Abstract
Mutations gef1, stp22, STP26, and STP27 in Saccharomyces cerevisiae were identified as suppressors of the temperature-sensitive α-factor receptor (mutation ste2-3) and arginine permease (mutation can1ts). These suppressors inhibited the elimination of misfolded receptors (synthesized at 34°C) as well as damaged surface receptors (shifted from 22 to 34°C). The stp22 mutation (allelic to vps23 [M. Babst and S. Emr, personal communication] and the STP26 mutation also caused missorting of carboxypeptidase Y, and ste2-3 was suppressed by mutations vps1, vps8, vps10, and vps28 but not by mutation vps3. In the stp22 mutant, both the mutant and the wild-type receptors (tagged with green fluorescent protein [GFP]) accumulated within an endosome-like compartment and were excluded from the vacuole. GFP-tagged Stp22p also accumulated in this compartment. Upon reaching the vacuole, cytoplasmic domains of both mutant and wild-type receptors appeared within the vacuolar lumen. Stp22p and Gef1p are similar to tumor susceptibility protein TSG101 and voltage-gated chloride channel, respectively. These results identify potential elements of plasma membrane quality control and indicate that cytoplasmic domains of membrane proteins are translocated into the vacuolar lumen.
Plasma membrane proteins link the interior of the cell with the extracellular environment. Removal of defective membrane proteins prevents the accumulation of damage that might otherwise compromise the ability of the cell to maintain electrochemical gradients, transport nutrients, and respond to sensory information. However, degradation of integral membrane proteins presents special problems for the cell because the internal and external sides of the protein are exposed to different biochemical environments. In eucaryotic cells, membrane proteins which have not folded or assembled properly are normally eliminated by the endoplasmic reticulum (ER) quality control process (25); however, examples of post-ER quality control are known (16, 31). Degradation of defective membrane proteins in the yeast vacuole has been recognized; however, the molecular details of this process are unknown. This report describes a genetic approach toward elucidating the steps that comprise the quality control of integral plasma membrane proteins.
Recent work with Saccharomyces cerevisiae has shown that temperature-sensitive forms of plasma membrane ATPase (Pma1p) (5) and α-factor receptors (19) are delivered directly to the vacuole, where they are degraded. In a previous report (19), we described a temperature-sensitive form of the yeast α-factor receptor (Ste2-3p) as a model for investigating the consequences of structural defects of integral plasma membrane proteins. Operationally, we define “misfolded receptors” as mutant receptors that are synthesized at the nonpermissive temperature, whereas “damaged cell surface receptors” are receptors that are exposed to the nonpermissive temperature only after they have been expressed at the cell surface. Misfolded receptors are delivered to the vacuole and then degraded without traversing the plasma membrane. Damaged cell surface receptors are removed from the plasma membrane more rapidly than undamaged wild-type receptors; internalized receptors are degraded in the vacuole. Interestingly, receptor domains located on opposite sides of the membrane are degraded in concert, and the degradation of the two sides of the receptor depends on the activity of vacuolar proteases. When receptor degradation is blocked, Ste2-3p accumulates as a series of high-molecular-weight species, suggesting a role for posttranslational modification in the elimination process. Both the α-factor and the a-factor receptors are also subject to ligand-induced endocytosis (7, 20, 44) and to ubiquitination (13, 42).
This report defines steps in the quality control of two different plasma membrane proteins and identifies molecular events associated with their delivery to the vacuole. Processes that operate on misfolded receptors and damaged cell surface receptors appeared to have common elements. A subset of these elements controlled the sorting of vacuolar enzymes, as was found for the quality control of plasma membrane ATPase (28). The cytoplasmic sides of both normal and mutant receptors were translocated into the vacuolar lumen. DNA sequence analysis of the cloned genes and phenotypic analysis of the mutants suggest roles for homologous proteins in mammals.
MATERIALS AND METHODS
Yeast strains.
Congenic yeast strains are listed in Table 1. The can1ts mutation in strain 3262-14-3 was identified by selecting for canavanine resistance at 34°C and screening for sensitivity at 22°C; the mutation segregated 2:2 in tetrad analysis in a cross with strain DJ215-7-1. Strains DJ283-7-1stp22Δ, DJ283-7-1vps3Δ, DJ283-7-1vps8Δ, DJ283-7-1vps10Δ, and DJ283-7-1vps28Δ were constructed by one-step gene disruption of strain DJ283-7-1 with plasmids pDJ225, pCKR70A (from C. Raymond), pCKR42 (from C. Raymond), pAAC220 (from T. Stevens), and pKKD28 (from S. Rieder and S. Emr), respectively. Strain DJ1323-14-1 was constructed by one-step gene disruption of a diploid strain (DJ262-10-1 × DJ1317-6-1) with plasmid pCKR3A (from C. Raymond), followed by tetrad analysis. Chromosomal gene disruptions were confirmed by PCR analysis or by immunoblotting (DJ283-7-1vps10Δ). Strain DJ147-1-2pep4Δura3 was a spontaneous 5-fluoro-orotic acid-resistant isolate of DJ147-1-2pep4Δ (19). Strains 3262-14-3, DJ120-6, DJ147-2-1, DJ178-2-2, DJ211-5-3, DJ256-2-1, and DJ256-2-1pep4Δ are described elsewhere (18, 19, 44). Other strains were constructed by standard genetic crosses.
TABLE 1.
Yeast strains used in this study
| Straina | Genotypeb |
|---|---|
| 381G | MATa cry1 ade2-1 his4-580 lys2 trp1 tyr1 SUP4-3ts |
| DJ120-6 | 381G bar1-1 leu2 TYR1+ |
| DJ147-1-2 | 381G leu2 ura3 TYR1+ |
| DJ147-1-2pep4Δura3 | 381G leu2 ura3 TYR1+pep4::ura3 |
| DJ178-2-2 | 381G bar1-1 leu2 TYR1+ste2-3 |
| DJ209-7-4 | 381G matΔ::LEU2 leu2 ste2-3; containing plasmid pDJ102 |
| DJ211-5-3 | 381G bar1-1 leu2 ura3 |
| DJ211-5-3stp22Δ | 381G bar1-1 leu2 ura3 stp22::TRP1 |
| DJ214-7-2 | 381G ste2-3 stp22-1 leu2 TYR1+ |
| DJ215-7-1 | 381G leu2 ste2-3 TYR1+ |
| DJ256-2-1 | 381G leu2 ura3 TYR1+ste2-3 |
| DJ256-2-1pep4Δ | 381G leu2 ura3 TYR1+ste2-3 pep4::URA3 |
| DJ262-10-1 | 381G ste2-3 leu2 ADE2+TYR1+can1ts |
| DJ272-4-4 | 381G MATα ste2-3 ura3 can1ts |
| DJ276-6-3 | 381G bar1-1 ste2-3 stp22-1 leu2 ura3 can1ts |
| DJ280-1-4 | 381G ste2-3 STP26-1 ura3 leu2 TYR1+can1ts |
| DJ281-6-2 | 381G ste2-3 STP27-1 ura3 TYR1+can1ts |
| DJ283-7-1 | 381G ste2-3 leu2 ura3 TYR1+bar1-1 can1ts |
| DJ283-7-1stp22Δ | 381G ste2-3 leu2 ura3 TYR1+bar1-1 can1ts stp22::TRP1 |
| DJ283-7-1vps3Δ | 381G ste2-3 leu2 ura3 TYR1+ bar1-1 can1ts vps3::LEU2 |
| DJ283-7-1vps8Δ | 381G ste2-3 leu2 ura3 TYR1+bar1-1 can1ts vps1::URA3 |
| DJ283-7-1vps10Δ | 381G ste2-3 leu2 ura3 TYR1+bar1-1 can1ts vps10::URA3 |
| DJ283-7-1vps28Δ | 381G ste2-3 leu2 ura3 TYR1+bar1-1 can1ts vps28::URA3 |
| DJ1200-4-4 | 381G stp24-1 ste2-3 ADE2+TYR1+ura3 |
| DJ1201-15-3 | 381G ste2-3 stp22::TRP1 leu2 TYR1+ura3 |
| DJ1317-6-1 | 381G MATα ste2-3 leu2 ura3 TYR1+can1ts stp22::TRP1 |
| DJ1323-15-3 | 381G ste2-3 leu2 ura3 TYR1+can1ts vps1::LEU2 |
| 1rB | 381G bar1-1 leu2 TYR1+ste2-3 stp22-1 |
| 2rA | 381G bar1-1 leu2 TYR1+ste2-3 stp22-2 |
| 19rB | 381G bar1-1 leu2 TYR1+ste2-3 stp23-1 |
| 25rA | 381G bar1-1 leu2 TYR1+ste2-3 stp24-1 |
| 27rB | 381G bar1-1 leu2 TYR1+ste2-3 STP26-1 |
| 31rA | 381G bar1-1 leu2 TYR1+ste2-3 STP27-1 |
| 70rB | 381G bar1-1 leu2 TYR1+ste2-3 stp25-1 |
| 3262-14-3 | 381G ADE2+ade6 TYR1+sup4-87 |
| 5504-23 | 381G ADE2+ade6 HIS4+LYS2+TRP1+TYR1+cyh2 |
All strains were congenic with strain 381G (11).
The designation 381G indicates that the genotype is the same as that for strain 381G, except for the markers shown. Mutation bar1-1 (47) inhibits α-factor degradation in a cell cultures. Temperature-sensitive mutation SUP4-3 suppresses amber mutations his4-580 and trp1 at 22°C, and sup4-87 is a nonsuppressing allele (18). ste2-10::LEU2 is a deletion of the STE2 gene. Mutation stp24-1 is an allele of the GEF1 gene (10).
