Abstract
Proline-rich macrocyclic peptides (PRMPs) are natural products present in geographically and phylogenetically dispersed marine sponges. The large diversity and low abundance of PRMPs in sponge metabolomes precludes isolation and structure elucidation of each individual PRMP congener. Here, using standards developed via biomimetic enzymatic synthesis of PRMPs, a mass spectrometry-based workflow to sequence PRMPs was developed and validated to reveal that the diversity of PRMPs in marine sponges is much greater than that has been realized by natural product isolation-based strategies. Findings are placed in the context of diversity-oriented transamidative macrocyclization of peptide substrates in sponge holobionts.
Keywords: chemoenzymatic synthesis, cyclopeptides, fragmentation, marine sponge
Graphical Abstract
By adopting an integrated approach of chemoenzymatic synthesis of standards and HCD-MSn fragmentation, we have sequenced proline-rich cyclopeptide natural products from marine sponges. Our MSn fragmentation workflow successfully differentiated between leucine and isoleucine residues in the cyclopeptides and can be used to sequence low abundance cyclic peptides from sub-gram amounts of sponge biomass.
Macrocyclization of ribosomal and non-ribosomal peptides is ubiquitous in natural product chemistry. Macrocyclization imparts pharmacologically desirable properties of stability and rigidity which reduces the entropic cost associated with target binding.[1] From neutrophil defensins and snake venom toxins to cyclic peptide natural products produced by bacteria, across all scales of life, macrocyclic peptides demonstrate potent bioactivities and serve various different biological roles.[2] Benthic marine invertebrates- sponges, are prolific sources of proline rich macrocyclic peptides (PRMPs).[3] PRMPs are detected in marine sponges without restriction of geographical location and are widespread among several sponge phyla. Cell growth inhibitory cytotoxic activities of sponge derived PRMPs has sustained interest in their development as drug candidates.[3] Unlike other natural product chemical classes, the abundance of which dominates the sponge metabolomes, PRMPs in sponges are present in trace amounts. For instance, in Stylissa spp. sponges the metabolomes of which are enriched in pyrrole imidazole alkaloids (PIAs),[4] PRMPs were isolated in 0.01% yields (PRMP weight/sponge dry weight).[5] Low abundance of PRMPs interspersed with much higher abundance other natural products in sponge extracts encumbers their structure elucidation using natural product isolation-based strategies. Here, we devise a mass spectrometry-based workflow to sequence sponge-derived PRMPs and validate the spectrometric sequencing using standards furnished by biomimetic enzymatic peptide macrocyclization.
We previously described the detection of PIAs in Stylissa and Axinella spp. sponges using LC/MS-based untargeted metabolomics.[6] Screening these LC/MS data using the DEREPLICATOR tool[7] hinted at the presence of peptidic natural products in Stylissa and Axinella spp. extracts consistent with prior reports describing the isolation of PRMPs from these sponge genera.[3] The low abundance of PRMPs and sub-gram amounts of dry sponge biomass which were available to us precluded a natural product isolation-based effort. Thus, we decided to pursue PRMP structure elucidation from mass spectrometry fragmentation data.
In a proline containing peptide, fragmentation preferentially occurs at the N-terminus of the Pro residue.[8] Sequencing PRMPs detected in sponge extracts thus began with the annotation of the NPro-XaaC dipeptide oxonium MS2 fragment ions per the inventory shown in Table S1. Proceeding from the NPro-XaaC dipeptide, to recover the PRMP sequence, we annotated mass shifts corresponding to proteinogenic amino acids till we reached the parent ion in the MS2 spectra. As shown in Fig. 1A for a PRMP detected in Axinella sp., PRMPs containing more than one proline residue yielded multiple NPro-XaaC dipeptide MS2 fragments. Progressing from each of these different dipeptide fragments, NPro-PheC (MS2 m/z 245) and NPro-GluC (MS2 m/z 227), the same cyclic sequence, cyclo(FFPEXWP), was recovered. Here, X denotes Leu/Ile residues that cannot be differentiated based on MS2 fragmentation alone. Identities of the amino acid constituents in cyclo(FFPEXWP), Phe, Pro, Glu, Leu/Ile, and Trp, were supported by the detection of the individual amino acyl immonium ions in the MS2 spectra (Fig. S1, Table S2).
Figure 1.
