Abstract
Influenza polymerase (FluPol) plays a key role in the viral infection cycle by transcribing and replicating the viral genome. FluPol is a multifunctional, heterotrimeric enzyme with cap-binding, endonuclease, RNA-dependent RNA polymerase and polyadenylation activities. It performs its functions in the context of the viral ribonucleoprotein particle (RNP), wherein the template viral RNA is coated by multiple copies of viral nucleoprotein. Moreover, it interacts with a number of host proteins that are essential cofactors and, consequently, adaptive mutations in the polymerase are required for crossing the avian–human species barrier. In this review, we show how mechanistic understanding of how FluPol performs its multiple functions has greatly advanced over the last decade through determination of high-resolution structures by X-ray crystallography and cryo-electron microscopy. These have revealed not only the detailed architecture of FluPol but highlighted the remarkably conformational flexibility that is inherent to its functioning as a dynamic RNA synthesis machine. Structural studies are also underpinning current attempts to develop next-generation anti-influenza drugs that directly target FluPol.
Influenza virus possesses a heterotrimeric RNA-dependent RNA polymerase (FluPol) composed of subunits PA (polymerase acidic protein, P3 in influenza C), PB1 (polymerase basic protein 1), and PB2 (polymerase basic protein 2) of total molecular weight of ∼270 kDa (Fig. 1A). Each of the negative-sense, single-stranded (ss) RNA genome segments of influenza viruses is packaged as a distinct ribonucleoprotein (RNP) by multiple copies of the viral nucleoproteins (NPs) together with one polymerase complex. The RNP is the functional context in which FluPol transcribes and replicates the viral RNA (vRNA). In the RNP, the polymerase is bound to the conserved and quasicomplementary 3′ and 5′ ends of the vRNA, jointly known as the “promoter,” which pseudocircularizes the ssRNA. Synthesis of viral mRNA (transcription) starts with “cap-snatching” of short, capped oligomers (10–15 nt in length) from nascent host Pol II transcripts, using the PB2 cap-binding and PA endonuclease activities of FluPol (Plotch et al. 1981). These oligomers prime template-directed RNA synthesis at the PB1 active site, following the typical polymerase sequence of initiation, elongation, termination, and recycling. However, in the case of FluPol transcription, termination involves stuttering at the 5′ proximal oligo(U) polyadenylation signal, thus generating the viral mRNA poly(A) tail (Poon et al. 1999). FluPol thus uses unique mechanisms to synthesize capped and polyadenylated viral mRNAs fully compatible with the downstream processing (e.g., transport, splicing, and translation) by the host gene expression machinery. Synthesis of unmodified genome copies (replication) occurs first by FluPol copying vRNA into complementary, positive-sense cRNA, with the cRNA cosynthetically being packaged with newly synthesized NP and FluPol into cRNPs. Subsequently, cRNPs replicate by a similar but not identical process into vRNPs that can be the template for further (secondary) transcription or exported from the nucleus and packaged into progeny virions. Replication differs from transcription in being unprimed, and termination occurs at the template 5′ extremity with read-through of the polyadenylation signal.
Figure 1.
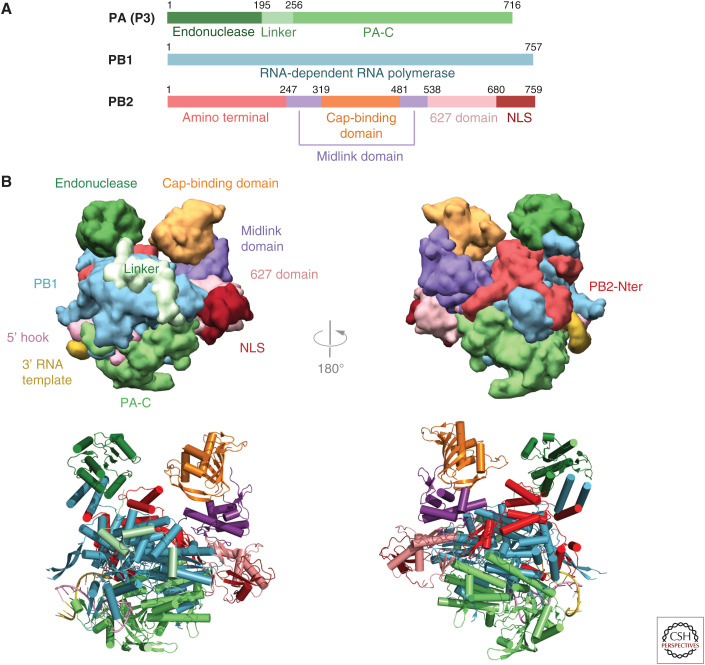
Overall structure of influenza polymerase. (A) Schematic of the three subunits showing major domains. Colors used throughout this review are PA-N endonuclease (dark green), PA linker (light green), PA-C (medium green), PB1 (cyan), PB2-N (red), PB2 midlink (purple), PB2 cap-binding domain (orange), PB2 627 domain (salmon), PB2 NLS domain (dark red). (B) Front and back views of influenza polymerase in the transcription active conformation in surface (top) and ribbon (bottom) representation showing the major domains colored as in A. Drawn from PDB: 4WSB.
It has always been a goal to understand in atomic resolution detail how FluPol carries out its unique functions, but solving the structure of the complete heterotrimer proved a tough nut to crack, largely because of the difficulty of producing sufficient amounts of pure, active polymerase. The first, atomic-resolution crystal structures of independently folded domains and subunit interfaces were published in 2007–2009 (Tarendeau et al. 2007, 2008; Guilligay et al. 2008; He et al. 2008; Obayashi et al. 2008; Dias et al. 2009; Sugiyama et al. 2009; Yuan et al. 2009; for review, see Ruigrok et al. 2010) and only in 2014 was the structure of the complete heterotrimer determined (Pflug et al. 2014; Reich et al. 2014). More recently, crystallographic and/or cryo-electron microscopy (cryo-EM) structures of the full-length heterotrimer have been obtained for influenza viruses of types A, B, C, and D in several functional states. Together, these structures give detailed insight into the mechanism of action of this remarkably dynamic, multifunctional RNA synthesis machine as well as underpinning new opportunities for anti-influenza drug development. Here we summarize the current understanding of the architecture of FluPol and outline its structure-based mechanism of action, particularly for transcription.
FluPol is a highly flexible enzyme, comprising a catalytic core to which are attached flexibly linked peripheral domains (Thierry et al. 2016). It therefore has multiple different interchangeable conformations, even though in a crystal, a particular conformation can be selectively trapped. Indeed the first promoter-bound crystal structures trapped what is now referred to as the “transcriptionally active” state, which will now be described. Unless otherwise stated, residue type and number will refer to the avian/human influenza A strains.
OVERALL STRUCTURE OF INFLUENZA POLYMERASE
Transcriptionally active FluPol has a U-shaped structure, with approximate height, width, and depth of 115 × 100 × 75 Å, respectively (Fig. 1B). The two protruding arms are capped by the PA-N endonuclease (Dias et al. 2009; Yuan et al. 2009) and PB2 cap-binding (Guilligay et al. 2008) domains, which face each other across a solvent channel. The endonuclease is held in place by one of its helices contributing to the helical bundle formed by the interface between the carboxyl terminus of PB1 and the amino terminus of PB2 (Sugiyama et al. 2009). The bottom of the U is formed by the large PA-C domain (He et al. 2008; Obayashi et al. 2008). The carboxy-terminal two-thirds of PB2 (PB2-C) form one side of the U and comprise the midlink, 627, and NLS (nuclear localization signal) domains (Tarendeau et al. 2007, 2008). The body of the trimer is formed by the catalytic core of PB1, which has the canonical right-handed polymerase architecture comprising fingers, palm, and thumb. On the thumb side, it is decorated by the amino-terminal third of PB2 (PB2-N), on the opposite side, by three consecutive short helices of the PA linker that wraps around PB1 and connects the PA-N endonuclease with PA-C. Apart from the two hydrophobic intersubunit interfaces between PA-C and PB1-Nter (He et al. 2008; Obayashi et al. 2008) and PB1-Cter and PB2-Nter (Sugiyama et al. 2009), which stably link the subunits in head-to-tail fashion, there are numerous other hydrophilic intersubunit interactions, which can be reversibly broken or formed, and flexible interdomain linkers that permit remarkable conformational flexibility (Thierry et al. 2016).
