Abstract
Mitochondria are highly dynamic, maternally inherited cytoplasmic organelles, which fulfill cellular energy demand through the oxidative phosphorylation system. Besides, they play an active role in calcium and damage-associated molecular patterns (DAMPs) signaling, amino acid, and lipid metabolism, and apoptosis. Thus, the maintenance of mitochondrial integrity and homeostasis is extremely critical, which is achieved through continual fusion and fission. Mitochondrial fusion allows the transfer of gene products between mitochondria for optimal functioning, especially under metabolic and environmental stress. On the other hand, fission is crucial for mitochondrial division and quality control. The imbalance between these two processes is associated with various ailments such as cancer, neurodegenerative and cardiovascular diseases. This review discusses the molecular mechanisms that control mitochondrial fusion and fission and how the disruption of mitochondrial dynamics manifests into various disease conditions.
Keywords: Mitochondria, dynamics, fusion, fission, diseases
Introduction
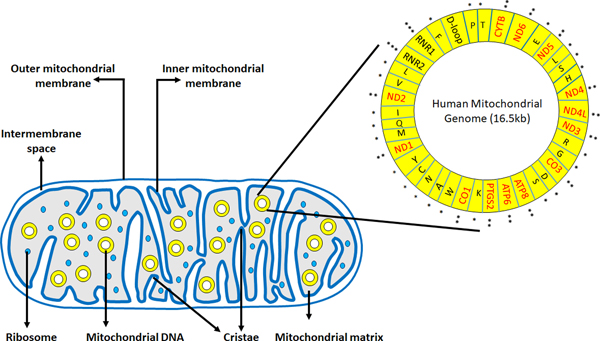
Mitochondria are highly dynamic double membrane-bound organelles found in most eukaryotic cells. They play critical roles in the cells, including energy production through oxidative phosphorylation (OXPHOS), integration of various metabolic pathways, regulation of apoptosis, and maintenance of calcium homeostasis (1). It is believed that mitochondria are the descendants of an ancient prokaryote that underwent an endosymbiotic event with early eukaryotes (2). A mitochondrion consists of an outer membrane, intermembrane space, inner membrane, and matrix (Figure 1). The outer membrane contains three integral protein families: translocase of outer mitochondrial membrane (TOMM) complex, sorting and assembly machinery (SAM) complex, and porins. The inner membrane is folded to form cristae, which contains the enzyme complexes responsible for oxidative phosphorylation. The mitochondrial matrix is the site where the tricarboxylic acid (TCA) cycle takes place. It also contains the mitochondrial genome, which is a 16.5 kb double-stranded closed circular DNA. The mitochondrial genome consists of 37 genes that encode 13 OXPHOS proteins, 2 ribosomal RNAs (12S and 16S rRNA), and 22 transfer RNAs (Figure 1). The majority of the mitochondrial proteins (~1500) are encoded by the nuclear genes and are imported into the mitochondria from the cytosol after their synthesis.
Figure 1.
Structure of a mitochondrion showing the outer mitochondrial membrane, inner mitochondrial membrane, intermembrane space and the mitochondrial matrix. Mitochondrial genome residing in the mitochondrial matrix has 37 genes coding for 13 OXPHOS proteins (in red, **), 2 ribosomal RNAs (12s rRNA, 16s rRNA,*), 22 tRNAs (***) and a regulatory D loop region.
Being a vital cellular organelle, maintenance of mitochondrial integrity and homeostasis is extremely critical. Therefore, mitochondria continuously undergo changes in their number and morphology through the processes of fusion and fission. This ‘mitochondrial dynamics’ helps maintain the pool of mitochondria within a cell and optimal OXPHOS activity by allowing efficient transport and distribution of mitochondrial content. Mitochondrial dynamics is linked to several cellular processes, including cell cycle, apoptosis, cell migration, mitophagy, and reactive oxygen species (ROS) production (3). Therefore, a fine balance between mitochondrial fusion and fission is crucial for cell survival and optimal functioning. Defects in mitochondrial dynamics are linked to many pathological conditions, including cancer, neurodegenerative, and cardiovascular diseases (4,5). This review discusses the molecular insights into the mitochondrial fusion and fission machinery and different disease conditions that develop due to the functional abnormalities in mitochondrial dynamics.
The fusion paradigm and the molecular insight
Mitochondrial fusion joins two mitochondria at the outer and inner membrane interfaces via three membrane GTPases, Mitofusin 1 (MFN1, human chromosome 3q26), Mitofusin 2 (MFN2, human chromosome 1p36), and Optic atrophy protein 1(OPA1, human chromosome 3q29). These proteins are members of the dynamin-related protein (DRP) superfamily. MFN 1 and 2 are homologs of the drosophila protein fuzzy onion (Fzo) (6), whereas OPA1 is the mammalian ortholog of Mgm1, which is a protein essential for the mitochondrial fusion in yeast (7). Increased fusion is triggered by treatments that inhibit protein synthesis, mammalian target of rapamycin (mTOR) inhibition-induced autophagy, and starvation (8, 9, 10).
Some mitochondria are unable to perform their respiratory function optimally because of mutations in mtDNA induced by reactive oxygen species (11). Therefore, mitochondrial fusion is vital as it allows the exchange of gene products and metabolites between the fusing mitochondria to enhance their overall respiratory function. Likewise, impediment of mitochondrial fusion is associated with the underperformance of mitochondrial function (12). However, a recent study demonstrated that promoting mitochondrial fusion in pancreatic adenocarcinoma cells by overexpression of MFN2 or inhibition of dynamin-related protein-1 (DRP1) led to increased mitophagy, which resulted in a loss of mitochondrial mass and reduced OXPHOS (13). Thus, the true significance of mitochondrial fusion is elusive and maybe context-dependent, requiring additional in-depth mechanistic investigations.
Outer mitochondrial membrane fusion: role of Mitofusins
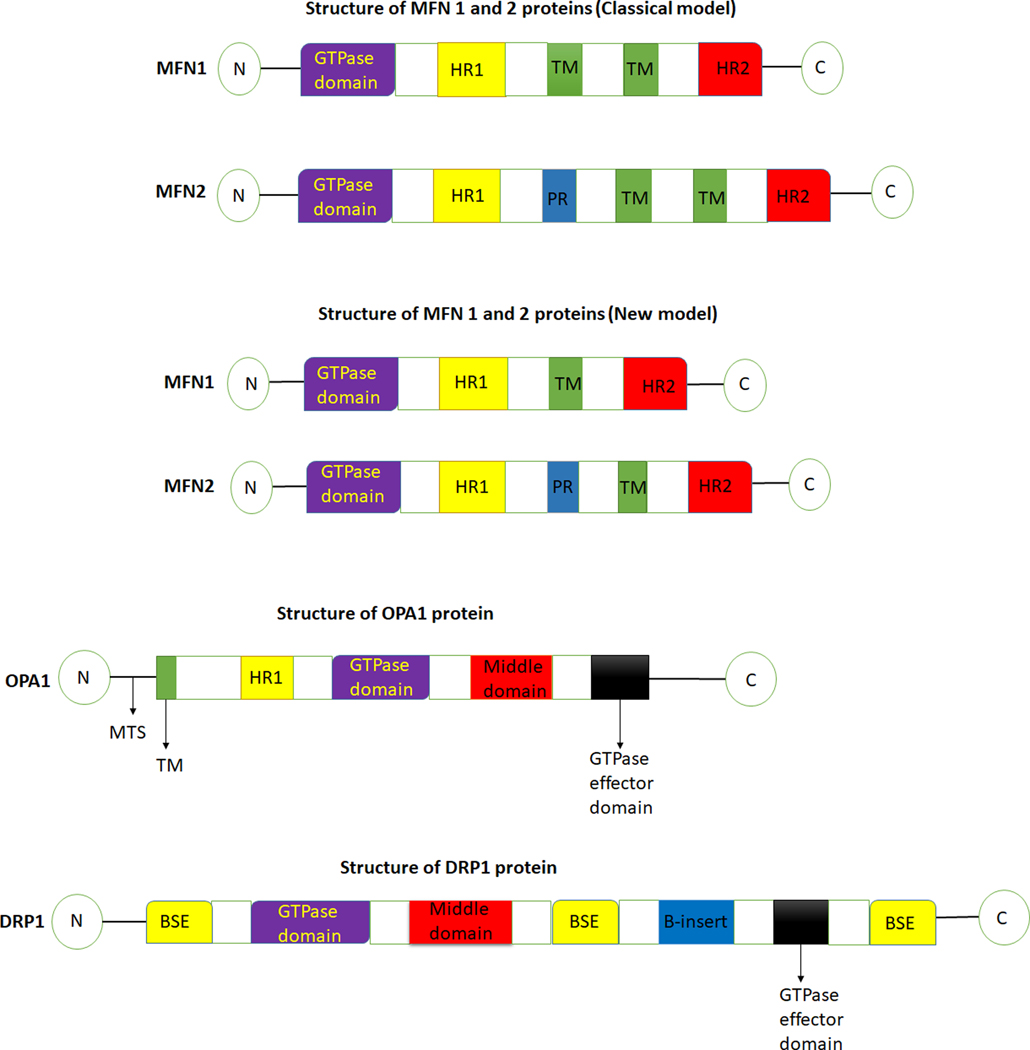
The outer mitochondrial membrane fusion is facilitated by two large membrane GTPases- Mitofusin 1 (MFN1) and Mitofusin 2 (MFN2), present on the outer mitochondrial membrane. MFN2 is also present on the endoplasmic reticulum (ER) membrane, where it tethers ER to mitochondria enabling the mitochondrial calcium uptake (14). MFN topology has been described using two models (Figure 2). The classical model of MFN topology suggests that MFNs have two transmembrane domains. However, a recent study has revealed that both MFN1 and MFN2 possess a single transmembrane domain that places the N-terminal GTPase and helical repeat 1 (HR1) domains in the cytoplasm and C-terminal HR2 domain in the mitochondrial intermembrane space (15).
