Abstract
Background
In Nigeria, decline in the sensitivity of Plasmodium falciparum to Artemisinin Combination Therapy (ACT) has prompted the unofficial use of chloroquine (CQ) for self-medication. This study was designed to determine the prevalence and distribution of CQ resistant/susceptible alleles of CQ resistance transporter (Pfcrt) and P. falciparum multidrug resistance gene 1 (Pfmdr1) in view of the possible re-introduction of CQ for malaria treatment.
Materials and methods
Four hundred and sixty six (466) P. falciparum positive samples were randomly collected from five states of northwest Nigeria. The samples were amplified using RT- PCR at codon 76 for Pfcrt and codon 86 for Pfmdr1. Data was analysed using chi-square, odds ratios and paired t-tests.
Results
Drug susceptible alleles (N86) were most prevalent in the study population (47.9%; 223/466), followed by the drug resistance alleles 86Y (28.3%; 132/466), followed by the drug susceptible alleles K76 (17.4%; 81/466), the resistant alleles 76T (12.4%; 58/466) and finally the mixed infection mutation K76T (3.6%; 17/466). Differences between the distributions of the Pfmdr1 and Pfcrt alleles were significant (P<0.05). There were significant differences (P<0.05) between N86 and 86Y alleles, but no significant differences between K76 and 76T alleles, including the prevalence of the various alleles across the different age groups.
Conclusion
The results of this study suggest the possibility of (re)introducing CQ for malaria treatment in north-western Nigeria and provide insight in the genetic background of P. falciparum in the study area.
1 Introduction
Malaria remains a major health problem in Sub-Saharan Africa [1] and the world’s most important parasitic infection, ranking among the major health and developmental challenges for developing countries [2,3]. Although there are encouraging reports that malaria morbidity and mortality are declining, there are globally still an estimated 207 million cases and 627 thousand deaths every year. The entire Nigerian population of >170 million is at risk of malaria, which is responsible for about 60% and 30% of outpatient visits and hospital admissions, respectively [4]. Emergence and spreading of clones of P. falciparum resistant to the most commonly available antimalarial drugs hinders effective control of the disease. Chloroquine (CQ) was the first antimalarial to be widely used in endemic countries, including Nigeria. Resistance to it was first documented in Thailand in the late 1950’s, and spread to Africa in the 1970s. It surfaced in Nigeria in the early 1980’s and continued to spread until 2005 when CQ was withdrawn. The Nigerian government subsequently changed its first line drug to the more expensive Artemisinin based Combination Therapy (ACT) and recommended that all fevers be treated presumptively with ACTs where confirmation could not be obtained [5]. However, since the detection of P. falciparum resistant to ACT in western Cambodia, malaria control and elimination efforts have become seriously threatened [6].
In different parts of Nigeria, P. falciparum resistance to CQ has prompted studies in the last decade [7-10]. While high resistance to CQ was reported in south-eastern Nigeria [8], most strains of P. falciparum were found to be fully sensitive to CQ in north-eastern Nigeria [7]. Erah et al. [11] showed that CQ is still effective in the treatment of uncomplicated malaria in Delta state.
Chloroquine resistant P. falciparum malaria is caused by mutations in two genes, the P. falciparum CQ resistance transporter (Pfcrt) and multidrug resistance transporter-1 (Pfmdr1), which are both located on the food vacuole of the parasite. While wild-type Pfmdr1 is thought to transport and accumulate CQ into the parasite food vacuole, mutations N86Y, S1034C, N1042D, and D1246Y abolish this transport, leading to reduced CQ sensitivity [12]. On the other hand, the CQ transporter Pfcrt is a stronger predictor of CQ resistance than Pfmdr1. Mutations on the Pfcrt K76T is directly linked with both in vitro and clinical resistance and is thus used as a biomarker of CQ resistance [13]. As polymorphisms of the mutations have also been linked to resistance or sensitivity to other antimalarial drugs, the withdrawal of drugs may have resulted in the re-emergence of sensitive genes as has been documented following CQ withdrawal. In Malawi, parasites carrying the CQ sensitive pfcrt 76, with 100% clinical efficacy, was reported just eight years after abandoning its use [14]. Two studies in Tanzania reported restoration of CQ-sensitive pfcrt 76 from 17.1% to 50.7% in five years [15] and from 48% to 89.6% in seven years [16]. In Kenya the restoration was slower than in Malawi and Tanzania, rising from 5% to 40% in 13 years [17].
This present study investigated the prevalence and distribution of pfcrt and pfmdr1 susceptible and resistance alleles in clinical isolates from five states of north-western Nigeria, for possible re-introduction of CQ for malaria treatment in that part of the country.
2 Materials and methods
2.1 Study area
The study was conducted in randomly selected States of north-western Nigeria (Figure 1). Mosquito breeding sites abound in the region due to farming, industrial and other human activities contributing to larval growth and development [18]. Malaria is meso- to hyper-endemic in the States and is seasonally transmitted, with the main peak of transmission from early June to late August and a minor peak from early October to mid November. These transmission periods correspond with rainfall patterns and resulting high populations of mosquitoes and intense P. falciparum transmission [19].
