In this study, Perez-Borrajero et al. set out to characterize PETISCO architecture and its interaction with RNA, together with its effector proteins TOST-1 and PID-1. Using biochemical and structural biology approaches, the authors found that PETISCO forms a dimer of tetramers, in which dimerization is mediated by both PID-3 and ERH-2. Crystal structures of the PID- 3/TOFU-6 and ERH-2/PID-3 subcomplexes reveal insights into PETISCO assembly, function, and subcellular localization. Using NMR spectroscopy, the authors also characterize the mutually exclusive interplay of ERH-2 with the two effector proteins TOST-1 and PID-1.
Keywords: piRNA, 21U RNA, Piwi, C. elegans, PETISCO, RRM domain, ERH-2, IFE-3, TOFU-6, PID-3, TOST-1, PID-1
Abstract
Piwi-interacting RNAs (piRNAs) constitute a class of small RNAs that bind PIWI proteins and are essential to repress transposable elements in the animal germline, thereby promoting genome stability and maintaining fertility. C. elegans piRNAs (21U RNAs) are transcribed individually from minigenes as precursors that require 5′ and 3′ processing. This process depends on the PETISCO complex, consisting of four proteins: IFE-3, TOFU-6, PID-3, and ERH-2. We used biochemical and structural biology approaches to characterize the PETISCO architecture and its interaction with RNA, together with its effector proteins TOST-1 and PID-1. These two proteins define different PETISCO functions: PID-1 governs 21U processing, whereas TOST-1 links PETISCO to an unknown process essential for early embryogenesis. Here, we show that PETISCO forms an octameric assembly with each subunit present in two copies. Determination of structures of the TOFU-6/PID-3 and PID-3/ERH-2 subcomplexes, supported by in vivo studies of subunit interaction mutants, allows us to propose a model for the formation of the TOFU-6/PID-3/ERH-2 core complex and its functionality in germ cells and early embryos. Using NMR spectroscopy, we demonstrate that TOST-1 and PID-1 bind to a common surface on ERH-2, located opposite its PID-3 binding site, explaining how PETISCO can mediate different cellular roles.
RNA molecules typically require processing after transcription before becoming fully functional. In eukaryotes, messenger RNAs (mRNAs) and many noncoding RNAs are spliced, capped, and polyadenylated by processing factors (Hocine et al. 2010). Transcripts can also be chemically modified, trimmed, or cleaved by ribonucleases, or extended by the addition of nontemplated nucleotides (Roundtree et al. 2017; Yu and Kim 2020). Such processing steps are crucial for activation, (de)stabilization, localization, and many other aspects relevant to RNA function.
Piwi-interacting RNAs (piRNAs) constitute one of the largest classes of noncoding RNA transcripts whose processing is only starting to be understood (Weick and Miska 2014; Ozata et al. 2019). piRNAs play a crucial role in the germline, where they act as specificity factors in genome defense pathways with transposable elements as major targets (Luteijn and Ketting 2013; Czech and Hannon 2016). The proteins guided by piRNAs, Piwi proteins, are an animal-specific subgroup of the Argonaute family. Piwi proteins are guided by piRNAs that bind to their target site, leading to either transcript cleavage or modification of chromatin, depending on the subclass of Piwi proteins involved (Luteijn and Ketting 2013). In either case, the sequence of the piRNAs bound by the Piwi proteins dictates the target specificity of the silencing process and is therefore crucial for function. Hence, the mechanism(s) that act in piRNA precursor selection and processing determines the specificity of Piwi proteins.
The precursors of piRNAs are single-stranded RNA transcripts. In Caenorhabditis elegans, these are produced from a multitude of miniature genes, each producing a small transcript of 27–30 nucleotides (Ruby et al. 2006; Gu et al. 2012). Mature piRNAs are bound by the Piwi protein PRG-1 and are typically 21 nucleotides long with a uracil base at the 5′ end. For these reasons, the piRNAs in C. elegans are often named 21U RNAs. To form mature 21U RNAs, the precursor transcripts are shortened at both ends, including the removal of the 5′ cap and trimming of the 3′ end (Ruby et al. 2006; Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008; Gu et al. 2012; Weick and Miska 2014; Tang et al. 2016). The vast majority of 21U RNAs stem from dedicated loci characterized by a specific sequence motif in their promoter termed the Ruby motif (Ruby et al. 2006; Cecere et al. 2012; Weick et al. 2014). Within the C. elegans genome, these loci are strongly clustered, suggesting that they may act in concert (Ruby et al. 2006). The transcription of these loci, including the termination of their transcription, bears hallmarks of small nuclear RNA (snRNA) biogenesis, suggesting that the C. elegans piRNA system has evolutionary connections to these noncoding snRNAs that play essential roles in splicing (Kasper et al. 2014; Beltran et al. 2019, 2021; Weng et al. 2019; Berkyurek et al. 2021).
Following the genetic identification of several 21U RNA processing genes (de Albuquerque et al. 2014; Goh et al. 2014), we and others identified a four-member protein complex required for 21U RNA biogenesis, which we named PETISCO (PID-3, ERH-2, TOFU-6, and IFE-3 small RNA complex) (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019). PETISCO interacts with and stabilizes 21U RNA precursors. Interestingly, PETISCO was shown to be additionally required for early embryogenesis, a function independent of 21U RNA biogenesis (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019). In this case, loss of PETISCO function leads to a so-called “maternal effect lethal” (Mel) phenotype, in which first-generation homozygous mutant animals develop normally, but their offspring arrest in embryogenesis. At the molecular level, low levels of the splice-leader transcript SL1, a small nuclear RNA (snRNA) involved in trans-splicing, were found to be bound by PETISCO. SL1-derived 21U RNAs have also been described, albeit at very low levels (Gu et al. 2012). Whether these findings relate to the Mel phenotype of PETISCO mutants is unclear, but they do strengthen the link between 21U RNAs and snRNAs in C. elegans. The two described functions of PETISCO are specified by two different effector proteins, PID-1 and TOST-1. PID-1:PETISCO mediates 21U RNA biogenesis (de Albuquerque et al. 2014), while TOST-1:PETISCO is required for early embryogenesis (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019).
The PETISCO subunits contain domains often present in RNA-binding proteins (Cordeiro Rodrigues et al. 2019). PID-3 and TOFU-6 are restricted to the nematode phylum and contain two domains. PID-3 has a predicted RNA recognition (RRM) domain and an Argonaute-related middle (MID) domain. TOFU-6 contains an RRM domain, an extended Tudor (eTudor) domain, and a C-terminal eIF4E interaction motif. IFE-3 is one of the five highly conserved C. elegans’ eIF4E homologs (Keiper et al. 2000) and binds to the C terminus of TOFU-6 (Cordeiro Rodrigues et al. 2019). Finally, ERH-2 is one of the two C. elegans paralogs of “enhancer of rudimentary” (Erh), a factor that is conserved throughout eukaryotes (Weng and Luo 2013). Erh was shown to participate in the RNA exosome-mediated degradation of meiotic RNAs in Schizosaccharomyces pombe (Sugiyama et al. 2016) and to facilitate miRNA processing in human cells (Fang and Bartel 2020; Hutter et al. 2020; Kwon et al. 2020).
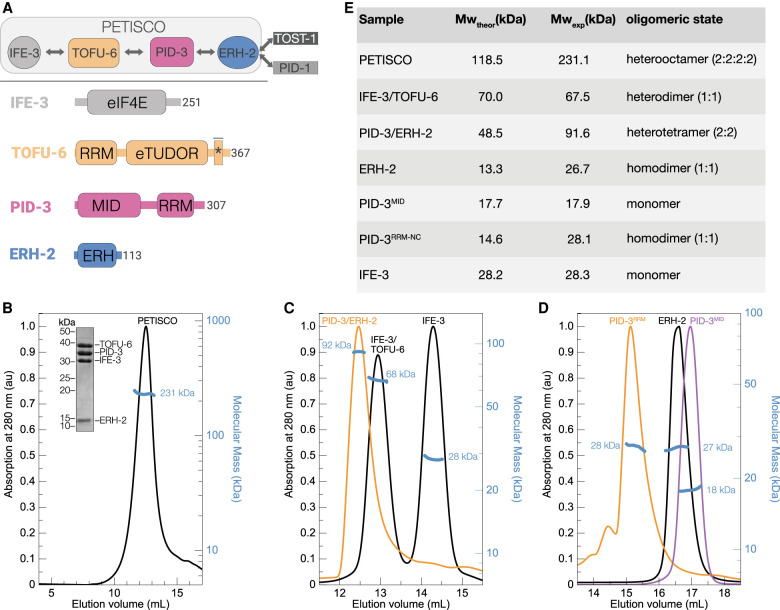
An approximate architecture of PETISCO was previously derived from yeast two-hybrid (Y2H) studies (Fig. 1A; Cordeiro Rodrigues et al. 2019). However, the structural basis of PETISCO assembly and its interaction with the effector proteins PID-1/TOST-1 and RNA substrates remain poorly understood, limiting our understanding of PETISCO function.
Figure 1.
PETISCO assembles into a hetero-octamer with 2:2:2:2 stoichiometry. (A, top) Schematic representation of the PETISCO topology and subunit interactions and binding of the effector proteins TOST-1 and PID-1. (Bottom) Domain organization of IFE-3, TOFU-6, PID-3, and ERH-2 from C. elegans. Rounded rectangles indicate predicted domains. The asterisk marks the position of the eIF4E interaction motif that mediates binding of TOFU-6 to IFE-3. (B–D) SEC-MALS chromatograms showing UV absorption at 280 nm and the calculated molecular mass in kilodaltons. The UV absorption signal was normalized to the highest peak. (B) SEC-MALS profile of PETISCO. The inset shows a Coomassie-stained SDS polyacrylamide gel of PETISCO after SEC. (C) SEC-MALS profiles of the IFE-3/TOFU-6, IFE-3 (black line), and PID-3/ERH-2 (orange line) samples. (D) SEC-MALS profiles of ERH-2 (black line), PID-3MID (purple line), and PID-3RRM (orange line) domains. (E) Summary of molecular masses and stoichiometries of PETISCO and its subunits.
Here, we studied PETISCO assembly using a bottom-up approach with purified proteins, interaction studies, and structural analyses. We found that PETISCO forms a dimer of tetramers, in which dimerization is mediated by both PID-3 and ERH-2. Crystal structures of the PID-3/TOFU-6 and ERH-2/PID-3 subcomplexes reveal insights into PETISCO assembly, function, and subcellular localization. Using NMR spectroscopy, we also characterize the mutually exclusive interplay of ERH-2 with the two effector proteins TOST-1 and PID-1. These results represent the first structural characterization of a piRNA biogenesis complex, and we start to reveal how PETISCO may execute its dual role in vivo.
