu A, Kari K, et al. Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients. Oral Dis. 2004;10:298‐305. [DOI] [PubMed] [Google Scholar]
- 114.Hoare A, Soto C, Rojas‐Celis V, Bravo D. Chronic inflammation as a link between periodontitis and carcinogenesis. Mediators Inflamm. 2019;2019:1029857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Choi J‐I, Seymour GJ. Vaccines against periodontitis: a forward‐looking review. J Periodontal Implant Sci. 2010;40:153‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology (Reading, Engl). 2003;149:279‐294. [DOI] [PubMed] [Google Scholar]
- 117.Socransky SS, Haffajee AD, Smith C, et al. Use of checkerboard DNA‐DNA hybridization to study complex microbial ecosystems. Oral Microbiol Immunol. 2004;19:352‐362. [DOI] [PubMed] [Google Scholar]
- 118.Leonardi R, Talic NF, Loreto C. MMP‐13 (collagenase 3) immunolocalisation during initial orthodontic tooth movement in rats. Acta Histochem. 2007;109:215‐220. [DOI] [PubMed] [Google Scholar]
- 119.Szkaradkiewicz A, Karpiński T. Microbiology of chronic periodontitis. J Biol Earth Sc. 2013;3:M14‐M20. [Google Scholar]
- 120.Multhoff G, Molls M, Radons J. Chronic inflammation in cancer development. Front Immunol. 2011;2:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Katz J, Wallet S, Cha S. Periodontal disease and the oral‐systemic connection: “is it all the RAGE?”. Quintessence Int. 2010;41:229‐237. [PubMed] [Google Scholar]
- 122.Yilmaz O, Verbeke P, Lamont RJ, Ojcius DM. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect Immun. 2006;74:703‐710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brauncajs M, Sakowska D, Krzemiński Z. Production of hydrogen peroxide by lactobacilli colonising the human oral cavity. Med Dosw Mikrobiol. 2001;53:331‐336. [Google Scholar]
- 124.Abranches J, Zeng L, Kajfasz JK, et al. Biology of oral streptococci. Microbiol Spectr. 2018;6(5):1–18. 10.1128/microbiolspec.GPP3-0042-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Karpiński TM. Role of oral microbiota in cancer development. Microorganisms. 2019;7:20. 10.3390/microorganisms7010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bhatt AP, Redinbo MR, Bultman SJ. The role of the microbiome in cancer development and therapy. CA Cancer J Clin. 2017;67:326‐344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Carbonero F, Benefiel AC, Alizadeh‐Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012;3:448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chelombitko MA. Role of reactive oxygen species in inflammation: a minireview. Moscow Univ Biol Sci Bull. 2018;73:199‐202. [Google Scholar]
- 129.Lin K‐Y, Chung C‐H, Ciou J‐S, et al. Molecular damage and responses of oral keratinocyte to hydrogen peroxide. BMC Oral Health. 2019;19:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Piao J‐Y, Lee HG, Kim S‐J, et al. Helicobacter pylori activates IL‐6‐STAT3 signaling in human gastric cancer cells: potential roles for reactive oxygen species. Helicobacter. 2016;21:405‐416. [DOI] [PubMed] [Google Scholar]
- 131.Shirazi MSR, Al‐Alo KZK, Al‐Yasiri MH, Lateef ZM, Ghasemian A. Microbiome dysbiosis and predominant bacterial species as human cancer biomarkers. J Gastrointest Cancer. 2020;51:725–728. 10.1007/s12029-019-00311-z [DOI] [PubMed] [Google Scholar]
- 132.Wagener FADTG, Carels CE, Lundvig DMS. Targeting the redox balance in inflammatory skin conditions. Int J Mol Sci. 2013;14:9126‐9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276‐285. [DOI] [PubMed] [Google Scholar]
- 134.Stornetta A, Guidolin V, Balbo S. Alcohol‐derived acetaldehyde exposure in the oral cavity. Cancers (Basel). 2018;10(1):20–47. 10.3390/cancers10010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. Identification of DNA adducts of acetaldehyde. Chem Res Toxicol. 2000;13:1149‐1157. [DOI] [PubMed] [Google Scholar]