Figure 1.
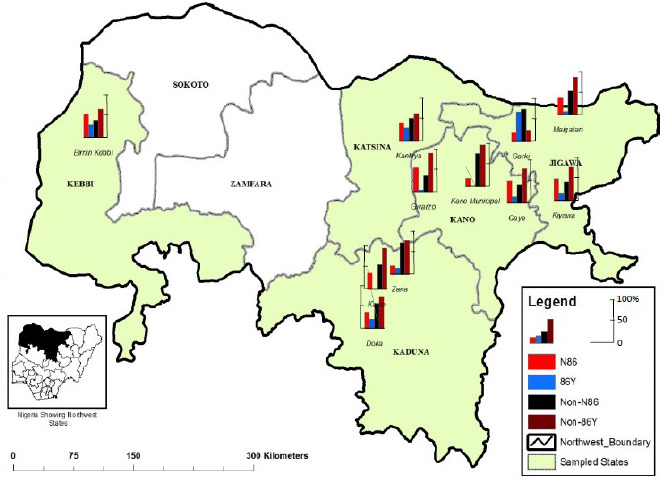
Distribution of P. falciparum pfmdr1 alleles in five states of northwest Nigeria. Map source: Cartography Laboratory, Bayero University, Kano. For data see also Supplementary file 1.
2.2 Blood collection and sample handling
For P. falciparum screening, 2-5 ml of venous blood was collected from hospital patients by clinicians or medical laboratory scientists after obtaining the patient’s consent and demographics. Blood was stored in sample bottles containing EDTA as anti-coagulant. Both thin and thick smears of each sample were Giemsa stained for microscopic analysis. Four hundred and sixty six (466) P. falciparum-positive samples were blotted on Whatman filter paper (24 cm) in quadruplets. These were allowed to dry and stored in a separate clean envelope for molecular analysis. A sterile paper punch (6 mm) was used to cut out the dry blood spots (DBS). DNA was extracted from the DBS using a modified Qiagen protocol (QIAamp) DNA blood kit (Qiagen, Hilden, Germany).
2.3 Real time polymerase chain reaction (RT-PCR)
RT-PCR was used to determine the susceptible and resistant alleles of Pfcrt at codon K76T and Pfmdr1 at codon N86Y in positive samples using specific primers adopted from the work of Ojurongbe et al. [20], and designed by Sysmex UK Ltd. The primers and probes used are shown in Table 1.
Table 1.
Primers, probes and melting temperatures for P. falciparum chloroquine resistance transporter (pfcrt) and P. falciparum multidrug resistance gene 1 (pfmdr1).
| Primers and probes used | Melting temperatures (°C) | |
|---|---|---|
| Pfcrt | F: 5’-CTTGTCTTGGTAAATGTGCTCA-3’ | Wild: 65.3 ± 0.4 |
| R:5’-GTTACCAATTTTGTTTAAAGTTCT-3’ | Mutant: 46.5 ± 0.2 | |
| SensorProbe 76: | ||
| SPTGTGTAATTGAAACAATTTTTGCTAA-3 | ||
| Pfmdr1 | F:5’-TGTATTATCAGGAGGAACATTACC-3’ | Wild: 51.8 ± 0.3 |
| R:5’- ACCACCAAACATAAATTAACGGA-3’ | Mutant: 56.5 ± 0.2 | |
| SensorProbe86: | ||
| ATTAATATCATCATAAATACATG 51 | ||
| AnchorProbe86: | ||
| TCTTTAATATTACACCAAACACAGATAT |
The samples were analysed for pfcrt alleles at codon 76 (lysine to threonine). The sensor probe labelled with fluorescein at the 3' end was designed to be perfectly complementary to the mutation site. An amplification primer iLC labelled with Cy5 on the third base from the 3' end was used as a reverse primer, which is extended during amplification. During fluorescence Resonance Energy Transfer (FRET), fluorescein, which is excited by the light source of the Rotor Gene instrument transfers its energy to the Cy5 incorporated into the PCR product working as anchor probe [21,22]. A specific melting temperature is then obtained for each genotype: a sensor probe spanning one mismatch could still hybridise to the target sequence but will melt off at lower temperature than a sensor probe with a perfect match [20].
The samples were also analysed for the pfmdr1 alleles at codon 86 (asparagine to tyrosine). Pfmdr1 allele’s hybridisation probes consisted of two different oligonucleotides that bind to an internal sequence amplified by forward and reverse primers. The sensor probe, labelled at the 3' end with FAM, is designed to match the mutation sites. The anchor probe, labelled at the 5' end with Cy5 and phosphorylated at the 3' end, to prevent extension by Taq polymerase, is designed to conserve sequences adjacent to the mutation sites. Both probes, localised on the same DNA strand, could hybridise in a head to tail arrangement, bringing the two fluorescent dyes into close proximity. During Fluorescence Resonance Energy Transfer (FRET), FAM was excited by the light source of the Rotor Gene instrument. The excitation energy was transferred to the acceptor fluorophore, Cy5, and the emitted fluorescence was measured continuously on the Rotor Gene channel during the melting phase.