Results
PETISCO forms an octameric assembly
PETISCO consists of the proteins IFE-3, TOFU-6, PID-3, and ERH-2. Previous Y2H experiments (Cordeiro Rodrigues et al. 2019) revealed a linear topology in which IFE-3 binds to TOFU-6, which in turn binds PID-3, which associates with ERH-2 (Fig. 1A; Supplemental Fig. 1A). To analyze the oligomeric state and stoichiometry of PETISCO, we recombinantly expressed and purified PETISCO components from bacterial cells and subjected the complex to size exclusion chromatography (SEC) coupled to multiangle light scattering (MALS) (Fig. 1B). The complete PETISCO complex eluted as a single peak and showed an average molecular mass of 231 kDa (Fig. 1B), twice the sum of the individual components (assuming a 1:1:1:1 complex, 116 kDa), suggesting that PETISCO forms a hetero-octameric assembly, a dimer of tetramers, with 2:2:2:2 stoichiometry (Fig. 1B). Next, we set out to determine which protein or domains mediate oligomerization. The linear topology of PETISCO suggested that the complex could be divided into IFE-3/TOFU-6 and PID-3/ERH-2 subcomplexes (Fig. 1A). We individually purified these and analyzed their molecular mass by SEC-MALS (Fig. 1C). In the case of IFE-3/TOFU-6, we measured an average mass of 69 kDa, in line with the calculated mass of a heterodimer. The PID-3/ERH-2 subcomplex, however, had an average mass of 92 kDa, consistent with that of a heterotetramer, suggesting that the PID-3/ERH-2 module mediates oligomerization. Since the human and fission yeast ERH orthologs have been shown to form homodimers (Wan et al. 2005; Xie et al. 2019; Hazra et al. 2020), we hypothesized that ERH-2 is responsible for dimerization. Therefore, we determined the oligomeric state of both ERH-2 and PID-3 separately. However, PID-3 full-length protein could not be prepared in a quality suitable for SEC-MALS, and therefore we analyzed the PID-3 MID and RRM domains individually, denoted here as PID-3MID and PID-3RRM, respectively. We determined average masses of 18, 28, and 27 kDa for PID-3MID, PID-3RRM-NC, and full-length ERH-2FL, respectively (Fig. 1D,E; Supplemental Fig. S1C). This suggests that PID-3MID is monomeric, whereas both PID-3RRM and ERH-2 are homodimers (Fig. 1E). We thus concluded that both PID-3RRM and ERH-2 contribute to PID-3/ERH-2 subcomplex dimerization, and through binding of two IFE-3/TOFU-6 subcomplexes, this results in the formation of the octameric PETISCO.
Crystal structure of the TOFU-6–PID-3 RRM core complex
The topological arrangement of PETISCO places TOFU-6 and PID-3 at the core, and we thus proceeded to narrow down their interacting regions. Our previous experiments indicated that the interaction between TOFU-6 and PID-3 is mediated by the RRM domains (denoted as TOFU-6RRM and PID-3RRM) (Fig. 1A; Supplemental Fig. S1A; Cordeiro Rodrigues et al. 2019). To better map which domains mediate the interaction between TOFU-6 and PID-3, we used a combination of pull-down experiments and SEC. We recombinantly coexpressed maltose binding protein (MBP)-tagged TOFU-6 constructs with glutathione-S-transferase (GST)-tagged PID-3 constructs. Both MBP and GST pull-downs revealed that the RRM domains of PID-3 and TOFU-6 mediate the interaction between the two proteins and that neither the eTudor domain of TOFU-6 (TOFU-6eTUDOR) nor the PID-3MID are required (Supplemental Fig. S2A). This is supported by SEC using purified TOFU-6RRM and PID-3RRM proteins (Supplemental Fig. S2B).
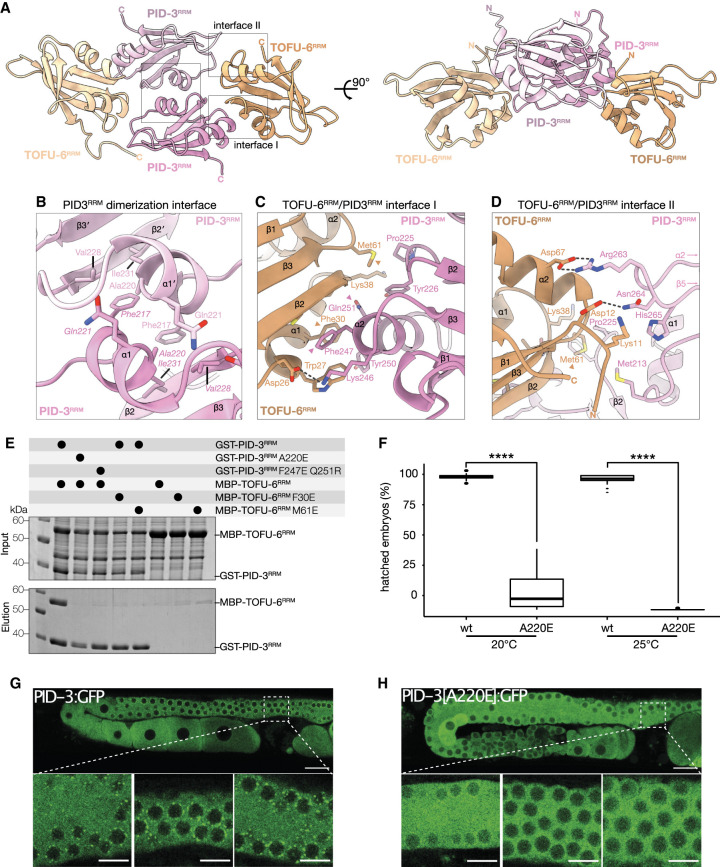
To gain structural insights into the TOFU-6RRM/PID-3RRM complex, we determined the structures of PID-3RRM and the TOFU-6RRM/PID-3RRM complex, at 1.8 Å and 1.7 Å resolutions, respectively (Fig. 2A; Supplemental Fig. S2C; Supplemental Table S1). Both structures are very similar. The PID-3RRM forms a homodimer in both cases, with slight differences in the relative orientation of the RRM domains (Supplemental Fig. S2C). Here, we will focus on the analysis of the TOFU-6RRM/PID-3RRM complex structure, containing one tetrameric TOFU-6RRM/PID-3RRM complex in the asymmetric unit (Fig. 2A), consistent with the SEC-MALS analysis (Supplemental Fig. S2D). The PID-3RRM adopts a canonical RRM fold with an antiparallel four-stranded β sheet packing opposite two α helices (α1 and α2) (Supplemental Fig. S2E). The TOFU-6RRM has a similar architecture but contains an additional fifth β-strand β4* located between the α2 and β4 elements (Supplemental Fig. S2F). The PID-3RRM dimerizes via the α1 helix by a combination of hydrophobic and polar interactions (Fig. 2A,B), reminiscent, for instance, of the dimer interface in HuR-RRM3 (Pabis et al. 2019; Ripin et al. 2019) and RBPMS (Teplova et al. 2016). Phe217 from one PID-3RRM protomer packs in a hydrophobic pocket created by Phe217, Ala220, Val228, and Ile231 from the other protomer. Moreover, Gln221 forms a hydrogen bond with the polypeptide backbone of the neighboring protomer (Fig. 2B).
Figure 2.
The TOFU-6 and PID-3 RRM domains form the PETISCO core. (A) Crystal structure of the PID-3RRM/TOFU-6RRM complex shown in cartoon representation in two orientations related by a 90° rotation about the horizontal axis. The two PID-3RRM protomers are shown in different shades of pink, while the two TOFU-6RRM protomers in different shades of orange. The N-terminal and C-terminal residues are highlighted. (B) Zoomed-in view of the homodimerization interface of PID-3RRM. Interacting residues are shown in stick representation and labeled. (C,D) Zoomed-in view of a representative set of residues at the PID-3/TOFU-6 interaction interface I (C) and interface II (D). Interacting residues are shown in stick representation as indicated. (E) Analysis of the effect of structure-guided mutations on the PID-3/TOFU-6 interaction by GST pull-down assays. Wild-type (WT) and mutant versions of MBP-tagged TOFU-6 constructs were coexpressed with WT and mutant versions of GST-tagged PID-3 constructs. Input and elution fractions were analyzed by SDS-PAGE followed by Coomassie staining. (F) Box plot showing the percentage of hatched embryos of pid-3::gfp(wt) and pid-3[a220e]::gfp animals grown at 20°C and 25°C. Progeny of 30 different mothers were analyzed for each condition, and development of at least 2600 eggs was scored. A two-sided t-test was used to assess significance. P-values are indicated in the graph. (G,H) Single-plane confocal micrographs of PID-3::GFP(WT) and PID-3[A220E]::GFP at 25°C. The boxes indicate the regions (above the spermatheca) from which three zoomed-in examples are shown below. Scale bars: overview, 20 µm; zoom-in, 8 µm.
Two TOFU-6 RRM domains bind on either side of the interfaces created through the PID-3RRM dimerization and contact both PID-3 protomers (Fig. 2A). The TOFU-6 α1 helix, β2 strand, and β3-α2 loop form a surface that interacts with the α1–β2 loop and α2 helix from the PID-3RRM (Interface I/protomer 1) by a combination of hydrophobic and polar interactions (Fig. 2C). The TOFU-6RRM interaction with the second PID-3RRM protomer involves electrostatic interactions between the N terminus and the α2 helix of TOFU-6RRM with the PID-3RRM α2-β5 loop (Interface II/protomer 2), featuring a salt bridge between TOFU-6 Asp67 and PID-3 Arg263, as well as a hydrogen bond between TOFU-6 Asp12 and PID-3 Asn264 (Fig. 2D).
Our structural analysis suggests that dimerization of the PID-3RRM is a prerequisite for TOFU-6RRM binding and that two interfaces at the two protomers contribute to the interaction. To test this prediction, we engineered a series of substitutions both at the PID-3 dimerization interface (PID-3RRM A220E) and at interface I (TOFU-6RRM F30E and M61E; PID-3RRM F247E/Q251R) and analyzed their impact on the interaction by bacterial coexpression pull-down experiments. Both PID-3 mutant proteins (A220E and F247E/Q251R) are folded (Supplemental Fig. S2G); however, the A220E mutation rendered PID-3RRM monomeric, while the F247E/Q251R mutant remained homodimeric as expected from the structural analysis (Supplemental Fig. S2H,I). Mutations at the PID-3RRM dimerization surface and interface I disrupted TOFU-6 binding, confirming that these sites contribute to a stable association with TOFU-6RRM (Fig. 2E).
Next, we tested the effect of the monomer-inducing A220E mutation in PID-3 in vivo. Using CRISPR-Cas9-mediated gene editing, we first created a strain expressing C-terminally GFP-tagged PID-3, such that in vivo expression could be monitored by fluorescence microscopy. We then introduced the A220E mutation and scored its effect on subcellular localization and embryonic viability. pid-3(a220e) animals showed a strong Mel phenotype, which was fully penetrant at 25°C (Fig. 2F), consistent with loss of PETISCO function. In addition, PID-3(A220E) did not form perinuclear foci (Fig. 2G,H), indicating that PID-3 dimerization is required for the previously described P granule localization of PETISCO (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019).