Plasmids.
pDJ379 is a yeast-integrating plasmid containing the URA3 gene, the bacterial Tetr determinant, and a fragment of the STE2 gene (codons 302 to 431) fused to the coding sequence of a mutant green fluorescent protein (GFP) gene (1). When pDJ379 is cleaved at the PstI site in the STE2 sequence and used to transform a ura3 strain, the resulting genetic duplication contains one STE2 gene fused to the GFP gene and a second STE2 copy that lacks the promoter and N-terminal coding sequence. Plasmid pDJ379 was constructed by ligating two PCR products. One PCR product, containing the STE2 gene, was synthesized by use of the DNA template, plasmid pDJ252 (44), and the primers GCGGCCCGCGGCTAAATTATTATTATCTTCAGTCCAGAA and GGCTCTAGACCCCAGCTTTAATGCGGTAGTTTA. The second PCR product, containing the GFP-coding sequence, resulted from a reaction containing the template, plasmid pBEX-1 (1), and the primers GCGCGGCCGTCTAGATTATTTGTATAGTTCATCCATGCCAT and GCGGCCGCGGTCGACGGTATGTGTAAAGGAGAAGAACTTTTCACTG. Both PCR products were digested with SacII and XbaI, and the purified fragments were ligated to yield plasmid pDJ320. pDJ320 was digested with NsiI, and the ends of the purified 5-kb fragment were ligated together to yield plasmid pDJ379. The PCR product of pBEX-1 (see above) digested with EagI and SalI and the PCR product of the STP22 gene (primers GACCTAGCTAGCACTAGTAATATGGAGACACATCG and TCACGCGTCGACCGATAACGGTGAGGTGATTCGTTG) digested with SalI and NheI were cloned between the EagI and NheI sites of episomal plasmid YEp24 to yield plasmid pDJ380 (URA3 STP22::GFP). The 3.3-kb BamHI-XhoI fragment containing the MATα locus from plasmid pJH64 (from J. Haber) was ligated with the 6.2-kb BamHI-SalI fragment of the centromere vector pYE(CEN3)30 to yield plasmid pDJ102.
Culture media.
Solid and liquid culture media are described elsewhere (19). Liquid cultures were grown in YM-1 medium unless indicated otherwise. For 35S labeling of cells, 0.1 mCi of Tran-35S label (ICN Radiochemicals) was used in 1 ml of minimal medium containing 1 mg of bovine serum albumin per ml and lacking methionine and cysteine.
Antisera and immunoblotting.
Polyclonal rabbit antisera were anti-Ste2p, specific for the C-terminal domain of the α-factor receptor (24); anti-carboxypeptidase Y (CPY) (from R. Gilmore); and anti-Vps10p (6). Mouse monoclonal anti-GFP serum was from Clontech (Palo Alto, Calif.). Immunoblotting methods were as described previously (14).
Isolation and genetic analysis of stp mutants.
Strain DJ178-2-2 was mutagenized with ethyl methanesulfonate to about 50% survival. Forty independent subcultures were plated for single colonies at 22°C and then screened for mating at 34°C. The purified isolates were considered further if they showed more than a 10-fold increase in mating efficiency (quantitative filter mating assay) and if they represented different subcultures of the original mutagenized population. Mutants (MATa ste2-3 stp) were crossed with strain DJ209-7-4 (matΔ ste2-3) containing plasmid pDJ102. The resulting diploids (MATa/matΔ ste2-3/ste2-3 stp/+) were subjected to tetrad analysis and cured of the plasmid to test for dominance in the quantitative mating assay. For complementation tests, recessive mutants (MATa ste2-3 stpX) were crossed with a tester strain (matΔ ste2-3 stpY) containing plasmid pDJ102 and cured of pDJ102. The STP26, STP27, and STE2 loci were unlinked, as judged by tetrad analysis of diploid strains (MATa/matΔ ste2-3/ste2-3 STP26/+ STP27/+ and MATa/matΔ ste2-3/+ STP/+) containing pDJ102. The genetic map position of stp22 was assigned by tetrad analysis with cross DJ214-7-2 × 5504-23; simple genetic distances were 3 cM to leu2 (58 parental ditype [PD], 0 nonparental ditype [NPD], 4 tetratype [T], 27 cM to his4 (29 PD, 0 NPD, 33 T), 37 cM to MAT (29 PD, 2 NPD, 41 T), and 2 cM to CEN3 (3 first-division and 69 second-division segregations with trp1).
Cloning and DNA sequence analysis of the STP22 and STP24 genes.
The STP22 gene was obtained from an existing plasmid library (32) that represented chromosome III as BamHI fragments cloned into plasmid vector YIp5. Plasmid pDJ166 contains the 2.3-kb EcoRI-BamHI and 3-kb BamHI-BglII fragments from plasmids C2G and D8B, respectively (32); it complemented stp22-1 and stp22-2 when integrated at the ura3 locus of strain DJ276-6-3. A deletion map of the insert (see Fig. 7B) was generated by subcloning selected fragments of pDJ166 into plasmid YCp50 and then testing for complementation of the mating, canavanine sensitivity, and 38°C growth phenotypes of strain DJ276-6-3. The 1.9-kb HindIII-SacI fragment was cloned into pUC19 (yielding plasmid pDJ223) and used as a template for double-stranded DNA sequencing with a Sequenase kit (U.S. Biochemicals). Overlapping sequences of both DNA strands were obtained. The correct junction of sequences from plasmids C2G and D8B was confirmed by partial sequencing of the 3.3-kb BglII fragment from a YCp50 yeast genomic library (41). Deletion allele stp22::TRP1 was constructed in plasmid pDJ225 by cloning a 1.4-kb BglII fragment containing TRP1 between the BamHI and BglII sites of pDJ223. One-step gene disruption of a diploid strain (confirmed by Southern blot analysis) followed by tetrad dissection indicated that STP22 is not essential.
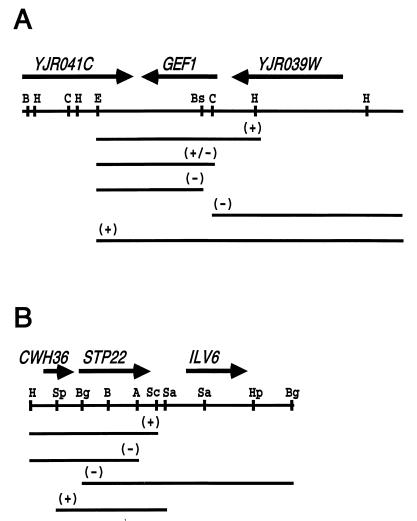
FIG. 7.
STP24 and STP22 cloning. (A) Restriction map of the plasmid insertion (pDJ266) that complements mutation stp24-1. (B) Restriction map of the plasmid insertion (pDJ166) that complements mutations stp22-1 and stp22-2. Arrows above maps indicate the position and direction of ORFs. Lines below the maps indicate subcloned fragments and their ability to complement the stp24 and stp22 mutants. Restriction enzymes: BamHI, B; HindIII, H; ClaI, C; EcoRI, E; BstEII, Bs; SpeI, Sp; BglII, Bg; AatII, A; SacI, Sc; and HpaI, Hp.
Plasmid pDJ266 (see Fig. 7A) from a YCp50 yeast genomic library (41) complemented mutation stp24-1 in strain DJ1200-4-4. It reversed the growth defect on pH 7 yeast extract-peptone-glycerol plates and the sterility and α-factor sensitivity phenotypes. Integration plasmid pDJ269 contained the 3.4-kb HindIII fragment cloned into YIp352. Strain DJ272-4-4 was transformed with pDJ269, crossed with strain DJ1200-4-4, and subjected to tetrad analysis. The site of integration was linked to stp24-1, since the Ura and growth phenotypes gave only PD asci (10 total); all MATa URA+ segregants were fertile. The deletion map (see Fig. 7A) was generated by digesting pDJ226 with restriction enzymes, religating, and transforming strain DJ1200-4-4. The DNA sequence of the deletion breakpoint in the partially complementing clone (see Fig. 7A) was contained in the GEF1 gene. The restriction map was consistent with GEF1.
The predicted amino acid sequences were compared to other known sequences by use of the Bestfit program (Genetics Computer Group, Inc., Madison, Wis.) with the standard default settings. Coiled-coil structures in Stp22p were predicted by the method of Lupas (29). As recommended previously (29), the extent of the coiled-coil structure was estimated by use of the 21-residue window, and the probability was estimated by use of the 28-residue window. The weighted and unweighted MTIDK matrices gave scores of 96 and 79%, respectively (residues 277 to 302), whereas the weighted and unweighted MTK matrices gave scores of only 53 and 45%, respectively. The TSG101 protein had scores of greater than 99% for all four matrices.
Fluorescence microscopy.
Cells were stained with FM4-64 (Molecular Probes) essentially as described previously (52). Cells growing exponentially at 30°C were collected by centrifugation, suspended in 0.5 ml of YM-1 medium containing 1 μl of FM4-64 (16 mM in dimethyl sulfoxide), incubated for 15 min at 30°C, collected, suspended in 0.5 ml of YM-1 medium, incubated for 1 h at 30°C, rinsed, suspended in 0.5 ml of phosphate-buffered saline, kept on ice, and examined by epifluorescence with a Nikon microscope. Photographic negatives were digitized with a laser densitometer (Molecular Dynamics).
α-Factor binding, sensitivity, and mating assays.