- 136.Vaca CE, Fang JL, Schweda EK. Studies of the reaction of acetaldehyde with deoxynucleosides. Chem Biol Interact. 1995;98:51‐67. [DOI] [PubMed] [Google Scholar]
- 137.Rajalakshmi TR, AravindhaBabu N, Shanmugam KT, Masthan KMK. DNA adducts—chemical addons. J Pharm Bioallied Sci. 2015;7:S197‐S199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Barros SP, Hefni E, Nepomuceno R, Offenbacher S, North K. Targeting epigenetic mechanisms in periodontal diseases. Periodontol 2000. 2018;78:174–184. [DOI] [PubMed] [Google Scholar]
- 139.Larsson L. Current concepts of epigenetics and its role in periodontitis. Curr Oral Health Rep. 2017;4:286‐293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Viana MB, Cardoso FP, Diniz MG, et al. Methylation pattern of IFN‐γ and IL‐10 genes in periodontal tissues. Immunobiology. 2011;216:936‐941. [DOI] [PubMed] [Google Scholar]
- 141.Planello AC, Singhania R, Kron KJ, et al. Pre‐neoplastic epigenetic disruption of transcriptional enhancers in chronic inflammation. Oncotarget. 2016;7:15772‐15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Irimie AI, Ciocan C, Gulei D, et al. Current insights into oral cancer epigenetics. Int J Mol Sci. 2018;19: 10.3390/ijms19030670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kang MK, Mehrazarin S, Park NH, Wang CY. Epigenetic gene regulation by histone demethylases: emerging role in oncogenesis and inflammation. Oral Dis. 2017;23:709‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Martins MD, Jiao Y, Larsson L, et al. Epigenetic modifications of histones in periodontal disease. J Dent Res. 2016;95:215‐222. [DOI] [PubMed] [Google Scholar]
- 145.Cantley MD, Bartold PM, Marino V, et al. Histone deacetylase inhibitors and periodontal bone loss. J Periodontal Res. 2011;46:697‐703. [DOI] [PubMed] [Google Scholar]
- 146.Francis M, Pandya M, Gopinathan G, et al. Histone methylation mechanisms modulate the inflammatory response of periodontal ligament progenitors. Stem Cells Dev. 2019;28:1015‐1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Li S, Liu X, Zhou Y, et al. Shared genetic and epigenetic mechanisms between chronic periodontitis and oral squamous cell carcinoma. Oral Oncol. 2018;86:216‐224. [DOI] [PubMed] [Google Scholar]
- 148.Andia DC, de Oliveira NFP, Casarin RCV, Casati MZ, Line SRP, de Souza AP. DNA methylation status of the IL8 gene promoter in aggressive periodontitis. J Periodontol. 2010;81:1336‐1341. [DOI] [PubMed] [Google Scholar]
- 149.Oliveira NFP, Damm GR, Andia DC, et al. DNA methylation status of the IL8 gene promoter in oral cells of smokers and non‐smokers with chronic periodontitis. J Clin Periodontol. 2009;36:719‐725. [DOI] [PubMed] [Google Scholar]
- 150.Ishida K, Kobayashi T, Ito S, et al. Interleukin‐6 gene promoter methylation in rheumatoid arthritis and chronic periodontitis. J Periodontol. 2012;83:917‐925. [DOI] [PubMed] [Google Scholar]
- 151.Gao D, Mittal V. Tumor microenvironment regulates epithelial‐mesenchymal transitions in metastasis. Expert Rev Anticancer Ther. 2012;12:857‐859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Barros SP, Fahimipour F, Tarran R, et al. Epigenetic reprogramming in periodontal disease: dynamic crosstalk with potential impact in oncogenesis. Periodontol 2000. 2020;82:157–172. [DOI] [PubMed] [Google Scholar]
- 153.Chen L, Deng H, Cui H, et al. Inflammatory responses and inflammation‐associated diseases in organs. Oncotarget. 2018;9:7204‐7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074‐1080. [DOI] [PubMed] [Google Scholar]
- 155.Eccleston A, Cesari F, Skipper M. Transcription and epigenetics. Nature. 2013;502:461. [DOI] [PubMed] [Google Scholar]