FUNCTIONAL DOMAINS
Cap-Binding Domain
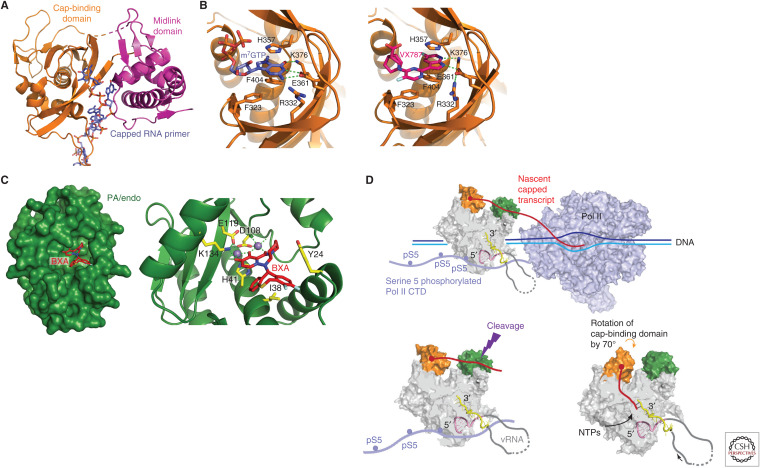
The cap-binding domain (residues 320–485 of PB2) (Fig. 2A,B) has a unique fold, distinct from cellular cap-binding proteins such as the translation initiation factor eIF4E or the nuclear cap-binding complex (CBC) (Guilligay et al. 2008). It does, however, preserve the aromatic sandwich formed by residues His357 and Phe404, as the means by which specificity for the cap-specific methylated guanosine (m7G) is enhanced compared to the unmethylated base (Fig. 2B, left). The isolated FluA cap-binding domain binds relatively weakly to the cap analog m7Gppp (Guilligay et al. 2008), and even more weakly for FluB (Xie et al. 2016), but in the heterotrimer context, additional interactions to the capped RNA physiological substrate are provided by the adjacent PB2 midlink domain (Fig. 2A; Pflug et al. 2018). Even so, it is not yet clear how influenza polymerase is able to preferentially pirate fully capped nascent Pol II transcripts in competition with the much higher affinity nuclear CBC that normally sequesters the cap early in transcription. A key insight emerging from the original crystal structures of the complete heterotrimer was that the cap-binding domain was flexibly linked to the polymerase core allowing its rotation, presumably stochastically, over a large angle (∼70°) (Fig. 2D; Reich et al. 2014; Pflug et al. 2018). One extreme position is consistent with the cap-bound nascent Pol II transcript reaching across to the endonuclease to be cleaved during cap-snatching. The other extreme position allows the cleaved primer to be directed into the RNA synthesis active site to initiate transcription (Pflug et al. 2018; Kouba et al. 2019). As for the endonuclease, the cap-binding domain is a promising target for anti-influenza drugs (Pautus et al. 2013; Mifsud et al. 2019). The most advanced compound to date is VX-787/JNJ63623872/pimodivir, which is very potent against FluA polymerase because of picomolar affinity for the cap-binding site (Fig. 2B, right; Clark et al. 2014). In contrast, it has no antiviral activity against FluB because it binds too weakly to the FluB cap-binding domain as a result of amino acid differences in the cap-binding site (Pflug et al. 2018). Interestingly, part of the potency of pimodivir derives from its binding to not only the cap-binding domain but also to the adjacent midlink domain, thus stabilizing a particular configuration of these domains that is transcriptionally inactive (see the section Conformational Flexibility and Dimer Formation; Ma et al. 2017; Pflug et al. 2018). However, resistant mutations to pimodivir (e.g., PB2 S324K/N/R, F325L, S337P, M431I, N510T) have been selected in vitro (Byrn et al. 2015; Ma et al. 2017; Pflug et al. 2018) or in treated patients (Trevejo et al. 2018; Finberg et al. 2019). Combination therapy with other anti-influenza drugs may be a solution to this problem (Mifsud et al. 2019).
Figure 2.
Cap-snatching. (A) Capped RNA primer (slate blue) binding to the PB2 cap-binding and midlink domains and descending into the PB1 active site to initiation transcription. Drawn from PDB: 6QCX. (B) Close-up of (left) cap-analog m7GTP (slate blue) and (right) VX787/pimodivir (pink) binding in the influenza A cap-binding domain showing conserved interacting residues (orange sticks). Drawn from PDB: 4CB4 and 6EUV, respectively. (C) Surface representation of influenza A endonuclease with bound baloxavir acid (BXA) in the active site, showing two divalent cations (purple spheres) and conserved interacting residues (yellow sticks). The substitution I38T confers partial resistance to the drug. Drawn from PDB: 6FS6. (D) Schematic of how cap-snatching is performed. FluPol (gray surface with cap-binding domain in orange and endonuclease in green) binds to the serine 5 phosphorylated (pS5) carboxy-terminal domain (CTD) of host polymerase II (Pol II, light blue surface). This gives FluPol access to emerging nascent capped transcripts (red) that bind to the cap-binding domain and are cleaved 10–15 nt downstream by the endonuclease. The cap-binding domain then rotates to direct the capped primer into the PB1 RNA synthesis active site to initiate transcription.
Endonuclease
The cap-snatching endonuclease (residues 1–196 of PA) is a member of the PD-(D/E)xK two-metal nuclease superfamily (Dias et al. 2009; Yuan et al. 2009). Conserved residues His41, Glu80, Asp108, and Glu119 chelate the two essential divalent cations (probably magnesium in vivo, but manganese works better in vitro), and Lys134 also plays an important catalytic role (Fig. 2C; Crepin et al. 2010; Kowalinski et al. 2012; Datta et al. 2013). The isolated endonuclease domain has weak affinity for RNA, which probably explains why there is still no structure available of an RNA substrate complex. However, in the context of the complete heterotrimer, the cis-acting cap-binding domain considerably enhances the affinity for the capped nascent Pol II transcript, which is the physiological substrate (Pflug et al. 2018). Because the endonuclease is essential to provide the capped primers for initiating transcription, endonuclease inhibitors that chelate the two-metal ions are potential anti-influenza drugs. Since the 1990s, many inhibitors have been described (Tomassini et al. 1994; DuBois et al. 2012; Kowalinski et al. 2012; Sagong et al. 2014; Stevaert and Naesens 2016; Mifsud et al. 2019) culminating in the discovery of baloxavir acid (BXA), which potently inhibits FluA and FluB endonuclease. Under the trade name Xofluza, a pro-drug of BXA is approved for clinical use in Japan and the United States (Noshi et al. 2018). However, an I38T mutation in the endonuclease active site, which can emerge during treatment, significantly reduces susceptibility to the drug (Omoto et al. 2018; Mifsud et al. 2019), and such resistant viruses can propagate between humans (Imai et al. 2020). Combination therapy with, for instance, Tamiflu is being tested as a potential means to get around this problem.