Methods for 3H-labeled α-factor binding were described previously (20). α-Factor sensitivity was assayed by a halo test. Four paper disks (1/4 in; Difco) containing α-factor were placed on a yeast extract-peptone-dextrose plate spread with 5 × 106 exponentially growing cells, incubated at 34°C, and scored for the size and clarity of the halos surrounding the disks. Disks contained 1.2, 2.5, 5, and 10 pmol of α-factor for BAR+ strains and 0.12, 0.25, 0.5, and 1 pmol for bar1 strains. Quantitative mating assays were done as described previously (18).
Canavanine sensitivity assay.
Quantitative determination of canavanine sensitivity was evaluated by determining the MIC breakpoint. Cells growing exponentially at 34°C in minimal medium without arginine were used to inoculate tubes (final concentration at A650, 0.01) containing different canavanine concentrations (including a no-canavanine control). After 20 h at 34°C with shaking, the culture density was determined spectrophotometrically. The MIC breakpoint was the concentration of canavanine that resulted in half-maximal growth.
Nucleotide sequence accession number.
The GenBank accession no. for the STP22 gene is AF004731.
RESULTS
Isolation of stp mutants.
Mutation ste2-3 leads to the substitution of threonine for alanine at position 52 in the first transmembrane segment of the α-factor receptor (19). At 22°C, ste2-3 mutant cells accumulate functional receptors on the plasma membrane, whereas at 34°C, the receptors are diverted to the vacuole and degraded. As a consequence, haploid MATa cells containing ste2-3 fail to mate with haploid MATα cells at 34°C. We isolated seven independent suppressor mutants (designated stp according to established nomenclature [22]) that improved mating activity in MATa ste2-3 cells at 34°C. Genetic analysis of stp mutations was complicated by the inability to detect ste2-3 phenotypes in MATα cells. This problem was overcome by exploiting the facts that haploid strains carrying a deletion of the MAT locus (matΔ) exhibit the a mating type and that matΔ strains with plasmid pDJ102 (MATα) exhibit the α mating type. In all cases, the suppressor phenotype showed 2:2 segregation in tetrad analysis (MATa ste2-3 stp × matΔ ste2-3), indicating a single chromosomal mutation. Five mutations, stp22-1, stp22-2, stp23-1, STP24-1, and STP25-1 (strains 1rB, 2rA, 19rB, 25rB, and 70rB, respectively), were recessive in diploid cells (MATa/matΔ ste2-3/ste2-3 stp/+). Complementation tests indicated that stp22-1 and stp22-2 were allelic, whereas stp23-1, stp24-1, and stp25-1 defined independent complementation groups. Mutations stp22-1 and stp22-2 also resulted in a growth defect at 38°C that cosegregated with the suppressor phenotype. Two mutations (STP26 and STP27) were dominant (strains 27rB and 31rA, respectively). In tetrad analysis, STP26 and STP27 were not linked to each other. None of the suppressor mutations was linked to the original ste2-3 mutation (see Materials and Methods).
Suppressor mutations affect quality control of plasma membrane proteins.
Three criteria were used to identify the suppressors that affect the quality control of α-factor receptors: (i) increased α-factor binding sites and enhanced α-factor sensitivity in the ste2-3 mutant, (ii) slower turnover of mutant receptor protein (Ste2-3p), and (iii) suppression of temperature-sensitive defects in a second, unrelated plasma membrane protein. Table 2 indicates the binding capacity of the cells relative to the wild-type control at four different temperatures. In the unsuppressed ste2-3 mutant, no α-factor binding was detected at 30°C or higher. The stp22, stp24, STP26, and STP27 mutants showed a significant increase in α-factor binding (Table 2), whereas no increase was detected for the stp23 and stp25 mutants. The suppressed mutants that showed more α-factor binding also exhibited enhanced α-factor sensitivity (Fig. 1). The stp23 and stp25 mutants exhibited very weak sensitivity to α-factor (data not shown). Thus, four of the suppressors (stp22, stp24, STP26, or STP27) satisfied the first criterion for determinants that affect quality control, whereas two of the suppressors did not (stp23 and stp25).
TABLE 2.
Accumulation and turnover of α-factor binding sites
| Strain | Relevant genotype | α-Factor bindinga (% of wild-type value) at the following °C:
|
% Surface sites remaining after 70 min at 34°C | |||
|---|---|---|---|---|---|---|
| 22 | 26 | 30 | 34 | |||
| DJ120-6 | STE2+ | 100 | 100 | 100 | 100 | 57 ± 7 |
| DJ178-2-2 | ste2-3 | 58 | 18 | <2 | <2 | 25 ± 2 |
| 1rB | ste2-3 stp22-1 | 62 | 31 | 12 | 5 | 42 ± 3 |
| 2rA | ste2-3 stp22-2 | 39 | 17 | 7 | ||
| 19rB | ste2-3 stp23-1 | 84 | 12 | <2 | <2 | |
| 25rA | ste2-3 stp24-1 | 54 | 33 | 22 | 5 | 39 ± 4 |
| 70rB | ste2-3 stp25-1 | 62 | 26 | 2 | <2 | |
| 27rB | ste2-3 STP26-1 | 54 | 34 | 27 | 4 | 38 ± 12 |
| 31rA | ste2-3 STP27-1 | 92 | 57 | 30 | 6 | 48 ± 13 |
Cells that had been cultured continuously at the temperatures indicated were assayed for α-factor binding. Wild-type control cells were assigned a relative binding value of 100%.
Cells that had been cultured at 22°C were shifted to 34°C and received cycloheximide. Errors are the standard deviations for three or four determinations.
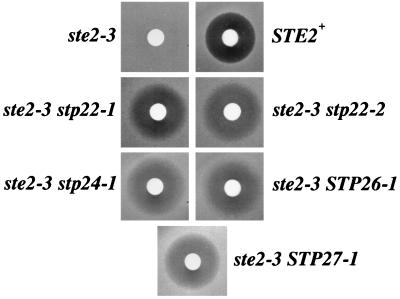
FIG. 1.
Suppressors restore α-factor sensitivity in the ste2-3 mutant. Halo assays indicated α-factor sensitivity of the suppressor mutants at 34°C. The unsuppressed ste2-3 strain (DJ178-2-2) showed no zone of growth inhibition surrounding the disk containing 0.5 pmol of α-factor, whereas a zone of growth inhibition was observed for the wild-type control strain, DJ120-6 (STE2+), and the suppressed ste2-3 strains, 1rB (ste2-3 stp22-1), 2rA (ste2-3 stp22-2), 25rA (ste2-3 stp24-1), 27rB (ste2-3 STP26-1), and 31rA (ste2-3 STP27-1).
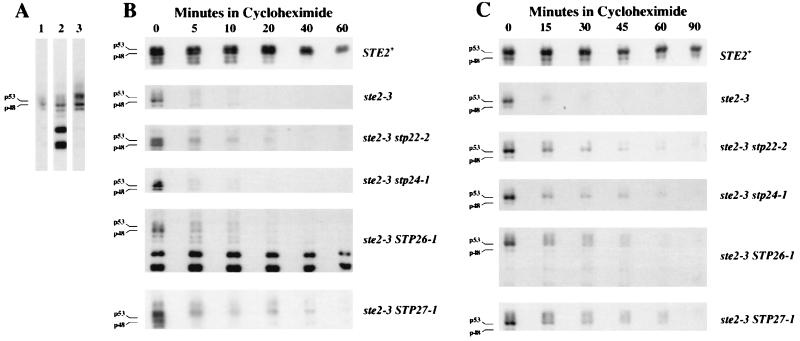
The increased α-factor binding activity observed for the stp22, stp24, STP26, and STP27 mutants potentially reflects either faster synthesis or slower degradation of receptors. The amount of receptor mRNA in the ste2-3 mutant cells was not affected by mutation stp22, stp24, STP26, or STP27 (data not shown). To evaluate the turnover of Ste2-3p in the suppressor mutants, we used immunoblot analysis to monitor the decay of Ste2-3p after protein synthesis had been blocked with cycloheximide. Quantitative analysis of the decay kinetics was complicated by the heterogeneity of Ste2-3p from the various suppressor mutants. The unsuppressed ste2-3 mutant (Fig. 2A, lane 1) produced p48 and p53 glycosylated forms similar to the wild-type proteins (19). Multiple larger receptor species were most prominent in the STP27 mutant (Fig. 2A, compare lanes 1 and 3). Apparent degradation intermediates were prominent only in the STP26 ste2-3 mutant (Fig. 2A, lane 2). Figure 2B shows the loss of Ste2p and Ste2-3p in cells that received cycloheximide after they had been cultured continuously at 34°C. For the suppressed mutants (stp22-2, stp24-1, STP26-1, and STP27-1), p53 and p48 disappeared at a rate similar to that in the unsuppressed ste2-3 mutant (DJ178-2-2), whereas the larger Ste2-3p species (i.e., larger than p53) decayed more slowly in three of the suppressed mutants (stp22-2, STP26-1, and STP27-1) than in the unsuppressed control (ste2-3). The slower decay of the higher-molecular-weight forms than of p48 and p53 is consistent with the rapid conversion in the suppressor mutants of p48 and p53 to other structural forms (presumably, different covalent modifications) that are degraded more slowly. Alternatively, the slowly degrading electrophoretic forms of the receptors may exist only in the suppressor mutants. We conclude that the stp22, STP26, and STP27 mutants satisfied the second criterion for defects in quality control. In this analysis, we were unable to detect a Ste2-3p species that decayed more slowly in the stp24-1 mutant; however, slower decay of Ste2-3p was evident when stp24 mutant cells were shifted from 22 to 34°C (see below).