Figure 2.
Schematic diagram of the structure of fusion (MFN1, MFN2, OPA1) and fission (DRP1) regulators. MFN1- Mitofusin 1, MFN2- Mitofusin 2, OPA1- Optic atrophy protein 1, DRP1- Dynamin related protein 1, TM- transmembrane, PR- proline rich, HR1- helical repeat 1, HR2- helical repeat 2, N- N terminal, C- C terminal, MTS- mitochondrial targeting sequence, BSE- bundle signaling element.
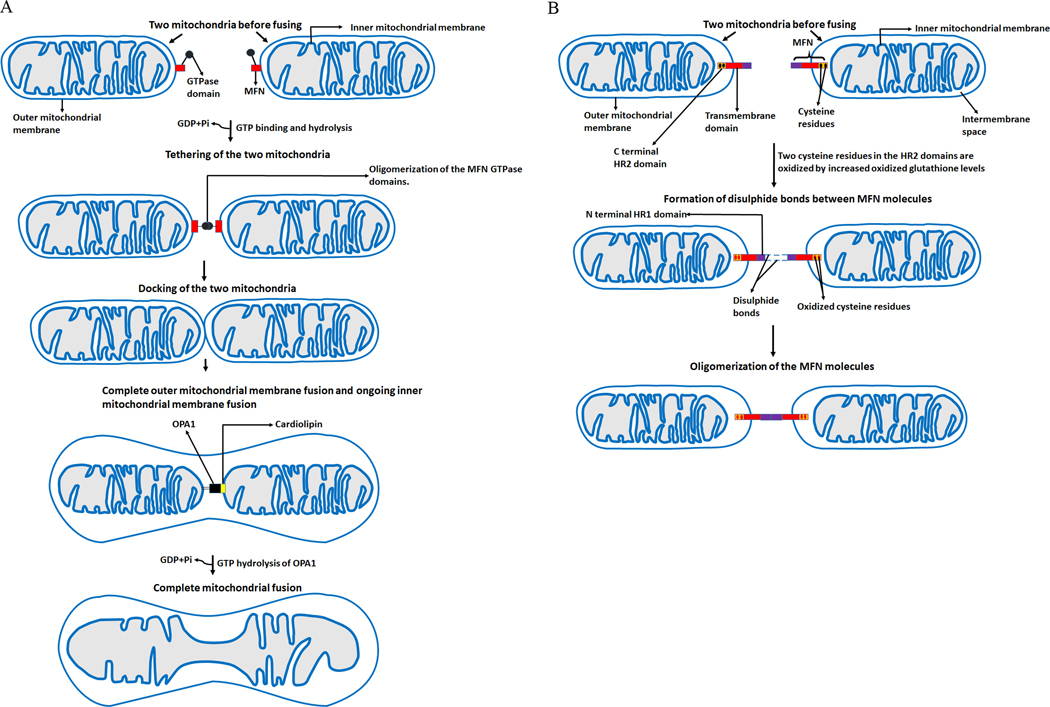
The primary function of Mitofusins is the promotion of docking and fusion of the mitochondrion. At present, a complete understanding of the molecular mechanisms involved in mitochondrial fusion is lacking. However, a couple of studies have proposed that tethering of two OMMs occurs through the oligomerization of the GTPase domains of MFNs, and GTP hydrolysis is required for this oligomerization (16, 17). Upon GTP binding and hydrolysis, a conformational change occurring in the GTPase domains leads to their oligomerization, facilitating the docking of the two mitochondria at the two outer membranes and their subsequent fusion (Figure 3a).
Figure 3.
The fusion paradigm. Depiction of various steps involved in mitochondrial fusion. (a) GTP binding and hydrolysis leads to a conformational change in the GTPase domains of MFNs resulting in their oligomerization. The oligomerization of the GTPase domains brings about the tethering of the two mitochondria, which facilitates their docking and fusion. (b) A new mechanism of the oligomerization of MFN molecules. Increased oxidized glutathione levels oxidizes two cysteine residues located in the HR2 domains of MFN molecules. This leads to the formation of disulphide bonds between MFN molecules and their oligomerization. MFNs- Mitofusins, OPA1- Optic atrophy protein 1, GTP- Guanosine triphosphate, GDP- Guanosine diphosphate, Pi- inorganic phosphate, HR2- helical repeat 2.
Based on the new MFNs topology suggesting only one transmembrane domain in human MFNs, another mechanism of the oligomerization of MFN molecules, critical for outer membrane fusion, has evolved (Figure 3b). Mattie et al. demonstrated that two cysteine residues located in the HR2 domains (which reside in the intermembrane space) can be oxidized by increased oxidized glutathione levels leading to the formation of disulfide bonds between MFN molecules and their oligomerization (15). They also showed that the addition of GSH to preformed GSSG-induced oligomers of MFN2 led to a reversal of MFN2 oligomer formation. This new mechanism indicates that redox signaling plays a vital role in OMM fusion. Further studies are needed to clarify the mechanistic basis of mitochondrial fusion and better understand the effects of aberrant redox signaling on fusion.
The activity of MFN 1 is regulated by phosphorylation, ubiquitination, and deacetylation. Phosphorylation of Mfn1 in the HR1 domain by the extracellular-signal-regulated kinase (ERK) inhibits mitochondrial fusion and promotes apoptosis (18). On the other hand, deacetylation of Mfn1 by histone deacetylase 6 (HDAC6) leads to its activation and promotion of fusion in conditions of glucose deprivation (19). In response to cellular stress, JNK phosphorylates MFN 2, leading to the recruitment of E3 ubiquitin ligase, which ubiquitinates MFN2 causing its proteasomal degradation. This degradation of MFN2 results in mitochondrial fragmentation and enhanced apoptotic cell death (20). Mfn2 can also be phosphorylated by PINK1, leading to its ubiquitination by Parkin and eventual mitophagy (21).
Inner mitochondrial membrane fusion: role of Optic atrophy protein 1 and Cardiolipin
Inner mitochondrial membrane (IMM) fusion is mediated by OPA1 and specific IMM lipid components, for example- cardiolipin (22). OPA1 is a complex protein that localizes to the inner mitochondrial membrane (Figure 3). It consists of eight isoforms which are products of alternative splicing and are characterized by different combinations of three small exons (4,4b and 5b) located in the N-terminal part of the molecule (23). One to three sites of proteolytic cleavage can be formed in different isoforms. The formation of these cleavage sites depends on the set of exons (24). These sites are usually termed S1, S2, and S3. Site S1 is present in all eight isoforms, whereas each S2 and S3 sites are present in four isoforms (24). All isoforms encode for a polypeptide containing an N-terminal mitochondrial targeting sequence (MTS) (Figure 2). The MTS is removed by the matrix processing protease during the import of the N-terminus into the matrix (4). The cleavage of the S1 site is mediated by OMA1. If the S1 site is left intact, the resulting long-form of OPA1 (L-OPA1) is anchored to the inner membrane, with most of the protein facing the intermembrane space (4). On the other hand, if the S1 site is cleaved by OMA1, a short form of Opa1 (S-OPA1) is produced. Cleavage at the S2 and S3 sites occurs constitutively, and this cleavage is mediated by YME1L (25, 26), while the cleavage of OPA1 isoforms at the S1 site occurs only under certain conditions associated with stress (27).