- 156.Ateia IM, Sutthiboonyapan P, Kamarajan P, et al. Treponema denticola increases MMP‐2 expression and activation in the periodontium via reversible DNA and histone modifications. Cell Microbiol. 2018;20:e12815 10.1111/cmi.12815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nwizu N, Wactawski‐Wende J, Genco RJ. Periodontal disease and cancer: epidemiologic studies and possible mechanisms. Periodontol 2000. 2020;83:213–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Y, Sun C, Song EJ, et al. Is periodontitis a risk indicator for gastrointestinal cancers? A meta‐analysis of cohort studies. J Clin Periodontol. 2020;47:134‐147. [DOI] [PubMed] [Google Scholar]
- 159.Yin X‐H, Wang Y‐D, Luo H, et al. Association between tooth loss and gastric cancer: a meta‐analysis of observational studies. PLoS One. 2016;11:e0149653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fan X, Alekseyenko AV, Wu J, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population‐based nested case‐control study. Gut. 2018;67:120‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jacob JA. Study links periodontal disease bacteria to pancreatic cancer risk. JAMA. 2016;315:2653‐2654. [DOI] [PubMed] [Google Scholar]
- 162.Michaud DS, Izard J, Wilhelm‐Benartzi CS, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764‐1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lo C‐H, Nguyen LH, Wu K, et al. Periodontal disease, tooth loss, and risk of serrated polyps and conventional adenomas. Cancer Prev Res (Phila Pa). 2020;13:699‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wu WM, Yang YS, Peng LH. Microbiota in the stomach: new insights. J Dig Dis. 2014;15:54‐61. [DOI] [PubMed] [Google Scholar]
- 165.Nardone G, Compare D. The human gastric microbiota: is it time to rethink the pathogenesis of stomach diseases? United European Gastroenterol J. 2015;3:255‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bruno G, Rocco G, Zaccari P, Porowska B, Mascellino MT, Severi C. Helicobacter pylori infection and gastric dysbiosis: can probiotics administration be useful to treat this condition? Can J Infect Dis Med Microbiol. 2018;2018:6237239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yuan X, Liu Y, Kong J, et al. Different frequencies of Porphyromonas gingivalis infection in cancers of the upper digestive tract. Cancer Lett. 2017;404:1‐7. [DOI] [PubMed] [Google Scholar]
- 168.Yamamura K, Baba Y, Miyake K, et al. Fusobacterium nucleatumin gastroenterological cancer: evaluation of measurement methods using quantitative polymerase chain reaction and a literature review. Oncol Lett. 2017;14:6373‐6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Boehm ET, Thon C, Kupcinskas J, et al. Fusobacterium nucleatum is associated with worse prognosis in Lauren’s diffuse type gastric cancer patients. Sci Rep. 2020;10:16240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nieminen MT, Listyarifah D, Hagström J, et al. Treponema denticola chymotrypsin‐like proteinase may contribute to orodigestive carcinogenesis through immunomodulation. Br J Cancer. 2018;118:428‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Thomas RM, Jobin C. Microbiota in pancreatic health and disease: the next frontier in microbiome research. Nat Rev Gastroenterol Hepatol. 2020;17:53‐64. [DOI] [PubMed] [Google Scholar]
- 172.Pushalkar S, Hundeyin M, Daley D, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gnanasekaran J, Binder Gallimidi A, Saba E, et al. Intracellular Porphyromonas gingivalis promotes the tumorigenic behavior of pancreatic carcinoma cells. Cancers (Basel). 2020;12:2331 10.3390/cancers12082331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Öğrendik M. Periodontal pathogens in the etiology of pancreatic cancer. Gastrointest Tumors. 2017;3:125‐127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Evans T, Faircloth M, Deery A, Thomas V, Turner A, Dalgleish A. Analysis of K‐ras gene mutations in human pancreatic cancer cell lines and in bile samples from patients with pancreatic and biliary cancers. Oncol Rep. 1997;4:1373‐1381. [DOI] [PubMed] [Google Scholar]