627-NLS Double Domain
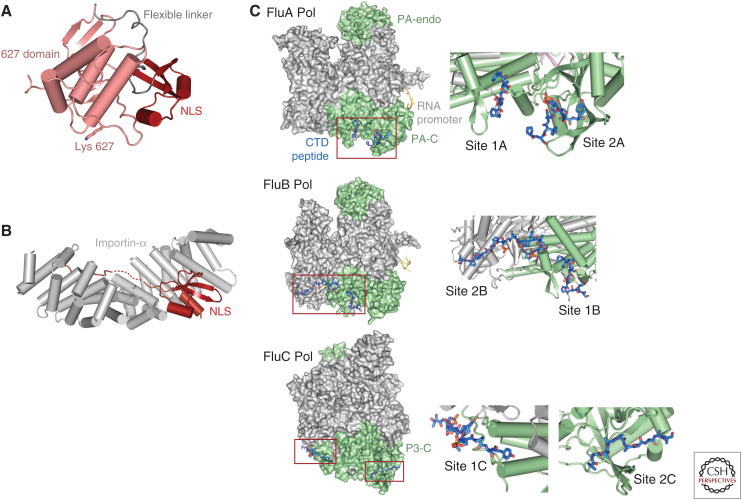
The 627-NLS double domain (PB2/538-757) forms the carboxy-terminal region of PB2 and is separable into two independently folded subdomains, the “627 domain” and the “NLS domain,” separated by a linker (PB2/679-684) (Fig. 3A; Tarendeau et al. 2008; Kuzuhara et al. 2009). The 627 domain (PB2/538-678) is so-called because it contains the host-specific residue 627 (see the subsection Host-Specific Mutations). The NLS domain (PB2/685-759) contains a flexible carboxy-terminal extension beyond PB2/736 that features the PB2 bipartite nuclear localization sequence with minor (737-RKR-739) and major (752-KRIR-755) basic motifs that are recognized and bound by the nuclear import adapter importin-α (Fig. 3B; Tarendeau et al. 2007; Pumroy et al. 2015). In crystal structures of the isolated double domain and in the “transcriptionally active” conformation of the trimer, the two folded subdomains are packed against each other (Fig. 3A), whereas in the so-called “apo”-conformation of the polymerase (in which the promoter is not bound, see the section Conformational Flexibility and Dimer Formation) the two domains are separated and the linker is extended (Hengrung et al. 2015; Thierry et al. 2016). In solution the equilibrium between the open and closed configurations of the 627-NLS double domain has been characterized by NMR, and it has been shown that the open configuration is required for importin-α binding (Delaforge et al. 2015). The 627-domain is not required for transcription in vitro (Nilsson et al. 2017), but in vivo it contributes to binding the Pol II CTD during cap-snatching (see the subsection The PA-C Domain and Host Pol II CTD Binding). The domain is also required for replication, perhaps for recruiting a second polymerase to package the nascent replicate (Nilsson et al. 2017).
Figure 3.
Host–protein interacting domains. (A) Ribbon diagram showing the closed state of the PB2 627-NLS double domain. (B) Binding of the PB2 NLS domain, which carries a bipartite nuclear localization signal (NLS) in its carboxy-terminal extremity, to nuclear import factor importin-α. Drawn from PDB: 2JDQ. (C) Serine 5 phosphorylated Pol II CTD binding to influenza A, B, and C polymerases. In each case, two distinct sites are observed.
Host-Specific Mutations
Both the 627 and NLS domains of PB2 are hotspots for host-specific residues that mutate during interspecies transmission (Peacock et al. 2019). First and foremost is residue 627 itself, which is almost invariably a glutamate in avian strains but needs to be a lysine for efficient polymerase activity in mammalian cells (Subbarao et al. 1993; Hatta et al. 2001). However, the combination of the avian Glu627 signature with additional substitutions G590S and Q591R in the same domain, as found in the 2009 pandemic A strain, also confers high activity in mammalian cells (Mehle and Doudna 2009). Consistently, these avian-to-human substitutions render the surface of the 627 domain more extensively positively charged in human compared to avian variants (Tarendeau et al. 2008; Mehle and Doudna 2009). The specific role of the different signatures at position 627 is still not understood mechanistically. However, a recent breakthrough has correlated characteristic differences between the avian and human versions of the host factor ANP32, which is important for vRNA to cRNA replication (Sugiyama et al. 2015), with host differences in the 627 domain signature (Long et al. 2016; Mistry et al. 2019). PB2 residues 701 and 702 of the NLS domain are also strongly implicated in host adaptation (Chin et al. 2017). Residue 701 is normally an aspartate in all strains but the D701N substitution is selected upon adaptation of highly pathogenic avian strains to mice (Li et al. 2005; Gabriel and Fodor 2014) and also in cases of avian to human or guinea pig transmission (de Jong et al. 2006; Steel et al. 2009). Although it has been suggested that the D701N substitution could affect the interaction with importin-α (Tarendeau et al. 2007), the exact role of these two residues remains to be elucidated.
The PA-C Domain and Host Pol II CTD Binding
The large PA carboxy-terminal domain (PA/258-713) has an important structural role in tightly binding the extreme amino terminus of PB1 and more generally buttressing the entire trimer. However, it also has specific functional roles in promoter binding (see the section Different Modes of Promoter Binding), in interacting with the carboxy-terminal domain (CTD) of host Pol II to enable cap-snatching, and in contributing to the recently discovered secondary 3′ end binding site (see the section Different Modes of Promoter Binding). Cap-snatching is thought to occur while Pol II is paused to allow transcript capping, soon after transcription initiation (Fig. 2D). At this stage, the 52 heptad repeats of the CTD are preferentially phosphorylated at the serine 5 position. Although it was shown some time ago that FluPol interacts, probably directly, with the serine 5 phosphorylated form of the CTD (Engelhardt and Fodor 2006), the structural basis for binding was only revealed recently for influenza A and B (Lukarska et al. 2017) and C (Serna Martin et al. 2018) polymerases (Fig. 3C). In each case, there is a different bipartite binding site, exclusively or partially on PA-C. For influenza A, there are two phosphoserine binding sites on PA-C with the phosphates bound respectively by conserved Lys635 and Arg638 (site 1A) and Lys289 and Arg449 (site 2A). Mutation of any one of these residues severely attenuates viral replication (Lukarska et al. 2017), suggesting that disruption of CTD binding by FluPol could be a novel antiviral strategy. However, the highly deleterious R638A mutation can be largely compensated by a second site C453R mutation that restores phosphoserine binding (Fodor et al. 2003; Lukarska et al. 2017). Interestingly, site 2A involves residues from the PA 550-loop, some of which are also implicated in interspecies adaptation (Lukarska et al. 2017). In influenza B, site 1 is conserved (and denoted site 1B) but site 2A is replaced by a distinct site 2B that crosses over from the PA subunit to the PB2 627 domain (Lukarska et al. 2017). For influenza C, both sites 1C and 2C are distinct from previously described sites. They are primarily on PA-C, although site 1C also involves PB1 residues (Serna Martin et al. 2018). These results show that FluPol binds multiple, not necessarily adjacent Pol II CTD repeats, with each individual interaction being only in the micromolar range but, through avidity and cooperativity, resulting in an overall high-affinity interaction. Finally, there is evidence that CTD binding across to the 627 domain stabilizes the transcriptionally active form of the polymerase (Serna Martin et al. 2018).