Currently, the precise mechanism of IMM fusion is not yet fully understood. A study by Song et al. revealed that a combination of S-OPA1 and L-OPA1 is required for IMM fusion to occur (26). Similarly, Ge et al. showed that L-OPA1 and S-OPA1 work together to stimulate fusion activity leading to efficient and fast membrane pore opening in liposomes (28). In contrast, some other studies have shown that only L-OPA1 is sufficient to promote IMM fusion (8, 29). Therefore, further studies are needed to confirm if S-OPA1 is dispensable for IMM fusion.
Cardiolipin (CL), a phospholipid and component of the inner mitochondrial membrane, is vital for IMM fusion. Ban et al. showed that the incubation of recombinant L-OPA1 with reconstituted CL-containing liposomes led to an interaction between L-OPA1 and CL, resulting in IMM fusion. Findings also revealed that when cardiolipin was removed from the liposomes, no membrane fusion was observed even in the presence of L-OPA1 on both sides of the membranes (30). They further demonstrated that GTP hydrolysis was needed for OPA1-mediated fusion and that fusion activity was absent in liposomes containing an OPA1 mutant lacking GTP hydrolysis activity. (30). These findings indicate that the binding of Cardiolipin to OPA1 and GTP hydrolysis of OPA-1 are crucial for IMM fusion. Additional studies are required to decipher the precise roles of cardiolipin in IMM fusion and delineating the underlying molecular mechanisms.
Apart from the proteolytic processing of Opa1, which is mediated by YME1L and OMA1, NAD-dependent deacetylase sirtuin-3 (SIRT3) deacetylates OPA1 at its GTPase effector domain (Lys926 and Lys931), elevating its GTPase activity and enhancing mitochondrial fusion (31).
Mitochondrial fission and the molecular insight
Mitochondrial fission is the process whereby a mitochondrion divides into two mitochondria (Figure 4). It plays different roles, including inheritance and partitioning of organelles during cell division, proper distribution of mitochondria, and cytochrome C release during apoptosis (32, 33, 34). Fission is also crucial for removing damaged organelles by mitophagy, and its loss results in an increase in the number of elongated mitochondria due to unbalanced fusion (35). On the other hand, disruption in the fusion process results in more fragmented mitochondria (36, 37). Although the precise reason for this is unknown, this could happen because of the need to maintain ATP supply in the cells. Fission in mammals is coordinated by a large dynamin-like GTPase, known as Dynamin-related protein 1 (DRP1, a.k.a.DNM1L, human chromosome 12p11). Other proteins involved in the fission process include Dynamin 2 (DNM2, a.k.a. DYN2, human chromosome 19p13.2), human mitochondrial dynamics proteins 49 (MID49, a.k.a.MIEF2, human chromosome 17p11) and 51 (MID51, a.k.a.MIEF1, human chromosome 22q13), mitochondrial fission 1 protein (FIS1, a.k.a. TTC11, human chromosome 7q22) and mitochondrial fission factor (MFF, a.k.a. C2orf33, human chromosome 2q36.3) (22, 35, 38, 39). DRP1 is a cytosolic protein recruited to the mitochondria by its adaptors present on the OMM, including MFF, MID49, MID51, and FIS1. DRP1 consists of four domains, an N-terminal GTPase domain followed by the middle domain, variable domain (or B-insert), and the GED in C-terminal (Figure 2).
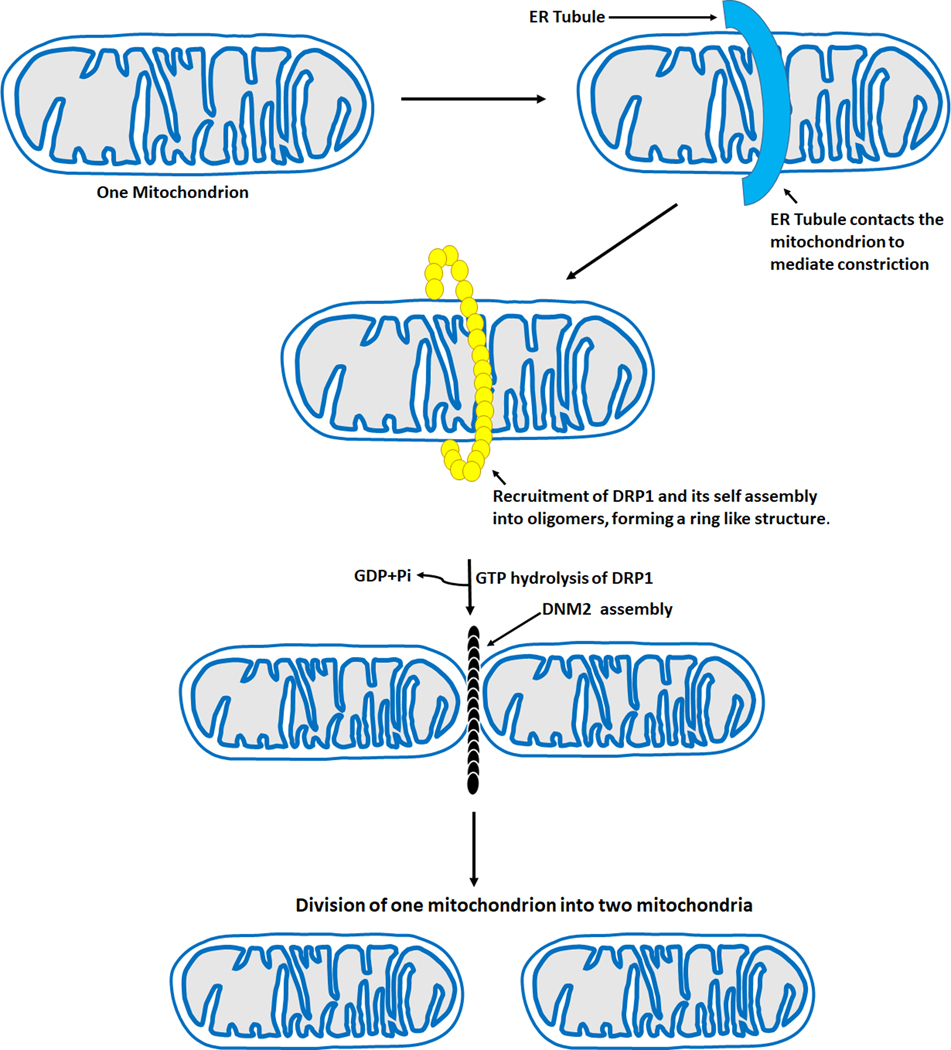
Figure 4.
Mitochondrial fission machinery. The steps involved in mitochondrial fission are as follows- ER tubules contact the mitochondria to mediate constriction before the recruitment of DRP1. After the recruitment of DRP1 to the OMM, it forms a ring like structure, GTP hydrolysis of DRP1 then occurs followed by the recruitment of DNM2 to the mitochondrion-constricted site, where it assembles and completes the division process leading to two daughter mitochondria. ER- Endoplasmic reticulum, DRP1- Dynamin related protein 1, DNM2- Dynamin 2, GTP- Guanosine triphosphate, GDP- Guanosine diphosphate, Pi- inorganic phosphate, OMM- Outer mitochondria membrane.
The initial step of mitochondrial division occurs at positions where the endoplasmic reticulum (ER) tubules contact the mitochondria to mediate constriction before Drp1 recruitment (40) (Figure 4). This ER-mediated mitochondrial constriction is required to decrease the average mitochondrial diameter from approximately 300–500 nm to around150 nm to allow Drp1-oligomeric ring formation (40). At the ER-mitochondria contact sites, the ER-bound inverted-formin 2 (INF2) cooperates with mitochondria anchored formin-binding Spire1C to regulate the actin assembly required for mitochondrial constriction before Drp1 recruitment and oligomerization (41, 42). Studies have shown that some proteins involved in regulating the actin cytoskeleton, for example, Arp2/3, cofilin, and cortactin, are also involved in mitochondrial fission (43, 44). Therefore, ER and actin are essential in the process and regulation of mitochondria division. However, it is not fully understood how the ER recognizes the sites for mitochondria constriction requiring more investigations to better understand the underlying mechanisms.