- 176.Barton CM, Staddon SL, Hughes CM, et al. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br J Cancer. 1991;64:1076‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Prassolov VS, Sakamoto H, Nishimura S, Terada M, Sugimura T. Activation of c‐Ki‐ras gene in human pancreatic cancer. Jpn J Cancer Res. 1985;76:792‐795. [PubMed] [Google Scholar]
- 178.Sun C‐H, Li B‐B, Wang B, et al. The role of Fusobacterium nucleatum in colorectal cancer: from carcinogenesis to clinical management. Chronic Dis Translat Med. 2019;5:178‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299‐306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E‐cadherin/β‐catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor‐immune microenvironment. Cell Host Microbe. 2013;14:207‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gur C, Ibrahim Y, Isaacson B, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Abed J, Emgård JEM, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor‐expressed Gal‐GalNAc. Cell Host Microbe. 2016;20:215‐225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ashare A, Stanford C, Hancock P, et al. Chronic liver disease impairs bacterial clearance in a human model of induced bacteremia. Clin Transl Sci. 2009;2:199‐205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387‐6392. [PubMed] [Google Scholar]
- 186.Zeng X‐T, Xia L‐Y, Zhang Y‐G, Li S, Leng W‐D, Kwong JSW. Periodontal disease and incident lung cancer risk: a meta‐analysis of cohort studies. J Periodontol. 2016;87:1158‐1164. [DOI] [PubMed] [Google Scholar]
- 187.Yan X, Yang M, Liu J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. 2015;5:3111‐3122. [PMC free article] [PubMed] [Google Scholar]
- 188.Pu CY, Seshadri M, Manuballa S, Yendamuri S. The oral microbiome and lung diseases. Curr Oral Health Rep. 2020;7:79‐86. [Google Scholar]
- 189.Yu G, Gail MH, Consonni D, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 2016;17:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bassis CM, Erb‐Downward JR, Dickson RP, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015;6:e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang D, Xing Y, Song X, Qian Y. The impact of lung microbiota dysbiosis on inflammation. Immunology. 2020;159:156‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Morris A, Beck JM, Schloss PD, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067‐1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Berger G, Wunderink RG. Lung microbiota: genuine or artifact? Isr Med Assoc J. 2013;15:731‐733. [PubMed] [Google Scholar]
- 194.Michaud DS, Liu Y, Meyer M, Giovannucci E, Joshipura K. Periodontal disease, tooth loss, and cancer risk in male health professionals: a prospective cohort study. Lancet Oncol. 2008;9:550‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hiraki A, Matsuo K, Suzuki T, Kawase T, Tajima K. Teeth loss and risk of cancer at 14 common sites in Japanese. Cancer Epidemiol Biomarkers Prev. 2008;17:1222‐1227. [DOI] [PubMed] [Google Scholar]
- 196.da Silva APB, Alluri LSC, Bissada NF, Gupta S. Association between oral pathogens and prostate cancer: building the relationship. Am J Clin Exp Urol. 2019;7:1‐10. [PMC free article] [PubMed] [Google Scholar]
- 197.Estemalik J, Demko C, Bissada NF, et al. Simultaneous detection of oral pathogens in subgingival plaque and prostatic fluid of men with periodontal and prostatic diseases. J Periodontol. 2017;88:823‐829. [DOI] [PubMed] [Google Scholar]
- 198.Nabil F, Bissada NA. Periodontal treatment improves prostate symptoms and lowers serum PSA in men with high PSA and chronic periodontitis. Dentistry. 2015;05(3):1–4. 10.4172/2161-1122.1000284 [DOI] [Google Scholar]