FIG. 2.
Effect of suppressor mutations on mutant receptor turnover. Immunoblots were probed with antiserum specific for the C-terminal domain of the α-factor receptor. (A) Variant forms of Ste2-3p. Cells were cultured continuously at 34°C without cycloheximide. Lane 1, ste2-3 control (strain DJ178-2-2); lane 2, ste2-3 STP26 mutant (strain 27rB); lane 3, ste2-3 STP27-1 mutant (strain 31rA). p48 and p53 indicate the positions of the two major glycosylated forms of the receptor. (B) Turnover of misfolded Ste2-3p. Cultures growing exponentially at 34°C received cycloheximide at time zero. Samples were removed periodically and processed for immunoblotting. (C) Turnover of damaged cell surface receptors. Cultures growing at 22°C were shifted to 34°C and received cycloheximide. At the times indicated, samples were withdrawn and analyzed for receptors. Relevant genotypes are indicated to the right of panels B and C. Strains were DJ120-6, 178-2-2, 2rA, 25rA, 27rB, and 31rA.
We tested whether stp mutants suppress temperature-sensitive defects in another plasma membrane protein, arginine permease (encoded by CAN1). A can1ts mutant was isolated. It was resistant to the toxic arginine analog canavanine at 34°C yet sensitive at 22°C. At 34°C, the CAN1+ control was sensitive to less than 1 μg of canavanine per ml, whereas the can1ts mutant was resistant to greater than 80 μg/ml (Table 3). The can1ts stp22 double mutant showed nearly wild-type sensitivity; the can1ts stp24, can1ts STP26, and can1ts STP27 mutants exhibited intermediate sensitivity. Drug sensitivity was not due to acquisition of an alternate canavanine transport pathway, since stp22-1 failed to suppress a nonconditional can1 allele (data not shown). stp22-1 also failed to suppress temperature-sensitive alleles of eight other genes (STE4, STE5, STE7, STE11, STE12, CDC4, CDC25, and CDC28) that do not encode integral membrane proteins (data not shown). Thus, the stp22, stp24, STP26, and STP27 mutants satisfied the third criterion for defective quality control of plasma membrane proteins.
TABLE 3.
Suppression of the arginine permease defect
| Relevant genotypea | Canavanine MIC breakpoints (μg/ml)b |
|---|---|
| CAN1+ | 0.6, 0.6, 0.4 |
| can1ts | >80, >80 |
| can1ts stp22-1 | 1.3, 1.4 |
| can1ts stp22-2 | 1.0, 0.8 |
| can1ts stp24-1 | 50, 56 |
| can1ts STP26-1 | 24, 24 |
| can1ts STP27-1 | 11, 18 |
The strains listed in Table 2 were crossed with strain DJ272-4-4 (MATa can1ts). The CAN1+ control was strain DJ178-2-2 assayed in triplicate. Two segregants containing the suppressor and the can1ts mutation were tested for canavanine sensitivity.
The MIC breakpoint was the canavanine concentration that caused 50% inhibition of growth at 34°C in liquid minimal medium lacking arginine.
Elimination of damaged cell surface receptors.
To test whether the suppressor mutations affect the elimination of damaged cell surface receptors, we cultured ste2-3 mutant cells at 22°C to allow cell surface accumulation of Ste2-3p and then shifted them to 34°C to induce the structural defect. Turnover of the receptor protein was evaluated by monitoring the decay of Ste2-3p after the addition of cycloheximide (Fig. 2C). As was observed for misfolded receptors, damaged Ste2-3p in the suppressor mutants accumulated higher-molecular-weight forms and decayed more slowly than Ste2-3p in the unsuppressed control. Curiously, the STP26 ste2-3 mutant (strain 27rB) did not accumulate significant levels of the lower-molecular-weight species that were observed during continuous growth at the restrictive temperature (compare Fig. 2B and C). The net rate at which the α-factor binding sites disappeared from the cell surface was evaluated with a radioactive ligand binding assay (19). Cells growing at 22°C received cycloheximide and were shifted to 34°C; α-factor binding activity was assayed immediately and after 70 min at 34°C (Table 2). More binding sites were lost in the unsuppressed ste2-3 mutant (DJ178-2-2) than in the wild-type control (DJ120-6). The ste2-3 mutants containing suppressor mutations stp22-1, stp24-1, STP26-1, and STP27-1 had intermediate values. In control experiments, the binding sites were lost when α-factor (10−8 M) was present during the 70-min period at 34°C (data not shown); thus, the binding sites that remained in the suppressor mutants (Table 2) were not a consequence of dead or inactive cells.
Vacuolar protein sorting mutants have the Stp phenotype.
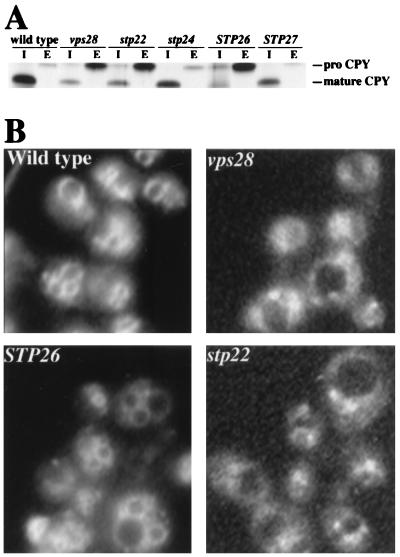
Since Ste2-3p is degraded in the vacuole (19), the set of quality control mutants is likely to include mutants with more general defects in the sorting of proteins to the vacuole. Vacuolar protein sorting (vps) mutants were originally identified by their ability to secrete the vacuolar enzyme CPY (21). Previous work (28) has shown that the degradation of defective plasma membrane ATPase is blocked in certain vps mutants. We tested for overlap between the Stp and Vps pathways. Processing of CPY to its mature form in the vacuole was evaluated in pulse-chase experiments. CPY was efficiently sorted to the vacuole in the wild type as well as in the stp24 and STP27 mutants (Fig. 3A). In contrast, the stp22 and STP26 mutants secreted the majority of CPY as the precursor, proCPY (similar to vps28Δ). The vps mutants have been assigned to six phenotypic classes (37); certain classes are distinguished by use of the fluorescent vital stain FM4-64 (52). FM4-64 is endocytosed by wild-type yeast cells and accumulates on the vacuolar surface; however, in class E vps mutants (e.g., vps28), the majority of the stain accumulates in an exaggerated endosome-like compartment (class E compartment) adjacent to the vacuole. By this criterion, the stp22 mutant (but not the STP26 mutant) is a class E mutant (Fig. 3B). The stp22 mutant also accumulated vacuolar ATPase in the class E compartment (data not shown), as was observed for other class E vps mutants (37). The ste2-3 vps double mutants representing four of the Vps classes were tested for α-factor sensitivity (Table 4). All of the vps mutations except vps3 suppressed the ste2-3 mutant. pep4 yielded no detectable suppression. Thus, quality control of plasma membrane proteins requires some but not all elements of the Vps pathway; other gene products (i.e., Stp24p and Stp27p) play roles in quality control distinct from vacuolar protein sorting.
FIG. 3.
Common elements of the quality control and Vps pathways. (A) Missorting of CPY. Cells were pulse-labeled for 10 min with Tran-35S label and chased for 30 min, and CPY was immunoprecipitated from the cells and culture supernatants as described previously (30). Mature CPY and proCPY were detected in the intracellular compartment (I) and in the extracellular compartment (E). Strains used were DJ283-7-1 (wild type), DJ283-7-1vps28Δ (vps28), DJ283-7-1stp22Δ (stp22), 25rA (stp24), 27rB (STP26), and 31rA (STP27). (B) FM4-64 staining (52). The STP26 mutant (strain 27rB) and the wild-type control (DJ283-7-1) gave normal vacuolar staining, whereas FM4-64 accumulated in the class E compartment of the stp22 and vps28 mutants (DJ283-7-1stp22Δ and DJ283-7-1vps28Δ, respectively).
TABLE 4.
Suppression of the ste2-3 mutant by vps mutations
| Strain | Relevant genotype | Vps class | Halo formationa |
|---|---|---|---|
| DJ147-1-2 | STE2+ | ++ (clear) | |
| DJ256-2-1 | ste2-3 | − | |
| DJ120-15-3 | ste2-3 stp22 | E | ++ (turbid) |
| DJ256-2-1pep4Δ | ste2-3 pep4 | − | |
| DJ1323-15-3 | ste2-3 vps1 | F | + (turbid) |
| DJ211-5-3 | bar1 STE2+ | ++ (clear) | |
| DJ211-5-3stp22Δ | bar1 STE2+stp22 | E | ++ (clear) |
| DJ283-7-1 | bar1 ste2-3 | − | |
| DJ283-7-1stp22Δ | bar1 ste2-3 stp22 | E | ++ (turbid) |
| DJ283-7-1vps3Δ | bar1 ste2-3 vps3 | D | − |
| DJ283-7-1vps8Δ | bar1 ste2-3 vps8 | A | ++ (turbid) |
| DJ283-7-1vps10Δ | bar1 ste2-3 vps10 | A | ++ (turbid) |
| DJ283-7-1vps28Δ | bar1 ste2-3 vps28 | E | ++ (turbid) |
Halos of growth inhibition surrounding α-factor disks at 34°C were compared with values for the wild-type control strains DJ147-1-2 and DJ211-5-3. Relative halos sizes are as follows: ++, wild-type control; +, intermediate; −, no halo. The clarity of the halo is indicated as clear or turbid.