After DRP1 is recruited to the OMM by MFF, MID 49, MID 51, and FIS1, it forms a ring-like structure around the mitochondrion, enhancing the pre-existing constriction of the mitochondrion (38). GTP hydrolysis of DRP1 then occurs, followed by the recruitment of DNM2 to the mitochondrion-constricted site, where it assembles and completes the division process (Figure 4). In contrast, another study has revealed that DNM2 is dispensable for mitochondrial fission and that DRP1 has constricting and severing abilities, which enables it to complete the fission process even in the absence of DNM2 (45). Further studies would help determine if complete mitochondrial fission can occur without DNM2.
Inner mitochondrial membrane (IMM) constriction is Ca2+-dependent and occurs at mitochondria–ER contact sites (46). The calcium release from the ER into the mitochondria leads to the constriction of the inner membrane compartment, resulting in IMM division before the recruitment of DRP1. Apart from playing a role in mitochondrial fusion, Cardiolipin (CL) also interacts with DRP1. This interaction drives the oligomerization of DRP1 and stimulation of its GTPase activity, which increases the constriction of liposome membranes (47, 48). Future investigations are needed to reveal how cardiolipin balances mitochondrial fusion and fission and what triggers its differential actions.
DRP1 is phosphorylated by cdk1/cyclin B kinase on serine (Ser) 585 during the process of mitosis, leading to its oligomerization and promotion of mitochondrial fission (49). Mitogen-activated protein kinase I (MAPK1) also plays a vital role in DRP-1 phosphorylation. MAPK1, also known as ERK2, phosphorylates DRP1 at Ser 616 to cause its activation, promoting mitochondrial fission (50). In contrast, protein kinase A phosphorylates DRP1 at Ser 637, leading to its retention in the cytoplasm, thereby inhibiting fission and sparing mitochondria from autophagic degradation during nutrient deprivation and cell death (10,51). Phosphorylation at Ser 637 is abrogated by calcineurin through dephosphorylation leading to its translocation to the mitochondria and mitochondrial fission (52).
SUMOylation, S-nitrosylation, O-GlcNAcylation, and Ubiquitination, can also regulate DRP1activity. SUMOylation of DRP1 by mitochondrial -associated protein ligase (MAPL) is triggered by the activation of apoptosis. After the SUMOylation of DRP1 by MAPL, DRP1 stabilizes ER- mitochondrial contact sites, calcium flux, cristae remodeling, and cytochrome c release (53). O-GlcNAcylation and S-nitrosylation of DRP1 increase its fission activity. O-GlcNAc transferase induces the translocation of Drp1 from the cytoplasm to mitochondria, enhancing fission (54). Comparably, S-nitrosylation of DRP1 by nitric oxide increases the GTPase activity of DRP1, inducing mitochondrial fission (55). Ubiquitination of DRP1 by Parkin results in its degradation by the proteasome-dependent pathway (56).
Pathobiological implications of the imbalance in mitochondrial fusion and fission
The processes of fusion and fission are critical for normal mitochondrial functioning, energetics, and movement. Reduction or excess of any of these processes results in an imbalance affecting mitochondrial function, ultimately resulting in various diseases, including neurodegenerative disorders, cardiovascular diseases, and cancer (Table 1), as discussed below:
Table 1–
A list of fusion and fission associated genes, their functions, reported alterations and associated diseases.
| Gene Name | Yeast Orthologue | Functions | Alterations | Diseases |
|---|---|---|---|---|
| MFN1 | Fzo1 | Outer mitochondrial membrane fusion | Mfn1 downregulation | Hepatocellular carcinoma (97, 98), Triple negative breast cancer (99) |
| MFN2 | Fzo1 | Outer mitochondrial membrane fusion | Heterozygous mutations in Mfn 2Mfn2 downregulation |
Charcot Marie Tooth Disease type 2A (4)Breast cancer (100) |
| OPA1 | Mgm1 | Inner mitochondrial membrane fusion | Mutations (mostly missense) | Dominant optic atrophy (68). |
| DRP1 | Dnm1 | Mitochondrial fission | Increased Drp1 phosphorylation levels. | Sporadic Parkinson’s disease (81) Diseases |
| DRP1 | Dnm1 | Mitochondrial fission | Increased phosphorylation of Drp1 at serine 616 Increased expression levels of Drp1 Increased Drp1 mRNA expression and protein levels Drp1 upregulation |
Pancreatic ductal adenocarcinoma (50), Glioblastoma (101). Left ventricular hypertrophy (85), Lung adenocarcinomas (102). Hepatocellular carcinoma (97). Triple negative breast cancer (99) |
Charcot-Marie-Tooth Disease Type 2A (CMT2A)
CMT2A is an autosomal dominant peripheral neuropathy characterized by sensory and motor loss in the distal limbs (57, 58). This disease occurs as a result of heterozygous mutations in MFN2. In contrast to demyelinating forms of CMT, CMT2A is an axonopathy, and in most cases, there is no change or a slight decrease in nerve conduction velocity (5). The disease is primarily characterized by muscle weakness, hyporeflexia, sensory loss in the lower limbs, and gait defects. These symptoms usually appear before the age of 10 in most patients (59, 60).
Most of the MFN2 mutations are missense mutations distributed throughout the entire ORF sequence with some enrichment in functional domains (60, 61, 62). The overexpression of T105M (63, 64) or R94W mutation (65) in transgenic mice resulted in various neurological phenotypes associated with clumping of mitochondria in neurons and sparsity of mitochondria in axons due to defective trafficking. The mitochondrial defects result in mitochondrial aggregation, dysmotility, fragmentation, and depolarization. Under normal conditions, MFN1 and MFN2 interact with Miro and Milton proteins that form the molecular complex linking MFN2 to kinesin motors. In CMT2A, the mutation in MFN2 affects this interaction leading to impairment in axonal mitochondrial transport. Some MFN2 mutations are also associated with optic atrophy, thereby overlapping with Dominant optic atrophy (DOA) and, in severe cases, resembling the DOA plus syndrome. These findings indicate MFN2 is crucial for proper neuronal functioning.
Dominant Optic Atrophy
Dominant Optic Atrophy (DOA) is the most common inherited optic neuropathy, with most cases caused by mutations in the optic atrophy 1 gene (OPA1). The disease is usually diagnosed in early childhood and characterized by a progressive bilateral loss of visual acuity, blue‐yellow dyschromatopsia or generalized color vision deficits, variable centrocecal, central or paracentral visual field defects, and temporal or diffuse optic nerve pallor with optic disc excavation (66). DOA shows variable expression, both between and within families, ranging from an asymptomatic state to blindness (67). Different OPA1 mutations have been reported, including missense, frameshift, nonsense, and deletion mutations. Of which missense mutations have the highest frequency (26%) (68). Although the main feature of DOA is optic nerve degeneration, some patients with OPA1 mutations also develop additional extra-ocular neurological features called DOA plus syndrome, which entails deafness, ataxia, peripheral neuropathy, myopathy, and progressive external ophthalmoplegia.
Mutations in the OPA1 gene lead to mitochondrial morphology and function impairments, leading to increased autophagy or apoptosis. A study by Ban et al. revealed that the expression of seven Opa1 pathogenic mutants associated with DOA or DOA plus in Opa1-null mouse embryonic fibroblasts (MEFs) did not restore tubular mitochondrial morphology, indicating a loss of mitochondrial fusion activity (69). Davies et al. also reported that fibroblasts taken from adult heterozygous Opa1 mutant mice had alterations in mitochondrial morphology, along with an increase in mitochondrial fission and fragmentation (70). Another study using mouse models of DOA revealed that retinal ganglion cells from adult heterozygous Opa1 mutant mice showed an increased number of autophagosomes than the Opa1 wild-type mice (71). These studies show that mutations in the OPA1 gene lead to defective mitochondrial fusion and aberrant mitochondrial functioning.