ERH-2 binds to a region upstream of the PID-3 RRM domain
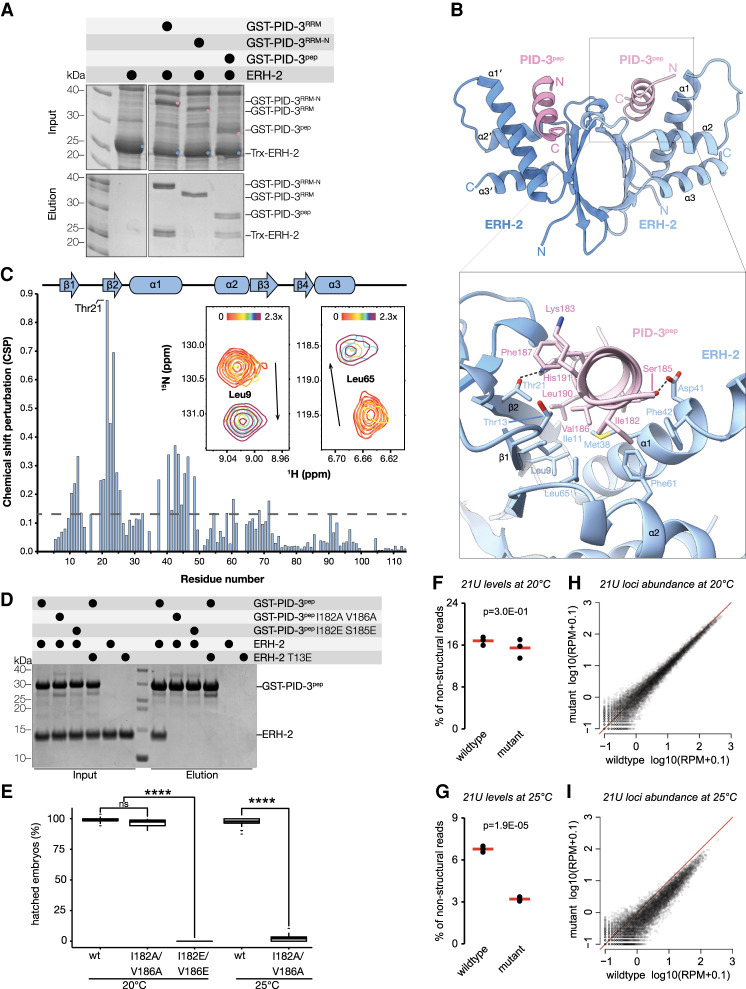
Next, we investigated the interaction between PID-3 and ERH-2. ERH-2 consists of an ERH domain followed by a C-terminal region of ∼15 amino acids (Fig. 1A). Previous experiments (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019) suggested a direct interaction between ERH-2 and PID-3, in particular between PID-3RRM and ERH-2 (Cordeiro Rodrigues et al. 2019). However, the PID-3 constructs used in the latter study contained additional N-terminal and C-terminal regions flanking the PID-3RRM (Supplemental Fig. S1A). To better define the ERH-2 binding site of PID-3, we used PID-3 constructs covering the RRM domain (PID-3RRM) or the RRM domain with an N-terminal extension (PID-3RRM-N) and performed bacterial coexpression pull-down experiments. While PID-3RRM-N pulled down ERH-2, PID-3RRM failed to bind ERH-2 (Fig. 3A). To rule out that the C-terminal extension downstream from the PID-3 RRM domain contributed to binding, we performed SEC experiments. We incubated PID-3RRM-N and PID-3RRM-C with ERH-2 and analyzed the mixtures by SEC to assess binding. This confirmed that PID-3RRM-N did but PID-3RRM-C did not interact with ERH-2 (Supplemental Fig. S3A,B). We then asked whether the N-terminal extension of the PID-3 RRM alone was sufficient for ERH-2 binding. We purified a GST-tagged PID-3 peptide (PID-3pep, residues 171–203) corresponding to this region and found that ERH-2 could indeed bind to GST-PID-3pep (Fig. 3A). To obtain quantitative insights into PID-3pep/ERH-2 interaction, we used isothermal titration calorimetry (ITC). We determined a dissociation constant (Kd) of 0.65 µM and a stoichiometry N∼1 (0.97) (Table 1; Supplemental Fig. S3C), consistent with the 2:2 stoichiometry observed in the case of the full-length PID-3/ERH-2 complex by SEC-MALS (Fig. 1C). Finally, we tested whether the PID-3pep binding specificity may explain our previous observation that PETISCO specifically incorporates ERH-2, and not its close paralog ERH-1 (36% sequence identity and 60% similarity) (Supplemental Fig. S3D; Cordeiro Rodrigues et al. 2019). ERH-1 did not bind PID-3pep in GST pull-down, SEC or, ITC experiments (Supplemental Fig. S3E–G), indicating that PID-3pep can discriminate between the two ERH paralogs. We concluded that the N-terminal extension of the PID-3RRM domain binds to ERH-2 and is likely responsible for the ERH-2 specificity of PETISCO.
Figure 3.
Structural and functional analysis of the PID-3/ERH-2 interaction. (A) GST pull-down assays. GST-tagged PID-3 constructs were coexpressed with Trx-tagged ERH-2 in bacterial cells and then subjected to coprecipitation using glutathione-coupled beads. Input and elution fractions were analyzed on SDS-PAGE gels with Coomassie brilliant blue staining. (B, top) The crystal structure of the ERH-2ΔC/PID-3pep complex is shown in cartoon representation. The two ERH-2 protomers are shown in different shades of blue, while the two PID-3 peptides are in different shades of pink. (Bottom) Zoomed-in view of the interaction between ERH-2 and PID-3pep. Interacting residues are shown in stick representation. (C) Binding of PID-3pep to ERH-2 monitored by NMR spectroscopy. (Inset) Upon incremental addition of PID-3pep, the 1H-15N amide signals of 15N-labeled ERH-2 move in slow exchange from their free (shown in red) to their bound state (in maroon) with a 2.3-fold molar excess of PID-3pep. Two representative zoomed-in regions of the overlaid spectra are shown (full view in Supplemental Fig. S5B). The arrows indicate the direction of the change in CSP from free to bound states. The CSP values determined for a saturated 1:3 ERH-2:PID-3pep complex are plotted below as a function of residue number. The most highly perturbed amino acid, Thr21, is indicated. The gray dashed line corresponds to the average CSP of all residues. The secondary structure elements as found in the crystal structure are shown above. (D) Mutational analysis of the PID-3/ERH-2 interface. GST-tagged PID-3 wild type and mutants were coexpressed with wild-type or mutant Trx-tagged ERH-2 in bacterial cells as described in A. (E) Box plot showing the percentage of hatched embryos of pid-3::gfp(wt) and pid-3[i182a;v186a]::gfp and pid-3[i182e;v186e]::gfp animals grown at 20 and 25°C. Progeny of 30 different mothers were analyzed for each condition, and development of at least 2600 eggs was scored. A two-sided t-test was used to assess significance. P-values are indicated in the graph. (F,G) Total 21U levels in wild-type and pid-3[i182a/v186a]-mutant embryos grown at 20°C (F) and 25°C (G) (n = 3). Group means are depicted by red lines and P-values are calculated using two-tailed unpaired t-test. (H,I) Scatter plots depicting the relative abundance of individual 21U loci in pid-3[i182a/v186a]-mutant versus wild-type embryos grown at 20°C (H) and 25°C (I). (RPM) Reads per million nonstructural sRNA reads.
Table 1.
Dissociation constants determined using ITC
Structural insights into the formation of the ERH-2/PID-3 complex
We next determined the crystal structures of a C-terminally truncated ERH-2 (residues 1–99, ERH-2ΔC) in the free and PID-3pep-bound states, at resolutions of 1.50 Å and 2.17 Å, respectively (Fig. 3B; Supplemental Table S1). The overall structure of ERH-2ΔC is similar to human ERH (Wan et al. 2005) and S. pombe Erh (Supplemental Fig. S4A; Xie et al. 2019). ERH-2ΔC adopts a mixed α-β-fold, with a four-stranded antiparallel β sheet that packs against three amphipathic α helices on the back side (Fig. 3B). The front side of the β sheet mediates dimerization that results in the formation of a pseudo-β-barrel structure. The structure of free ERH-2ΔC is very similar to PID-3pep-bound ERH-2ΔC, with the exception that the loop connecting helices α1 and α2 is not visible in the electron density, suggesting that it becomes ordered upon PID-3pep binding (Supplemental Fig. S4B).
PID-3pep is unfolded in solution (Supplemental Fig. S4C) but forms an amphipathic α helix when bound to ERH-2ΔC (Fig. 3B; Supplemental Fig. S4D). PID-3pep shows well-ordered electron density for residues 177–193 (chain C) and 179–193 (chain D) and occupies a similar surface on ERH as DGCR8 in the human ERH/DGCR8peptide complex (Kwon et al. 2020), and Mmi1 in the fission yeast Erh1/Mmi1peptide complex (Supplemental Fig. S4A; Xie et al. 2019). However, while both Mmi1 and DGCR8 bind as extended peptides lacking defined secondary structure, PID-3pep binds as an α helix (Supplemental Fig. S4A). The hydrophobic interface of the amphipathic PID-3pep helix is formed by Ile182, Val186, Phe187, Val189, and Leu190 and points toward a hydrophobic groove in ERH-2 formed by strands β1, β2, as well as the α1 helix (Fig. 3B; Supplemental Fig. S4E). In addition to the hydrophobic interactions, hydrogen bonds formed between PID-3pep Ser185 and ERH-2 Asp41, as well as between PID-3pep His191 and ERH-2 Thr21 further contribute to the affinity and specificity of the interaction.
In a complementary approach, we used NMR spectroscopy and assigned the backbone chemical shifts (1H-15N, Cα, and Cβ) of free full-length ERH-2. We used these chemical shifts to determine the secondary structural elements of ERH-2, which were fully consistent with the crystal structure, and also showed a disordered C terminus (residues 104–113) (Supplemental Fig. S5A). In addition, the loop region between α1 and α2 encompassing residues 46–55 was found to contain two sets of amide peaks. This indicates the presence of two well-defined alternative conformations that exchange in the millisecond timescale and longer and explains the lack of order in the crystal structure of free ERH-2ΔC (Supplemental Fig. 4B).
We then monitored the interaction between PID-3pep and ERH-2. Unlabeled PID-3pep was titrated into 15N-labeled ERH-2, and the positions of the amide chemical shifts were measured at each titration point by 1H-15N HSQC experiments (Fig. 3C; Supplemental Fig. S5B). We observed binding in the slow exchange regime, whereby peaks disappear from one location and reappear at a new position corresponding to the bound complex. This indicates a strong interaction with a dissociation constant in the low micromolar to nanomolar range, in agreement with the ITC data (Supplemental Fig. S3C). We next assigned the chemical shifts of the ERH-2/PID-3pep complex and calculated the chemical shift perturbations (CSPs). The ERH-2 residues most affected by the interaction with PID-3pep were located in β2, with Thr21, Trp22, and Gly23 exhibiting the largest CSPs (Fig. 3C). Mapping of the amide CSPs onto the crystal structure of ERH-2ΔC was consistent with the binding interface seen in the crystal structure (Supplemental Fig. S5C). Taken together, the crystal structure and NMR data provide a clear view for the basis of the ERH-2/PID-3 interaction.
Mutational analysis of the ERH-2/PID-3 interface
To test the relevance of residues found at the complex interface, we used GST-pull-down experiments with purified proteins. Mutation of PID-3 Ile182 and Ser185 to glutamate residues (I182E and S185E), completely abrogated binding of PID-3pep to ERH-2 (Fig. 3D), and even milder substitutions of PID-3 Ile182 and Val186 to alanine (I182A and V186A) were sufficient to abolish binding (Fig. 3D). Mutation of Thr13 in ERH-2, which lines the hydrophobic pocket (Fig. 3B), to glutamate (T13E), also abrogated binding to PID-3pep (Fig. 3D). In vivo, PID-3 (I182E and V186E) worms displayed a full Mel phenotype at 20°C (Fig. 3E), while the milder PID-3 (I182A and V186A) mutations showed a Mel phenotype only at 25°C. Finally, at the subcellular level, loss of ERH-2 binding did not affect the localization of PID-3 (Supplemental Fig. S5D).
Figure 4.