2.4 Ethical clearance
Scientific and Ethical permit/clearance was obtained from the Ministries of Health/Hospital Management Board (MOH/HMB) of Kano, Kaduna, Katsina, Kebbi, and Jigawa States before commencing the research. Written/informed consent was obtained from patients prior to recruitment into the study. Consent from children was provided by the parents/guardians.
3 Results
A total of four hundred and sixty six (466) P. falciparum-positive samples were amplified at codon 76 for the pfcrt gene and at codon 86 for the pfmdr1gene. Drug susceptible alleles (N86) were more prevalent (47.9%; 223/466) in the study population, followed by the drug resistance alleles 86Y(28.3%; 132/466), followed by the drug susceptible K76 (17.4%; 81/466), the resistant 76T (12.4%; 58/466) and finally the mixed infection mutation K76T(3.6%; 17/466). There was a significant difference (P≤0.05) between the geographical distributions of the Pfmdr1 and the Pfcrt alleles (Table 2).
Table 2.
Geographical distribution of pfcrt and pfmdr1 alleles in the study population.
| State | # Examined | Pfcrt | Pfmdr1 | |||
|---|---|---|---|---|---|---|
| K76 | 76T | K76T | N86 | 86Y | ||
| Kaduna | 53 | 17 (32.1) | 10 (18.9) | 3 (5.67) | 18 (34.0) | 10 (18.9) |
| Jigawa | 43 | 8 (18.6) | 11 (25.6) | 5 (11.6) | 17 (39.5) | 14 (32.6) |
| Kano | 53 | 13 (24.5) | 9 (17.0) | 1 (1.9) | 28 (52.8) | 6 (11.3) |
| Kebbi | 135 | 10 (7.4) | 7 (5.2) | 2 (1.5) | 78 (57.8) | 42 (31.1) |
| Katsina | 182 | 33 (18.1) | 21 (11.5) | 6 (3.3) | 82 (45.1) | 60 (33.0) |
| Total | 466 | 385 (82.6) | 58 (12.4) | 17 (3.6) | 223 (47.9) | 132 (28.3) |
| Chi-square | 19.3 | 16.5 | 6.3 | 11.7 | 12.7 | |
| df | 4 | 4 | 4 | 4 | 4 | |
| p value | 0.001 | 0.002 | 0.181ns | 0.020 | 0.013 | |
Based on gender both K76 and 76T were more frequently found in females (18.8%; 52/276, and 12.4%; 58/276, respectively). The mixed mutation infection K76T was more common in males (4.2%; 8/190). The results also showed that both N86 and 86Y were more commonly found in males (47.9 %; 91/190 and 32.1%; 61/190, respectively). Overall there was no significant difference between the sexes as well as between the alleles (Table 3).
Table 3.
Distribution of pfcrt and pfmdr1 alleles according to gender.
| Gender | # Examined | Pfcrt | Pfmdr1 | |||
|---|---|---|---|---|---|---|
| K76 | 76T | K76T | N86 | 86Y | ||
| Male | 190 | 29 (15.3) | 22 (11.6) | 8 (4.2) | 91 (47.9) | 61 (32.1) |
| Female | 276 | 52 (18.8) | 36 (13.0) | 9 (3.3) | 132 (47.8) | 71 (25.7) |
| Total | 466 | 81 (17.4) | 58 (12.4) | 17 (3.6) | 223 (47.9) | 132 (28.3) |
| Chi-square | 1.00 | 0.22 | 0.29 | 0.000 | 2.26 | |
| df | 1 | 1 | 1 | 1 | 1 | |
| p value | 0.32ns | 0.64ns | 0.59ns | 0.99ns | 0.13ns | |
| Odds ratio | 0.78 | 0.87 | 1.30 | 1.00 | 1.37 | |
| 95% CI | 0.47-1.27 | 0.49-1.54 | 0.49-3.44 | 0.69-1.45 | 0.91-2.05 | |
Age wise, K76 was found more often in the age group 6-15 yrs (21.8%; 52/78), 76T was highest in the age group 26-40 yrs (17.0%; 17/100) while the mixed mutation infection (K76T) was higher in the 16-25 yrs age group 6.9% (6/87). Also N86 was higher among the age group of > 40 yrs of age (52.8%; 28/53) while 86Y was higher among the age group 1-5 yrs (31.1%; 46/148). But like for gender, there was no significant difference between the ages the alleles (Table 4).
Table 4.