Receptor localization.
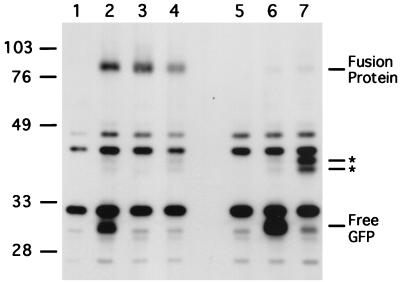
Previous experiments indicated that both wild-type and ste2-3 mutant receptors are delivered to the vacuole, where they are degraded, since receptor degradation is blocked in the pep4 mutant (7, 19, 44). The relative distribution of receptors between the plasma membrane and intracellular compartments was evaluated by subcellular fractionation (19). When cells are cultured at 34°C and the membranes are resolved on Renografin density gradients, Ste2p occurs in plasma membrane fractions, whereas Ste2-3p occurs in fractions containing membranes from intracellular compartments. In the present study, receptors tagged with GFP were used to clarify the distribution of receptors among the various intracellular compartments. The GFP-coding sequence was inserted after the last codon of STE2, and a single copy of the chimeric gene under the control of the native STE2 promoter was integrated at the STE2 locus. The fusion protein was functional in that strains expressing only the wild-type fusion protein (Ste2p-GFP) exhibited full α-factor sensitivity, whereas the mutant fusion protein (Ste2-3p–GFP) led to temperature-sensitive α-factor responsiveness. Immunoblot analysis of the strain expressing Ste2p-GFP (Fig. 4, compare lane 2 with control lane 1) revealed the full-length fusion protein (80 kDa) as well as other, smaller proteins that contained GFP. The electrophoretic mobility of the smallest species (30 kDa) was within the range of published values for free GFP (9). The absence of the 30-kDa species in pep4 mutant cells (Fig. 4, compare lanes 2 and 3) suggested that it forms when the fusion protein is cleaved within the lumen of the vacuole. GFP forms a compact protease-resistant structure (34) that is likely to persist in the vacuole. The approximately 40-kDa fragments were consistent with a polypeptide containing GFP and the C-terminal cytosolic domain of the receptor. As expected, anti-Ste2p antiserum detected the 80- and 40-kDa (but not 30-kDa) forms (data not shown). In contrast to Ste2p-GFP, essentially all of the mutant fusion protein was cleaved in the cytoplasmic domain, generating free GFP (Fig. 4, compare lane 6 with control lane 5).
FIG. 4.
Cleavage of GFP from the cytoplasmic domain of the receptor requires PEP4 and STP22. GFP was fused to the C terminus of Ste2p and Ste2-3p by transformation with integrating plasmid pDJ379. Cells that had been cultured at 30°C were subjected to immunoblot analysis with anti-GFP antiserum. Controls containing no GFP protein were STE2+ (lane 1) and ste2-3 (lane 5) that had not been transformed with pDJ379. These lanes show the positions of nonspecific protein species. Strains transformed with pDJ379 were STE2+ (lane 2), STE2+ pep4 (lane 3), STE2+ stp22Δ (lane 4), ste2-3 (lane 6), and ste2-3 stp22Δ (lane 7). Molecular masses (in kilodaltons) of marker proteins are indicated at the left. Predicted positions for receptor-GFP fusion protein, free GFP, and novel fragments (asterisks) are indicated at the right. Strains were DJ211-5-3, DJ211-5-3stp22Δ, DJ147-1-2pep4Δ, DJ283-7-1, and DJ283-7-1stp22Δ
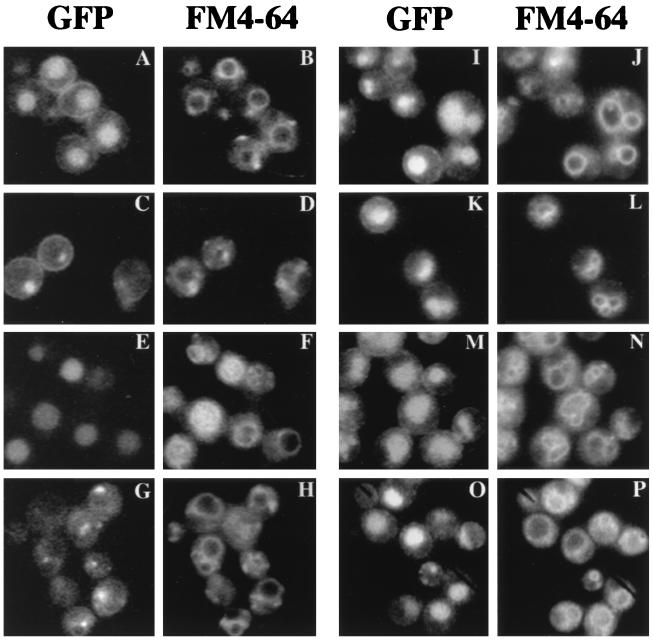
The ability of the fusion protein to reach the vacuole was also evaluated by comparing fluorescent signals from GFP and FM4-64 (Fig. 5). GFP appeared within the vacuolar lumen as well as on the cell surface (Fig. 5A and B), consistent with PEP4-dependent formation of free GFP (Fig. 4, lanes 2 and 3). In contrast to the situation with Ste2p-GFP, essentially all of the GFP fluorescence arising from the mutant Ste2-3p–GFP accumulated within the vacuole (Fig. 5E and F), consistent with the predominance of free GFP in this mutant (Fig. 4, lane 6). All of the stp mutants showed an altered subcellular distribution of Ste2-3p–GFP. For both Ste2p-GFP (Fig. 5C and D) and Ste2-3p–GFP (Fig. 5G and H), traffic to the vacuole was blocked in the stp22 mutant. GFP fluorescence was concentrated in a subset of the internal compartments that were stained with FM4-64. Moreover, the appearance of free GFP on the immunoblot was blocked (Fig. 4, compare lanes 4 and 7 with lanes 2 and 6). These results are consistent with the larger proteolytic fragments forming in the class E compartment. Degradation of membrane proteins in the class E compartment has been established previously (reviewed in reference 21). In stp24, STP26, and STP27 mutants, the distribution of GFP for Ste2-3p–GFP (Fig. 5I through N) was similar to the distribution for Ste2p-GFP (Fig. 5A). The relatively faint fluorescence on the surface of stp22 mutant cells (Fig. 5G) probably reflects the fewer α-factor binding sites in ste2-3 stp22 mutants (Table 2) and the fact that stp22 suppressed the ste2-3::GFP mutant more weakly than the ste2-3 mutant in the α-factor halo test (data not shown).
FIG. 5.
Subcellular localization of GFP-tagged receptors. Cells that had been transformed with integrating plasmid pDJ379 were cultured at 30°C and stained with FM4-64. Fluorescent images indicate GFP (A, C, E, G, I, K, M, and O) and FM4-64 (B, D, F, H, J, L, N, and P). The relevant genotypes were STE2+ (A and B), STE2+ stp22Δ (C and D), ste2-3 (E and F), ste2-3 stp22 (G and H), ste2-3 stp24-1 (I and J), ste2-3 STP26-1 (K and L), ste2-3 STP27-1 (M and N), and STE2+ pep4 (O and P). Strains were DJ211-5-3, DJ211-5-3stp22Δ, DJ147-1-2pep4Δ, DJ283-7-1, DJ283-7-1stp22Δ, DJ1200-4-4, DJ280-1-4, and DJ281-6-2.
According to two different criteria, the cytoplasmic domain of the receptor is translocated into the lumen of the vacuole. First, PEP4-dependent cleavage occurs near the junction of Ste2p and GFP (Fig. 4, lanes 2 and 3). Second, in the pep4 mutant strain, essentially all GFP is associated with the fusion protein (Fig. 4, lane 3), yet most of the fluorescence is inside the vacuole (Fig. 5O and P). These observations provide an explanation for our earlier finding that the N-terminal and C-terminal domains of Ste2p are degraded simultaneously (19).
Stp22p localization.
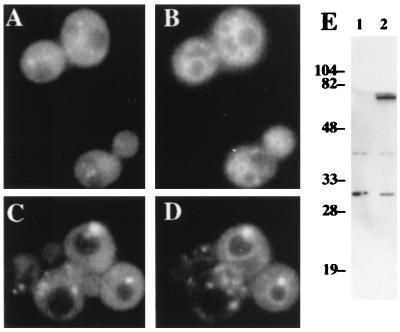
We examined the intracellular localization of Stp22p that had been tagged with GFP at its C terminus. The chimeric gene was expressed under the control of the native STP22 promoter on a multiple-copy plasmid. The plasmid reversed the ability of the stp22::TRP1 mutation to suppress ste2-3 and can1ts (data not shown). The size (72 kDa) of the fusion protein was consistent with the predicted value (70 kDa), as judged by immunoblot analysis (Fig. 6E); no other protein products containing GFP were detected. The intracellular distribution of Stp22p was inferred by comparing the fluorescence of GFP and FM4-64. In wild-type cells, Stp22p-GFP exhibited a disperse pattern but was excluded from the vacuole. Some of the fluorescence was concentrated in punctate foci (Fig. 6A and B). However, in vps28Δ cells, the bulk of Stp22p-GFP was concentrated in class E compartments together with FM4-64, and the remaining Stp22p-GFP was concentrated in structures that did not stain with FM4-64 (Fig. 6C and D). The class E Vps phenotype of the stp22 mutant and the accumulation of Stp22p in class E compartments suggest that Stp22p plays a direct role in the function of the prevacuole. Other class E VPS gene products show a similar distribution in a class E vps genetic background (3, 36).