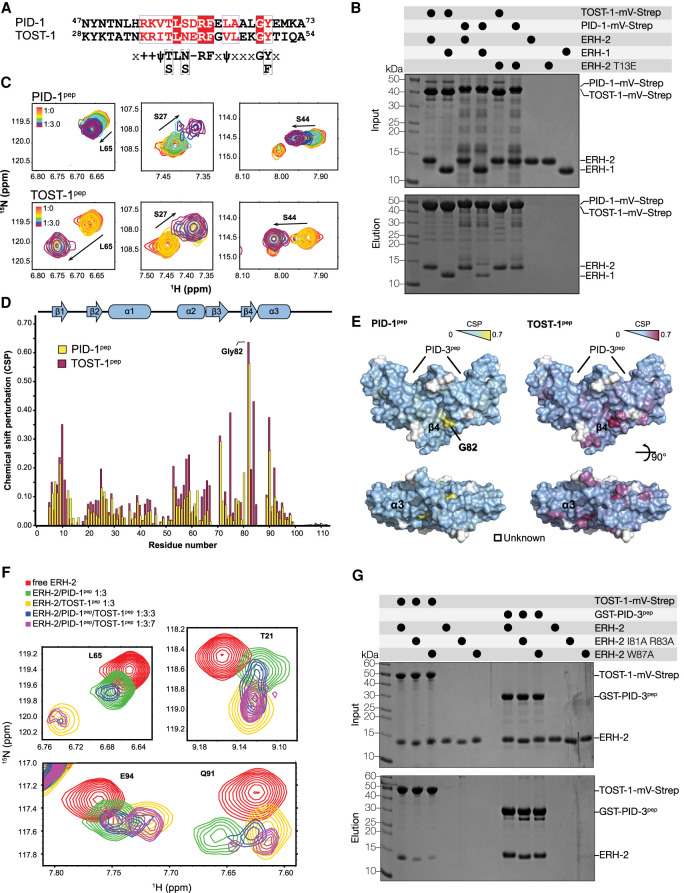
PID-1pep and TOST-1pep bind to ERH-2 through a common interface. (A) Sequence alignment of C. elegans PID-1 and TOST-1 corresponding to the conserved motif previously determined (Cordeiro Rodrigues et al. 2019). (B) Analysis of the interaction between TOST-1/PID-1 with ERH-1/ERH-2 by streptavidin pull-down assays. Purified, recombinant TOST-1 or PID-1 mVenus-Strep fusion proteins were used as baits and ERH-1 and ERH-2 as prey. Input and elution fractions were analyzed by SDS-PAGE followed by Coomassie staining. (C) PID-1pep and TOST-1pep bind to ERH-2 in a similar fashion. NMR-monitored titrations of 15N-labeled ERH-2 upon addition of unlabeled TOST-1 and PID-1 peptides, as indicated (full view in Supplemental Fig. S6). (D) The bound ERH-2/peptide complexes (1:3 molar ratios) were assigned and the extent of the CSPs quantified. The peptides produced changes in similar directions (indicated by the arrows) and generally included the same set of perturbed amino acids. (E) Mapping of the CSP values onto the dimeric structure of ERH-2 (PDB: 7O6N) shows that the most affected residues lie in strand β4 and adjacent helix α3, opposite the interface bound by PID-3pep. (F) Interplay of TOST-1pep and PID-1pep binding to ERH-2. Comparison of 15N-labeled ERH-2 moieties in its free form (red), bound to PID-1pep (green, 1:3 molar ratio), TOST-1pep (yellow, 1:3), in the presence of equimolar amounts of both peptides (blue, 1:3:3), or in the presence of excess TOST-1pep (magenta, 1:3:7). (G) Pull-down assays with purified recombinant wild-type and mutant versions of ERH-2 as prey and TOST-1-mVenus-Strep as bait (streptavidin pull-down) or GST-PID-3pep as bait (GST pull-down). Input and elution fractions were analyzed by SDS-PAGE followed by Coomassie staining.
To analyze the effect of the PID-3 (I182A and V186A) mutations on 21U RNA biogenesis, we sequenced mature piRNAs at permissive (20°C) and restrictive (25°C) temperatures in wild-type and mutant animals. This revealed a temperature-sensitive effect on overall 21U RNA levels (Fig. 3F,G). This was found to affect all 21U RNA species (Fig. 3H,I). We note that elevated temperatures also reduced 21U RNA levels in wild-type animals, but to a lesser extent. The reason behind this effect is currently unclear.
These results strongly support the in vivo relevance of the structures that we determined but also suggest that additional interactions between ERH-2 and PID-3 may exist within the full PETISCO complex.
ERH-2 interacts with the effector proteins TOST-1 and PID-1 through a common interface
PID-1 and TOST-1 share a common sequence motif with which they interact with ERH-2, as previously assessed through Y2H and MS analysis (Fig. 4A; Cordeiro Rodrigues et al. 2019). To understand the interplay of ERH-2 with these proteins, we purified full-length TOST-1 and PID-1, C-terminally tagged with monomeric Venus StrepII tag (mV-Strep), and probed their binding to ERH-2 and ERH-1. Both PID-1 and TOST-1 bound ERH-2 (Fig. 4B), but unlike PID-3, they also interacted with ERH-1 (Fig. 4B). We noted as well that TOST-1 was a more efficient bait than PID-1 (Fig. 4B), suggesting that it binds more strongly to ERH-2 and ERH-1 than PID-1.
To define the binding interface at the residue level, we analyzed the interaction of PID-1 and TOST-1 with ERH-2 using NMR spectroscopy. We titrated peptides corresponding to the conserved region of TOST-1 (residues 28–53, TOST-1pep) and PID-1 (residues 47–74, PID-1pep) into 15N-labeled full-length ERH-2 to monitor amide CSPs (Fig. 4C; Supplemental Fig, S6A,B). The changes occurred in slow-intermediate exchange for TOST-1pep, and in intermediate exchange for PID-1pep (Fig. 4C). This indicates binding constants in the micromolar range for both peptides, with PID-1pep binding being weaker than TOST-1pep.
To determine where PID-1pep and TOST-1pep bind ERH-2, we assigned the backbone chemical shifts of the respective complexes and calculated their amide CSPs (Fig. 4D). The changes observed occurred in similar ERH-2 residues and mostly in the same direction, indicating that both peptides bound in analogous fashion and to the same surface of ERH-2 (Fig. 4C). Both peptides caused the largest perturbations at the interface formed by strand β4, including the highly perturbed amino acids Ile81, Gly82, and Arg83, as well as the adjacent α3 helix, including Trp87 (Fig. 4D,E). Mapping of the largest CSPs on the surface of ERH-2 showed that the interface bound by TOST-1/PID-1 laid opposite the interface used for PETISCO binding through PID-3pep (Fig. 4E). Of note, we observed only one set of peaks for both the ERH-2 homodimer (except for the flexible loop), as well as the protein-peptide complexes. This suggests a 2:2 stoichiometry for protein to effector peptide, as any other scenario would break the symmetry required to observe only one species in solution.
We then explored the PID-1 and TOST-1 interplay on ERH-2 and compared spectra of free ERH-2 and in the presence of PID-1pep, TOST-1pep, as well as mixtures of both. When the peptides were present at the same concentrations, we observed chemical shifts at intermediate locations between that of the ERH-2/TOST-1pep and ERH-2/PID-1pep complexes (Fig. 4F). However, we cannot establish at the microscopic level whether this mixed state arises from rapid exchange between the two complexes in the NMR timescale or from association of both TOST-1 and PID-1 peptides, one at each available ERH-2 site. Nevertheless, once TOST-1pep was present in excess, we observed chemical shifts consistent with that of an ERH-2/TOST-1pep complex only (Fig. 4F; Supplemental Fig. S7). Since the peptides associate at the same interface and with comparable dissociation constants, we concluded that PID-1 and TOST-1 compete for ERH-2 binding, and their relative concentrations will determine which of the two complexes will be favored. Despite the larger size of the ERH-2/TOST-1FL complex (60 kDa), we detected very similar amide CSPs upon addition of full-length TOST-1 (TOST-1FL) to 15N-labeled ERH-2, indicating that the shorter peptide recapitulates full-length TOST-1 binding (Supplemental Fig. S8).
Figure 5.
PID-3 and TOST-1 associate with ERH-2 simultaneously. (A) Analysis of the interaction between TOST-1 with PETISCO components. Pull-down experiments with purified, recombinant TOST-1-mVenus-Strep fusion as bait with the indicated preys. Input and elution fractions were analyzed by SDS-PAGE followed by Coomassie staining. (B) Interactions between ERH-2, TOST-1, and PID-3 were assessed by SEC. (Left panel) Purified proteins were incubated alone or in the indicated mixtures in 1:1 ratio and subjected to SEC. (Right panel) Analysis of the peak fraction from SEC by SDS-PAGE and Coomassie staining. (C) The formation of a ternary ERH-2/TOST-1pep/PID-3pep complex shown by NMR spectroscopy. Zoomed-in views of overlaid 1H-15N HSQCs of ERH-2 in its free form (red), in complex with TOST-1pep (blue, 1:4 molar ratio), in complex with PID-3pep (orange, 1:3 molar ratio), or in the presence of both (green, 1:4:1.5 ERH-2:TOST-1pep:PID-3pep). Residues Thr21 (T21), Glu26 (E26), and Gly82 (G82) are indicated, with their positions color-coded according to the same scheme. The black arrows highlight amide peaks present in the ternary complex that do not correspond to either the ERH-2/TOST-1pep or ERH-2/PID-3pep subcomplexes.
To validate the PID-1/TOST-1 binding interface observed by NMR, we designed two ERH-2 mutants (I81A/R83A and W87A) and performed pull-down experiments with purified proteins. Both ERH-2 mutants, in particular W87A, showed a weaker interaction with TOST-1. (Fig. 4G). These mutations in ERH-2 did not affect PID-3pep binding, and conversely, the mutation in ERH-2 (T13E) that disrupted association with PID-3 did not significantly affect TOST-1 binding (Fig. 4B), consistent with their binding at opposite sides.
Analysis of the interplay between TOST-1 and PID-3 upon binding to ERH-2
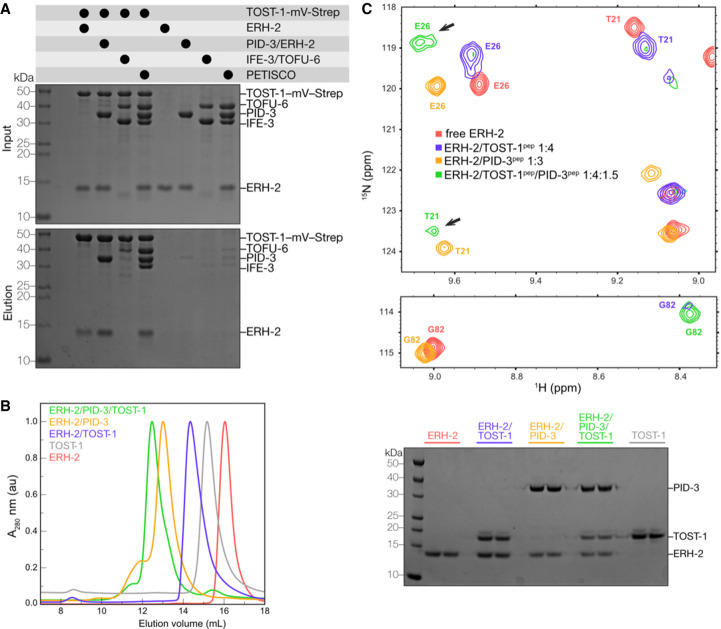
Our analysis suggested that ERH-2 could simultaneously bind TOST-1 and PID-3. To test this, we performed pull-down assays with TOST-1-mVenus-Strep as bait and ERH-2 alone, as part of an ERH-2/PID-3 subcomplex, or with full PETISCO as prey. We found that TOST-1 pulled down ERH-2 in all conditions (Fig. 5A). In addition, we observed that more ERH-2 is pulled down when ERH-2 is in complex with PID-3 or as part of PETISCO, indicating that the presence of PID-3 might facilitate the binding to TOST-1. The formation of a trimeric complex was supported by SEC experiments with purified full-length proteins, as a sample containing ERH-2, TOST-1, and PID-3 elutes at smaller elution volumes than the dimeric subcomplexes (Fig. 5B).