Distribution of pfcrt and pfmdr1 alleles according to age.
| Age (yrs) | # Examined | Pfcrt | Pfmdr1 | |||
|---|---|---|---|---|---|---|
| K76 | 76T | K76T | N86 | 86Y | ||
| 1-5 | 148 | 25 (16.9) | 16 (10.8) | 4 (2.7) | 70 (47.3) | 46 (31.1) |
| 6-15 | 78 | 17 (21.8) | 11 (14.1) | 3 (3.8) | 36 (46.2) | 22 (28.2) |
| 16-25 | 87 | 17 (19.5) | 13 (14.9) | 6 (6.9) | 40 (46.0) | 24 (27.6) |
| 26-40 | 100 | 13 (13.0) | 17 (17.0) | 3 (3.0) | 49 (49.0) | 24 (24.0) |
| >40 | 53 | 9 (17.0) | 1 (1.9) | 1 (1.9) | 28 (52.8) | 16 (30.2) |
| Total | 466 | 81 (17.4) | 58 (12.4) | 17 (3.6) | 223 (47.9) | 132 (28.3) |
| Chi square | 2.708 | 8.383 | 3.584 | 0.81 | 1.590 | |
| df | 4 | 4 | 4 | 4 | 4 | |
| p value | 0.61ns | 0.08ns | 0.47ns | 0.94ns | 0.81ns | |
Both the drug resistance and susceptible alleles of the pfmdr1 and pfcrt genes were widely distributed in the study area. Pfmdr1 drug susceptible alleles (N86) were most prevalent in Kebbi State (57.8%; 78/135) and least so in Kaduna State (34.0%; 18/53). The pfmdr1drug resistance alleles were most commonly found in Katsina State (33.0%; 60/182) and least so in Kano State (11.3%; 6/53) (Table 2 and Figure 1; data shown in Supplementary file 1).
Pfcrt drug susceptible alleles K76 were most prevalent in Kaduna State (32.1%; 17/53), and least so in Kebbi State (7.4%; 10/135). The pfcrt drug resistance alleles 76T were more common in Jigawa State (25.6%; 11/43) and least common in Kebbi State (5.2%; 7/135) (Table 2). K76T mixed infection mutation were most prevalent in the following Jigawa State (11.6%; 5/43) and again least so in Kebbi State (1.5%; 2/135) (Table 2 and Figure 2).
Figure 2.
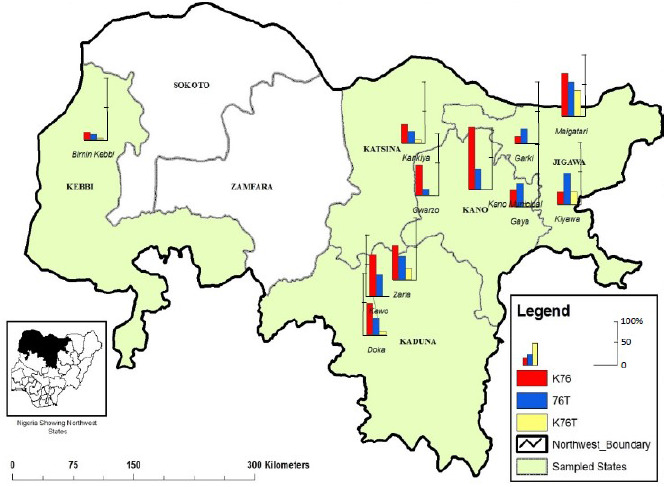
Distribution of P. falciparum pfcrt alleles in five states of northwest Nigeria. Map source: Cartography Laboratory, Bayero University Kano. For data see also Supplementary file 1.
4 Discussion
The high prevalence of malaria in most parts of Nigeria, coupled with the sole reliance on ACT treatment and the withdrawal of CQ since 2005, could have resulted in the selection and propagation of ACT resistance and re-emergence of CQ susceptible alleles. This study also showed that the CQ susceptible alleles of the two genes pfmdr1and pfcrt were prevalent (47.9% and 17.4%, respectively) in the study population, which could be the result of withdrawal of CQ for malaria treatment or drug pressure that is mounting on artemether-lumefantrine (AL). It further suggests that CQ can be used for malaria treatment since pfcrt, K76, and pfmdr1, N86, do not confer resistance to CQ [23,24].
The occurrence of mutant parasites with pfcrt T76 (28.3%) and pfmdr1Y86 (12.4%) is a threat to some of the ACTs in use, suggesting drug pressure. This was shown in an in vitro study conducted in southeast Nigeria, which demonstrated an association between the T76 mutation and decreased susceptibility to artemether [25]. In addition, an increasing trend for K76 will create a future problem for ACT use because it has been seen in recrudescent samples after AL use [26]. The mixed mutation, pfcrt K76T, is directly linked with both in vitro and clinical resistance and is thus used as a biomarker of CQ resistance [13]. Another report, from Ibadan, Nigeria, suggested an association between the pfcrt T76 and pfmdr1 Y86 alleles in CQ-resistant isolates [27].
The fact that the drug resistance alleles 86Y were higher in males (32.1%) than in females (25.7%) implies that males stand a greater chance of developing drug resistance than females. Also the higher occurrences of the drug susceptible alleles N86 in males suggest that CQ may be used to cure the disease in that gender. Moreover the higher presence of pfcrt K76T mixed infection alleles in males (4.2%) further buttressed the fact above that males are more likely to develop drug resistance because the presence of K76T mutation is pre-condition for the parasite to develop multidrug resistance property against CQ [28].