DIFFERENT MODES OF PROMOTER BINDING
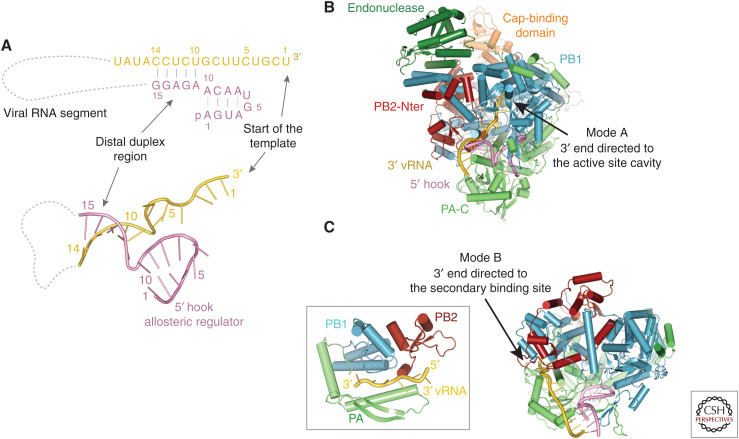
The vRNA promoter is composed of the conserved, quasicomplementary pyrimidine-rich 3′ (nucleotides 1–12) and purine-rich 5′ (nucleotides 1–13) termini (Fig. 4A). The isolated promoter RNA can form an irregular base-paired duplex, but this is not how it binds to the polymerase. Indeed, it is unlikely that the full “panhandle” duplex has any role in influenza replication, although it may bind as such to the innate immune RNA sensor RIG-I (Lee et al. 2016). When bound to the polymerase, nucleotides 1–10 of the 5′ end form an intramolecular stem-loop, the so-called “hook,” with four base-pairs, two of them noncanonical A-A base-pairs (A1:A10 and A4:A7) (Fig. 4A; Pflug et al. 2014). This compact RNA structure, which forms with both vRNA and cRNA (Thierry et al. 2016), binds with high affinity in a specific pocket in the interface between the PA and PB1 subunits. The 5′ hook is secured by a latch formed by the so-called PA-arch and PB1 β-hairpin, thus becoming an integral part of the polymerase structure (Fig. 4B; Pflug et al. 2014). This places the loop of the hook (nucleotides 5–6) close to the polymerase active site. Indeed, 5′-hook binding is critical for correctly structuring critical peptides (e.g., the fingertips) on the RNA synthesis active site and thus acts as an allosteric activator.
Figure 4.
Promoter binding. (A) Schematic of the vRNA promoter structure that comprises the conserved 5′ (pink) and 3′ (yellow) extremities of each genomic vRNA segment. Nucleotides 1–10 of the 5′ end form a stem-loop structure called the “hook,” which binds tightly to the polymerase and acts as an allosteric regulator of polymerase function. Nucleotides 1–9 of the 3′ end are single-stranded and flexible. The distal duplex region is formed by interstrand base-pairs. The cRNA promoter forms a similar structure, but the single-stranded region of the 3′ end is 2 nt longer. (B) Mode A promoter binding showing the 5′ hook bound in a pocket between the PB1 and PA-C domains and the 3′ end of the template entering the PB1 RNA synthesis site. (C) Mode B promoter binding showing the vRNA 3′ end binding in the “secondary 3′-end binding site.”
The proximal vRNA 3′ end nucleotides 1–9 (or 1–11 for cRNA), where RNA synthesis initiates, are single-stranded and flexible, allowing different modes of binding to FluPol depending on the polymerase functional state. In the pre-initiation (resting) state (i.e., in the absence of ongoing RNA synthesis), the vRNA 3′ end is particularly mobile (Robb et al. 2019) and has been observed to take up three positions. The first corresponds to the 3′ end entering through the template entrance tunnel into the PB1 RNA synthesis active site (Fig. 4B; Reich et al. 2017). This has been designated as mode A promoter binding (Peng et al. 2019). Alternatively, for FluB polymerase, the 3′ end has been observed on the external surface of the polymerase, interacting with the PB1 β-ribbon (Reich et al. 2014). More recently, it has been found that the 3′ end nucleotides 1–5 can bind sequence specifically in a “secondary 3′-end binding site” between PA-C and the PB1 thumb, which has been called mode B promoter binding (Fig. 4C; Fan et al. 2019; Peng et al. 2019; Wandzik et al. 2020). In the case of cRNA, the longer single-stranded 3′ end is considerably less flexible (Robb et al. 2019) and preferentially bound in the secondary 3′-end binding site (i.e., mode B) (Fan et al. 2019; Peng et al. 2019). Importantly, the secondary 3′-end binding is mutually exclusive with the homodimer interface (Fan et al. 2019). This suggests that dimerization with a second “activating” polymerase in trans might promote transfer of the cRNA 3′ end to the active site, thus initiating cRNA to vRNA replication, although the detailed molecular mechanism of such regulation of replication remains unknown.
The distal parts of the promoter 5′ and 3′ ends typically make four canonical base-pairs with each other (e.g., 5′ nucleotides 11–14 base-pair with 3′ nucleotides 10–13 for vRNA, and 5′ nucleotides 11–14 base-pair with 3′ nucleotides 12–15 for cRNA) (Fig. 4A). This short duplex projects away from the polymerase. Stabilization of the interstrand base-pairing between the promoter ends is one way the polymerase maintains the pseudocircularity of the viral RNA. However, even in the absence of this base-pairing (which must be broken once RNA synthesis starts and the template begins to translocate), the existence of the high affinity 5′ hook and secondary 3′-end binding site provide a second means by which the polymerase can remain bound to both genome segment ends. Indeed, at least for transcription, there is now strong evidence that during the complete cycle of viral mRNA synthesis, the polymerase never lets go of either end of the vRNA (see the section Model for Complete Transcription Cycle).
CONFORMATIONAL FLEXIBILITY AND DIMER FORMATION
As noted above, FluPol comprises a relatively stable core comprising PA-C, PB1, and PB2-N with flexibly linked peripheral PA-N and PB2-C domains that can be repacked in very distinct ways or simply left dangling, depending on the functional state of the polymerase (Fig. 5). As described above, during cap-snatching, the PB2 cap-binding domain rotates in situ by ∼70° (Fig. 2D). Even more dramatically, crystal structures of FluC (Hengrung et al. 2015), FluB (Thierry et al. 2016), and FluA (Fan et al. 2019) have trapped the so-called “apo” state (i.e., in the absence of any promoter RNA or with only the cRNA 5′ hook bound in the case of FluB), in which the PA endonuclease and PB2-C midlink, cap-binding, 627-, and NLS domains have all translated and/or rotated by significant amounts (Fig. 5C,D). For instance, in the “transcriptionally active” polymerase conformation, the 627-NLS double domain is in the closed state and packs against PB1, distant from the endonuclease (Fig. 5B). However, in the “apo”-conformation, the double domain is relocated and opens, the NLS domain moiety binding to the endonuclease, which itself rotates in situ by ∼45°. The carboxy-terminal NLS motif containing an extension to the NLS domain refolds into an α-helix that also binds to the endonuclease (Hengrung et al. 2015; Thierry et al. 2016; Fan et al. 2019). The functional role of the “apo” state has yet to be elucidated, although it is presumed to be transcriptionally inactive. There is also strong evidence that the cap-binding inhibitor pimodivir stabilizes the “apo” state (Ma et al. 2017; Pflug et al. 2018). In solution, the polymerase is in dynamic equilibrium between several different configurations, as is apparent from recent cryo-EM single-particle reconstructions that typically resolve several different 3D structural classes on the same grid (Kouba et al. 2019; Peng et al. 2019).
Figure 5.
Polymerase conformational flexibility. (A) Schematic showing the FluPol core, comprising PA-C, PB1, and PB2-N, to which are flexibly attached the peripheral PA endonuclease and PB2 midlink, cap, 627-, and NLS domains. (B) Surface representation of domain arrangement in the transcriptionally active state with bound full vRNA promoter. Drawn from PDB: 4WSB. (C) Domain arrangement in the transcriptionally inactive state of FluB polymerase with bound cRNA 5′ end only. Drawn from PDB: 5EPI. (D) Domain arrangement in the transcriptionally inactive state of FluC polymerase with no bound RNA (apo). Similar to C, but endonuclease and 627-domains orientated differently. Drawn from PDB: 5D9A. (E) Symmetric dimeric form of influenza A polymerase as observed in the crystal. The conformation of each monomer is as in D. Drawn from PDB: 6QNW.