- 199.Joshi N, Bissada NF, Bodner D, et al. Association between periodontal disease and prostate‐specific antigen levels in chronic prostatitis patients. J Periodontol. 2010;81:864‐869. [DOI] [PubMed] [Google Scholar]
- 200.Dennis LK, Lynch CF, Torner JC. Epidemiologic association between prostatitis and prostate cancer. Urology. 2002;60:78‐83. [DOI] [PubMed] [Google Scholar]
- 201.Stark T, Livas L, Kyprianou N. Inflammation in prostate cancer progression and therapeutic targeting. Transl Androl Urol. 2015;4:455‐463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sfreddo CS, Maier J, De David SC, Susin C, Moreira CHC. Periodontitis and breast cancer: a case‐control study. Community Dent Oral Epidemiol. 2017;45:545‐551. [DOI] [PubMed] [Google Scholar]
- 203.Shao J, Wu L, Leng W‐D, et al. Periodontal disease and breast cancer: a meta‐analysis of 1,73,162 participants. Front Oncol. 2018;8:601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mai X, LaMonte MJ, Hovey KM, et al. Periodontal disease severity and cancer risk in postmenopausal women: the Buffalo OsteoPerio Study. Cancer Causes Control. 2016;27:217‐228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mai X, Genco RJ, LaMonte MJ, et al. Periodontal pathogens and risk of incident cancer in postmenopausal females: the Buffalo OsteoPerio study. J Periodontol. 2016;87:257‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Parhi L, Alon‐Maimon T, Sol A, et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. 2020;11:3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Arora M, Weuve J, Fall K, Pedersen NL, Mucci LA. An exploration of shared genetic risk factors between periodontal disease and cancers: a prospective co‐twin study. Am J Epidemiol. 2010;171:253‐259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun. 2004;72:2272‐2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fischer LA, Demerath E, Bittner‐Eddy P, Costalonga M. Placental colonization with periodontal pathogens: the potential missing link. Am J Obstet Gynecol. 2019;221:383‐392.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Tuominen H, Rautava S, Syrjänen S, Collado MC, Rautava J. HPV infection and bacterial microbiota in the placenta, uterine cervix and oral mucosa. Sci Rep. 2018;8:9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chim SSC, Shing TKF, Hung ECW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482‐490. [DOI] [PubMed] [Google Scholar]
- 213.Gao L, Kang M, Zhang MJ, et al. Polymicrobial periodontal disease triggers a wide radius of effect and unique virome. NPJ Biofilms Microbiomes. 2020;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Radaic A, de Jesus MB, Kapila YL. Bacterial anti‐microbial peptides and nano‐sized drug delivery systems: the state of the art toward improved bacteriocins. J Control Release. 2020;321:100‐118. 10.1016/j.jconrel.2020.02.001 [DOI] [PubMed] [Google Scholar]
- 215.Tornesello AL, Borrelli A, Buonaguro L, Buonaguro FM, Tornesello ML. Antimicrobial peptides as anticancer agents: functional properties and biological activities. Molecules. 2020;25:2850. 10.3390/molecules25122850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shin JM, Ateia I, Paulus JR, et al. Antimicrobial nisin acts against saliva derived multi‐species biofilms without cytotoxicity to human oral cells. Front Microbiol. 2015;6:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Joo NE, Ritchie K, Kamarajan P, Miao D, Kapila YL. Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC1. Cancer Med. 2012;1:295‐305. 10.1002/cam4.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kamarajan P, Hayami T, Matte B, et al. Nisin ZP, a bacteriocin and food preservative, inhibits head and neck cancer tumorigenesis and prolongs survival. PLoS One. 2015;10:e0131008. 10.1371/journal.pone.0131008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shin JM, Gwak JW, Kamarajan P, Fenno JC, Rickard AH, Kapila YL. Biomedical applications of nisin. J Appl Microbiol. 2016;120:1449‐1465. 10.1111/jam.13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Baker MA, Maloy WL, Zasloff M, Jacob LS. Anticancer efficacy of Magainin2 and analogue peptides. Cancer Res. 1993;53:3052‐3057. [PubMed] [Google Scholar]