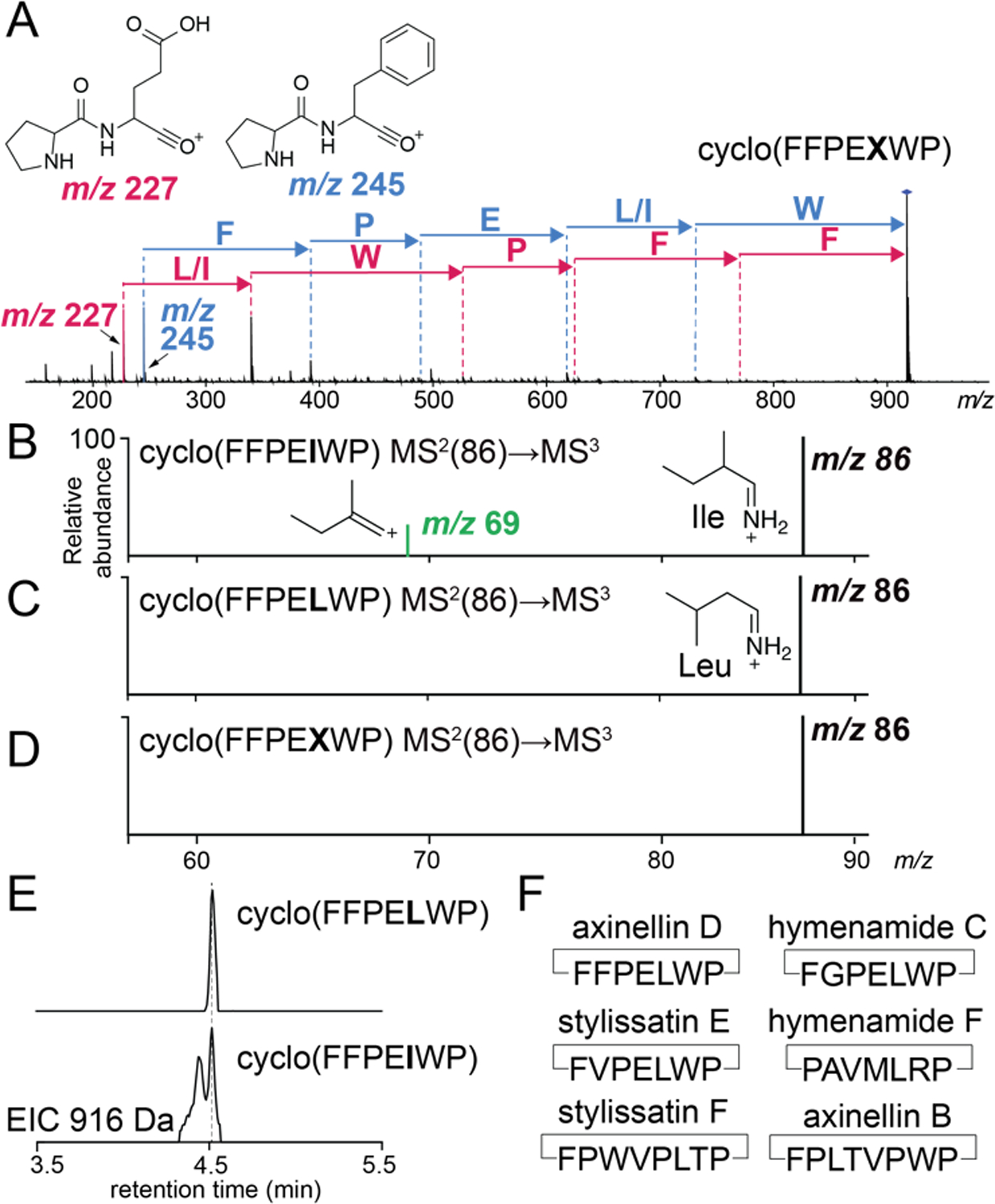
(A) MS2 spectra for cyclo(FFPEXWP). MS2(86)→MS3 spectra for the (B) Ile-derived MS2 m/z 86 precursor ion (cyclo(FFPEIWP) standard), (C) Leu-derived MS2 m/z 86 precursor ion (cyclo(FFPELWP) standard), and the (D) Leu/Ile-derived MS2 m/z 86 precursor ion (cyclo(FFPEXWP) PRMP). (E) Extracted ion chromatograms (EICs) showing co-injection of cyclo(FFPELWP) (top) and cyclo(FFPEIWP) (bottom) standards with the sponge extract. (F) Inventory of single Leu/Ile containing PRMPs sequenced in this study.
To resolve Leu and Ile residues, we developed standards for the two possible PRMPs– cyclo(FFPELWP) and cyclo(FFPEIWP). For this, synthetic linear peptides FFPELWPFQA and FFPEIWPFQA were obtained. The C-terminal FQA tripeptide was proteolyzed and transamidatively macrocyclized products were furnished by the plant peptidase PCY1.[9] Upon fragmentation of the cyclo(FFPELWP) and cyclo(FFPEIWP) products, the MS2 immonium ion for Leu/Ile, m/z 86, was isolated and fragmented. Consistent with prior reports,[10] fragmentation of the Ile-derived MS2 m/z 86 immonium ion generated a characteristic MS3 m/z 69 product ion in high abundance, while the MS3 m/z 69 product ion was detected in lesser abundance when the MS2 m/z 86 precursor ion was derived from Leu. By modulating the MS3 fragmentation energy (Fig. S2), we generated an unambiguous binary result in which the MS3 m/z 69 product ion was observed only for Ile (Fig. 1B), but not observed for Leu (Fig. 1C). Adopting the thus optimized methodology for the cyclo(FFPEXWP) PRMP detected in the sponge extract, we annotated X as Leu (Fig. 1D). Validation of the mass spectrometry-based result was obtained by spiking the sponge extract with enzymatically synthesized cyclo(FFPELWP) and cyclo(FFPEIWP) standards; the cyclo(FFPELWP) standard coeluted with the sponge-derived PRMP (Fig. 1E) which we term as axinellin D. PRMP congeners detected in this study are highlighted in Table S3.