Both pfmdr drug susceptible alleles N86 and 86Y were prevalent across the various age groups with no clear demarcation. This may be caused by the withdrawal of CQ for malaria treatment or it could be due to a mounting pressure on AL or both. Also the pfcrt drug susceptible alleles K76 and drug resistant alleles 76T and K76T mixed infections alleles were higher among different age groups, suggesting similar pressure on the drugs. These results further suggest possible drug pressure among the age groups that resulted in declined drug susceptibility or that the effect of CQ withdrawal for about a decade resulted in the CQ susceptibility increase. Although there are no previously documented data in the study area to support these findings, studies have shown that removal of CQ for the treatment of P. falciparum or the pressure from AL that has been used eventually lead to replacement of pfmdr1 resistance genes by susceptible parasite populations [29,30]. The findings are consistent with observations from Malawi [29], Kenya [30] and Tanzania [31], where the withdrawal of CQ resulted in the rapid spread of a CQ susceptible pfcrt K76 population. In Malawi, recovery of the susceptible pfcrt K76 from <15% to 100% within 13 years of CQ withdrawal was reported by Mang’era et al. [29].
The high distribution level of both drug susceptible and drug resistance alleles observed in this study showed that either CQ remained widely used at the community level even 10 years after its official withdrawal or that drug pressure is mounting on AL, or both. This might have been caused by poor education of drug retailers during the change over period, or simply due high cost of ACT, that made patients to continue using the much cheaper CQ. As there is no documented record, it is difficult to determine whether the CQ-susceptible resurgence is due to back mutations in the CQ-resistant alleles or the expansion of surviving CQ-susceptible reservoir populations. However re-expansion appears to be more common in Africa, where transmission rates are higher and naturally immune individuals are more common than in southeast Asia where CQ-resistant alleles appear to have gone to fixation in many areas [29].
Pfmdr1 drug susceptible alleles N86 were more prevalent Kebbi state (57.8%), followed by Kano state (52.8%), Katsina (45.1%), Jigawa state (39.5%) and Kaduna state (33.96%). These results showed that there has been strong selection for CQ-susceptible parasites after the nationwide replacement of CQ with AL in 2005. The return of CQ-susceptible alleles in a country like Nigeria which is endemic for malaria, can be considered as a positive development toward possible replacement of expensive AL with a safe and cheaper drug (CQ).
The lower distribution of pfmdr1 drug resistance alleles 86Y across the states, 33.0% in Katsina, 32.56% in Jigawa, 31.1% in Kebbi, 18.9% in Kaduna and 11.32% in Kano, suggests recovery of the wild type alleles, as the same scenario was observed for pfmdr1 N86Y, where there was a decline in the prevalence of the mutant from 46% to 28% from 2005 to 2010 and an increase in the prevalence of the wild type strains N86 from 77% to 86% in Ghana [32]. Similar observations have been made in other studies in Africa where K76T and N86Y were investigated concurrently [33]. This is also an indication of a gradual gain of stability of these genotypes in the population similar to what was observed by Nzila’s group in Kenya [17].
As the drug susceptible allele K76 was more prevalent in almost all the locations than the drug resistance allele 76T (Figure 2) this suggests either a gradual recovery of the susceptible alleles over the period since the CQ withdrawal, or it could be due partial withdrawal of CQ for P. falciparum treatment, or both. In similar studies like in coastal Tanzania, the prevalence of mutant alleles of pfcrt decreased after only 2.5 years of CQ withdrawal [34]; Malawi showed 100% recovery of the mutant alleles after a decade of non-use CQ [14].
In addition, an increasing trend for K76 will create a future problem for ACT use as it has been observed in recrudescent samples after AL use [26]. Finally, factors such as farmland activities, especially irrigation farming, might also have contributed to increased transmission and the higher prevalence of resistance alleles obtained in Jigawa and Kaduna States. Mohammed et al [31] suggested that factors such as differences in malaria transmission pattern and intensity may also play a role.
5 Conclusions and recommendation
This study concludes that pfcrt and pfmdr1 resistant alleles are prevalent in the study area but the prevalence is neither influenced by gender nor by age. The study provided insight into the genetic background of P. falciparum parasites prevalent in north-western Nigeria. The possibility of using CQ in future as a combination therapy with other short acting drugs with different pharmacokinetic and pharmacodynamic profiles will be an additional antimalarial option. Since CQ-resistant alleles are relatively rare, the drug may be introduced as prophylaxis for malaria risk groups, such as children and pregnant women. There is also the need for continuous monitoring of the molecular markers to be able to establish a trend of drug resistance or susceptibility in this part of Nigeria.
Supporting information
Table 1.