Parkinson’s disease
Parkinson’s disease (PD) is one of the most common neurodegenerative disorders, with most cases being sporadic while about 10–15% of patients having a family history. Clinical features include tremors, akinesia or bradykinesia, progressive rigidity, and postural instability. One of the significant neuropathological findings in Parkinson’s disease is the loss of pigmented dopaminergic neurons of the substantia nigra pars compacta, which occurs due to dopamine deficit in the striatum. The dopaminergic neurons from the substantia nigra are vulnerable to changes in mitochondrial dynamics. One reason for this is that these neurons have lower basal mitochondrial content than other midbrain neurons (72). Imbalance in mitochondrial dynamics and mitochondrial dysfunction resulting from mtDNA defects are associated with PD (73, 74, 75, 76). Sporadic Parkinson’s disease (sPD) cybrid cells exhibit dysfunctional mitochondria due to mtDNA defects, abnormal mitochondrial localization, and an increased fragmentation pattern (77, 78, 79). Similarly, increased mitochondrial fragmentation has been linked to Huntington’s disease (80). Santos et al. revealed that the increase in mitochondrial fission in sPD was due to OPA1 long isoform cleavage and increased mitochondrial DRP1 phosphorylation levels, which led to mitochondrial fragmentation (81). The knockdown of DRP1 in sPD cybrid cells led to increased mitochondrial interconnectivity, elongation, and membrane potential, while ROS production was decreased, indicating that DRP1 inhibition could alleviate mitochondrial deficits in sporadic cases of Parkinson disease (81).
Atherosclerosis
Atherosclerosis is a chronic and progressive vascular disease characterized by narrowing of an artery due to the formation of a plaque containing inflammatory cells, oxidized- LDL, and myofibroblasts (82). One of the factors promoting the development of this disease is the activation of the vascular smooth muscle cells (VSMCs). The activation of VSMCs by Platelet-derived growth factors (PDGF) is associated with decreased MFN2 levels and mitochondrial fragmentation. Conversely, inhibition of fission with Mitochondrial division inhibitor 1 (Mdivi-1) has also been associated with decreased PDGF- induced mitochondrial fragmentation and reduction of VSMCs proliferation (83). Similarly, the inhibition of Drp1 has been shown to reduce endothelial dysfunction and atherosclerosis in apolipoprotein E (ApoE) knockout diabetic mice (84). These findings indicate that maintaining the balance between fusion and fission is vital in preventing atherosclerosis progression.
Left ventricular hypertrophy
Left ventricular (LV) hypertrophy is a condition characterized by either enlargement of the left ventricular cavity or thickening of the heart’s left ventricular wall or both. LV hypertrophy can lead to an increased risk of arrhythmias and heart failure. A study by Javadov et al. showed that in a cell model of phenylephrine-induced cardiomyocyte hypertrophy, the expression of Drp1 was increased while Mfn2 levels were reduced (85). Using a pressure overload animal model, the inhibition of mitochondrial fission by Mdivi-1 led to abnormal cardiac mitophagy inhibition, induced angiogenesis by increasing expression of CD31 and VEGF, and decreased the expression of anti-angiogenic factors, preventing collagen deposition. These effects of Mdivi-1 led to the amelioration of left ventricular dysfunction (86), suggesting that increased mitochondrial fragmentation and mitophagy might be associated with LV hypertrophy development.
Carcinogenesis
The imbalance in mitochondrial dynamics contributes significantly to cancer development and metastatic progression. Most studies investigating mitochondrial morphology in tumor cells have reported that increased fission promotes tumorigenesis (13, 50, 87, 88). Several cancer cell lines, which have been transformed with oncogenes, have more fragmented mitochondria than their non-transformed control lines (13, 50, 87). One pathway associated with mitochondrial fission in cancer is the MAPK pathway, activated in cancer cells by excess signaling from growth factors or oncogenic mutations like BRAFV600E or RASG12V. MAPK activation causes the phosphorylation of DRP1 at Ser 616, leading to its activation and subsequent increase in mitochondrial fission. The inhibition of DRP1 or promotion of fusion reduces cancer cell growth by inducing cell death (13, 50, 87, 88). A study from our laboratory has demonstrated that increased fusion led to the growth suppression of breast cancer cells. Overexpression of SH3GL2, an SH3 domain-containing cytosolic protein, upon translocation to the mitochondria, led to increased MFN2 expression and reduced proliferation and invasion of breast cancer cells (89).
Since fission is critical during mitosis to assure equal segregation of mitochondrial contents between daughter cells (90), its loss possibly induces cell death and cellular dysfunction due to replicative stress and mitotic defects affecting genomic integrity (91). A recent study proposed that increased mitophagy due to increased fusion might be another mechanism of reduced cancer growth. Yu et al. revealed that inhibition of fission in pancreatic cancer cells led to increased fusion, which resulted in increased mitophagy, reduced mitochondrial mass, and decreased mitochondrial respiration, leading to a reduction in ATP production and a decrease in tumor growth (13).
Increased mitochondrial fusion has also been reported to promote cancer growth (92, 93). According to Humphries et al., treatment of breast cancer cells with leflunomide, a potent activator of mitochondrial fusion proteins, overcame inhibitory effects of fission on migration, signaling, and metastasis (93). Similarly, mining existing datasets for breast cancer showed increased expression of genes associated with mitochondrial fission, which correlated with improved survival in human breast cancer (93). Since increased mitochondrial fusion and fission have been reported to drive tumor growth and vice versa, more mechanistic studies are needed to have a comprehensive understanding of how an imbalance in mitochondrial dynamics causes cancer development and progression.
Increased DRP1 levels also promote metastasis. Zhao et al. showed that invasive breast cancer cell lines have more fragmented mitochondria, higher total and phosphorylated DRP1S616, and lower MFN1 levels in comparison with a non-metastatic cell line (87). Increased mitochondrial fragmentation via activated DRP1 or MFN silencing also resulted in metabolic repositioning to the peripheral cytoskeleton’s lamellipodia, providing a concentrated energy source to power tumor cell motility and invasion (87). Another factor that stimulates migration and invasion in cancer cells is hypoxia. Wan et al. showed that hypoxia led to increased DRP1 expression and mitochondrial fission in glioblastoma U251 cells. They also showed that exogenously expressed GFP-DRP1 enhanced hypoxia-induced migration of U251 cells and mitochondrial division inhibitor-1 (Mdivi-1), a selective dynamin-related protein 1 (DRP1) inhibitor, efficiently attenuated hypoxia-induced migration of U251 cells (88). These studies strongly suggest that increased fission facilitates cancer metastasis.
High-mobility group box 1 (HMGB1) is a cellular response signaling protein released by immune cells following cell death or injury. It is also released into the tumor microenvironment following cell stress or necrosis. Increased HMGB1 levels stimulate pancreatic tumor cell proliferation and ATP production, and its inhibition led to a decrease in tumor growth (94). HMGB1 binds to the receptor for advanced glycation end products (RAGE), a transmembrane receptor overexpressed in cancer. The binding of HMGB1 to RAGE led to the activation of RAGE and increased phosphorylation of Erk1/2, leading to increased mitochondrial ATP production (94). A recent study revealed that HMGB1 protein led to the activation of Erk1/2, which led to the phosphorylation of DRP1, thereby leading to increased tumor growth (95). HMGB1 has also been associated with high glucose levels. Huang et al. showed that culturing human umbilical vein endothelial cells (HUVECs) under high glucose conditions led to increased secretion of HMGB1 (96). Since diabetes is associated with an increased risk of malignancies, HMGB1 may play a role in driving tumor growth and metastasis due to its effects on Erk1/2 activation and DRP-1 phosphorylation. More studies are required to investigate the link between hyperglycemia and imbalance in mitochondrial dynamics as a contributor to tumor growth.