Employing the now validated mass spectrometry workflow which could differentiate between Leu and Ile residues, we additionally sequenced the PRMPs stylissatin E cyclo(FVPELWP) and stylissatin F cyclo(FPWVPLTP) from Stylissa sp. specimens used in this study (Fig. 1F, S3–6, Table S3). We also detected the presence of the PRMPs cyclo(FGPELWP), cyclo(PAVMLRP), and cyclo(FPLTVPWP) in Axinella sp. extracts (Fig. S7–12). Primary sequences of these PRMPs correspond to the previously described hymenamide C, hymenamide F, and axinellin B. The presence of axinellin B serves as an internal control as it has been reported from an Axinella sponge previously.[11] However, hymenamides C and F were isolated from Hymeniacidon sp. sponges (Table S3).[12] Hymeniacidon sp. sponges belong to a different phylogenetic order (Suberitidia) as compared to Axinella sp. (order Axinellida) within the Demospongiae class of marine sponges.
Abovementioned workflow is applicable when a single Leu/Ile is present in the peptide sequence. When more than one Leu/Ile residues are present, multiple side chains will contribute to MS2 m/z 86 ion which will complicate the MS2(86)→MS3-based Leu/Ile distinction. This scenario presents itself for the cyclo(FYSX1AX2P) PRMP that we detected in Axinella sp.; Xn denote Leu/Ile residues. As before, the PRMP was sequenced starting from the NPro-PheC MS2 m/z 245 fragment ion (Fig. 2A, S13). Next, the ion corresponding to the fragment PFYSX1A, MS2 m/z 679 (Fig. 2A) was isolated and the MS3 m/z 86 Leu/Ile immonium ion was generated. Here, only a single residue, X1, contributes to the production of the MS3 m/z 86 product ion. Further fragmentation of the MS3 m/z 86 ion did not generate the characteristic MS4 m/z 69 ion (Fig. 2B), denoting that X1 was Leu.
Figure 2.
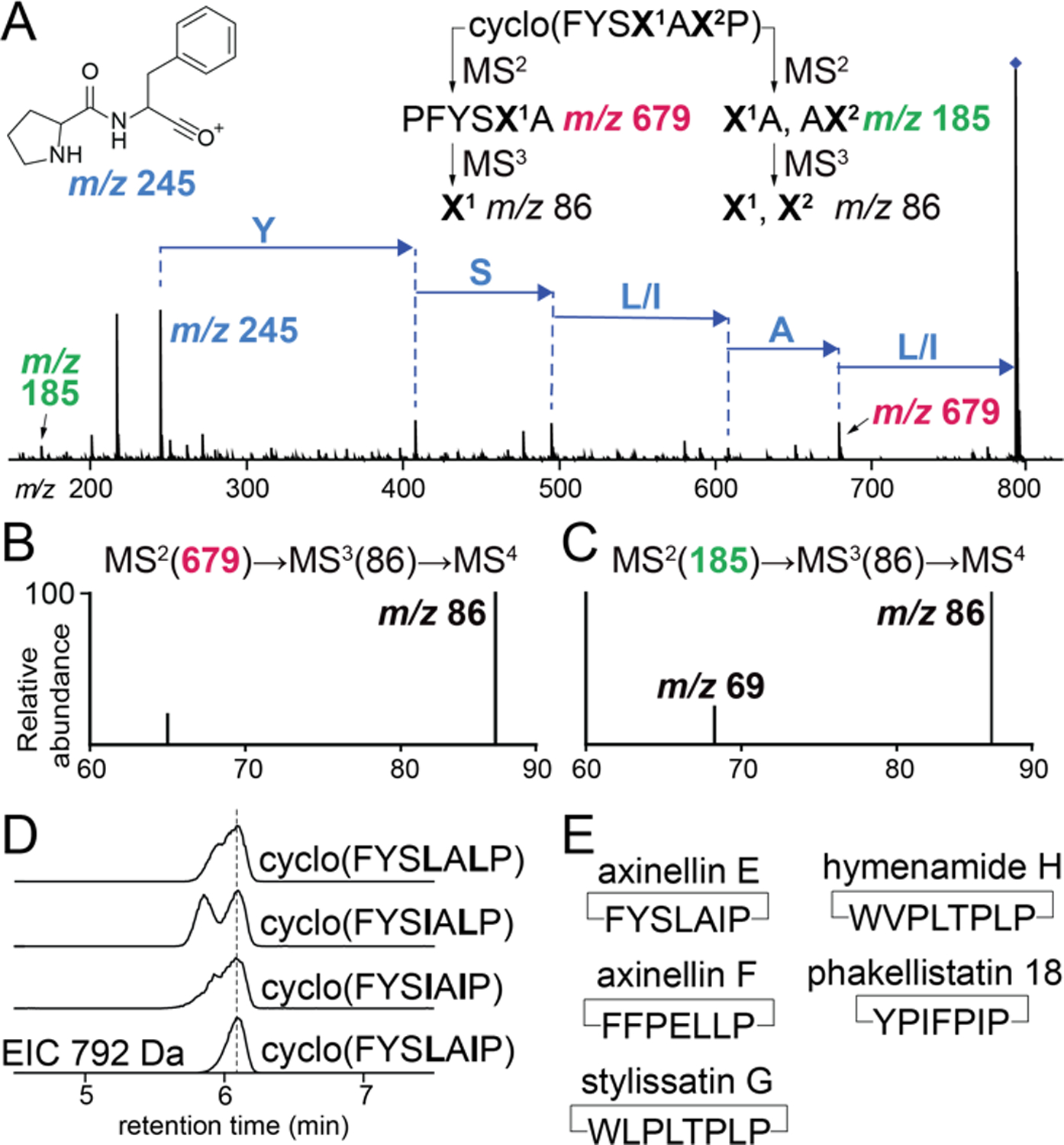
(A) MS2 spectra for the PRMP cyclo(FYSX1AX2P) detected in Axinella sp. where X corresponds to Leu/Ile. MSn fragmentation strategies to query X1 and X2 identities are illustrated. (B) Confirmation of X1 as Leu as MS4 m/z 69 product ion is not detected. (C) Confirmation of X2 as Ile as MS4 m/z 69 product ion is detected. (D) EICs showing co-injection of enzymatically synthesized PRMP standards with the sponge extract. (E) Inventory of multiple Leu/Ile containing PRMPs sequenced in this study.