Distribution of Pfcrt alleles in the senatorial districts of Kano State.
| Senatorial Districts | Number Examined | Pfcrt K76 | Pfcrt 76T | Pfcrt K76T | |||
|---|---|---|---|---|---|---|---|
| K76 | Non-K76 | 76T | Non-76T | K76T | Non-K76T | ||
| N-S (Gaya) | 17 | 5 (29.4) | 12 (70.6) | 1 (5.9) | 16 (94.1) | 0 (0.0) | 17 (100.0) |
| N-W (Gwarzo) | 31 | 5 (16.1) | 26 (83.9) | 7 (22.6) | 24 (77.4) | 1 (3.2) | 30 (96.8) |
| N-C (Municipal) | 5 | 3 (60.0) | 2 (40.0) | 1 (20.0) | 4 (80.0) | 0 (0.0) | 5 (100.0) |
| Total | 53 | 13 (24.53) | 40 (75.47) | 9 (16.98) | 44 (83.02) | 1 (1.89) | 52 (98.11) |
| Chi square | 4.799 | 2.207 | 0.723 | ||||
| df | 2 | 2 | 2 | ||||
| P value | 0.091ns | 0.332ns | 0.697ns | ||||
Table 2.
Distribution of Pfmdr1 alleles in the senatorial districts of Kano State.
| Senatorial Districts | Number Examined | Pfdmr1 N86 | Pfdmr1 86Y | ||
|---|---|---|---|---|---|
| N86 | Non-N86 | 86Y | Non-86Y | ||
| N-S (Gaya) | 17 | 10 (58.8) | 7 (41.2) | 1 (5.9) | 16 (94.1) |
| N-W (Gwarzo) | 31 | 17 (54.8) | 14 (45.2) | 5 (16.1) | 26 (83.9) |
| N-C (Municipal) | 5 | 1 (20.0) | 4 (80.0) | 0 (0.0) | 5 (100.0) |
| Total | 53 | 28 (52.83) | 25 (47.17) | 6 (11.32) | 47 (88.68) |
| Chi square | 2.458 | 1.853 | |||
| df | 2 | 2 | |||
| P value | 0.293ns | 0.396ns | |||
Table 3.
Distribution of pfcrt alleles in the senatorial districts of Jigawa State.
| Senatorial Districts | Number Examined | Pfcrt K76 | Pfcrt 76T | Pfcrt K76T | |||
|---|---|---|---|---|---|---|---|
| K76 | Non-K76 | 76T | Non-76T | K76T | Non-K76T | ||
| N-W (Garki) | 14 | 1 (7.1) | 13 (92.9) | 2 (14.3) | 12 (85.7) | 0 (0.0) | 14 (100.0) |
| N-C (Kiyawa) | 17 | 2 (11.8) | 15 (88.2) | 5 (29.4) | 12 (70.6) | 2 (11.8) | 15 (88.2) |
| N-E (Maigatari) | 12 | 5 (41.7) | 7 (58.3) | 4 (33.3) | 8 (66.7) | 3 (25.0) | 9 (75.0) |
| Total | 43 | 8 (18.60) | 35 (81.40) | 11 (25.58) | 32 (74.42) | 5 (11.63) | 38 (88.37) |
| Chisquare | 5.954 | 1.448 | 3.931 | ||||
| df | 2 | 2 | 2 | ||||
| P value | 0.051ns | 0.485ns | 0.140ns | ||||
Table 4.
Distribution of pfmdr 1 alleles in the senatorial districts of Jigawa State.
| Senatorial Districts | Number Examined | Pfdmr1 N86 | Pfdmr1 86Y | ||
|---|---|---|---|---|---|
| N86 | Non-N86 | 86Y | Non-86Y | ||
| N-W (Garki) | 14 | 3 (21.4) | 11 (78.6) | 10 (71.4) | 4 (28.6) |
| N-C (Kiyawa) | 17 | 9 (52.9) | 8 (47.1) | 3 (17.6) | 14 (82.4) |
| N-E (Maigatari) | 12 | 5 (41.7) | 7 (58.3) | 1 (8.3) | 11 (91.7) |
| Total | 43 | 17 (39.53) | 26 (60.47) | 14 (32.56) | 29 (67.44) |
| Chisquare | 3.221 | 14.562 | |||
| df | 2 | 2 | |||
| P value | 0.200ns | 0.001** | |||
Table 5.
Distribution of pfcrt alleles in the senatorial districts of Kaduna State.
| Senatorial Districts | Number Examined | Pfcrt K76 | Pfcrt 76T | Pfcrt K76T | |||
|---|---|---|---|---|---|---|---|
| K76 | Non-K76 | 76T | Non-76T | K76T | Non-K76T | ||
| N-S (Doka) | 30 | 9 (30.0) | 21 (70.0) | 5 (16.7) | 25 (83.3) | 1 (3.3) | 29 (96.7) |
| N-W (Zaria) | 18 | 6 (33.3) | 12 (66.7) | 4 (22.2) | 14 (77.8) | 2 (11.1) | 16 (88.9) |
| N-C (Kawo) | 5 | 2 (40.0) | 3 (60.0) | 1 (20.0) | 4 (80.0) | 0 (0.0) | 5 (100.0) |
| Total | 53 | 17 (32.08) | 36 (67.92 | 10 (18.87) | 43 (81.13) | 3 (5.66) | 50 (94.34) |
| Chisquare | 0.217 | 0.231 | 1.606 | ||||
| df | 2 | 2 | 2 | ||||
| P value | 0.897ns | 0.891ns | 0.448ns | ||||
Table 6.