To obtain quantitative insights, we performed ITC measurements. Both TOST-1FL and TOST-1pep bound to ERH-2 with a dissociation constant of 13 µM, and ERH-2 had a higher affinity to TOST-1pep than to PID-1pep, both consistent with the NMR experiments (Table 1; Supplemental Figs. S8, S9). The difference in binding affinities for TOST-1pep and PID-1pep was more pronounced for ERH-1, with the dissociation constant differing by more than a factor of five. However, we note that in vivo ERH-1 has not been detected in complex with TOST-1 or PID-1.
Similar binding affinities between ERH-2 and TOST-1pep were obtained when the experiments were performed in the presence of PID-3pep (Table 1; Supplemental Fig. S9).
We also used NMR spectroscopy to monitor the interplay of TOST-1pep and PID-3pep on ERH-2. Upon addition of PID-3pep to a saturated ERH-2/TOST-1pep complex, we observed changes in the amide positions of 15N-labeled ERH-2 to locations consistent with the formation of an ERH-2/PID-3pep complex. However, signature chemical shifts corresponding to TOST-1-bound ERH-2 remained, suggesting the formation of a ternary complex (Fig. 5C; full view in Supplemental Fig. S10). Consistent with this idea, we observed the appearance of new peaks not present in either the ERH-2/TOST-1pep or ERH-2/PID-3pep subcomplexes (Fig. 5C).
Interestingly, some amide peaks diagnostic of the ERH-2/TOST-1pep interaction became stronger in the presence of PID-3pep (Fig. 5C), suggesting that PID-3pep stabilizes the interaction of TOST-1pep and ERH-2. Given that the two binding sites are connected through a central β sheet, this synergy in binding could be explained by an allosteric mechanism. Taken together, the pull-down, SEC, ITC, and NMR results clearly show that both PID-3pep and TOST-1pep can simultaneously associate with ERH-2 to form a trimeric complex and that the association of ERH-2 with PID-3pep facilitates its interaction with TOST-1pep.
A composite structural model of the PETISCO core
In the ERH-2ΔC/PID-3pep subcomplex, PID-3pep contains residues 177–193 and in the PID-3RRM/TOFU-6RRM subcomplex, the PID-3RRM domain contains residues 196–274. Hence, only two connecting residues between PID-3pep and PID-3RRM are unaccounted for. This allowed us to combine both structures into a composite model (Supplemental Fig. S11A). Using structure calculation protocols with slow simulated annealing, we generated a structure ensemble of 20 structures. We fixed atoms present in the crystal structures except the linker region connecting PID-3pep and PID-3RRM, allowing the two subcomplexes to sample different relative orientations. In the resulting ensemble, ERH-2 is positioned above the PID-3RRM domain through the interaction with PID-3pep (average RMSD of the Cα atoms of 6 Å ± 1 Å) (Fig. 6A; Supplemental Fig. S11B). Because ERH-2 is bound by two PID-3pep moieties, this restricts the flexibility of ERH-2 with respect to the RRM domain core.
Figure 6.
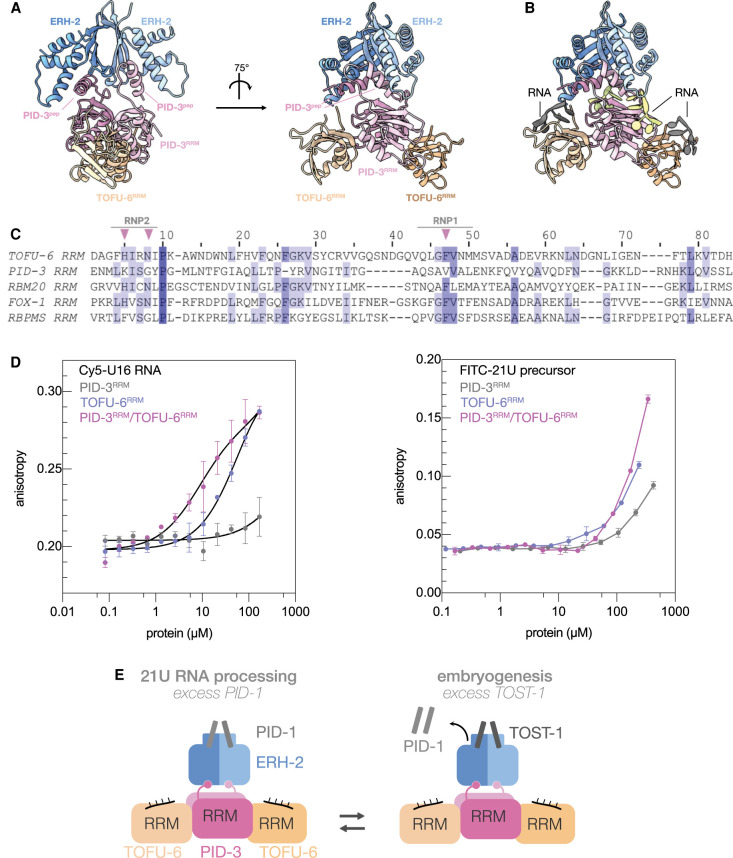
Modeling of the PETISCO core complex. (A) Modeling of the PETISCO core complex by combining the PID-3RRM/TOFU-6RRM and ERH-2ΔC/PID-3pep subcomplexes. The core complex is shown in two different orientations. The PID-3RRM/TOFU-6RRM complex is colored as in Figure 2, ERH-2ΔC/PID-3pep as in Figure 3 (B) Modeling of RNAs on the four RRMs of the PID-3RRM/TOFU-6RRM complex. The positions of the four modeled RNA molecules are shown in ribbon representation. RNAs on the TOFU-6RRM and PID-3RRM are shown in gray and green, respectively. (C) Structure-based multiple sequence alignment of PID-3RRM and TOFU-6RRM in comparison with well-characterized RNA-binding RRMs of RBM20, FOX-1, and RBPMS. The RNP1 and RNP2 motifs contributing to RNA binding are indicated and differences are highlighted with pink triangles. (D) Fluorescence anisotropy binding assay using a Cy5-U16-mer RNA (left panel) and a FITC-28-mer RNA (right panel). Values are presented as the average, and error bars correspond to the standard deviation of three (n = 3) technical replicates. The solid lines for TOFU-6RRM and PID-3RRM/TOFU-6RRM in the left panel represent the fit of a single-binding site model to the data, whereas for PID-3RRM and all three curves in the right panel, the solid lines only serve as a guide for the eye. (E) Schematic model of the PETISCO core complex (TOFU-6RRM/PID-3RRM/ERH-2) and its interaction with the effector proteins PID-1 and TOST-1. TOFU-6RRM/PID-3RRM form a tetrameric core and ERH-2 binds to a region upstream of the PID-3RRM. TOST-1 and PID-1 bind to a common surface on ERH-2, located opposite to its PID-3 binding site, and thereby specify PETISCO function.
In addition, we modeled RNA onto the PID-3RRM/TOFU-6RRM complex guided by known RRM-RNA structures (Auweter et al. 2006; Teplova et al. 2016; Upadhyay and Mackereth 2020), and we noticed that, while the RNA-binding surface of the TOFU-6RRM would be accessible, the RNA-binding surface of the PID-3RRM would be occupied by the ERH-2ΔC/PID-3pep module (Fig. 6B), suggesting that TOFU-6 is more likely to bind RNA than PID-3.
To investigate the potential RNA-binding properties of the TOFU-6RRM/PID-3RRM complex in more detail, we compared the RRM domains of both TOFU-6 and PID-3 with well-characterized RRMs using structure-based sequence alignments (Pei et al. 2008). RRM domains bind single-stranded RNA molecules via the outer β-sheet surface with the contribution of two conserved motifs called RNP1 and RNP2, located in the β3 and β1 strands, respectively (Maris et al. 2005). The residues critical for RNA binding, located in RNP1 and RNP2, are retained in TOFU-6RRM, whereas the PID-3RRM shows several differences, including the absence of aromatic residues in RNP1 and RNP2 (Fig. 6C).
We next tested RNA binding of PID-3RRM, TOFU-6RRM, and the PID-3RRM/TOFU-6RRM complex experimentally using fluorescence anisotropy assays using two different probes: (1) a Cy5-labeled 16-mer oligo(U) RNA, and (2) a FITC-labeled 28-mer RNA based on the 21U-3372 precursor (AUUGCAUCUAAAGUUGAUUGAAGAGUUA) containing all four nucleotides and with no predicted secondary structure (Cecere et al. 2012). Consistent with the modeling and sequence analysis, the PID-3RRM showed the lowest binding affinity for both RNAs (Fig. 6D). We were able to fit the data to a single binding site model for the poly(U) probe and derive binding constants for the TOFU-6RRM (Kd of ∼60 µM) and the tetrameric PID-3RRM/TOFU-6RRM complex (Kd of ∼10 µM). The higher RNA binding affinity of the PID-3RRM/TOFU-6RRM complex could result from additional contribution from the PID-3RRM, or from other, currently unknown effects. We also note that the 21U precursor-related probe was bound weaker than the poly(U) RNA. Presently, we have no explanation for this observation, but we note that the PETISCO complex also has additional RNA binding domains—for example, IFE-3, which can bind 5′-cap structures (Peter et al. 2015)—that may well affect overall RNA binding by PETISCO.
Discussion
PETISCO—oligomeric state
We show that PETISCO is an octameric protein complex, consisting of two copies each of IFE-3, TOFU-6, PID-3, and ERH-2, with a mass of ∼231 kDa. We found that the PID-3RRM/TOFU-6RRM complex binds a model RNA substrate with higher affinity than the isolated TOFU-6RRM, suggesting that oligomerization enhances RNA binding properties, but the functional consequence of PETISCO dimerization for piRNA processing remains to be determined. However, there are interesting parallels with other RNA processing complexes. First, it was recently demonstrated that the yeast and human THO (TREX) complexes involved in the transcription and export of RNA also associate into higher-order oligomers (Pühringer et al. 2020; Schuller et al. 2020). Whereas the yeast THO complex forms a dimer, the human THO complex forms a tetramer (Pühringer et al. 2020; Schuller et al. 2020). Although the functional consequence of oligomerization remains to be shown experimentally, it was hypothesized that dimerization of the yeast THO complex plays a crucial role in preventing R-loop formation during transcription of mRNA by RNA polymerase II (Schuller et al. 2020). In addition, dimerization is required for the function of the Drosophila melanogaster SFiNX complex, which facilitates cotranscriptional gene silencing downstream from the piRNA-PIWI complex. Dimerization of SFiNX is promoted through the interaction with the dynein light chain protein “Cut up”/LC8, which forms a homodimer and binds a short linear motif present in one of the SFiNX subunits. At the functional level, dimerization of SFiNX is required for the nucleic acid-stimulated formation of biomolecular condensates in vitro and heterochromatin formation at piRNA target loci in vivo (Schnabl et al. 2021). In the case of SFiNX, dimerization thus plays a direct role in the formation of condensates. Since the proteins that participate in the processing of piRNAs also localize in biomolecular condensates, the P granules, dimerization might also play a role in modulating potential phase separation properties of PETISCO.