Next, the cyclo(FYSLALP), cyclo(FYSIALP), cyclo(FYSLAIP), and cyclo(FYSIAIP) standards were generated by PCY1-mediated macrocyclization of the corresponding -FQA appended linear peptides. Fragmenting cyclo(FYSIALP) and cyclo(FYSIAIP) using the MS2(679)→MS3(86)→MS4 procedure yielded the characteristic MS4 m/z 69 ion while fragmentation of cyclo(FYSLALP) and cyclo(FYSLAIP) did not (Fig. S14). To query the identity of X2 in the PRMP cyclo(FYSX1AX2P), the MS2 m/z 185 ion was isolated. This MS2 ion can be generated by the X1A, or the AX2 dipeptides. The MS2(185)→MS3(86)→MS4 fragmentation scheme demonstrated the production of the characteristic MS4 m/z 69 ion (Fig. 2C). As X1 was Leu (and hence the X1A fragment could not contribute to the production of the MS4 m/z 69 product ion), these data denote that X2 was Ile. Analogous MS2(185)→MS3(86)→MS4 fragmentation for the cyclo(FYSLAIP) standard generated the MS4 m/z 69 ion while the cyclo(FYSLALP) standard did not (Fig. S15). Validation of the mass spectrometry-based PRMP sequencing scheme was obtained by spiking the sponge extract with the PCY1 generated standards; the cyclo(FYSLAIP) standard coeluted with the sponge PRMP which is termed axinellin E (Fig. 2D, Table S3).
Using the workflow developed above, we additionally sequenced the multiple Leu/Ile containing PRMPs axinellin F cyclo(FFPELLP) from Axinella sp., and stylissatin G cyclo(WLPLTPLP) from Stylissa sp. (Fig. 2E, Fig. S16–19, Table S3). In addition to these PRMPs, we detected the presence of the previously described PRMPs hymenamide H[13]- cyclo(WVPLTPLP) and phakellistatin 18[14]- cyclo(YPIFPIP) from Stylissa sp. (Fig. 2E, Fig. S20–23). Hymenamide H was described from Hymeniacidon sp. (order Suberitidia) while phakellistatin 18 was described from Phakellia fusca (order Axinellida), sponges belonging to orders different from Stylissa sp. (order Scopalinida). Together with the detection of hymenamide C in Axinella sp., it is evident that PRMP congeners are shared among phylogenetically different sponges.