Distribution of pfmdr1 alleles in the senatorial districts of Kaduna State.
| Senatorial Districts | Number Examined | Pfdmr1 N86 | Pfdmr1 86Y | ||
|---|---|---|---|---|---|
| N86 | Non-N86 | 86Y | Non-86Y | ||
| N-S (Doka) | 30 | 12 (40.0) | 18 (60.0) | 7 (23.3) | 23 (76.7) |
| N-W (Zaria) | 18 | 4 (22.2) | 14 (77.8) | 3 (16.7) | 15 (83.3) |
| N-C (Kawo) | 5 | 2 (40.0) | 3 (60.0) | 0 (0.0) | 5 (100.0) |
| Total | 53 | 18 (33.96) | 35 (66.04) | 10 (18.87) | 43 (81.13) |
| Chisquare | 1.675 | 1.611 | |||
| df | 2 | 2 | |||
| P value | 0.433ns | 0.447ns | |||
6 Acknowledgement
The authors gratefully acknowledge the support received from Dr. Abdullahi Sulaiman.
7 Competing interests
The authors declare that they have no competing interests.
References
- 1.Kebede S, Aseffa A, Medhin G, Berhe N et al. Re-evaluation of microscopy confirmed Plasmodium falciparum and Plasmodium vivax malaria by nested PCR detection in southern Ethiopia. Malar. J. 2014;13:48. doi: 10.1186/1475-2875-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sachs J, Malaney P. The economic and social burden of malaria. Nature. 2002415:680–685.:7. doi: 10.1038/415680a. [DOI] [PubMed] [Google Scholar]
- 3.Breman J. The ears of the hippopotamus: manifestations, determinants, and estimates of the malaria burden. Am. J. Trop. Med. Hyg. 2001;64:1–11. doi: 10.4269/ajtmh.2001.64.1. [DOI] [PubMed] [Google Scholar]
- 4.President's Malaria Initiative. Malaria operational plan 2013. The United States Agency for International Development (USAID) 2001. p. 59.
- 5.Federal Ministry of Health (FMOH). Abuja, Nigeria: National Antimalarial Treatment Policy. FMOH, National malaria and Vector Control Division 2005. [Google Scholar]
- 6.World Health Organization. World Health Organization; Geneva: Global report on antimalarial drug efficacy and drug resistance, 2000–2010. [Google Scholar]
- 7.Molta NB. Susceptibility of Plasmodium falciparum to malarial drugs in north-eastern Nigeria. Trans. R. Soc. Trop. Med. Hyg. 1995;89:422–425. [Google Scholar]
- 8.Umotong AB, Ezedinachi EN, Okerengwo AA, Usanga EA et al. Correlation between in vivo and in vitro response of chloroquine resistant Plasmodium falciparum in Calabar, south-eastern Nigeria. Acta Trop. 1991;49:119–125. doi: 10.1016/0001-706x(91)90059-s. [DOI] [PubMed] [Google Scholar]
- 9.Salako LA. An African perspective. World Health. 1998;3:24–25. [Google Scholar]
- 10.Sowunmi A, Salako LA, Walker O, Ogundahunsi OA. Clinical efficacy of mefloquine in children suffering from CRPF in Nigeria. Trans. R. Soc. Trop. Med. Hyg. 1990;84:761–764. doi: 10.1016/0035-9203(90)90067-o. [DOI] [PubMed] [Google Scholar]
- 11.Patrick O, Erah GA, Augustine OO. Plasmodium falciparum malaria resistance to chloroquine in five communities in southern Nigeria. Afr. J. Biotechnol. 2003;2:384–389. [Google Scholar]
- 12.Koenderink JB, Kavishe RA, Rijpma SR, Russel FG. The ABCs of multidrug resistance in malaria. Trends Parasitol. 2010;26:440–446. doi: 10.1016/j.pt.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Wellems TE, Plowe CV. Chloroquine-resistant malaria. J. Infect. Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- 14.Kublin JG, Cortese JF, Njunju EM, Mukadam RA et al. Re-emergence of chloroquine sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 15.Alifrangis M, Lusingu JP, Mmbando B, Dalgaard MB et al. Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the P. falciparum dihydropteroate synthase gene. Am. J. Trop. Med. Hyg. 2009;80:523–527. [PubMed] [Google Scholar]
- 16.Malmberg M, Ngasala B, Ferreira PE, Larsson E et al. Temporal trends of molecular markers associated with artemether -lumefantrine tolerance/resistance in Bagamoyo district. Tanzania. Malar. J., 2013;12:103. doi: 10.1186/1475-2875-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwai L, Ochong E., Abdirahman A, Kiara SM et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar. J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Afrane YA, Lawson BW, Brenya R, Kruppa T et al. The ecology of mosquitoes in an irrigated vegetable farm in Kumasi, Ghana: abundance, productivity and survivorship. Parasit. Vectors. 