Structural studies of human A/H3N2, avian A/H5N1, and avian A/H7N9 polymerases show that in the absence of full promoter binding they form homodimers (Fig. 5E; Chang et al. 2015; Fan et al. 2019; Wandzik et al. 2020). However, similar dimers have not yet been reported for bat FluA or influenza B, C, and D polymerases. The extensive FluA dimer interface is twofold symmetric and involves core peptides from all three subunits. These include PA loops 292–297 and 348–356, each interacting with their counterparts in the second monomer, PB1 helix 567–581 and PB2 loop 71–76. In the H3N2 and H5N1 crystal structures, the polymerases forming the dimer are both in the “apo” conformation, whereas in the dimeric H3N2 cryoEM structure only the core is visible, the peripheral domains being disordered. Full promoter binding leads to monomerization of the polymerase (Chang et al. 2015; Fan et al. 2019), and the observed dimerization is also incompatible with the promoter 3′-end binding in the secondary site (Fan et al. 2019). Interestingly, in a recent study, mutations PB1/K577G, PB2/G74R, or PA/E349K, all of which weaken the observed dimer interface, were selected that could compensate for a replication defect arising from a mismatched PB2 segment from another strain (Chen et al. 2019). Indeed, numerous reports implicate a second “trans-acting” polymerase in the process of replication, particularly cRNA to vRNA synthesis (York et al. 2013; Chen et al. 2019; Fan et al. 2019). However, the exact nature of the interaction between the two polymerases (e.g., whether symmetric or asymmetric) and the mechanistic role of the trans-acting polymerase (e.g., releasing the cRNA 3′ end from the secondary site, template realignment during initiation of vRNA synthesis, and/or nucleating encapsidation of the nascent replicate into an RNP) remains elusive.
MECHANISM OF RNA SYNTHESIS
Active Site and Nucleotide Addition Cycle
The PB1 subunit contains the conserved catalytic core, comprising right-handed fingers, palm, and thumb domains (Fig. 6). Six characteristic motifs (A–F) with key conserved residues, together with two divalent magnesium cations (denoted A and B), control the template-directed nucleotide addition reaction (Fig. 7). The motifs are contained in the palm domain and the fingertips, a loop that extends from the fingers toward the thumb domain. Recent structures of the actively transcribing polymerase reveal in detail how these motifs, and other parts of the influenza polymerase core, interact with the primer, template, and incoming nucleotide during the nucleotide addition cycle (Kouba et al. 2019). Nucleotides are selected for addition to the growing product by their Watson–Crick complementarity to the template base at the so-called +1 position. Nucleotide addition comprises sequential steps of initial NTP binding (Fig. 7B), active site closure, and catalysis with release of pyrophosphate (Fig. 7C). Subsequently, concerted product/template duplex translocation leaves the +1 position vacant for the next incoming NTP (Fig. 7D).
Figure 6.
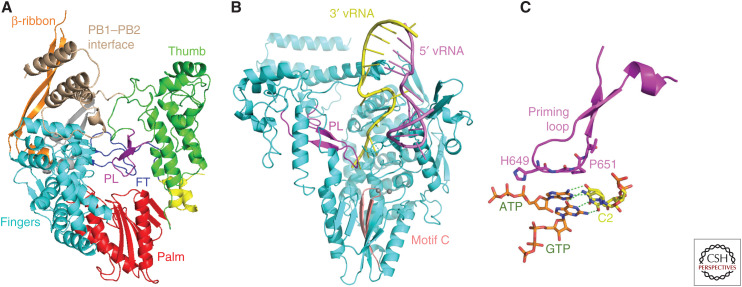
PB1 RNA synthesis active site. (A) Ribbon diagram showing the conserved core polymerase architecture of the PB1 subunit comprising a right-handed arrangement of fingers (cyan), palm (red), and thumb (green). The fingertips loop (FT, blue) and priming loop (PL, magenta) occupy the central cavity where RNA synthesis occurs (see text). Additional PB1 specific features include the β-ribbon (orange), and the carboxy-terminal helices (wheat) that form a strong interface with PB2-N. (B) Mode A vRNA promoter binding to PB1 showing the template 3′ end, where RNA synthesis initiates, in the active site in close proximity to the tip of the priming loop (PL, magenta). Motif C (rose), a β-hairpin in the palm domain that carries two key metal binding aspartates and the two catalytic magnesium ions (gray spheres) are highlighted (see also Fig. 7). (C) Model of how the priming loop might promote vRNA to cRNA replication initiation by stabilizing the first phosphor-transfer reaction that requires ATP and GTP to assemble opposite U1 and C2 of the template, respectively.
Figure 7.
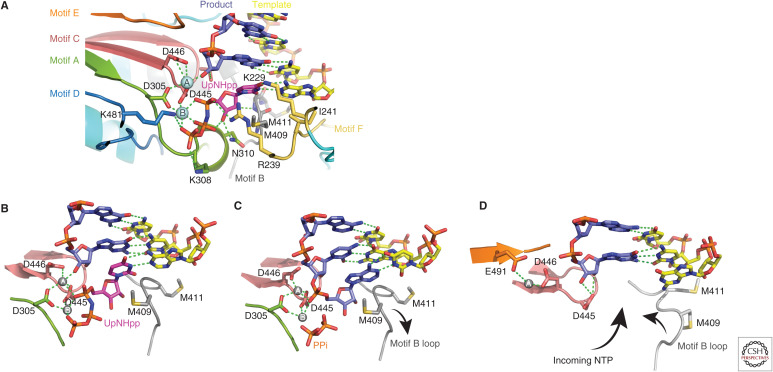
Nucleotide addition cycle. (A) Details of the PB1 catalytic site showing key residues associated with the conserved polymerase motifs A–F and the two catalytic magnesium ions A and B (gray spheres). The primer/product RNA is in blue and the template strand in yellow. An incoming NTP, here a nonhydrolyzable UTP analog (UpNHpp), is shown in the precatalytic state. (B) First precatalytic step in the nucleotide addition cycle with incoming NTP stabilized by stacking on motif B Met409 (same structure as in A). (C) Postcatalytic, pretranslocation step in which the nucleotide has been added onto the product releasing pyrophosphate (PPi). (D) Translocation step, in which the template–product duplex has moved forward one position, bringing the next unpaired template base opposite the empty NTP binding site. Translocation may be assisted by flipping of the motif B loop into an alternative configuration.
The template enters the active site by undergoing a characteristic kink at the +1 to +2 junction, with the +1 base held in place by a stacking interaction with Ile241 of motif F. A complementary NTP, entering through the NTP entrance tunnel, induces the motif B methionine-rich loop (409–GMMMGMF-) to flip conformation, allowing Met409 to stack on the incoming base (Fig. 7B). Prior to NTP binding, the active site is in an “open” configuration with metal A coordinated motif C Asp445 and motif E Glu490 in a side location, distinct from its position during catalysis. Upon NTP binding, which brings the metal B, the three conserved aspartates, Asp444 and Asp445 from motif C and Asp305 from motif A, coordinate both the metals in the “closed” catalytic conformation. In this conformation, key active site residues (e.g., Arg239 [motif F], Lys308 [motif A], Ser444 [motif C], Lys481 [motif D], and the two magnesium ions) position the receiving nucleotide ribose and the substrate NTP triphosphate in the precise geometry required for catalysis. Upon nucleotide incorporation, the translocation of the template–product is probably helped by the mobile methionine-rich loop, which serves as the Brownian ratchet/power stroke mechanism, together with pyrophosphate release. Translocation leaves the incoming nucleotide +1 position vacant for another round of the addition cycle. Direct inhibition of the RNA synthesis activity or incorporation of mutagenic nucleotides is an obvious antiviral strategy. However, although some active nucleoside analogs have been identified, such as T705 (favipiravir) (Furuta et al. 2017) and N4-hydroxycytidine (Toots et al. 2019), they are not in clinical use.