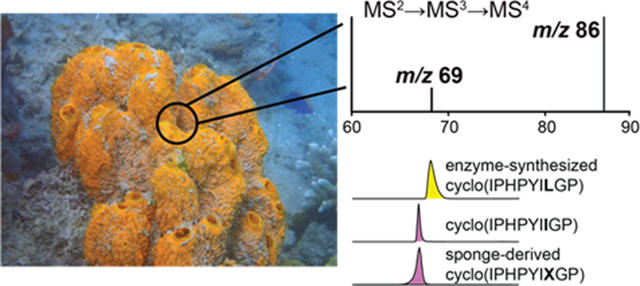
Unlike axinellin F, cyclo(FFPELLP), where Ile residues occur next to Leu/Ile residues in a PRMP sequence, mass spectrometry alone may be insufficient to provide unambiguous results. This scenario presented itself during the sequencing of the PRMP cyclo(X1PHPYX2X3GP) that we detected in Axinella sp. As before, the PRMP was sequenced starting from the NPro-HisC (m/z 235), NPro-TyrC (m/z 261), and NPro-X1C (m/z 211) dipeptide MS2 ions (Fig. 3A). Using PCY1 and appropriately designed linear peptide substrates, we generated standards for all eight possible Leu/Ile combinations for cyclo(X1PHPYX2X3GP). Residue X1 was annotated as Ile by isolating the MS2 m/z 211 ion corresponding to the PX1 dipeptide and following the MS2(211)→MS3(86)→MS4 fragmentation scheme (Fig. 3B). Annotation of X1 as Ile was supported by analogous fragmentation of the cyclo(LPHPYX2X3GP) and cyclo(IPHPYX2X3GP) standards (Fig. S24–25). Similarly, identity of X2 as Ile was decided by isolating the MS2 m/z 608 ion corresponding to the PHPYX2 pentapeptide and following the MS2(608)→MS3(86)→MS4 fragmentation scheme (Fig. 3C) and comparing the MS4 spectra to cyclo(IPHPYLX3GP) and cyclo(IPHPYIX3GP) standards (Fig. S26). Because X2 was Ile (which generates the MS4 m/z 69 diagnostic ion) and no MS2 fragment could be detected which contained only X3 without X1 or X2, identity of X3 could not be determined by mass spectral fragmentation. To annotate X3, retention times for the cyclo(IPHPYILGP) (X1,X2=Ile; X3=Leu) and cyclo(IPHPYIIGP) (X1,X2=Ile; X3=Ile) standards were compared to reveal the sponge PRMP as cyclo(IPHPYIIGP) which we term axinellin G (Fig. 3D, Table S3). Among the inventory of PRMPs, cyclo-nonapeptides such as axinellin G are relatively rare (Table S3). Overall, we demonstrate the utility of enzymatic synthesis of cyclic peptide standards for discovery and sequencing of sponge PRMPs. At present, we cannot determine the cis/trans conformation of the Pro residues.
Figure 3.
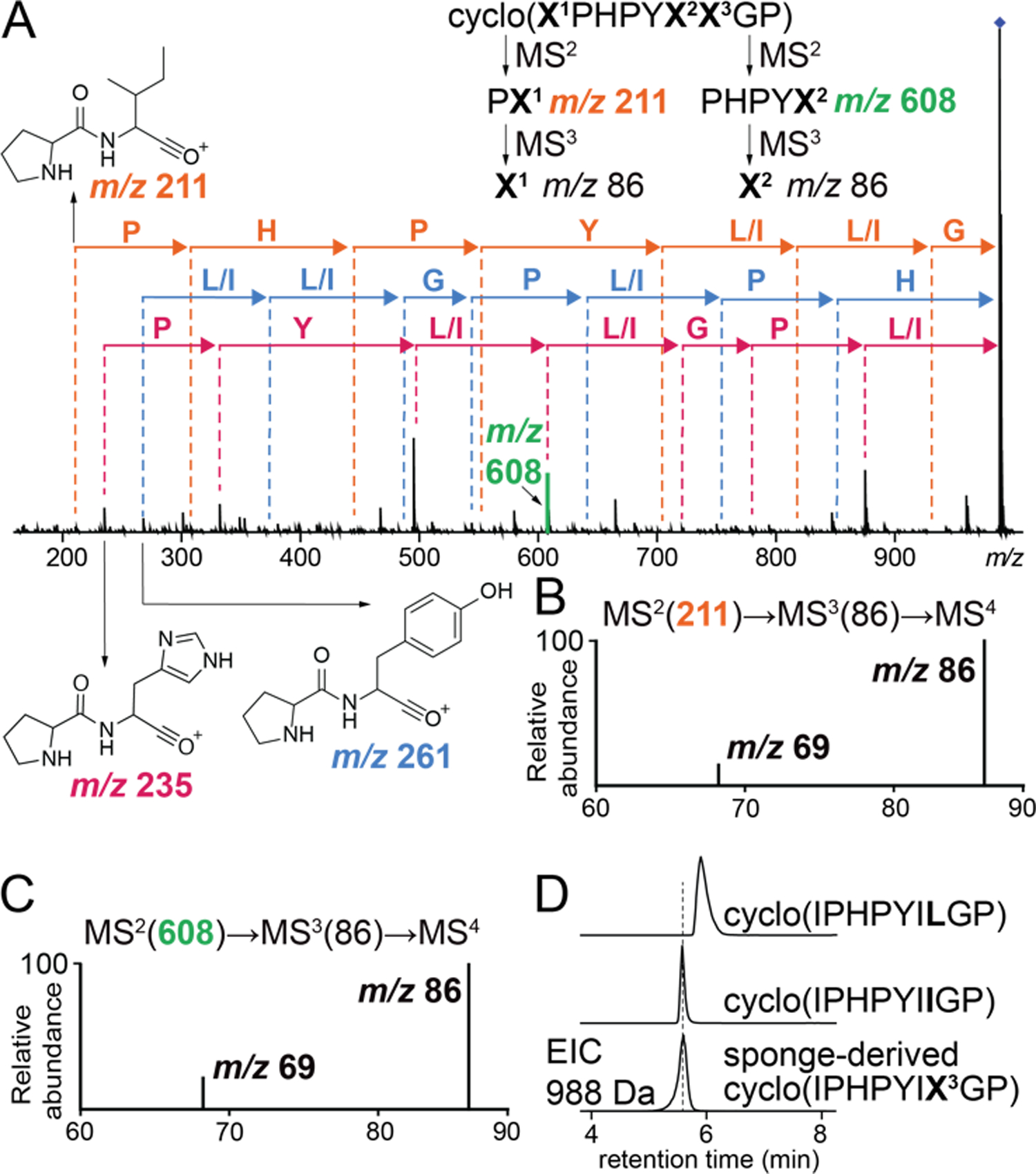
(A) Sequencing of the cyclo(X1PHPYX2X3GP) based on MS2 spectra. Note that three Pro residues in this octapeptide generate three different NPro-XaaC MS2 dipeptide product ions. Fragmentation strategies to query identities of X1 and X2 residues are illustrated. Annotation of (B) X1 and (C) X2 residues as Ile by the detection of the MS4 m/z 69 product ion. (D) Annotation of X3 as Ile by retention time comparison of the sponge detected PRMP against enzymatically synthesized cyclo(IPHPYILGP) and cyclo(IPHPYIIGP) standards.