Conclusion and Future perspectives
It appears from the above discussion that mitochondrial fusion and fission play an indispensable role in determining cell fate by maintaining the functional homeostasis of mitochondria. Fission is essential for maintaining the mitochondrial number and proper distribution in the daughter cells, while fusion ensures optimal mitochondrial activity by allowing the exchange of contents between fusing mitochondria. Imbalance in mitochondrial dynamics results in alteration of the mitochondrial number, morphology, and functioning, leading to the development of various diseases, including cancer. At present, the knowledge of molecular events involved in mitochondrial fusion and fission is premature, and more in-depth mechanistic studies are required. Studies are also warranted to better understand how fusion and fission processes are linked to mitophagy, a crucial phenomenon associated with various mitochondrial disorders and human cancers. In addition, it is also important to enhance our knowledge on the crosstalk between various oncogenic signaling and mitochondrial dynamics regulatory pathways. Considering the emerging role of mitochondrial dynamics in determining cellular fate, mitochondria hold great promise in developing novel treatment strategies and biomarkers for disease diagnosis and treatment. Next-generation sequencing of genome, transcriptome, or proteome can identify molecular heterogeneity and relevant pathways associated with the changes in the mitochondrial dynamics in a patient-specific manner. Functional characterization of these alterations utilizing appropriate humanized model systems and clinical validation may open up new avenues for therapeutic interventions.
Acknowledgments:
This work was supported by the National Institutes of Health/National Cancer Institute [CA185490, CA224306 (to AP Singh)] and [CA204801, CA231925 (to S Singh)] and the University of South Alabama Mitchell Cancer Institute. S Dasgupta acknowledges generous support from the University of South Alabama and Mitchell Cancer Institute.
Abbreviations
- TOMM
Translocase of the outer mitochondrial membrane
- SAM
Sorting and assembly machinery complex
- MFN1
Mitofusin 1
- MFN2
Mitofusin 2
- OPA1
Optic atrophy protein 1
- DRP1
Dynamin related protein 1
- Fzo
Fuzzy onions
- HR
Helical repeat
- MTS
Mitochondrial targeting sequence
- DNM2
Dynamin 2
- MID49
Mitochondrial dynamics protein 49
- MID51
Mitochondrial dynamics protein 51
- FIS1
Mitochondrial fission 1 protein
- MFF
Mitochondrial fission factor
- INF2
Inverted formin 2
- CMT2A
Charcot- Marie-Tooth Disease Type 2A
- DOA
Dominant optic atrophy
- sPD
Sporadic Parkinson Disease
- HMGB1
High mobility group box 1
- RAGE
Receptor for advanced glycation end products
Footnotes
Conflict of Interest: The authors declare no conflicts of interests
References
- 1.Srinivasan S, Guha M, Kashina A, and Avadhani NG (2017) Mitochondrial dysfunction and mitochondrial dynamics-The cancer connection. Biochim Biophys Acta Bioenerg 1858, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archibald JM (2015) Endosymbiosis and Eukaryotic Cell Evolution. Curr Biol 25, R911–921 [DOI] [PubMed] [Google Scholar]
- 3.Ma Y, Wang L, and Jia R. (2020) The role of mitochondrial dynamics in human cancers. Am J Cancer Res 10, 1278–1293 [PMC free article] [PubMed] [Google Scholar]
- 4.Chan DC (2020) Mitochondrial Dynamics and Its Involvement in Disease. Annu Rev Pathol 15, 235–259 [DOI] [PubMed] [Google Scholar]
- 5.Vásquez-Trincado C, García-Carvajal I, Pennanen C, Parra V, Hill JA, Rothermel BA, and Lavandero S. (2016) Mitochondrial dynamics, mitophagy and cardiovascular disease. J Physiol 594, 509–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hales KG, and Fuller MT (1997) Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121–129 [DOI] [PubMed] [Google Scholar]
- 7.Sesaki H, Southard SM, Yaffe MP, and Jensen RE (2003) Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol Biol Cell 14, 2342–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, Ehses S, Krause F, Chan DC, Alexander C, Bauer C, Youle R, Langer T, and Martinou JC (2009) SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J 28, 1589–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rambold AS, Kostelecky B, Elia N, and Lippincott-Schwartz J. (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A 108, 10190–10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes LC, Di Benedetto G, and Scorrano L. (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13, 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westermann B. (2012) Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta 1817, 1833–1838 [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Chomyn A, and Chan DC (2005) Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 280, 26185–26192 [DOI] [PubMed] [Google Scholar]
- 13.Yu M, Nguyen ND, Huang Y, Lin D, Fujimoto TN, Molkentine JM, Deorukhkar A, Kang Y, San Lucas FA, Fernandes CJ, Koay EJ, Gupta S, Ying H, Koong AC, Herman JM, Fleming JB, Maitra A, and Taniguchi CM (2019) Mitochondrial fusion exploits a therapeutic vulnerability of pancreatic cancer. JCI Insight 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Brito OM, and Scorrano L. (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 [DOI] [PubMed] [Google Scholar]
- 15.Mattie S, Riemer J, Wideman JG, and McBride HM (2018) A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J Cell Biol 217, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi Y, Yan L, Yu C, Guo X, Zhou X, Hu X, Huang X, Rao Z, Lou Z, and Hu J. (2016) Structures of human mitofusin 1 provide insight into mitochondrial tethering. J Cell Biol 215, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao YL, Meng S, Chen Y, Feng JX, Gu DD, Yu B, Li YJ, Yang JY, Liao S, Chan DC, and Gao S. (2017) MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 542, 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pyakurel A, Savoia C, Hess D, and Scorrano L. (2015) Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol Cell 58, 244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JY, Kapur M, Li M, Choi MC, Choi S, Kim HJ, Kim I, Lee E, Taylor JP, and Yao TP (2014) MFN1 deacetylation activates adaptive mitochondrial fusion and protects metabolically challenged mitochondria. J Cell Sci 127, 4954–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, and Weissman AM (2012) Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell 47, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, and Dorn GW (2013) PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340, 471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tilokani L, Nagashima S, Paupe V, and Prudent J. (2018) Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem 62, 341–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olichon A, Elachouri G, Baricault L, Delettre C, Belenguer P, and Lenaers G. (2007) OPA1 alternate splicing uncouples an evolutionary conserved function in mitochondrial fusion from a vertebrate restricted function in apoptosis. Cell Death Differ 14, 682–692 [DOI] [PubMed] [Google Scholar]
- 24.Patrushev MV, Mazunin IO, Vinogradova EN, and Kamenski PA (2015) Mitochondrial Fission and Fusion. Biochemistry (Mosc) 80, 1457–1464 [DOI] [PubMed] [Google Scholar]
- 25.Griparic L, Kanazawa T, and van der Bliek AM (2007) Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol 178, 757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Z, Chen H, Fiket M, Alexander C, and Chan DC (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol 178, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baricault L, Ségui B, Guégand L, Olichon A, Valette A, Larminat F, and Lenaers G. (2007) OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp Cell Res 313, 3800–3808 [DOI] [PubMed] [Google Scholar]
- 28.Ge Y, Shi X, Boopathy S, McDonald J, Smith AW, and Chao LH (2020) Two forms of Opa1 cooperate to complete fusion of the mitochondrial inner-membrane. Elife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihara N, Fujita Y, Oka T, and Mihara K. (2006) Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J 25, 2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ban T, Ishihara T, Kohno H, Saita S, Ichimura A, Maenaka K, Oka T, Mihara K, and Ishihara N. (2017) Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat Cell Biol 19, 856–863 [DOI] [PubMed] [Google Scholar]