Priming Loop
A particular feature of viral RNA polymerases that perform unprimed RNA synthesis, as FluPol does during replication, is the so-called priming loop (PB1 residues 631–660), which partially fills the active site cavity (Fig. 6A–C). Its role is to provide a stable platform, formed by the conserved tip residues 649-HGP, to promote the assembly of the vRNA to cRNA replication initiation state in which ATP as the priming nucleotide in the +2 position and GTP in the +1 position need to line up opposite terminal template nucleotides U1 and C2, respectively, to form pppApG in the first catalytic step (Fig. 6C; Te Velthuis et al. 2016). However, once this role is fulfilled, the priming loop blocks translocation of the product-template duplex and has to be retracted. The priming loop has been shown to be less important for cRNA to vRNA replication (Te Velthuis et al. 2016). This is because initiation of cRNA to vRNA replication occurs internally opposite nucleotides 4-UC of the cRNA template followed by backtracking of the template and realignment of the formed pppApG opposite 1-UC (Deng et al. 2006; Fan et al. 2019). In the case of transcription, the resting position of the priming loop is incompatible with hybridization of the capped primer with the template during transcription initiation. The priming loop therefore has to progressively retract in the early steps of transcription, as has been recently visualized (Kouba et al. 2019) (see the next section).
MODEL FOR COMPLETE TRANSCRIPTION CYCLE
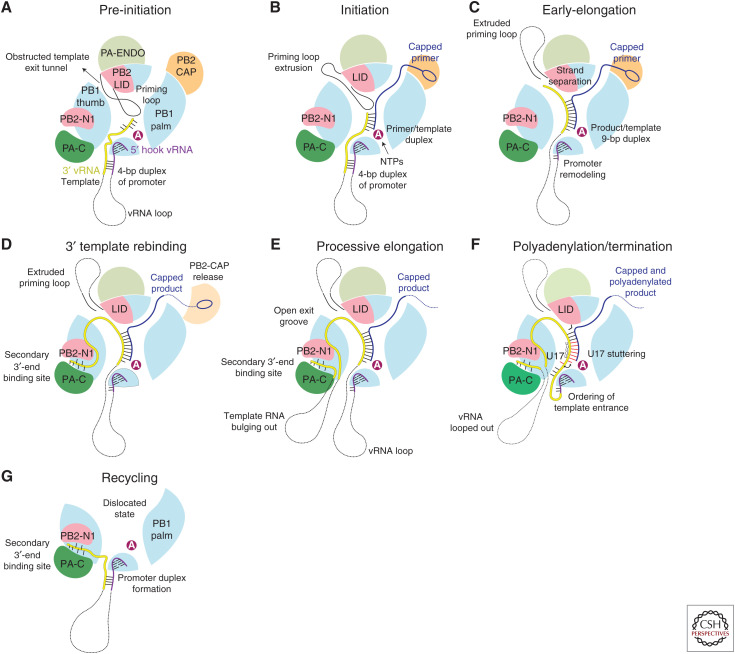
Recently, a combination of X-ray crystallography and cryo-electron microscopy has enabled snapshots to be determined of key steps in transcription encompassing initiation, elongation, termination/poly(A) tail generation, and recycling, leading to the following model of the complete transcription cycle (Kouba et al. 2019; Wandzik et al. 2020). In this model, the 5′ and 3′ extremities of the template remain bound to the polymerase throughout (Fig. 8).
Figure 8.
Model for the complete transcription cycle (see text).
In the pre-initiation state with mode A promoter binding, the template 3′ end can flip in and out of the active site cavity (Fig. 8A). Transcription initiates when the 3′ end of the capped primer hybridizes with the 3′ end of the template and this requires partial withdrawal of the priming loop (Fig. 8B). Typically a primer ending in AG-3′ will base-pair with template 3′-UC with the next template base G3 at the +1 position awaiting a CTP as the first nucleotide to be incorporated into the product. After successful initiation, through subsequent nucleotide addition and template translocation, the template–product RNA duplex grows in the active site cavity up to 9 bp, necessitating further retraction of the priming loop (Kouba et al. 2019). At this point, the duplex abuts against the so-called PB2-lid domain, forcing strand separation (Fig. 8C). A specific α-helix of the lid domain faces the growing product–template duplex and conserved Tyr205 changes rotamer to stack on the template base of the last base-pair (Kouba et al. 2019). The separated template strand navigates into a narrow template exit tunnel, which is only open after the full retraction of the priming loop, while the product strand bulges out in the solvent channel between the endonuclease and cap-binding domain (Kouba et al. 2019). In the initiation to elongation transition, concomitant changes occur to the promoter. After a few rounds of nucleotide addition and template translocation, the base-pairs in the promoter duplex must disrupt, resulting in remodeling of the template entry pathway and clamping down of the PB1 β-ribbon that separates the incoming template from the tightly bound 5′ end (Kouba et al. 2019). As processive elongation proceeds, the outgoing template extremity eventually reaches the secondary 3′-end binding site, where it tightly binds, thus preventing it from becoming lost and degraded (Fig. 8D; Wandzik et al. 2020). Subsequently the outgoing template will bulge out to form a growing loop while at the same time the ingoing template loop shrinks (Fig. 8E). Once the template is translocated so that there is only a taut connect between the 5′ hook and the active site, stuttering occurs on the 5′ proximal oligo(U) motif, involving template U17 flipping in and out of the +1 active site position, to generate the poly(A) tail (Fig. 8F). After product disassociation by a mechanism still to be elucidated, further conformational changes involving opening of the polymerase release the template from the active site cavity. This allows reformation of the promoter in the mode B configuration with the 3′ template end still bound in the secondary site (Fig. 8G). Subsequently, upon reclosure of the polymerase, promoter mode A configuration can be reestablished and initiation of the next transcription cycle begin (Fig. 8A).
CONCLUDING REMARKS
Recent high-resolution structural analysis of multiple functional states of FluPol has led to remarkable advances in our understanding of how the viral transcription-replication machine functions. This is particularly true of cap-dependent transcription, although our understanding of vRNA to cRNA and cRNA to vRNA replication is less well-advanced as it likely involves more than one polymerase. However, most structural studies done so far use recombinant polymerase and do not include NP. Although many polymerase functions can be recapitulated in vitro without NP, notably cap-dependent transcription, the next and considerable challenge is to extend detailed structural studies, first, to the RNP context, to understand the dynamic interplay between polymerase and NP, and second, an even greater challenge, to observe the RNP at work in the nuclear context. With regard to the first challenge, a very recent publication uses cryo-electron microscopy to propose a “processive helical track” mechanism of how, within the RNP, antiparallel NP-RNA strands slide past each other during transcription (Coloma et al. 2020). This model is perfectly consistent with the polymerase-centric mechanism of transcription shown in Figure 8 (Wandzik et al. 2020).
ACKNOWLEDGMENTS
The authors thank all past and current members and collaborators of the Cusack group for their contributions to advancing the structure-based mechanistic understanding of influenza polymerase. This work was supported by the French National Research Agency (ANR) grant (ANR-18-CE11-0028) to S.C. T.K. holds a fellowship from the European Molecular Biology Laboratory (EMBL) Interdisciplinary Postdocs (EI3POD) initiative cofunded by Marie Skłodowska-Curie grant No. 664726.