FIG. 6.
Subcellular localization of GFP-tagged Stp22p. Strains were transformed with episomal plasmid pDJ380 (STP22::GFP). Fluorescent images of wild-type cells (A and B) and vps28 mutant cells (C and D) indicate GFP (A and C) and FM4-64 (B and D). Immunoblot analysis with anti-GFP antiserum (E) was performed on cells transformed with vector plasmid YEp24 (lane 1) or with pDJ380 (lane 2). Numbers at left are kilodaltons. Strains were DJ283-7-1 and DJ283-7-1vps28Δ.
Cloning of the STP22 and STP24 genes.
The STP24 gene was cloned by complementation. A plasmid that complemented stp24-1 was identified from a random genomic library. The insert depicted in Fig. 7A complemented two phenotypes of the stp24-1 mutant: suppression of ste2-3 and failure to grow on yeast extract-peptone-glycerol plates at pH 7. A subcloned fragment of the insert (pDJ269) was integrated at a site that was linked to stp24 in tetrad analysis. A partial DNA sequence was used to locate the clone in the Yeast Genome Database (Stanford University). The restriction map of STP24 was identical to that of the GEF1 gene, which encodes a voltage-gated chloride channel homolog (8, 10) (Fig. 7A). Gef1p may play a role in regulating electrochemical gradients required for the transport of defective membrane proteins to the vacuole.
The STP22 gene was cloned by testing specific fragments of chromosome III (32) for complementation of the stp22-1 mutation. In tetrad analysis, the stp22-1 mutation mapped to chromosome III between LEU2 and the centromere. Plasmids containing this region of chromosome III complemented stp22-1 only when the 1.7-kb SpeI-SacI segment was present (Fig. 7B). The published chromosome III sequence (33) shows no open reading frame (ORF) spanning this region (i.e., containing the essential BglII and AatII sites). Our sequence of the 1.7-kb SpeI-SacI fragment differs from the published sequence at several positions and defines an ORF of 385 codons. This ORF contains the 119-codon ORF YCL008C from the published sequence. The overlapping portions of our sequence (nucleotides 1 to 789) and another published sequence (48) (GenBank accession no. S61879) are identical. The cloned sequence was used to construct a deletion allele of STP22 that was marked with the TRP1 gene. When this allele was introduced into the STP22 locus on chromosome III, the resulting mutant (stp22::TRP1) was viable and indistinguishable from stp22-1 and stp22-2 mutants.
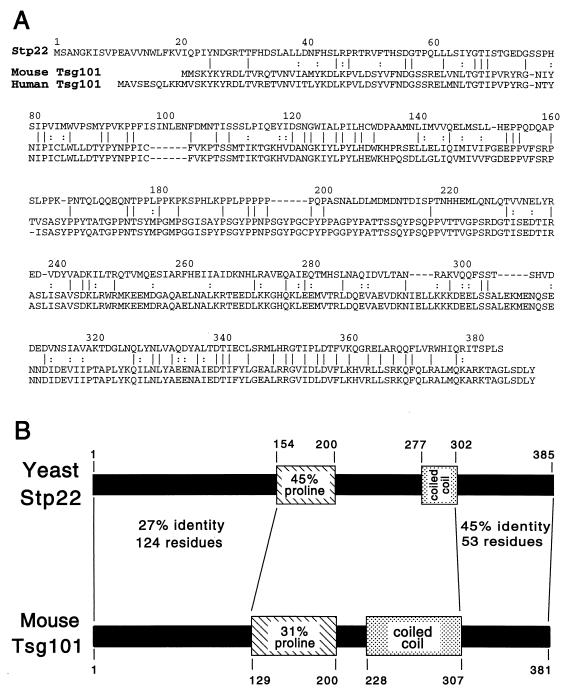
Stp22p shows significant homology to the tumor susceptibility protein TSG101 (27) (Fig. 8A). Bestfit alignment of Stp22p gave 25% identity with respect to mouse TSG101 and 24% identity with respect to human TSG101. Control alignments to the randomized sequences gave scores that differed by 33 standard deviations in both cases. The strongest homology between Stp22p and mouse TSG101 was within the C-terminal portion (45% identity). Stp22p and the mammalian TSG101 proteins are similar in length and contain proline-rich domains in comparable positions (Fig. 8B). In TSG101, the proline-rich sequence is followed by a sequence predicted to form a coiled-coil structure (27). Stp22p also shows a significant probability of forming a coiled coil in the corresponding position, although the values for the size and the probability of the predicted structure are lower than the values predicted for TSG101. STP22 is completely identical to VPS23 (2a). STP22 is 76% identical to a 376-codon ORF (GenBank accession no. 1870128) from the related yeast Saccharomyces pastorianus.
FIG. 8.
Comparison of the predicted Stp22p and TSG101 protein sequences. (A) Bestfit alignment of predicted sequences for Stp22p and mouse TSG101. Identical (vertical lines) and similar (colons) residues are indicated. Human TSG101 is also shown. (B) Common structural features of yeast Stp22p and mouse TSG101. Both proteins contain a proline-rich domain (hatched) and a sequence with a high probability of forming a coiled coil (stippled). The Bestfit alignments are indicated for the overlapping portions of the N-terminal and C-terminal domains. The Bestfit alignment of the entire polypeptide chains indicates 25% identity and 46% similarity.
DISCUSSION
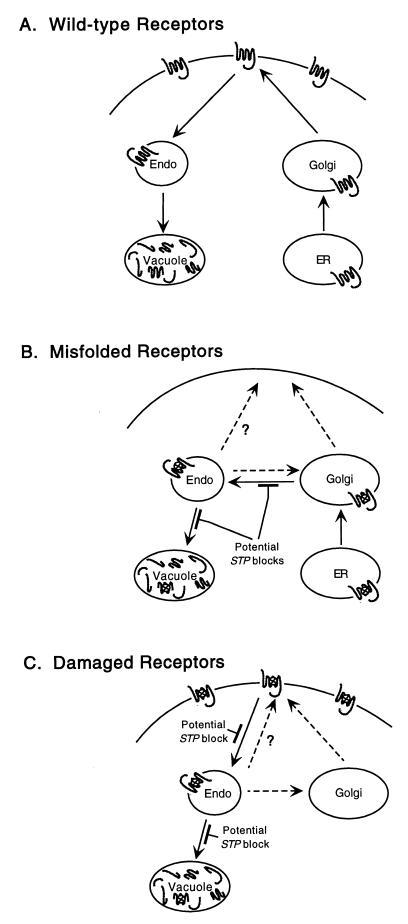
We have shown previously (19) that both misfolded α-factor receptors and receptors damaged in the plasma membrane are eliminated by a quality control process. Quality control is likely to involve three steps: recognition of the structural defect, transport to the vacuole, and proteolytic degradation. In this study, we used a genetic approach to identify steps in the quality control process. A gene was assumed to play a role in quality control if mutations in the gene suppressed temperature-sensitive defects in two unrelated plasma membrane proteins, the α-factor receptor and the arginine permease. Mutations in the four STP genes that we identified slowed the elimination of both misfolded and damaged Ste2-3p, suggesting that the processes that eliminate misfolded and damaged receptors have common elements. Working models for the trafficking of wild-type, misfolded, and damaged receptors are summarized in Fig. 9. We provide evidence indicating that cytoplasmic surfaces of plasma membrane proteins are degraded within the vacuolar lumen.
FIG. 9.
Working models for quality control of α-factor receptors. (A) Newly synthesized wild-type receptors are sorted efficiently to the plasma membrane. Turnover results from endocytosis and degradation in the vacuole. Endo, endosomal compartments. (B) Misfolded receptors are diverted from the secretory pathway in the Golgi complex and enter the Vps pathway. They are delivered to the endosomal compartment. When quality control is blocked, the defective receptors follow one of three potential routes to the plasma membrane (broken arrows). (C) Damaged receptors are eliminated from the plasma membrane following a shift to a nonpermissive temperature, and they are delivered to the same endosomal compartment as misfolded receptors.
A subset of the quality control elements that we identified belongs to the vacuolar protein sorting pathway. Two of the four STP mutants missorted CPY, and certain vps mutations (vps1, vps8, vps10, and vps28 but not vps3) suppressed ste2-3. Although the pep4 mutant blocks Ste2-3p degradation (19), the pep4 ste2-3 mutant remained unresponsive to α-factor. This result indicates that vps mutations suppress ste2-3 because of protein sorting defects and not because of a protease-deficient vacuole. The inability of vps3 to suppress ste2-3 suggests that some factors required for sorting CPY and assembling vacuolar ATPase (38) are not required for sorting Ste2-3p to the vacuole. Mutant forms of plasma membrane ATPase are also diverted to the vacuole (5); vps mutants, as well as non-vps mutants (sop), restore delivery to the cell surface (28).
Misfolded Ste2-3p is apparently recognized in the Golgi complex. In mammalian cells, misfolded and unassembled membrane proteins are usually retained in the ER (25); however, partially assembled T-cell receptors are degraded in the lysosome, suggesting that they are sorted in the Golgi complex (31). Most rhodopsin mutants that lead to the disease retinitis pigmentosa exhibit protein folding defects and ER retention; they influence the packing of the transmembrane helices and the tertiary structure of the intradiscal domain (17, 23). ER retention of misfolded Ste2-3p may represent a minor quality control pathway in that deletion of the CNE1 gene (encoding the ER chaperone calnexin) leads to partial suppression of ste2-3 (35). However, since Ste2-3p is stable in pep4 mutants (19) and since vps mutants that block Golgi-to-vacuole protein traffic prevent Ste2-3p from reaching the vacuole, the major quality control mechanism apparently recognizes misfolded Ste2-3p in the Golgi complex. Vps10p serves as a Golgi receptor that targets soluble CPY to the vacuole (6, 30), and it has been suggested that Vps10p serves as a receptor for misfolded soluble proteins as well (15). Our finding that vps10 suppresses ste2-3 (Table 4) raises the possibility that Vps10p also recognizes misfolded membrane proteins in the Golgi complex; however, we cannot exclude an indirect role for Vps10p.