Amino acid residues reported to date for sponge PRMPs are proteinogenic amino acids in the L- configuration (to the best of our knowledge the only exception is Trp derived L-kynurenine in phakefustatins A–C[15]). It is thus conceivable that sponge-derived PRMPs are ribosomally synthesized and posttranslationally modified peptides[16] and that the PCY1 catalyzed proteolysis of a C-terminal recognition sequence (RS in Fig. 4A; PCY1 RS is FQA[9]) and intramolecular transamidative macrocyclization mimics their physiological biosynthetic route. Peptide macrocyclization by prolyl oligopeptidases and cyanobactin macrocyclases is well established.[16] A recurring feature in biosynthesis of macrocyclic peptides is the requirement of a proline residue or a Ser/Thr/Cys-derived azol(in)e heterocycle immediately upstream of the RS (shaded green in Fig. 4A).[9, 17] Organizing the sponge PRMP octa- and heptapeptide sequences containing a single Pro residue such that the Pro is at the C-terminus leads to the observation that the transamidating N-terminal residues in PRMP linear peptides are hydrophobic amino acids with Trp, Phe, and Tyr predominating (Fig. 4B). Further organizing PRMP sequences containing multiple Pro residues in this sequence alignment then identifies position 2 to be a site for hypervariability.
Figure 4.
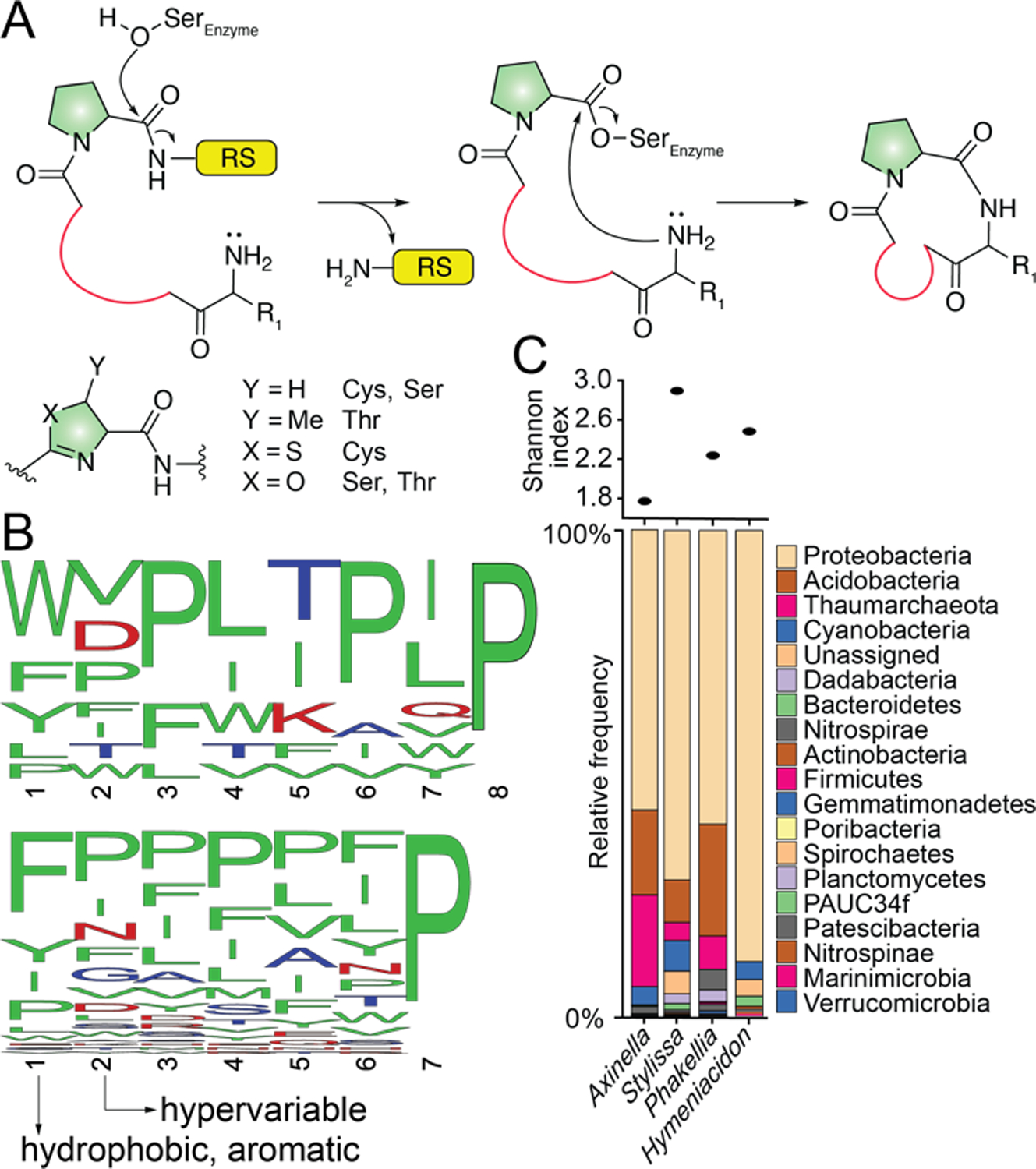
(A) Intramolecular transamidative peptide cyclization catalyzed by serine proteases such as PCY1. (B) WebLogo representation of PRMP octa- (top) and heptapeptides (bottom) with a C-terminal Pro. (C) Representative phylum-level microbiome architectures for Axinella,[6] Stylissa,[6] Phakellia,[20] and Hymeniacidon[20] spp. sponges. Microbiome diversity is denoted by Shannon indices. Shannon indices for HMA sponges are typically greater than 4.
It is as yet unknown whether the sponge host or a bacterial symbiont produces PRMPs in sponge holobionts. To date, natural products that have been shown to be produced by bacterial symbionts within marine sponges are all derived from high microbial abundance (HMA) sponges.[18] As microbial diversity correlates with microbial abundance for sponge microbiomes that are not dominated by cyanobacteria,[19] PRMP harboring sponges of the genera- Axinella, Stylissa, Phakellia, and Hymeniacidon spp. are all LMA sponges (Fig. 4C).
The diversity of natural products in marine sponges is much greater than that has been realized.[21] Here, with the sequencing of 12 macrolactam cyclopeptide sequences of which seven were novel PRMP sequences including a rare cyclononapeptide (Table S3), we demonstrate that inventorying natural product diversity using mass spectrometry and supplementing structural information using enzymatic synthesis can rapidly expand the natural product chemical space without recourse to sacrificing large amounts of sponge biomass which is otherwise necessary for the isolation of low abundance molecules.