Finally, the human ERH homolog has recently been shown to support the microprocessor complex during microRNA (miRNA) processing (Fang and Bartel 2020; Hutter et al. 2020; Kwon et al. 2020). Microprocessor is a trimeric complex consisting of one copy of DROSHA and two copies of DGCR8, and dimerization of DGCR8 is necessary for miRNA processing (Faller et al. 2007). The dimeric ERH protein binds to a short linear motif located in the N-terminal region of DGCR8 and is thought to additionally contribute to DGCR8 dimerization (Kwon et al. 2020). At the functional level, oligomerization facilitates processing of suboptimal miRNA hairpins located in clusters (Fang and Bartel 2020; Hutter et al. 2020). In PETISCO, we found a similar architecture: PID-3 dimerizes through its RRM domain and is supported by the binding of ERH-2 to a motif upstream of the RRM (Fig. 6E). Within PETISCO, ERH-2 not only reinforces dimerization but also binds the effector proteins TOST-1 and PID-1 at a surface opposite of PID-3. Similarly, SAFB has been shown to bind human ERH (Drakouli et al. 2017) and help process suboptimal miRNAs (Fang and Bartel 2020; Hutter et al. 2020). It thus appears that ERH homologs may act as signal integrators to control RNA processing. Because human ERH stimulates processing of suboptimal miRNA precursors, we hypothesize that PETISCO dimerization may likewise stimulate 21U RNA processing. However, dissecting whether these effects stem from dimerization or PETISCO integrity will be difficult, given the inter-dependence of PID-3 dimerization and TOFU-6 binding.
Subcellular localization of PETISCO
We found that loss of ERH-2 binding by PID-3 does not affect its subcellular distribution over the cytoplasm and P granules. However, we show that dimerization of PID-3 is a prerequisite for TOFU-6 binding and for localization of PID-3 to P granules. Previous work from Zeng et al. (2019) showed that an allele of tofu-6(ust95) that results in partial deletion of the eTudor domain specifically affected TOFU-6 P granule localization and 21U RNA production but not embryonic viability. Together, these results strongly imply that TOFU-6 plays an important role in P granule localization. Interestingly, the depletion of PID-3 by RNAi leads to loss of TOFU-6 from P granules (Zeng et al. 2019), suggesting that PID-3 can somehow enhance TOFU-6's ability to mediate this function. Whether this relates to dimerization or to other effects cannot be resolved at this stage.
Interplay between TOST-1 and PID-1
We found that TOST-1 can outcompete PID-1 for PETISCO binding when present in excess. Is this relevant for PETISCO functionality in vivo? A conclusive answer to this question will require an experiment in which the onset of expression of PID-1 and TOST-1 in the various stages of germ cell development can be assessed. Due to the stability of the fluorescent tags, and their large size with respect to the proteins of interest, standard localization studies under steady-state conditions (Zeng et al. 2019) are not suitable to resolve this. However, we do know that PID-1 does not have a role in the embryo and that 21U RNAs are already expressed at early stages, as judged from PRG-1 expression patterns (Batista et al. 2008; Wang and Reinke 2008). At the same time, germ cells do not show any defects in tost-1 mutants, while it is present in and required for the development of embryos (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019). Interestingly, in tost-1 mutants, some increase in 21U RNA levels have been reported (Cordeiro Rodrigues et al. 2019; Zeng et al. 2019), indicating a competition between the two PETISCO functions. Everything considered, it seems likely that PID-1-bound PETISCO assemblies are present in germ cells when TOST-1 starts to be up-regulated and that the PID-1-TOST-1 exchange that we describe is relevant to establish a good balance between PETISCO functions in vivo.
Evolutionary aspects
Evolutionary analysis of PETISCO components revealed that PID-3, TOFU-6, TOST-1, and PID-1 are restricted to nematodes, while ERH (ERH-2) proteins and eIF4E proteins (IFE-3) are present throughout eukaryotes (Cordeiro Rodrigues et al. 2019). C. elegans contains two ERH paralogs, ERH-1 and ERH-2. Strikingly, PID-3 can only interact with ERH-2, suggesting that ERH-2 has adopted a specialized function in PETISCO. Homology modeling of ERH-1 and superposition with our ERH-2 structure revealed that PID-3 binding residues Ile11, Thr13, and Gly20 in ERH-2 are not conserved in ERH-1 (Leu8, Pro10, and Ser20) (Supplemental Figs. S3D, S12A). Especially, ERH-1 Ser20 sterically interferes with PID-3 binding and we demonstrated that mutation of Thr13 in ERH-2 abrogates PID-3 binding. Interestingly, TOST-1 and PID-1 can, at least in vitro, interact with both ERH-1 and ERH-2. Furthermore, TOST-1 is present in nematode species that only contain one ERH paralog (Cordeiro Rodrigues et al. 2019), suggesting that in these species the single ERH protein may execute both PETISCO-related (bound by TOST-1) and PETISCO-unrelated activities.
Interestingly, there are also several nematode species that contain PID-3 but no ERH-2 paralog. Even though in C. elegans PID-3 does not bind ERH-1, it would be interesting to see whether PID-3 can interact with ERH-1 in those species, possibly representing an ancestor complex to what we now see in C. elegans.
Analysis of sequence conservation revealed that the most conserved feature of the PID-3RRM domain is the surface that mediates homodimerization. In contrast, the residues involved in TOFU-6 binding are less well conserved (Supplemental Fig. S12B). On the other hand, most residues of the TOFU-6RRM involved in PID-3RRM binding are well conserved (Supplemental Fig. S12C). This is consistent with our previous evolutionary analysis, which revealed that PID-3 is more widespread than TOFU-6 in the nematode phylum (Cordeiro Rodrigues et al. 2019). It might be that the TOFU-6 binding interface's evolutionary pressure is reduced compared with the PID-3 dimerization interface, or that TOFU-6 is replaced with another protein, possibly also an RRM domain protein, in species that have PID-3 but no TOFU-6.
These aspects clearly show that ERH is a nexus around which many RNA processing reactions concentrate and is amenable to significant variation in interacting proteins. The many parallels between ERH function in miRNA processing and PETISCO show that small RNA biogenesis pathways represent one domain of RNA processing that exploits ERH as an interaction platform. However, given the conservation of ERH also in species lacking small RNAs, such as S. cerevisiae, and the role of ERH in RNA decay in S. pombe, it is clear that ERH proteins fill a niche in RNA processing that is much more general (Weng and Luo 2013).
Materials and methods
Protein production
The genes coding for PETISCO subunits (IFE-3, TOFU-6, PID-3, and ERH-2), TOST-1, PID-1, and ERH-1 were cloned into modified pET vectors using ligation-independent cloning. All proteins were produced as an N-terminal His-tagged fusion protein with varying fusion partners. IFE-3, ERH-1, ERH-2, PID-1, and TOST-1 contained a His6-Trx-3C tag, and TOFU-6 a His10-MBP-3C tag, and PID-3 a His6-GST-3C tag. Addition of 3C protease allowed cleaving of this His fusion protein tag from the protein of interest. Proteins were produced in the E. coli BL21(DE3) derivative strain in terrific broth medium with the respective antibiotics. Briefly, cells were grown at 37°C, and when the culture reached an optical density (OD) at 600 nm of 2–3, the temperature was reduced to 18°C. After 2 h at 18°C, 0.2 mM IPTG was added to induce protein production for 12–16 h overnight.
To reconstitute PETISCO, single PETISCO subunits were coexpressed from individual plasmids with different antibiotic resistance markers. IFE-3 (His6-Trx-3C tag; ampicillin resistance) was coexpressed with TOFU-6 (His10-MBP-3C tag; kanamycin resistance), and PID-3 (His6-GST-3C tag; streptomycin resistance) with ERH-2 (His6-Trx-3C tag; ampicillin resistance). Cell pellets expressing the IFE-3/TOFU-6 and PID-3/ERH-2 subcomplexes were mixed in a 1:1 ratio, resuspended in lysis buffer (25 mM Tris/HCl, 50 mM NaPO4, 250 mM NaCl, 10% [v/v] glycerol, 5 mM 2-mercaptoethanol at pH 7.5), and lysed by sonication. PETISCO was purified by immobilized metal affinity chromatography (IMAC) using Ni2+-chelating beads (HisTrap FF; GE Healthcare). The HisTrap FF column was washed with 20 column volumes of lysis buffer, and proteins were eluted with 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 500 mM imidazole, 10% (v/v) glycerol, and 5 mM 2-mercaptoethanol. PETISCO was dialyzed against 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, and 5 mM 2-mercaptoethanol, and the His fusion tags were cleaved by the addition of 3C protease during dialysis. The His fusion tag and His-tagged 3C protease were removed by a second IMAC step. PETISCO was subsequently purified using Heparin affinity chromatography (HiTrap Heparin HP, GE Healthcare) and size exclusion chromatography using an S200 increase 10/300 column (GE Healthcare) in a buffer containing 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, and 2 mM DTT. All steps were performed on ice or at 4°C.
Size exclusion chromatography (SEC) assay
Purified proteins were incubated alone or in different combinations as indicated, in concentrations between 20 and 40 µM (total volume of 200 µL) in SEC buffer (20 mM Tris/HCl at pH 7.5, 150 mM NaCl, 2 mM DTT). Samples were incubated for 1 h on ice to allow complex formation. Complex formation was assayed by comparing the elution volumes in SEC on either a Superdex 200 Increase 10/300 or Superdex 200 Increase 3.2/300 (GE Healthcare) column. The SEC peak fractions were analyzed by SDS-PAGE and visualized by Coomassie brilliant blue staining.
Size exclusion chromatography coupled to multiangle light scattering (SEC-MALS)
The molecular mass and the oligomeric state of PETISCO, its subunits, and other proteins in solution were determined by size exclusion chromatography (SEC) coupled to multiangle light scattering (MALS). Individual proteins or protein complexes were analyzed at concentrations between 2 and 5 mg/mL in a buffer consisting of 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 2 mM DTT. A Superdex 200 Increase 10/300 GL column (GE Healthcare Life Sciences) was connected to a 1260 Infinity HPLC system (Agilent Technologies) coupled to a MiniDawn Treos detector (Wyatt Technologies) with a laser emitting at 690 nm. An RI-101 detector (Shodex) was used for refractive index measurement. Data analysis was performed using the Astra 7 software package (Wyatt Technologies).
ERH-2ΔC and PID-3pep production and purification
ERH-2ΔC containing residues 1–99 of C. elegans ERH-2 was cloned into a modified pET-vector with a 6xHis-Thioredoxin (Trx) tag followed by a 3C protease cleavage site. PID-3pep containing residues 171–203 of C. elegans PID-3 was cloned into a modified pET-vector with a 6xHis-GST tag followed by a 3C protease cleavage site. Cell growth and protein production were performed as described above. ERH-2ΔC and PID-3pep were purified by immobilized metal affinity chromatography using a Ni2+ matrix. The His-Trx and His-GST tags were cleaved with His-tagged 3C protease, and both the tags and 3C protease were removed in a reverse IMAC step. ERH-2ΔC and PID-3pep were present in the flow-through and were concentrated using ultrafiltration and subjected to size exclusion chromatography on a Superdex 75 (16/600) column equilibrated with 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, and 5 mM 2-mercaptoethanol. After size exclusion chromatography, ERH-2ΔC was concentrated to 15 mg/mland PID-3pep to 5 mg/ml and stored at −80°C until further use.