When the trafficking of misfolded Ste2-3p to the vacuole is blocked in stp mutants, a significant portion of Ste2-3p is directed to the plasma membrane. Potential pathways for the trafficking of misfolded Ste2-3p to the plasma membrane are depicted in Fig. 9B. It is not surprising that the vps1 mutant permits the accumulation of misfolded Ste2-3p on the plasma membrane, since it blocks vacuolar protein traffic from the Golgi complex to the prevacuole (21). In unsuppressed cells, misfolded Ste2-3p is apparently diverted from the Golgi complex to the Vps pathway, and the loss of Vps1p allows Ste2-3p to progress to the cell surface. The mechanism by which class E vps mutants (stp22 and vps28) suppress ste2-3 is less obvious. Two models account for suppression. First, when exit from the prevacuole is blocked, Ste2-3p may follow an alternate pathway to the cell surface. Second, defective prevacuoles may block the exit of Ste2-3p from the Golgi complex by backing up Golgi-to-vacuole traffic; in this case, the mechanism of suppression would be similar to that proposed for vps1.
The stp mutants that inhibited the elimination of misfolded Ste2-3p also inhibited the removal of damaged Ste2-3p from the cell surface. Therefore, the quality control pathways that eliminate damaged receptors and misfolded receptors apparently have common elements. Two models account for the ability of vps mutants (stp22 and STP26) to retard the exit of damaged receptors from the cell surface (Fig. 9C). First, late blocks in the pathway may cause the endocytic pathway to back up, inhibiting indirectly the exit of damaged Ste2-3p from the cell surface. Second, when endosome-to-vacuole traffic is inhibited, endocytosed Ste2-3p may become available to a recycling pathway, thus slowing the net rate at which Ste2-3p disappears from the plasma membrane. Davis et al. (7) offer these models to explain the ability of the class E vps2 mutant to inhibit the endocytosis of a-factor receptors (Ste3p). Non-vps mutants (stp24 and STP27) may block a step before the quality control and Vps pathways join or may affect a late event in quality control that is specific to defective membrane proteins.
Considerable evidence suggests that the Golgi-to-vacuole pathway and the endocytic pathway intersect at a common organelle. In class E vps mutants, a-factor receptors and FM4-64 accumulate in the class E compartment (7, 36, 39, 52), suggesting that endocytic and vacuolar protein sorting pathways merge at the prevacuole. Moreover, at low temperatures, endocytosed α-factor accumulates in intermediate compartments that cofractionate with partially processed CPY (53). ypt51 mutants (same as vps21) inhibit both α-factor endocytosis and the Vps pathway (45), and special alleles of YPT51 lead to large subcellular compartments that are positive for both Ste2p and CPY (46). The behavior of the ypt7 mutant also suggests the convergence of endocytic and vacuolar protein sorting pathways, as internalized α-factor is degraded in a prevacuolar compartment by PEP4-dependent proteases. Our results are consistent with a common prevacuolar organelle, since both Ste2p-GFP and Ste2-3p–GFP accumulate in similar structures in the stp22 mutant (Fig. 5), even though Ste2p traverses the plasma membrane during its turnover and misfolded Ste2-3p does not (19).
The cytoplasmic side of the α-factor receptor is apparently degraded within the vacuolar lumen. Consistent with this finding, an earlier study (55) found that the cytoplasmic domain of the Golgi membrane protein, Kex2p, is also transported to the vacuolar lumen during its turnover. Our earlier results with Ste2p indicated that a PEP4-dependent process simultaneously degrades the N-terminal and C-terminal domains of the receptor (19). This observation was consistent with two models: (i) both sides of the receptor enter the vacuolar lumen before they are degraded, or (ii) degradation of the luminal side renders the opposite side susceptible to cytosolic proteases. The present study favors the first model, since GFP fused to the C terminus of the receptor appeared within the vacuole. The accumulation of GFP in the vacuole did not appear to reflect translocation after GFP had been cleaved from the fusion protein, since GFP accumulated in the vacuole of the pep4 mutant even though GFP was not cleaved from the C-terminal sequences of the receptor. Our findings are consistent with a recent proposal for degradation of the mammalian epidermal growth factor (EGF) receptor. The endocytosed EGF receptor accumulates on vesicles contained within a compartment designated the multivesicular body (MVB); MVBs are believed to fuse with the lysosome, resulting in delivery of the EGF receptor complex to the lysosomal lumen (7a). Conceivably, Ste2p is sorted to vesicles contained within a late endosomal compartment as well. Although evidence for MVBs in yeast is somewhat limited, structures resembling MVBs accumulate in vps18 mutants (40), 3-phosphorylated phosphoinositides in the cytoplasmic leaflet of endosomal membranes are apparently sequestered into vesicles that enter the vacuolar lumen (56) and, during autophagy, cytoplasmic proteins are thought to enter the vacuole when the outer membrane of a double-membrane vesicle fuses with the vacuolar membrane (49).
The similarity of Stp22p to mammalian TSG101 suggests that membrane protein traffic or lysosomal function may be linked to cancer. The TSG101 gene is implicated in tumor susceptibility, since functional disruption of this gene in mouse NIH 3T3 cells causes cellular transformation and since the transformed cells form metastatic tumors when transferred to nude mice. Zhong et al. (58) found TSG101 only in the cytoplasm of human retinoblastoma cells. Xie et al. (57) found that TSG101 is prominent both in the cytoplasm and in the nucleus of mouse NIH 3T3 cells and that its localization is cell-cycle dependent—in interphase cells, it occurs in both the nucleus and the Golgi complex, whereas in mitotic cells, some TSG101 associates with mitotic spindles and centrosomes. Stp22p plays no obvious role in cell division, since stp22Δ mutant cells divide and respond to α-factor normally. GFP-tagged Stp22p occurred diffusely and as distinct foci in the cytoplasm of wild-type cells. We cannot determine whether a minor pool of Stp22p resides within the nucleus. In the class E vps28 mutant, Stp22p was concentrated in the class E compartment. Since this compartment also accumulated misfolded Ste2-3p, it is possible that Stp22p plays a direct role in the trafficking of misfolded membrane proteins. Stp22p and TSG101 proteins may contain similar protein-protein contacts, as they both contain proline-rich and coiled-coil domains. Proline-rich sequences potentially associate with profilin and SH3 domains. Several elements of the actin cytoskeleton contain SH3 domains, and genetic evidence links the yeast actin cytoskeleton to endocytosis (26). Many growth factor receptors are down-regulated by endocytosis, and the failure to down-regulate EGF receptors leads to enhanced EGF-dependent proliferation (54).
Quality control mutants that remain competent for sorting vacuolar proteases provide clues for understanding the regulatory events that are unique to the quality control pathway. The product of the GEF1 gene (identical to STP24) shows homology to the family of voltage-gated chloride channels (10). This homology apparently reflects the enzymatic activity of the protein, since gef1 mutants are suppressed when they express the CLC-0 gene from Torpedo marmorata (8). Gef1p localizes to late or post-Golgi vesicles (8, 12) and influences the homeostasis of iron and other cations (8, 10). Chloride channels potentially play a role in endosome function. In a transfected-cell model, the cystic fibrosis transmembrane conductance regulator influences chloride-dependent rates of endosomal fusion (4). Chloride ions also affect the initial rate of acidification of MVBs (51). In a mathematical model, voltage-gated chloride channels control endosomal pH and eliminate the dependence of luminal pH on the size and shape of the organelle (43); however, there is as yet no direct evidence for these channels in endosomes. Endosomes play roles both in recycling membrane proteins to the cell surface and in sorting the proteins to the lysosome (50). It is possible that Gef1p influences quality control by affecting the balance between recycling and sorting. Moreover, since recycling and sorting endosomes are very different in shape, their relative abilities to maintain proton gradients (or other ion gradients) in the absence of Gef1p may differ. Together, these results suggest that Gef1p limits the specific functions of endosomal or late Golgi compartments involved in quality control. Other yeast gene products implicated in quality control are the Golgi Ca+2 ATPase (encoded by PMR1), which limits the secretion of certain heterologous gene products (2), and a synaptojanin homolog (encoded by SOP2), which has been implicated in synaptic vesicle endocytosis and recycling (28).
ACKNOWLEDGMENTS
We thank S. Emr, R. Gilmore, J. Haber, J. Konopka, C. Raymond, S. Rieder, and T. Stevens for providing antisera and plasmids. We also thank M. Babst and S. Emr for communicating unpublished results and Ayce Yesilaltay, Aidan Hennigan, and Jodi Hirschman for comments on the manuscript.
This investigation was supported by grant VM-31 from the American Cancer Society and by Public Health Service research grant GM34719 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Anderson M T, Tjioe I M, Lorincz M C, Parks D R, Herzenberg L A, Nolan G P, Herzenberg L A. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell using engineered green fluorescent proteins. Proc Natl Acad Sci USA. 1996;93:8508–8511. doi: 10.1073/pnas.93.16.8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antebi A, Fink G R. The yeast Ca2+ ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992;3:633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Babst, M., and S. Emr. Personal communication.