Supplementary Material
Acknowledgements
The authors acknowledge support from the National Science Foundation (NSF, CHE-2004030) and National Institutes of Health (NIH, R35GM142882) to V.A., and support from the Georgia Institute of Technology’s Systems Mass Spectrometry Core Facility. The authors also thank Dr. S. K. Nair for the gift of PCY1 expression plasmid.
Footnotes
Supporting information for this article can be found under https://onlinelibrary.wiley.com
References
- [1].a) Zorzi A, Deyle K, Heinis C, Curr. Opin. Chem. Biol 2017, 38, 24–29; [DOI] [PubMed] [Google Scholar]; b) Driggers EM, Hale SP, Lee J, Terrett NK, Nat. Rev. Drug Discov 2008, 7, 608–624. [DOI] [PubMed] [Google Scholar]
- [2].Andrew TB, Cayla MM, Lokey RS, Curr. Top. Med. Chem 2013, 13, 821–836. [DOI] [PubMed] [Google Scholar]
- [3].Fang W-Y, Dahiya R, Qin H-L, Mourya R, Maharaj S, Mar. Drugs 2016, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Al-Mourabit A, Zancanella MA, Tilvi S, Romo D, Nat. Prod. Rep 2011, 28, 1229–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].a) Schmidt G, Grube A, Köck M, Eur. J. Org. Chem 2007, 2007, 4103–4110; [Google Scholar]; b) Cychon C, Köck M, J. Nat. Prod 2010, 73, 738–742; [DOI] [PubMed] [Google Scholar]; c) Wang X, Morinaka BI, Molinski TF, J. Nat. Prod 2014, 77, 625–630; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Arai M, Yamano Y, Fujita M, Setiawan A, Kobayashi M, Bioorg. Med. Chem. Lett 2012, 22, 1818–1821. [DOI] [PubMed] [Google Scholar]
- [6].Mohanty I, Moore SG, Yi D, Biggs JS, Gaul DA, Garg N, Agarwal V, ACS Chem. Biol 2020, 15, 2185–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mohimani H, Gurevich A, Shlemov A, Mikheenko A, Korobeynikov A, Cao L, Shcherbin E, Nothias L-F, Dorrestein PC, Pevzner PA, Nat. Commun 2018, 9, 4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Breci LA, Tabb DL, Yates JR, Wysocki VH, Anal. Chem 2003, 75, 1963–1971; [DOI] [PubMed] [Google Scholar]; b) Kapp EA, Schütz F, Reid GE, Eddes JS, Moritz RL, O’Hair RAJ, Speed TP, Simpson RJ, Anal. Chem 2003, 75, 6251–6264. [DOI] [PubMed] [Google Scholar]
- [9].Ludewig H, Czekster CM, Oueis E, Munday ES, Arshad M, Synowsky SA, Bent AF, Naismith JH, ACS Chem. Biol 2018, 13, 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Xiao Y, Vecchi MM, Wen D, Anal. Chem 2016, 88, 10757–10766; [DOI] [PubMed] [Google Scholar]; b) Moyer TB, Parsley NC, Sadecki PW, Schug WJ, Hicks LM, Nat. Prod. Rep 2021, 38, 489–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Randazzo A, Dal Piaz F, Orrù S, Debitus C, Roussakis C, Pucci P, Gomez-Paloma L, Eur. J. Org. Chem 1998, 1998, 2659–2665. [Google Scholar]