- 221.Lehmann J, Retz M, Sidhu SS, et al. Antitumor activity of the antimicrobial peptide magainin II against bladder cancer cell lines. Eur Urol. 2006;50:141‐147. [DOI] [PubMed] [Google Scholar]
- 222.Mader JS, Mookherjee N, Hancock REW, Bleackley RC. The human host defense peptide LL‐37 induces apoptosis in a calpain and apoptosis‐inducing factor‐dependent manner involving Bax activity. Mol Cancer Res. 2009;7:689‐702. [DOI] [PubMed] [Google Scholar]
- 223.Okumura K, Itoh A, Isogai E, et al. C‐terminal domain of human CAP18 antimicrobial peptide induces apoptosis in oral squamous cell carcinoma SAS‐H1 cells. Cancer Lett. 2004;212:185‐194. [DOI] [PubMed] [Google Scholar]
- 224.Ren SX, Cheng ASL, To KF, et al. Host immune defense peptide LL‐37 activates caspase‐independent apoptosis and suppresses colon cancer. Cancer Res. 2012;72:6512‐6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Bevers EM, Comfurius P, Zwaal RF. Regulatory mechanisms in maintenance and modulation of transmembrane lipid asymmetry: pathophysiological implications. Lupus. 1996;5:480‐487. [DOI] [PubMed] [Google Scholar]
- 226.Wodlej C, Riedl S, Rinner B, et al. Interaction of two antitumor peptides with membrane lipids—influence of phosphatidylserine and cholesterol on specificity for melanoma cells. PLoS One. 2019;14:e0211187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ran S, Downes A, Thorpe PE. Increased exposure of anionic phospholipids on the surface of tumor blood vessels. Cancer Res. 2002;62:6132‐6140. [PubMed] [Google Scholar]
- 228.Beloribi‐Djefaflia S, Vasseur S, Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lei J, Sun L, Huang S, et al. The antimicrobial peptides and their potential clinical applications. Am J Transl Res. 2019;11:3919‐3931. [PMC free article] [PubMed] [Google Scholar]
- 230.Scocchi M, Mardirossian M, Runti G, Benincasa M. Non–membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr Top Med Chem. 2016;16:76‐88. [DOI] [PubMed] [Google Scholar]
- 231.Gkeka P, Sarkisov L. Interactions of phospholipid bilayers with several classes of amphiphilic alpha‐helical peptides: insights from coarse‐grained molecular dynamics simulations. J Phys Chem B. 2010;114:826‐839. [DOI] [PubMed] [Google Scholar]
- 232.Radaic A, Matins‐de‐Souza D. The state of the art of nanopsychiatry for schizophrenia diagnostics and treatment. Nanomedicine. 2020;28:102222. 10.1016/j.nano.2020.102222 [DOI] [PubMed] [Google Scholar]
- 233.Radaic A, de Jesus MB. Solid lipid nanoparticles release DNA upon endosomal acidification in human embryonic kidney cells. Nanotechnology. 2018;29:315102. 10.1088/1361-6528/aac447 [DOI] [PubMed] [Google Scholar]
- 234.Radaic A, Barbosa LRS, Jaime C, Kapila YL, Pessine FBT, de Jesus MB. How lipid cores affect lipid nanoparticles as drug and gene delivery systems. In: Iglič A, Kulkarni CV, Rappolt M, eds. Advances in Biomembranes and Lipid Self‐Assembly, Vol. 24. Cambridge, MA: Academic Press; 2016:1–42. 10.1016/bs.abl.2016.04.001 [DOI] [Google Scholar]
- 235.Goudarzi F, Asadi A, Afsharpour M, Jamadi RH. In vitro characterization and evaluation of the cytotoxicity effects of nisin and nisin‐loaded PLA‐PEG‐PLA nanoparticles on gastrointestinal (AGS and KYSE‐30), hepatic (HepG2) and blood (K562) cancer cell lines. AAPS PharmSciTech. 2018;19:1554‐1566. [DOI] [PubMed] [Google Scholar]
- 236.Niemirowicz K, Prokop I, Wilczewska AZ, et al. Magnetic nanoparticles enhance the anticancer activity of cathelicidin LL‐37 peptide against colon cancer cells. Int J Nanomedicine. 2015;10:3843‐3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lilly DM, Stillwell RH. Probiotics: growth‐promoting factors produced by microorganisms. Science. 1965;147:747‐748. [DOI] [PubMed] [Google Scholar]