2012;5:233. doi: 10.1186/1756-3305-5-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization-MPAC. Malaria Policy Advisory Committee to the WHO: Conclusions and recommendations of March 2013 meeting. Malar. J. 2013;12:213. doi: 10.1186/1475-2875-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojurongbe O, Ogungbamigbe TO, Fagbenro-Beyioku AF, Fendel R et al. Rapid detection of Pfcrt and Pfmdr1 mutations in Plasmodium falciparum isolates by FRET and in vivo response to chloroquine among children from Osogbo, Nigeria. Malar. J. 2007;6:41. doi: 10.1186/1475-2875-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kearns AM, Guiver M, James V, King J. Development and evaluation of a real-time quantitative PCR for the detection of human cytomegalovirus. J. Virol. Methods. 2001;95:121–131. doi: 10.1016/s0166-0934(01)00307-x. [DOI] [PubMed] [Google Scholar]
- 22.De Monbrison F, Raynaud D, Latour-Fondanaiche C, Staal A et al. Real-time PCR for chloroquine sensitivity assay and for pfmdr1-pfcrt single nucleotide polymorphisms in Plasmodium falciparum. J. Microbiol. Methods. 2003;54:391–401. doi: 10.1016/s0167-7012(03)00086-1. [DOI] [PubMed] [Google Scholar]
- 23.Sutar SK, Gupta B, Ranjit M, Kar SK et al. Sequence analysis of coding DNA fragments of pfcrtand pfmdr-1 genes in Plasmodium falciparum isolates from Odisha, India. Mem. Inst. Oswaldo Cruz. 2011;106:78–84. doi: 10.1590/s0074-02762011000100013. [DOI] [PubMed] [Google Scholar]
- 24.Bin-Dajem SM, Al-Sheikh AA, Bohol MF, Alhawi M et al. A Detecting mutation in Pfcrt and Pfmdr1genes among Plasmodium falciparum isolates from Saudi Arabia bypyrosequencing. Parasitol. Res. 2011;109:291–296. doi: 10.1007/s00436-011-2251-5. [DOI] [PubMed] [Google Scholar]
- 25.Bustamante C, Folarin OA, Gbotosho GO, Batista CN et al. In-vitro reduced susceptibility to Artemether in P. falciparum and its association with polymorphisms on transporter genes. J. Infect. Dis. 2012;206:324–332. doi: 10.1093/infdis/jis359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisowath C, Strömberg J, Mårtensson A, Msellem M et al. In vivo selection of Plasmodium falciparum pfmdr1 86 N coding alleles by Artemether-lumefantrine (Coartem). J. infect. Dis. 2005;191:10141017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 27.Happi CT, Gbotosho GO, Folarin OA, Sowunmi A et al. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob. Agents Chemother. 2009;53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittra P, Vinayak S, Chandawat H, Das MK et al. Progressive increase in point mutations associated with chloroquine resistance in Plasmodium falciparum isolates from India. J. Infect. Dis. 2006;193:1304–1312. doi: 10.1086/502979. [DOI] [PubMed] [Google Scholar]
- 29.Mang’era CM, Mbai FN, Omedo IA, Mireji PO et al. Changes in genotypes of Plasmodium falciparum human malaria parasite following withdrawal of chloroquine in Tiwi, Kenya. Acta Trop. 2012;123:202–207. doi: 10.1016/j.actatropica.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Laufer MK, Takala-Harrison S, Dzinjalamala FK, Stine OC et al. Return of chloroquine-susceptible falciparum malaria in Malawi was there expansion of diverse susceptible parasites. J. Infect. Dis. 2010;202:801–808. doi: 10.1086/655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammed A, Arnold N, Akili K, Alphaxard M et al. Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar. J. 2013;12:415. doi: 10.1186/1475-2875-12-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duah NO, Wilson MD, Ghansah A, Abuaku B et al. Mutations in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance genes, and treatment out come in Ghanaian children with uncomplicated malaria. J. Trop. Pediatric. 2007;53:27–31. doi: 10.1093/tropej/fml076. [DOI] [PubMed] [Google Scholar]
- 33.Wurtz N, Fall B, Pascual A, Diawara S et al. Prevalence of molecular markers of Plasmodium falciparum drug resistance in Dakar, Senegal. Malar. J. 2012;11:197. doi: 10.1186/1475-2875-11-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Temu EA, Kimani I, Tuno N, Kawada H. Monitoring chloroquine resistance using Plasmodium falciparum parasites isolated from wild mosquitoes in Tanzania. Am. J. Trop. Med. Hyg. 2006;75:1182–1187. [PubMed] [Google Scholar]