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
- Byrn RA, Jones SM, Bennett HB, Bral C, Clark MP, Jacobs MD, Kwong AD, Ledeboer MW, Leeman JR, McNeil CF, et al. 2015. Preclinical activity of VX-787, a first-in-class, orally bioavailable inhibitor of the influenza virus polymerase PB2 subunit. Antimicrob Agents Chemother 59: 1569–1582. 10.1128/AAC.04623-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Sun D, Liang H, Wang J, Li J, Guo L, Wang X, Guan C, Boruah BM, Yuan L, et al. 2015. Cryo-EM structure of influenza virus RNA polymerase complex at 4.3 Å resolution. Mol Cell 57: 925–935. 10.1016/j.molcel.2014.12.031 [DOI] [PubMed] [Google Scholar]
- Chen KY, Santos Afonso ED, Enouf V, Isel C, Naffakh N. 2019. Influenza virus polymerase subunits co-evolve to ensure proper levels of dimerization of the heterotrimer. PLoS Pathog 15: e1008034. 10.1371/journal.ppat.1008034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AWH, Leong NKC, Nicholls JM, Poon LLM. 2017. Characterization of influenza A viruses with polymorphism in PB2 residues 701 and 702. Sci Rep 7: 11361. 10.1038/s41598-017-11625-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MP, Ledeboer MW, Davies I, Byrn RA, Jones SM, Perola E, Tsai A, Jacobs M, Nti-Addae K, Bandarage UK, et al. 2014. Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (VX-787) of influenza PB2. J Med Chem 57: 6668–6678. 10.1021/jm5007275 [DOI] [PubMed] [Google Scholar]
- Coloma R, Arranz R, de la Rosa-Trevín JM, Sorzano COS, Munier S, Carlero D, Naffakh N, Ortín J, Martín-Benito J.2020. Structural insights into influenza A virus ribonucleoprotein s reveal a processive helical track as transcription mechanism. Nat Microbiol 10.1038/s41564-020-0675-3 [DOI] [PubMed] [Google Scholar]
- Crepin T, Dias A, Palencia A, Swale C, Cusack S, Ruigrok RW. 2010. Mutational and metal binding analysis of the endonuclease domain of the influenza virus polymerase PA subunit. J Virol 84: 9096–9104. 10.1128/JVI.00995-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Wolkerstorfer A, Szolar OH, Cusack S, Klumpp K. 2013. Characterization of PA-N terminal domain of Influenza A polymerase reveals sequence specific RNA cleavage. Nucleic Acids Res 41: 8289–8299. 10.1093/nar/gkt603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12: 1203–1207. 10.1038/nm1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaforge E, Milles S, Bouvignies G, Bouvier D, Boivin S, Salvi N, Maurin D, Martel A, Round A, Lemke EA, et al. 2015. Large-scale conformational dynamics control H5N1 influenza polymerase PB2 binding to importin α. J Am Chem Soc 137: 15122–15134. 10.1021/jacs.5b07765 [DOI] [PubMed] [Google Scholar]
- Deng T, Vreede FT, Brownlee GG. 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J Virol 80: 2337–2348. 10.1128/JVI.80.5.2337-2348.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias A, Bouvier D, Crépin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458: 914–918. 10.1038/nature07745 [DOI] [PubMed] [Google Scholar]
- DuBois RM, Slavish PJ, Baughman BM, Yun MK, Bao J, Webby RJ, Webb TR, White SW. 2012. Structural and biochemical basis for development of influenza virus inhibitors targeting the PA endonuclease. PLoS Pathog 8: e1002830. 10.1371/journal.ppat.1002830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt OG, Fodor E. 2006. Functional association between viral and cellular transcription during influenza virus infection. Rev Med Virol 16: 329–345. 10.1002/rmv.512 [DOI] [PubMed] [Google Scholar]
- Fan H, Walker AP, Carrique L, Keown JR, Serna Martin I, Karia D, Sharps J, Hengrung N, Pardon E, Steyaert J, et al. 2019. Structures of influenza A virus RNA polymerase offer insight into viral genome replication. Nature 573: 287–290. 10.1038/s41586-019-1530-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finberg RW, Lanno R, Anderson D, Fleischhackl R, van Duijnhoven W, Kauffman RS, Kosoglou T, Vingerhoets J, Leopold L. 2019. Phase 2b study of pimodivir (JNJ-63623872) as monotherapy or in combination with oseltamivir for treatment of acute uncomplicated seasonal influenza A: TOPAZ trial. J Infect Dis 219: 1026–1034. 10.1093/infdis/jiy547 [DOI] [PubMed] [Google Scholar]
- Fodor E, Mingay LJ, Crow M, Deng T, Brownlee GG. 2003. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase promotes the generation of defective interfering RNAs. J Virol 77: 5017–5020. 10.1128/JVI.77.8.5017-5020.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Komeno T, Nakamura T. 2017. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci 93: 449–463. 10.2183/pjab.93.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Fodor E. 2014. Molecular determinants of pathogenicity in the polymerase complex. Curr Top Microbiol Immunol 385: 35–60. [DOI] [PubMed] [Google Scholar]
- Guilligay D, Tarendeau F, Resa-Infante P, Coloma R, Crepin T, Sehr P, Lewis J, Ruigrok RW, Ortin J, Hart DJ, et al. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15: 500–506. 10.1038/nsmb.1421 [DOI] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293: 1840–1842. 10.1126/science.1062882 [DOI] [PubMed] [Google Scholar]
- He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, et al. 2008. Crystal structure of the polymerase PAC–PB1N complex from an avian influenza H5N1 virus. Nature 454: 1123–1126. 10.1038/nature07120 [DOI] [PubMed] [Google Scholar]
- Hengrung N, El Omari K, Serna Martin I, Vreede FT, Cusack S, Rambo RP, Vonrhein C, Bricogne G, Stuart DI, Grimes JM, et al. 2015. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 527: 114–117. 10.1038/nature15525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Yamashita M, Sakai-Tagawa Y, Iwatsuki-Horimoto K, Kiso M, Murakami J, Yasuhara A, Takada K, Ito M, Nakajima N, et al. 2020. Influenza A variants with reduced susceptibility to baloxavir isolated from Japanese patients are fit and transmit through respiratory droplets. Nat Microbiol 5: 27–33. 10.1038/s41564-019-0609-0 [DOI] [PubMed] [Google Scholar]
- Kouba T, Drncová P, Cusack S. 2019. Structural snapshots of actively transcribing influenza polymerase. Nat Struct Mol Biol 26: 460–470. 10.1038/s41594-019-0232-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Zubieta C, Wolkerstorfer A, Szolar OH, Ruigrok RW, Cusack S. 2012. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog 8: e1002831. 10.1371/journal.ppat.1002831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuhara T, Kise D, Yoshida H, Horita T, Murazaki Y, Nishimura A, Echigo N, Utsunomiya H, Tsuge H. 2009. Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue. J Biol Chem 284: 6855–6860. 10.1074/jbc.C800224200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MK, Kim HE, Park EB, Lee J, Kim KH, Lim K, Yum S, Lee YH, Kang SJ, Lee JH, et al. 2016. Structural features of influenza A virus panhandle RNA enabling the activation of RIG-I independently of 5′-triphosphate. Nucleic Acids Res 44: 8407–8416. 10.1093/nar/gkw525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen H, Jiao P, Deng G, Tian G, Li Y, Hoffmann E, Webster RG, Matsuoka Y, Yu K. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol 79: 12058–12064. 10.1128/JVI.79.18.12058-12064.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Giotis ES, Moncorge O, Frise R, Mistry B, James J, Morisson M, Iqbal M, Vignal A, Skinner MA, et al. 2016. Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 529: 101–104. 10.1038/nature16474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukarska M, Fournier G, Pflug A, Resa-Infante P, Reich S, Naffakh N, Cusack S. 2017. Structural basis of an essential interaction between influenza polymerase and Pol II CTD. Nature 541: 117–121. 10.1038/nature20594 [DOI] [PubMed] [Google Scholar]
- Ma X, Xie L, Wartchow C, Warne R, Xu Y, Rivkin A, Tully D, Shia S, Uehara K, Baldwin DM, et al. 2017. Structural basis for therapeutic inhibition of influenza A polymerase PB2 subunit. Sci Rep 7: 9385. 10.1038/s41598-017-09538-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci 106: 21312–21316. 10.1073/pnas.0911915106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud EJ, Hayden FG, Hurt AC. 2019. Antivirals targeting the polymerase complex of influenza viruses. Antiviral Res 169: 104545. 10.1016/j.antiviral.2019.104545 [DOI] [PubMed] [Google Scholar]