PID-3RRM and TOFU-6RRM production and purification
TOFU-6RRM (residues 1–99) and PID-3RRM (residues 196–274) were cloned into a modified pET-vector containing His10-MBP-3C and His6-GST-3C tags, respectively. TOFU-6RRM and PID-3RRM were expressed individually as described for ERH-2ΔC and PID-3pep. To purify the PID-3RRM/TOFU-6RRM complex, the cultures expressing individual PID-3RRM and TOFU-6RRM proteins were mixed before cell harvest. The PID-3RRM/TOFU-6RRM complex was purified by immobilized metal affinity chromatography using a Ni2+ matrix, followed by cation exchange chromatography and size exclusion chromatography on a Superdex 200 (16/600) column (GE Healthcare) equilibrated in 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, and 5 mM 2-mercaptoethanol.
Crystallization
All crystallization trials were performed using a vapor diffusion setup by mixing the protein complex and crystallization solution in a 1:1 and 2:1 ratio. For ERH-2ΔC, the best diffracting crystals grew in condition A2 from the Morpheus screen (Molecular Dimensions). For crystallization of the ERH-2ΔC/PID-3pep complex, these were mixed in a 1:1 ratio. The best diffracting crystals grew in condition A5 from the JCSG+ (Molecular Dimensions). The best crystals of PID-3RRM were obtained in condition C5 from the Morpheus screen (Molecular Dimensions). Crystals were harvested and directly frozen in liquid nitrogen before data collection at 100 K. Crystals from ERH-2ΔC, ERH-2ΔC/PID-3pep, and the PID-3RRM grew in conditions that did not require further cryoprotection and were therefore directly frozen in liquid nitrogen before data collection at 100 K.
Initial small crystals for the PID-3RRM/TOFU-6RRM complex were obtained in several commercial screens in conditions containing sodium acetate buffer (pH 4–5) and various precipitants and additives. Through several rounds of microseeding, we obtained larger crystals. The best crystals grew in 0.1 M sodium acetate (pH 5.0), 200 mM NaCl, and 17% (v/v) PEG3350. Crystals were soaked with a mother liquor containing 300 mM NaI instead of 200 mM NaCl, and with 25% (v/v) glycerol for cryoprotection, and then frozen in liquid nitrogen.
Data processing, phase determination, refinement, and modeling
All data were processed with Xia2/Dials (Winter et al. 2013) within CCP4i2 (Potterton et al. 2018). For ERH-2ΔC, the phases were determined by molecular replacement using the human ERH structure (PDB 2nml) as model, while in the case of the ERH-2ΔC/PID-3pep complex, we used the C. elegans ERH-2ΔC structure (PDB 7O6L). The phases of the PID-3RRM/TOFU-6RRM complex were solved by single isomorphous replacement with anomalous scattering (SIRAS) phasing from iodide with Autosol from Phenix (Terwilliger et al. 2009). The mean figure of merit over all resolution shells had a value of 0.30 and an estimated map correlation coefficient value of 40 ± 11. The phases for the PID-3RRM were determined by molecular replacement using the C. elegans PID-3RRM domain from the PID-3RRM/TOFU-6RRM complex structure (PDB 7OCZ) as a model. The models were automatically built using Buccaneer (Cowtan 2006) or Autobuild (Terwilliger et al. 2008), manually completed with COOT (Emsley et al. 2010), and refined with phenix.refine (Afonine et al. 2012) and refmac5 (Kovalevskiy et al. 2018). PDB REDO (Joosten et al. 2012) and MolProbity (Williams et al. 2018) were used for validation. Data collection, phasing, and refinement statistics are listed in Supplemental Table S1. Molecular graphics of the structures were created using UCSF ChimeraX (Goddard et al. 2018).
Pull-down assays with purified proteins
For interaction studies with purified proteins, appropriate protein mixtures (bait 10–20 µM, prey in 1.2-fold molar excess) were incubated in 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 10% (v/v) glycerol, 0.05% (v/v) NP40, 1 mM DTT for 30 min at 4°C. The protein mixtures were then incubated with the indicated beads—glutathione sepharose beads (Cube Biotech), amylose sepharose beads (New England Biolabs), and Strep-Tactin XT beads (IBA)—for 2 h. After incubation, the beads were washed three times with 0.2 mL of incubation buffer, and the retained material was eluted with 0.05 mL incubation buffer supplemented with 20 mM of reduced glutathione, 20 mM maltose, or 50 mM biotin. Input material and eluates were analyzed by SDS-PAGE and Coomassie brilliant blue staining.
Coexpression pull-down assays
For interaction studies by the coexpression copurification strategy, two plasmids containing the gene of interest and different antibiotic resistance markers were cotransformed into BL21(DE3) derivative strains to allow coexpression. Cells were grown in TB medium at 37°C, and when the culture reached an OD at 600 nm of 2–3, the temperature was reduced to 18°C. After 2 h at 18°C, 0.2 mM IPTG was added to induce protein production for 12–16 h overnight. Cell pellets were resuspended in 2 ml of lysis buffer (50 mM sodium phosphate, 20 mM Tris/HCl, 250 mM NaCl, 10 mM Imidazole, 10% [v/v] glycerol, 0.05% [v/v] NP-40, 5 mM β-mercaptoethano at pH 8.0) per gram of wet cell mass. Cells were lysed by ultrasonic disintegration, and insoluble material was removed by centrifugation at 21,000×g for 10 min at 4°C. For GST pull-downs, 500 µL of supernatant was applied to 20 µL of glutathione-coupled resin (Cube Biotech); for MBP pull-downs, 500 µL of supernatant was applied to 20 µL of amylose resin (New England Biolabs) and incubated for 2 h at 4°C. Subsequently, the resin was washed three times with 500 µL of lysis buffer. The proteins were eluted in 50 µL of lysis buffer supplemented with 20 mM reduced glutathione or 20 mM maltose in the case of glutathione or amylose beads, respectively. Input material and eluates were analyzed by SDS-PAGE and Coomassie brilliant blue staining.
Circular dichroism spectroscopy
Far-UV CD spectra were measured using a Chirascan plus spectrometer (Applied Photophysics) and a 0.5-mm path-length cuvette at 22°C. Samples were prepared at 15 µM in 5 mM sodium phosphate, 80 mM NaCl (pH 8.0). At least three spectra were collected for each sample at 180–260 nm with 1-nm steps and averaged.
Multiple sequence alignments
Multiple sequence alignments were generated using MUSCLE (Edgar 2004), structure-based multiple sequence alignments using PROMALS3D (Pei et al. 2008). For the visualization of multiple sequence alignments, Jalview 2.0 was used (Waterhouse et al. 2009).
Isothermal titration calorimetry
All ITC experiments were carried out at 25°C using the PEAQ-ITC isothermal titration calorimeter (Malvern). The data were processed and curves were fitted using the PEAQ-ITC software. Before the measurements, all samples were dialyzed simultaneously against 1 L of sample buffer (50 mM Tris, 250 mM NaCl, 1 mM TCEP at pH 7.50) to ensure complete buffer matching. The proteins and peptides were concentrated to the values listed in Table 2.
Table 2.
Protein and peptide concentrations in ITC experiments
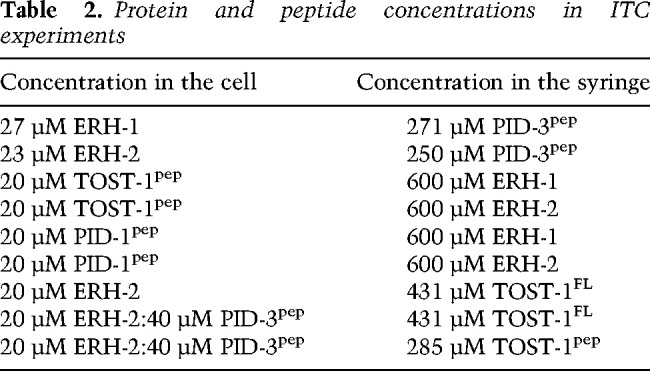
In each case, 2 µL of the titrant per injection were added to 200 µL of reactant cell solution. Due to difficulties in accurately measuring the TOST-1pep concentration (low extinction coefficient at 280 nm), this value was floated and the N value fixed at the known stoichiometry of 1. The heats of dilution were obtained by repeating the experiments with sample buffer in the calorimeter cell, while keeping all other experimental parameters identical. These dilution heats were fit to a straight line by linear regression and subtracted from the heats of binding. The baseline-corrected binding isotherms were then fit to a 1:1 model to give the reported thermodynamic parameters. For the purposes of comparing binding of TOST-1 in the presence and absence of PID-3pep, the heats of association were assumed to derive solely from TOST-1 binding, as we did not detect binding between TOST-1 and PID-3 peptides (not shown). For multiple measurements, the averages and population standard deviations of the fit results (Kd, ΔH°bind, and −TΔS°bind) are reported.
Fluorescence anisotropy experiments
The RNAs used for anisotropy experiments were purchased from Ella Biotech. The pre-piRNA is based on the following 21U-3372 precursor sequences: 5′-Cy5-U16 (Cy5-UUUUUUUUUUUUUUUU) and 5′-FITC-pre-piRNA (FITC-AUUGCAUCUAAAGUUGAUUGAAGAGUUA).
The fluorescently labeled RNAs were used at a concentration of 100 nM and incubated with increasing concentrations of protein in a volume of 20 µL in a buffer containing 10 mM Tris/HCl (pH 7.5), 50 mM KCl, and 0.5 mM EDTA for at least 2 h at 15°C. The anisotropy was recorded on a FS5 Spectrofluorometer (Edinburgh Instruments) with the following settings: for Cy5, excitation 649 nm, bandwidth 3 nm; emission 675 nm, and bandwidth 3 nm; for FITC, excitation 495 nm, bandwidth 3 nm; emission 519 nm, and bandwidth 3 nm. The anisotropy was measured five times in 30 sec, and the average anisotropy value was calculated. The data were fitted with a single binding site model using Graphpad Prism.
Modeling
A homology model of C. elegans ERH-1 was generated using the I-TASSER server (Yang et al. 2015) with default settings and the C. elegans ERH-2 structure (PDB 7OCZ) as a template.
We generated a model of the PETISCO core complex containing ERH-2(1–113) PID-3(179–274) TOFU-6(RRM) by combining the coordinates of the crystal structures of ERH-2(3–99)/PID-3(177–193/179–193) and PID0-3(198–274/196–274)/TOFU-6(7–99/7–91). A template structure of the complex was generated in cns-1.2 (Brunger 2007) by adding the missing linker residues (194–197 and 194–195) followed by a short energy minimization. To explore the structural flexibility of this core complex, we generated an ensemble of 20 structures from the energy-minimized template by randomly choosing 10 different Cα pairs of the two subcomplexes and ran a simulated annealing structure calculation protocol (Linge et al. 2003) with one distance restraint for each of the chosen pairs adding 7 Å to or subtracting 7 Å from the distance in the template structure, respectively. In the structure calculation, all residues observed in the crystal structures where fixed (Simon et al. 2010) except for eight linker residues (PID3 192–199) connecting the two subcomplexes. The resulting ensemble of structures exhibits an average root mean square deviation of the Cα atoms of 6 Å ± 1 Å to the template structure.