- 238.Guarner F, Schaafsma GJ. Probiotics. Int J Food Microbiol. 1998;39:237‐238. [DOI] [PubMed] [Google Scholar]
- 239.Morelli L, Capurso L. FAO/WHO guidelines on probiotics: 10 years later. J Clin Gastroenterol. 2012;46(Suppl):S1‐2. [DOI] [PubMed] [Google Scholar]
- 240.Nguyen T, Brody H, Lin G‐H, et al. Probiotics, including nisin‐based probiotics, improve clinical and microbial outcomes relevant to oral and systemic diseases. Periodontol 2000. 2020;82:173–185. [DOI] [PubMed] [Google Scholar]
- 241.Górska A, Przystupski D, Niemczura MJ, Kulbacka J. Probiotic bacteria: a promising tool in cancer prevention and therapy. Curr Microbiol. 2019;76:939‐949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Thirabunyanon M, Hongwittayakorn P. Potential probiotic lactic acid bacteria of human origin induce antiproliferation of colon cancer cells via synergic actions in adhesion to cancer cells and short‐chain fatty acid bioproduction. Appl Biochem Biotechnol. 2013;169:511‐525. [DOI] [PubMed] [Google Scholar]
- 243.Appleyard CB, Cruz ML, Isidro AA, Arthur JC, Jobin C, De Simone C. Pretreatment with the probiotic VSL#3 delays transition from inflammation to dysplasia in a rat model of colitis‐associated cancer. Am J Physiol Gastrointest Liver Physiol. 2011;301:G1004‐G1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chapman TM, Plosker GL, Figgitt DP. VSL#3 probiotic mixture: a review of its use in chronic inflammatory bowel diseases. Drugs. 2006;66:1371‐1387. [DOI] [PubMed] [Google Scholar]
- 245.Zheng D‐W, Li R‐Q, An J‐X, et al. Prebiotics‐encapsulated probiotic spores regulate gut microbiota and suppress colon cancer. Adv Mater. 2020:32:e2004529. [DOI] [PubMed] [Google Scholar]
- 246.Javanmard A, Ashtari S, Sabet B, et al. Probiotics and their role in gastrointestinal cancers prevention and treatment; an overview. Gastroenterol Hepatol Bed Bench. 2018;11:284‐295. [PMC free article] [PubMed] [Google Scholar]
- 247.Maksylewicz A, Bysiek A, Lagosz KB, et al. BET bromodomain inhibitors suppress inflammatory activation of gingival fibroblasts and epithelial cells from periodontitis patients. Front Immunol. 2019;10:933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457‐463. [DOI] [PubMed] [Google Scholar]
- 249.Meng S, Zhang L, Tang Y, et al. BET inhibitor JQ1 blocks inflammation and bone destruction. J Dent Res. 2014;93:657‐662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Ivanov M, Barragan I, Ingelman‐Sundberg M. Epigenetic mechanisms of importance for drug treatment. Trends Pharmacol Sci. 2014;35:384‐396. [DOI] [PubMed] [Google Scholar]
- 251.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067‐1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Belkina AC, Denis GV. BET domain co‐regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12:465‐477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Nicodeme E, Jeffrey KL, Schaefer U, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Chan CH, Fang C, Yarilina A, Prinjha RK, Qiao Y, Ivashkiv LB. BET bromodomain inhibition suppresses transcriptional responses to cytokine‐Jak‐STAT signaling in a gene‐specific manner in human monocytes. Eur J Immunol. 2015;45:287‐297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Bandukwala HS, Gagnon J, Togher S, et al. Selective inhibition of CD4+ T‐cell cytokine production and autoimmunity by BET protein and c‐Myc inhibitors. Proc Natl Acad Sci USA. 2012;109:14532‐14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sun Y, Wang Y, Toubai T, et al. BET bromodomain inhibition suppresses graft‐versus‐host disease after allogeneic bone marrow transplantation in mice. Blood. 2015;125:2724‐2728. [DOI] [PMC free article] [PubMed] [Google Scholar]