- 31.Samant SA, Zhang HJ, Hong Z, Pillai VB, Sundaresan NR, Wolfgeher D, Archer SL, Chan DC, and Gupta MP (2014) SIRT3 deacetylates and activates OPA1 to regulate mitochondrial dynamics during stress. Mol Cell Biol 34, 807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westermann B. (2010) Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11, 872–884 [DOI] [PubMed] [Google Scholar]
- 33.Elgass K, Pakay J, Ryan MT, and Palmer CS (2013) Recent advances into the understanding of mitochondrial fission. Biochim Biophys Acta 1833, 150–161 [DOI] [PubMed] [Google Scholar]
- 34.Detmer SA, and Chan DC (2007) Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol 8, 870–879 [DOI] [PubMed] [Google Scholar]
- 35.Chan DC (2006) Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol 22, 79–99 [DOI] [PubMed] [Google Scholar]
- 36.Machiela E, Liontis T, Dues DJ, Rudich PD, Traa A, Wyman L, Kaufman C, Cooper JF, Lew L, Nadarajan S, Senchuk MM, and Van Raamsdonk JM (2020) Disruption of mitochondrial dynamics increases stress resistance through activation of multiple stress response pathways. FASEB J 34, 8475–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanazawa T, Zappaterra MD, Hasegawa A, Wright AP, Newman-Smith ED, Buttle KF, McDonald K, Mannella CA, and van der Bliek AM (2008) The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet 4, e1000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smirnova E, Griparic L, Shurland DL, and van der Bliek AM (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12, 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan DC (2012) Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet 46, 265–287 [DOI] [PubMed] [Google Scholar]
- 40.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, and Voeltz GK (2011) ER tubules mark sites of mitochondrial division. Science 334, 358–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, and Lippincott-Schwartz J. (2015) A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Korobova F, Ramabhadran V, and Higgs HN (2013) An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Xu S, Roelofs BA, Boyman L, Lederer WJ, Sesaki H, and Karbowski M. (2015) Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J Cell Biol 208, 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehklau K, Hoffmann L, Gurniak CB, Ott M, Witke W, Scorrano L, Culmsee C, and Rust MB (2017) Cofilin1-dependent actin dynamics control DRP1-mediated mitochondrial fission. Cell Death Dis 8, e3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamerkar SC, Kraus F, Sharpe AJ, Pucadyil TJ, and Ryan MT (2018) Dynamin-related protein 1 has membrane constricting and severing abilities sufficient for mitochondrial and peroxisomal fission. Nat Commun 9, 5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chakrabarti R, Ji WK, Stan RV, de Juan Sanz J, Ryan TA, and Higgs HN (2018) INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol 217, 251–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basañez G, Terrones O, and Martinou JC (2014) Specific interaction with cardiolipin triggers functional activation of Dynamin-Related Protein 1. PLoS One 9, e102738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, and Ramachandran R. (2014) A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell 25, 1905–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taguchi N, Ishihara N, Jofuku A, Oka T, and Mihara K. (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 282, 11521–11529 [DOI] [PubMed] [Google Scholar]
- 50.Kashatus JA, Nascimento A, Myers LJ, Sher A, Byrne FL, Hoehn KL, Counter CM, and Kashatus DF (2015) Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol Cell 57, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cribbs JT, and Strack S. (2007) Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 8, 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, and Scorrano L. (2008) Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A 105, 15803–15808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prudent J, Zunino R, Sugiura A, Mattie S, Shore GC, and McBride HM (2015) MAPL SUMOylation of Drp1 Stabilizes an ER/Mitochondrial Platform Required for Cell Death. Mol Cell 59, 941–955 [DOI] [PubMed] [Google Scholar]
- 54.Gawlowski T, Suarez J, Scott B, Torres-Gonzalez M, Wang H, Schwappacher R, Han X, Yates JR, Hoshijima M, and Dillmann W. (2012) Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-β-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J Biol Chem 287, 30024–30034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho DH, Nakamura T, Fang J, Cieplak P, Godzik A, Gu Z, and Lipton SA (2009) S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science 324, 102–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Song P, Du L, Tian W, Yue W, Liu M, Li D, Wang B, Zhu Y, Cao C, Zhou J, and Chen Q (2011) Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J Biol Chem 286, 11649–11658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuppia G, Rizzo F, Riboldi G, Del Bo R, Nizzardo M, Simone C, Comi GP, Bresolin N, and Corti S. (2015) MFN2-related neuropathies: Clinical features, molecular pathogenesis and therapeutic perspectives. J Neurol Sci 356, 7–18 [DOI] [PubMed] [Google Scholar]
- 58.Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, Zappia M, Nelis E, Patitucci A, Senderek J, Parman Y, Evgrafov O, Jonghe PD, Takahashi Y, Tsuji S, Pericak-Vance MA, Quattrone A, Battaloglu E, Polyakov AV, Timmerman V, Schröder JM, Vance JM, and Battologlu E. (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet 36, 449–451 [DOI] [PubMed] [Google Scholar]
- 59.Lawson VH, Graham BV, and Flanigan KM (2005) Clinical and electrophysiologic features of CMT2A with mutations in the mitofusin 2 gene. Neurology 65, 197–204 [DOI] [PubMed] [Google Scholar]
- 60.Verhoeven K, Claeys KG, Züchner S, Schröder JM, Weis J, Ceuterick C, Jordanova A, Nelis E, De Vriendt E, Van Hul M, Seeman P, Mazanec R, Saifi GM, Szigeti K, Mancias P, Butler IJ, Kochanski A, Ryniewicz B, De Bleecker J, Van den Bergh P, Verellen C, Van Coster R, Goemans N, Auer-Grumbach M, Robberecht W, Milic Rasic V, Nevo Y, Tournev I, Guergueltcheva V, Roelens F, Vieregge P, Vinci P, Moreno MT, Christen HJ, Shy ME, Lupski JR, Vance JM, De Jonghe P, and Timmerman V. (2006) MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain 129, 2093–2102 [DOI] [PubMed] [Google Scholar]
- 61.Bombelli F, Stojkovic T, Dubourg O, Echaniz-Laguna A, Tardieu S, Larcher K, Amati-Bonneau P, Latour P, Vignal O, Cazeneuve C, Brice A, and Leguern E. (2014) Charcot-Marie-Tooth disease type 2A: from typical to rare phenotypic and genotypic features. JAMA Neurol 71, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 62.Feely SM, Laura M, Siskind CE, Sottile S, Davis M, Gibbons VS, Reilly MM, and Shy ME (2011) MFN2 mutations cause severe phenotypes in most patients with CMT2A. Neurology 76, 1690–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bannerman P, Burns T, Xu J, Miers L, and Pleasure D. (2016) Mice Hemizygous for a Pathogenic Mitofusin-2 Allele Exhibit Hind Limb/Foot Gait Deficits and Phenotypic Perturbations in Nerve and Muscle. PLoS One 11, e0167573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Detmer SA, Vande Velde C, Cleveland DW, and Chan DC (2008) Hindlimb gait defects due to motor axon loss and reduced distal muscles in a transgenic mouse model of Charcot-Marie-Tooth type 2A. Hum Mol Genet 17, 367–375 [DOI] [PubMed] [Google Scholar]
- 65.Cartoni R, Arnaud E, Médard JJ, Poirot O, Courvoisier DS, Chrast R, and Martinou JC (2010) Expression of mitofusin 2(R94Q) in a transgenic mouse leads to Charcot-Marie-Tooth neuropathy type 2A. Brain 133, 1460–1469 [DOI] [PubMed] [Google Scholar]
- 66.Lenaers G, Hamel C, Delettre C, Amati-Bonneau P, Procaccio V, Bonneau D, Reynier P, and Milea D. (2012) Dominant optic atrophy. Orphanet J Rare Dis 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertholet AM, Delerue T, Millet AM, Moulis MF, David C, Daloyau M, Arnauné-Pelloquin L, Davezac N, Mils V, Miquel MC, Rojo M, and Belenguer P. (2016) Mitochondrial fusion/fission dynamics in neurodegeneration and neuronal plasticity. Neurobiol Dis 90, 3–19 [DOI] [PubMed] [Google Scholar]
- 68.Ferré M, Caignard A, Milea D, Leruez S, Cassereau J, Chevrollier A, Amati-Bonneau P, Verny C, Bonneau D, Procaccio V, and Reynier P. (2015) Improved locus-specific database for OPA1 mutations allows inclusion of advanced clinical data. Hum Mutat 36, 20–25 [DOI] [PubMed] [Google Scholar]