- Mistry B, Long JS, Schreyer J, Staller E, Sanchez-David RY, Barclay WS. 2019. Elucidating the interactions between influenza virus polymerase and host factor ANP32A. J Virol 94: e01353-19. 10.1128/JVI.01353-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson BE, Te Velthuis AJW, Fodor E. 2017. Role of the PB2 627 domain in influenza A virus polymerase function. J Virol 91: e02467-16. 10.1128/JVI.02467-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noshi T, Kitano M, Taniguchi K, Yamamoto A, Omoto S, Baba K, Hashimoto T, Ishida K, Kushima Y, Hattori K, et al. 2018. In vitro characterization of baloxavir acid, a first-in-class cap-dependent endonuclease inhibitor of the influenza virus polymerase PA subunit. Antiviral Res 160: 109–117. 10.1016/j.antiviral.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454: 1127–1131. 10.1038/nature07225 [DOI] [PubMed] [Google Scholar]
- Omoto S, Speranzini V, Hashimoto T, Noshi T, Yamaguchi H, Kawai M, Kawaguchi K, Uehara T, Shishido T, Naito A, et al. 2018. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci Rep 8: 9633. 10.1038/s41598-018-27890-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautus S, Sehr P, Lewis J, Fortuné A, Wolkerstorfer A, Szolar O, Guilligay D, Lunardi T, Decout JL, Cusack S. 2013. New 7-methylguanine derivatives targeting the influenza polymerase PB2 cap-binding domain. J Med Chem 56: 8915–8930. 10.1021/jm401369y [DOI] [PubMed] [Google Scholar]
- Peacock TP, Sheppard CM, Staller E, Barclay WS. 2019. Host determinants of influenza RNA synthesis. Annu Rev Virol 6: 215–233. 10.1146/annurev-virology-092917-043339 [DOI] [PubMed] [Google Scholar]
- Peng Q, Liu Y, Peng R, Wang M, Yang W, Song H, Chen Y, Liu S, Han M, Zhang X, et al. 2019. Structural insight into RNA synthesis by influenza D polymerase. Nat Microbiol 4: 1750–1759. 10.1038/s41564-019-0487-5 [DOI] [PubMed] [Google Scholar]
- Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516: 355–360. 10.1038/nature14008 [DOI] [PubMed] [Google Scholar]
- Pflug A, Gaudon S, Resa-Infante P, Lethier M, Reich S, Schulze WM, Cusack S. 2018. Capped RNA primer binding to influenza polymerase and implications for the mechanism of cap-binding inhibitors. Nucleic Acids Res 46: 956–971. 10.1093/nar/gkx1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch SJ, Bouloy M, Ulmanen I, Krug RM. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23: 847–858. 10.1016/0092-8674(81)90449-9 [DOI] [PubMed] [Google Scholar]
- Poon LL, Pritlove DC, Fodor E, Brownlee GG. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol 73: 3473–3476. 10.1128/JVI.73.4.3473-3476.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, Ke S, Hart DJ, Zachariae U, Cingolani G. 2015. Molecular determinants for nuclear import of influenza A PB2 by importin α isoforms 3 and 7. Structure 23: 374–384. 10.1016/j.str.2014.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crépin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, et al. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516: 361–366. 10.1038/nature14009 [DOI] [PubMed] [Google Scholar]
- Reich S, Guilligay D, Cusack S. 2017. An in vitro fluorescence based study of initiation of RNA synthesis by influenza B polymerase. Nucleic Acids Res 45: 3353–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb NC, Te Velthuis AJW, Fodor E, Kapanidis AN. 2019. Real-time analysis of single influenza virus replication complexes reveals large promoter-dependent differences in initiation dynamics. Nucleic Acids Res 47: 6466–6477. 10.1093/nar/gkz313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RW, Crépin T, Hart DJ, Cusack S. 2010. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr Opin Struct Biol 20: 104–113. 10.1016/j.sbi.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Sagong HY, Bauman JD, Patel D, Das K, Arnold E, LaVoie EJ. 2014. Phenyl substituted 4-hydroxypyridazin-3(2H)-ones and 5-hydroxypyrimidin-4(3H)-ones: inhibitors of influenza A endonuclease. J Med Chem 57: 8086–8098. 10.1021/jm500958x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna Martin I, Hengrung N, Renner M, Sharps J, Martínez-Alonso M, Masiulis S, Grimes JM, Fodor E. 2018. A mechanism for the activation of the influenza virus transcriptase. Mol Cell 70: 1101–1110.e4. 10.1016/j.molcel.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Mubareka S, Palese P. 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 5: e1000252. 10.1371/journal.ppat.1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaert A, Naesens L. 2016. The influenza virus polymerase complex: an update on its structure, functions, and significance for antiviral drug design. Med Res Rev 36: 1127–1173. 10.1002/med.21401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol 67: 1761–1764. 10.1128/JVI.67.4.1761-1764.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama K, Obayashi E, Kawaguchi A, Suzuki Y, Tame JR, Nagata K, Park SY. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J 28: 1803–1811. 10.1038/emboj.2009.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama K, Kawaguchi A, Okuwaki M, Nagata K. 2015. pp32 and APRIL are host cell-derived regulators of influenza virus RNA synthesis from cRNA. Elife 4: e08939. 10.7554/eLife.08939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, et al. 2007. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol 14: 229–233. 10.1038/nsmb1212 [DOI] [PubMed] [Google Scholar]
- Tarendeau F, Crepin T, Guilligay D, Ruigrok RW, Cusack S, Hart DJ. 2008. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog 4: e1000136. 10.1371/journal.ppat.1000136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis AJ, Robb NC, Kapanidis AN, Fodor E. 2016. The role of the priming loop in influenza A virus RNA synthesis. Nat Microbiol 1: 16029. 10.1038/nmicrobiol.2016.29 [DOI] [PubMed] [Google Scholar]
- Thierry E, Guilligay D, Kosinski J, Bock T, Gaudon S, Round A, Pflug A, Hengrung N, El Omari K, Baudin F, et al. 2016. Influenza polymerase can adopt an alternative configuration involving a radical repacking of PB2 domains. Mol Cell 61: 125–137. 10.1016/j.molcel.2015.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomassini J, Selnick H, Davies ME, Armstrong ME, Baldwin J, Bourgeois M, Hastings J, Hazuda D, Lewis J, McClements W, et al. 1994. Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob Agents Chemother 38: 2827–2837. 10.1128/AAC.38.12.2827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toots M, Yoon JJ, Cox RM, Hart M, Sticher ZM, Makhsous N, Plesker R, Barrena AH, Reddy PG, Mitchell DG, et al. 2019. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci Transl Med 11: eaax5866. 10.1126/scitranslmed.aax5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevejo JM, Asmal M, Vingerhoets J, Polo R, Robertson S, Jiang Y, Kieffer TL, Leopold L. 2018. Pimodivir treatment in adult volunteers experimentally inoculated with live influenza virus: a Phase IIa, randomized, double-blind, placebo-controlled study. Antiviral Ther 23: 335–344. 10.3851/IMP3212 [DOI] [PubMed] [Google Scholar]
- Wandzik JM, Kouba T, Karuppasamy M, Pflug A, Drncova P, Provaznik J, Azevedo N, Cusack S. 2020. A structure-based model for the complete transcription cycle of influenza polymerase. Cell 10.1016/j.cell.2020.03.061 [DOI] [PubMed] [Google Scholar]
- Xie L, Wartchow C, Shia S, Uehara K, Steffek M, Warne R, Sutton J, Muiru GT, Leonard VH, Bussiere DE, et al. 2016. Molecular basis of mRNA cap recognition by influenza B polymerase PB2 subunit. J Biol Chem 291: 363–370. 10.1074/jbc.M115.693051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York A, Hengrung N, Vreede FT, Huiskonen JT, Fodor E. 2013. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc Natl Acad Sci 110: E4238–E4245. 10.1073/pnas.1315068110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Bartlam M, Lou Z, Chen S, Zhou J, He X, Lv Z, Ge R, Li X, Deng T, et al. 2009. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature 458: 909–913. 10.1038/nature07720 [DOI] [PubMed] [Google Scholar]