- [12].a) Tsuda M, Shigemori H, Mikami Y, Kobayashi J. i., Tetrahedron 1993, 49, 6785–6796; [Google Scholar]; b) Kobayashi J. i., Nakamura T, Tsuda M, Tetrahedron 1996, 52, 6355–6360. [Google Scholar]
- [13].Tsuda M, Sasaki T, Kobayashi J. i., Tetrahedron 1994, 50, 4667–4680. [Google Scholar]
- [14].Zhang H-J, Yi Y-H, Yang G-J, Hu M-Y, Cao G-D, Yang F, Lin H-W, J. Nat. Prod 2010, 73, 650–655. [DOI] [PubMed] [Google Scholar]
- [15].Wu Y, Liao H, Liu L-Y, Sun F, Chen H-F, Jiao W-H, Zhu H-R, Yang F, Huang G, Zeng D-Q, Zhou M, Wang S-P, Lin H-W, Org. Lett 2020, 22, 6703–6708. [DOI] [PubMed] [Google Scholar]
- [16].Montalbán-López M, Scott TA, Ramesh S, Rahman IR, van Heel AJ, Viel JH, Bandarian V, Dittmann E, Genilloud O, Goto Y, Grande Burgos MJ, Hill C, Kim S, Koehnke J, Latham JA, Link AJ, Martínez B, Nair SK, Nicolet Y, Rebuffat S, Sahl H-G, Sareen D, Schmidt EW, Schmitt L, Severinov K, Süssmuth RD, Truman AW, Wang H, Weng J-K, van Wezel GP, Zhang Q, Zhong J, Piel J, Mitchell DA, Kuipers OP, van der Donk WA, Nat. Prod. Rep 2021, 38, 130–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Sarkar S, Gu W, Schmidt EW, ACS Catal. 2020, 10, 7146–7153; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Luo H, Hong S-Y, Sgambelluri RM, Angelos E, Li X, Walton Jonathan D., Chem. Biol 2014, 21, 1610–1617; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sardar D, Lin Z, Schmidt Eric W., Chem. Biol 2015, 22, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Morita M, Schmidt EW, Nat. Prod. Rep 2018, 35, 357–378; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) McCauley EP, Piña IC, Thompson AD, Bashir K, Weinberg M, Kurz SL, Crews P, J. Antibiot 2020, 73, 504–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Moitinho-Silva L, Steinert G, Nielsen S, Hardoim CCP, Wu Y-C, McCormack GP, López-Legentil S, Marchant R, Webster N, Thomas T, Hentschel U, Front. Microbiol 2017, 8, 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Moitinho-Silva L, Nielsen S, Thomas T, Bell JJ, Vicente J, Björk JR, Montoya JM, Olson JB, Reveillaud J, Steindler L, Pineda M-C, Webster NS, Ilan M, Erwin PM, Lopez-Legentil S, Amir A, Gonzalez A, Ackermann GL, Knight R, Schupp PJ, Simister RL, Thacker RW, Costa R, Hill RT, Ravasi T, Hentschel U, Cerrano C, Astudillo-Garcia C, Taylor MW, Easson C, Sipkema D, Steinert G, Liu F, Feng G, Li Z, Kotoulas G, Polymenakou P, Dailianis T, McCormack GP, Marra MV, GigaScience 2017, 6, 1–7. [Google Scholar]
- [21].a) Reverter M, Rohde S, Parchemin C, Tapissier-Bontemps N, Schupp PJ, Front. Mar. Sci 2020, 7, 1062; [Google Scholar]; b) Paul VJ, Freeman CJ, Agarwal V, Integr. Comp. Biol 2019, 59, 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