- 3.Babst M, Wendland B, Estepa E J, Emr S D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biwersi J, Emans N, Verkman A S. Cystic fibrosis transmembrane conductance regulator activation stimulates endosome fusion in vivo. Proc Natl Acad Sci USA. 1996;93:12484–12489. doi: 10.1073/pnas.93.22.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang A, Fink G R. Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper A A, Stevens T H. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis N G, Horecka J L, Sprague G F., Jr cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J Cell Biol. 1993;122:53–65. doi: 10.1083/jcb.122.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Futter C E, Pearse A, Hewlett L J, Hopkins C R. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaxiola R A, Yuan D S, Klausner R D, Fink G R. The yeast CLC chloride channel functions in cation homeostasis. Proc Natl Acad Sci USA. 1998;95:4046–4050. doi: 10.1073/pnas.95.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerdes H-H, Kaether C. Green fluorescent protein: applications in cell biology. FEBS Lett. 1996;389:44–47. doi: 10.1016/0014-5793(96)00586-8. [DOI] [PubMed] [Google Scholar]
- 10.Greene J R, Brown N H, DiDomenico B J, Kaplan J, Eide D J. The GEF1 gene of Saccharomyces cerevisiae encodes an integral membrane protein; mutations in which have effects on respiration and iron-limited growth. Mol Gen Genet. 1993;241:542–553. doi: 10.1007/BF00279896. [DOI] [PubMed] [Google Scholar]
- 11.Hartwell L H. Mutants of Saccharomyces cerevisiae unresponsive to cell division control by polypeptide mating hormone. J Cell Biol. 1980;85:811–822. doi: 10.1083/jcb.85.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hechenberger M, Schwappach B, Fischer W N, Frommer W B, Jentsch T J, Steinmeyer K. A family of putative chloride channels from Arabidopsis and functional complementation of a yeast strain with a CLC gene disruption. J Biol Chem. 1996;271:33632–33638. doi: 10.1074/jbc.271.52.33632. [DOI] [PubMed] [Google Scholar]
- 13.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 14.Hirschman J, DeZutter G, Simonds W, Jenness D D. The Gβγ complex of the yeast pheromone response pathway: subcellular fractionation and protein-protein interaction. J Biol Chem. 1997;272:240–248. doi: 10.1074/jbc.272.1.240. [DOI] [PubMed] [Google Scholar]
- 15.Hong E, Davidson A R, Kaiser C A. A pathway for targeting soluble misfolded proteins to the yeast vacuole. J Cell Biol. 1996;135:623–633. doi: 10.1083/jcb.135.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu V W, Yuan L C, Nuchtern J G, Lippincott-Schwartz J, Hammerling G J, Klausner R D. A recycling pathway between the endoplasmic reticulum and the Golgi apparatus for retention of unassembled MHC class I molecules. Nature. 1991;352:441–444. doi: 10.1038/352441a0. [DOI] [PubMed] [Google Scholar]
- 17.Hwa J, Garriga P, Liu X, Khorana H G. Structure and function in rhodopsin: packing of the helices in the transmembrane domain and folding to a tertiary structure in the intradiscal domain are coupled. Proc Natl Acad Sci USA. 1997;94:10571–10576. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenness D D, Goldman B S, Hartwell L H. Saccharomyces cerevisiae mutants unresponsive to α-factor pheromone: α-factor binding and extragenic suppression. Mol Cell Biol. 1987;7:1311–1319. doi: 10.1128/mcb.7.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenness D D, Li Y, Tipper C, Spatrick S. Elimination of defective α-factor pheromone receptors. Mol Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenness D D, Spatrick P. Down regulation of the α-factor pheromone receptor in S. cerevisiae. Cell. 1986;46:345–353. doi: 10.1016/0092-8674(86)90655-0. [DOI] [PubMed] [Google Scholar]
- 21.Jones E W, Webb G C, Hiller M A. Biogenesis and function of the yeast vacuole. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. III. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 363–470. [Google Scholar]
- 22.Katz M E, Ferguson J, Reed S I. Temperature-sensitive lethal pseudorevertants of ste mutations in Saccharomyces cerevisiae. Genetics. 1987;115:627–636. doi: 10.1093/genetics/115.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushal S, Khorana H G. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry. 1994;33:6121–6128. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 24.Konopka J B, Jenness D D, Hartwell L H. The C-terminus of the S. cerevisiae α-pheromone receptor mediates an adaptive response to pheromone. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- 25.Kopito R R. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 26.Kubler E, Riezman H. Actin and fimbrin are required for the internalization step of endocytosis. EMBO J. 1993;12:2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Cohen S N. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell. 1996;85:319–329. doi: 10.1016/s0092-8674(00)81111-3. [DOI] [PubMed] [Google Scholar]
- 28.Luo W, Chang A. Novel genes involved in endosomal traffic in yeast revealed by suppression of a targeting-defective plasma membrane ATPase mutant. J Cell Biol. 1997;138:731–746. doi: 10.1083/jcb.138.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lupas A. Predictions and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 30.Marcusson E G, Horazdovsky B F, Cereghino J L, Gharakhanian E, Emr S D. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 31.Minami Y, Weissman A M, Samelson L E, Klausner R D. Building a multichain receptor: synthesis, degradation, and assembly of the T-cell antigen receptor. Proc Natl Acad Sci USA. 1987;84:2688–2692. doi: 10.1073/pnas.84.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newlon C S, Lipchitz L R, Collins I, Deshpande A, Devenish R J, Green R P, Klein H L, Palzkill T G, Ren R B, Synn S, Woody S T. Analysis of a circular derivative of Saccharomyces cerevisiae chromosome III: a physical map and identification and localization of ARS elements. Genetics. 1991;129:343–357. doi: 10.1093/genetics/129.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliver S G, van der Aart Q J, Agostoni-Carbone M L, Aigle M, Alberghina L, Alexandraki D, Antoine G, Anwar R, Ballesta J P, Benit P, Berben G, Bergantino E, Biteau N, Bolle P A, Bolotinfukuhara M, Brown A, Brown A J P, Buhler J M, Carcano C, Carignani G, Cederberg H, Chanet R, Contreras R, Crouzet M, Daignanfornier B, Defoor E, Delgado M, Demolder J, Doira C, Dubois E, Dujon B, Dusterhoft A. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 34.Ormö M, Cubitt A B, Kallio K, Gross L A, Tsien R Y, Remington S J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 35.Parlati F, Dominguez M, Bergeron J J M, Thomas D Y. Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J Biol Chem. 1995;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- 36.Piper R C, Cooper A A, Yang H, Stevens T H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raymond C K, Howaldstevenson I, Vater C A, Stevens T H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class-E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raymond C K, O’Hara P J, Eichinger G, Rothman J H, Stevens T H. Molecular analysis of the yeast VPS3 gene and the role of its product in vacuolar protein sorting and vacuolar segregation during the cell cycle. J Cell Biol. 1990;111:877–892. doi: 10.1083/jcb.111.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rieder S E, Banta L M, Kohrer K, McCaffery J M, Emr S D. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieder S E, Emr S D. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 42.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rybak S L, Lanni F, Murphy R F. Theoretical considerations on the role of membrane potential in the regulation of endosomal pH. Biophys J. 1997;73:674–687. doi: 10.1016/S0006-3495(97)78102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schandel K A, Jenness D D. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singerkruger B, Stenmark H, Dusterhoft A, Philippsen P, Yoo J S, Gallwitz D, Zerial M. Role of three rab5-like Gtpases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J Cell Biol. 1994;125:283–298. doi: 10.1083/jcb.125.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singerkruger B, Stenmark H, Zerial M. Yeast Ypt51p and mammalian Rab5: counterparts with similar function in the early endocytic pathway. J Cell Sci. 1995;108:3509–3521. doi: 10.1242/jcs.108.11.3509. [DOI] [PubMed] [Google Scholar]
- 47.Sprague G F, Jr, Herskowitz I. Control of yeast cell type by the mating type locus. I. Identification and control of expression of the a-specific gene BAR1. J Mol Biol. 1981;153:305–321. doi: 10.1016/0022-2836(81)90280-1. [DOI] [PubMed] [Google Scholar]
- 48.Symington L S, Brown A, Oliver S G, Greenwell P, Petes T D. Genetic analysis of a meiotic recombination hotspot on chromosome III of Saccharomyces cerevisiae. Genetics. 1991;128:717–727. doi: 10.1093/genetics/128.4.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trowbridge I S, Collawn J F, Hopkins C R. Signal-dependent membrane protein trafficking in the endocytic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 51.Van Dyke R W. Proton pump-generated electrochemical gradients in rat liver multivesicular bodies. Quantitation and effects of chloride. J Biol Chem. 1988;263:2603–2611. [PubMed] [Google Scholar]
- 52.Vida T A, Emr S D. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vida T A, Huyer G, Emr S D. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vieira A V, Lamaze C, Schmid S L. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- 55.Wilcox C A, Redding K, Wright R, Fuller R S. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wurmser A E, Emr S D. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie W, Li L, Cohen S N. Cell cycle-dependent subcellular localization of the TSG101 protein and mitotic and nuclear abnormalities associated with TSG101 deficiency. Proc Natl Acad Sci USA. 1998;95:1595–1600. doi: 10.1073/pnas.95.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong Q, Chen C F, Chen Y, Chen P L, Lee W H. Identification of cellular TSG101 protein in multiple human breast cancer cell lines. Cancer Res. 1997;57:4225–4228. [PubMed] [Google Scholar]