- 69.Ban T, Heymann JA, Song Z, Hinshaw JE, and Chan DC (2010) OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum Mol Genet 19, 2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davies VJ, Hollins AJ, Piechota MJ, Yip W, Davies JR, White KE, Nicols PP, Boulton ME, and Votruba M. (2007) Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum Mol Genet 16, 1307–1318 [DOI] [PubMed] [Google Scholar]
- 71.White KE, Davies VJ, Hogan VE, Piechota MJ, Nichols PP, Turnbull DM, and Votruba M. (2009) OPA1 deficiency associated with increased autophagy in retinal ganglion cells in a murine model of dominant optic atrophy. Invest Ophthalmol Vis Sci 50, 2567–2571 [DOI] [PubMed] [Google Scholar]
- 72.Liang CL, Wang TT, Luby-Phelps K, and German DC (2007) Mitochondria mass is low in mouse substantia nigra dopamine neurons: implications for Parkinson’s disease. Exp Neurol 203, 370–380 [DOI] [PubMed] [Google Scholar]
- 73.Su B, Wang X, Zheng L, Perry G, Smith MA, and Zhu X. (2010) Abnormal mitochondrial dynamics and neurodegenerative diseases. Biochim Biophys Acta 1802, 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arduíno DM, Esteves AR, and Cardoso SM (2011) Mitochondrial fusion/fission, transport and autophagy in Parkinson’s disease: when mitochondria get nasty. Parkinsons Dis 2011, 767230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trimmer PA, Swerdlow RH, Parks JK, Keeney P, Bennett JP, Miller SW, Davis RE, and Parker WD (2000) Abnormal mitochondrial morphology in sporadic Parkinson’s and Alzheimer’s disease cybrid cell lines. Exp Neurol 162, 37–50 [DOI] [PubMed] [Google Scholar]
- 76.Smigrodzki R, Parks J, and Parker WD (2004) High frequency of mitochondrial complex I mutations in Parkinson’s disease and aging. Neurobiol Aging 25, 1273–1281 [DOI] [PubMed] [Google Scholar]
- 77.Arduíno DM, Esteves AR, Cortes L, Silva DF, Patel B, Grazina M, Swerdlow RH, Oliveira CR, and Cardoso SM (2012) Mitochondrial metabolism in Parkinson’s disease impairs quality control autophagy by hampering microtubule-dependent traffic. Hum Mol Genet 21, 4680–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esteves AR, Arduíno DM, Swerdlow RH, Oliveira CR, and Cardoso SM (2010) Microtubule depolymerization potentiates alpha-synuclein oligomerization. Front Aging Neurosci 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Esteves AR, Gozes I, and Cardoso SM (2014) The rescue of microtubule-dependent traffic recovers mitochondrial function in Parkinson’s disease. Biochim Biophys Acta 1842, 7–21 [DOI] [PubMed] [Google Scholar]
- 80.Reddy PH (2014) Increased mitochondrial fission and neuronal dysfunction in Huntington’s disease: implications for molecular inhibitors of excessive mitochondrial fission. Drug Discov Today 19, 951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos D, Esteves AR, Silva DF, Januário C, and Cardoso SM (2015) The Impact of Mitochondrial Fusion and Fission Modulation in Sporadic Parkinson’s Disease. Mol Neurobiol 52, 573–586 [DOI] [PubMed] [Google Scholar]
- 82.Gimbrone MA, and García-Cardeña G. (2016) Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res 118, 620–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salabei JK, and Hill BG (2013) Mitochondrial fission induced by platelet-derived growth factor regulates vascular smooth muscle cell bioenergetics and cell proliferation. Redox Biol 1, 542–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Q, Zhang M, Torres G, Wu S, Ouyang C, Xie Z, and Zou MH (2017) Metformin Suppresses Diabetes-Accelerated Atherosclerosis via the Inhibition of Drp1-Mediated Mitochondrial Fission. Diabetes 66, 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Javadov S, Rajapurohitam V, Kilić A, Hunter JC, Zeidan A, Said Faruq N, Escobales N, and Karmazyn M. (2011) Expression of mitochondrial fusion-fission proteins during post-infarction remodeling: the effect of NHE-1 inhibition. Basic Res Cardiol 106, 99–109 [DOI] [PubMed] [Google Scholar]
- 86.Givvimani S, Munjal C, Tyagi N, Sen U, Metreveli N, and Tyagi SC (2012) Mitochondrial division/mitophagy inhibitor (Mdivi) ameliorates pressure overload induced heart failure. PLoS One 7, e32388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao J, Zhang J, Yu M, Xie Y, Huang Y, Wolff DW, Abel PW, and Tu Y. (2013) Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 32, 4814–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wan YY, Zhang JF, Yang ZJ, Jiang LP, Wei YF, Lai QN, Wang JB, Xin HB, and Han XJ (2014) Involvement of Drp1 in hypoxia-induced migration of human glioblastoma U251 cells. Oncol Rep 32, 619–626 [DOI] [PubMed] [Google Scholar]
- 89.Kannan A, Wells RB, Sivakumar S, Komatsu S, Singh KP, Samten B, Philley JV, Sauter ER, Ikebe M, Idell S, Gupta S, and Dasgupta S. (2016) Mitochondrial Reprogramming Regulates Breast Cancer Progression. Clin Cancer Res 22, 3348–3360 [DOI] [PubMed] [Google Scholar]
- 90.Qian W, Wang J, and Van Houten B. (2013) The role of dynamin-related protein 1 in cancer growth: a promising therapeutic target? Expert Opin Ther Targets 17, 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maycotte P, Marín-Hernández A, Goyri-Aguirre M, Anaya-Ruiz M, Reyes-Leyva J, and Cortés-Hernández P. (2017) Mitochondrial dynamics and cancer. Tumour Biol 39, 1010428317698391 [DOI] [PubMed] [Google Scholar]
- 92.Li M, Wang L, Wang Y, Zhang S, Zhou G, Lieshout R, Ma B, Liu J, Qu C, Verstegen MMA, Sprengers D, Kwekkeboom J, van der Laan LJW, Cao W, Peppelenbosch MP, and Pan Q. (2020) Mitochondrial Fusion Via OPA1 and MFN1 Supports Liver Tumor Cell Metabolism and Growth. Cells 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Humphries BA, Cutter AC, Buschhaus JM, Chen YC, Qyli T, Palagama DSW, Eckley S, Robison TH, Bevoor A, Chiang B, Haley HR, Sahoo S, Spinosa PC, Neale DB, Boppisetti J, Sahoo D, Ghosh P, Lahann J, Ross BD, Yoon E, Luker KE, and Luker GD (2020) Enhanced mitochondrial fission suppresses signaling and metastasis in triple-negative breast cancer. Breast Cancer Res 22, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kang R, Tang D, Schapiro NE, Loux T, Livesey KM, Billiar TR, Wang H, Van Houten B, Lotze MT, and Zeh HJ (2014) The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 33, 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang CY, Chiang SF, Chen WT, Ke TW, Chen TW, You YS, Lin CY, and Chao KSC (2018) HMGB1 promotes ERK-mediated mitochondrial Drp1 phosphorylation for chemoresistance through RAGE in colorectal cancer. Cell Death Dis 9, 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang X, Jiang J, Huang L, Ren Q, Gao X, and Yu S. (2020) Ropivacaine Prevents the Activation of the NLRP3 Inflammasome Caused by High Glucose in HUVECs. ACS Omega 5, 23413–23419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Z, Li TE, Chen M, Xu D, Zhu Y, Hu BY, Lin ZF, Pan JJ, Wang X, Wu C, Zheng Y, Lu L, Jia HL, Gao S, Dong QZ, and Qin LX (2020) MFN1-dependent alteration of mitochondrial dynamics drives hepatocellular carcinoma metastasis by glucose metabolic reprogramming. Br J Cancer 122, 209–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen L, Zhang J, Lyu Z, Chen Y, Ji X, Cao H, Jin M, Zhu J, Yang J, Ling R, Xing J, Ren T, and Lyu Y. (2018) Positive feedback loop between mitochondrial fission and Notch signaling promotes survivin-mediated survival of TNBC cells. Cell Death Dis 9, 1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang Q, Zhan L, Cao H, Li J, Lyu Y, Guo X, Zhang J, Ji L, Ren T, An J, Liu B, Nie Y, and Xing J. (2016) Increased mitochondrial fission promotes autophagy and hepatocellular carcinoma cell survival through the ROS-modulated coordinated regulation of the NFKB and TP53 pathways. Autophagy 12, 999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng CT, Kuo CY, Ouyang C, Li CF, Chung Y, Chan DC, Kung HJ, and Ann DK (2016) Metabolic Stress-Induced Phosphorylation of KAP1 Ser473 Blocks Mitochondrial Fusion in Breast Cancer Cells. Cancer Res 76, 5006–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xie Q, Wu Q, Horbinski CM, Flavahan WA, Yang K, Zhou W, Dombrowski SM, Huang Z, Fang X, Shi Y, Ferguson AN, Kashatus DF, Bao S, and Rich JN (2015) Mitochondrial control by DRP1 in brain tumor initiating cells. Nat Neurosci 18, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rehman J, Zhang HJ, Toth PT, Zhang Y, Marsboom G, Hong Z, Salgia R, Husain AN, Wietholt C, and Archer SL (2012) Inhibition of mitochondrial fission prevents cell cycle progression in lung cancer. FASEB J 26, 2175–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]