Production of fractionally deuterated 13C/15N-labeled ERH-2
The gene encoding full-length C. elegans ERH-2 (NCBI gene ID 185323) was cloned into pET28a (Addgene 69864-3), containing an N-terminal His6 tag followed by a TEV protease cleavage site (MGSSHHHHHHSSGENLYFQGHMAS) using NheI and BamHI restrictions sites. Due to the size of the dimeric ERH-2 protein (∼27 kDa), fractional deuteration in combination with stable isotope labeling was used for recombinant protein expression in Escherichia coli BL21 (λDE3) to allow NMR triple resonance experiments for backbone assignments (13Cα, 13Cβ, and 15NH). A starter 50-mL culture was grown for 16 h at 37°C with shaking at 200 rpm, using M9 minimal medium (dissolved in H2O) supplemented with 2 g/L 12C-glucose and 0.5 g/L 14NH4Cl. The cells were collected by centrifugation and resuspended in 500 mL of M9 minimal medium dissolved in ∼98% D2O supplemented with 2 g/L 13C-glucose and 0.5 g/L 15NH4Cl, followed by incubation for 16 h at 37°C. The resulting culture was used to inoculate additional 2.5 L of minimal M9/D2O/13C/15N medium, to a starting OD600 of ∼0.3. After growth at 37°C, protein expression was induced at an OD600 of ∼1 with 2 mM IPTG, and the culture was allowed to shake for 16 h at 20 °C. Subsequently, the cells were collected by centrifugation, resuspended in lysis buffer (20 mM Na2HPO4, 20 mM imidazole, 1 M NaCl, 1 M urea, 10% [v/v] glycerol at pH 7.4), and frozen at −20°C until use. The cell pellet was then thawed, and the lysis buffer supplemented with lysozyme (Genaxxon Bioscience) and protease inhibitor cocktail (Roche) according to instructions. After 15-min incubation on ice, the mixture was homogenized (Microfluidizer M-110L fluid processor, Microfluidics Corp.) by passaging four times. Unless otherwise noted, all purification steps were performed at room temperature. The lysate was cleared by centrifugation and applied to tandem fast-flow Ni+2-NTA HisTrap (GE Healthcare) columns. These were washed with 20 column volumes of lysis buffer. His-tagged ERH-2 was eluted in one step with ∼35 mL of 20 mM Na2HPO4, 500 mM imidazole, and 0.5 M NaCl (pH 7.4), and dialyzed against 2 L of TEV cleavage buffer (50 mM Tris, 250 mM NaCl, 1 mM DTT at pH 7.5) for 30 min at room temperature. At this point, ∼1 mg of TEV protease was added, and ERH-2 was allowed to continue dialyzing in its presence for 16 h at 4°C. The resulting cleaved product contained five nonnative residues at the N terminus and was further purified using a second HisTrap, followed by size exclusion chromatography column (Superdex 75 16/600, GE Healthcare) equilibrated with NMR buffer (50 mM Tris, 250 mM NaCl, 1 mM TCEP at pH 7.50). The appropriate pure fractions were pooled, and the protein was concentrated using Amicon 3 kDa MWCO centrifugal filters (Merck Millipore). The protein concentration was calculated using the measured absorbance at 280 nm under native conditions and assuming an extinction coefficient of 19940 M−1 cm−1.
NMR spectroscopy
NMR experiments were performed at 25°C using a Bruker Avance III 800-MHz spectrometer equipped with a cryoprobe. The ERH-2 sample was concentrated to 200–600 µM and contained 0.02% sodium azide, 10% D2O (as a locking agent), and 90% NMR buffer. For backbone assignments and titrations, 2D and 3D transverse relaxation optimized spectroscopy (TROSY) pulse sequences with 2H-decoupling and apodization weighted sampling were used (Pervushin et al. 1997; Salzmann et al. 1998; Simon and Köstler 2019). The resulting spectra were processed and analyzed using NMRPipe (Delaglio et al. 1995) and NMR-FAM Sparky (Lee et al. 2015), respectively. Ninety percent of all amides were assigned. The resulting chemical shifts (1HN, 15N, 13Cα, and 13Cβ) were deposited under BMRB 50914 and used as input for secondary structure prediction using the program MICS (Supplemental Fig. S5A; Shen and Bax 2012). To assign the peptide-bound ERH-2 complexes, NMR-monitored titrations were used in combination with TROSY-HNCA experiments to track the new positions of the amide peaks (Salzmann et al. 1998).
Peptides and NMR-monitored titrations
The lyophilized TOST-1 and PID-1 peptides were purchased from Proteogenix at 95% purity (for details, see Table 3). Approximately 10 mg of each peptide was resuspended in ∼0.8 mL of NMR buffer and further dialyzed using Pur-A-Lyzer dialysis containers (Sigma) with a MWCO of 1 kDa, against an additional 2 L of NMR buffer to ensure complete buffer match. The peptide concentration was typically ∼2 mM, calculated using the measured absorbance at 280 nm under native conditions and assuming extinction coefficients of 2980 M−1 cm−1 for both TOST-1 and PID-1 peptides. The peptides were added in a stepwise manner to labeled ERH-2 at a concentration of ∼200 μM, and the changes in chemical shifts were monitored with TROSY-HSQC experiments at each titration point. The extent of amide 1H-15N chemical shift perturbation (CSP) in free versus saturated peptide-bound ERH-2 were calculated according to Williamson (2013) to compensate for frequency bandwidth differences between 15N and 1H dimensions. The resulting values were plotted as a function of residue number.
Table 3.
Peptides used in this study
Worm methods
Worms were cultivated at 20°C on OP50 plates. For experiments at 25°C, animals were grown for 48 h at 25°C. The strains used were as follows: N2, C. elegans wild isolate; RFK1344, pid-3(xf271[pid-3::gfp]) I; RFK1346, pid-3(xf272[pid-3[A220E]::gfp]) I; RFK1348, pid-3(xf273[pid-3[I182A/V186A]::gfp]) I; and RFK1349, pid-3(xf274[pid-3[I182E/V186E]::gfp]) I.
CRISPR/Cas9 genome editing
Protospacer sequences were chosen using CRISPOR (http://crispor.tefor.net) cloned in pRK2412 by site-directed, ligase-independent mutagenesis (SLIM). For the pid-3 gene, the following alleles were used: xf271 (5′-CAAATTGGGGCACATTTAAG[AGG]-3′), xf272 (5′-GATCCCATTGACACGATATG[GGG]-3′), xf273 (5′-CTTCAACAGCGAAGACATCA[AGG]-3′), and xf274 (5′-CTTCAACAGCGAAGACATCA[AGG]-3′).
A PCR product from plasmid pDD282 (Addgene plasmid 66823) was used as a donor template for insertion of gfp. For creation of point mutations, the following Ultramer DNA oligodeoxynucleotides from Integrated DNA Technologies were used as a donor template: xf271 (TGGGAAAGCCGATCAGACTTTCGAAAGTCTCCTCTGGAGCATCGGGAGCCTCAGGAGCATCGATGAGTAAAGGAGAAGAATTGTTCACTGGAGTTGTCCCAATCCTCGTCGAGCTCGACGGAGACGTCAACGGACACAAGTTCTCCGTCTCCGGAGAGGGAGAGGGAGACGCCACCTACGGAAAGCTCACCCTCAAGTTCATCTGCACCACCGGAAAGCTCCCAGTCCCATGGCCAACCCTCGTCACCACCTTCTGCTACGGAGTCCAATGCTTCTCCCGTTACCCAGACCACATGAAGCGTCACGACTTCTTCAAGTCCGCCATGCCAGAGGGATACGTCCAAGAGCGTACCATCTTCTTCAAGGTAAGTTTAAACATATATATACTAACTACTGATTATTTAAATTTTCAGGACGACGGAAACTACAAGACCCGTGCCGAGGTCAAGTTCGAGGGAGACACCCTCGTCAACCGTATCGAGCTCAAGGTAAGTTTAAACAGTTCGGTACTAACTAACCATACATATTTAAATTTTCAGGGAATCGACTTCAAGGAGGACGGAAACATCCTCGGACACAAGCTCGAGTACAACTACAACTCCCACAACGTCTACATCATGGCCGACAAGCAAAAGAACGGAATCAAGGTCAACTTCAAGGTAAGTTTAAACATGATTTTACTAACTAACTAATCTGATTTAAATTTTCAGATCCGTCACAACATCGAGGACGGATCCGTCCAACTCGCCGACCACTACCAACAAAACACCCCAATCGGAGACGGACCAGTCCTCCTCCCAGACAACCACTACCTCTCCACCCAATCCGCCCTCTCCAAGGACCCAAACGAGAAGCGTGACCACATGGTCCTCCTCGAGTTCGTCACCGCCGCCGGAATCACCCACGGAATGGACGAGCTCTACAAGTAAATGTGCCCCAATTTGCTTATAAGTTATTTTTTTCT), xf272 (GTGGATATCCGGGGATGCTCAACACGTTCGGCATCGAGCAATTGCTCACTCCATATCGTGTCAATGGGATCACCATCACCGGCGCCCAG), xf273 (CGACGTTCGACAAGGTTCTCTTCAACAGCGAAGACGCTAAAGATTCTGCCTTCAAAGTTCTGCATGCCGAAGAAGAGCCGAGAGGTGCGGATCAGGAGAA), and xf274 (CGACGTTCGACAAGGTTCTCTTCAACAGCGAAGACGAGAAAGATTCTGAGTTCAAAGTTCTGCATGCCGAAGAAGAGCCGAGAGGTGCGGATCAGGAGAA).
In all cases, dpy-10 coconversion was used. All alleles were outcrossed twice before experiments.
Mel phenotype
For RFK1344, RFK1346, and RFK1348, 30 young adult worms were singled and left for 24 h at 20°C or overnight at 25°C. Afterward, mothers were removed and the eggs were counted, and after 2 d, developed animals were scored. RFK1349 is Mel (maternal embryonic lethal) at 20°C; therefore, progeny of heterozygous mothers were singled and genotyped. At least 2200 eggs from at least 23 worms were counted for each condition.
Microscopy
Young adult worms or young gravid adults grown at 25°C were washed in M9 buffer and paralyzed with 60 mM sodium azide in M9. As soon as worms stopped moving, they were imaged on a TCS SP5 Leica confocal microscope at 25°C. Images were processed using Fiji and Adobe illustrator.
21U RNA sequencing
NGS library preparation was performed with a NEXTflex small RNA-seq kit V3, following step A to step G of BioScientific's standard protocol (V19.01), with a starting amount of 30 ng, amplified in 21 PCR cycles, purified by running on an 8% TBE gel, and size-selected in the range of 144–163 nt. The libraries were profiled using a high-sensitivity DNA kit on a 2100 Bioanalyzer (Agilent Technologies) and quantified using the dsDNA HS assay kit in a Qubit 2.0 fluorometer (Life Technologies). All samples were pooled in equimolar ratio and sequenced on a NextSeq 500/550 Flowcell SR for 75 cycles plus seven cycles for the index read.
The raw sequence reads in FastQ format were cleaned from adapter sequences and size-selected for 18- to 40-base-long inserts (plus eight random adapter bases) using cutadapt v.2.4 (http:// cutadapt.readthedocs.org) with parameters “-a AGATCGGAAGAGCACACGTCT -m 26 -M 48.” Read alignment to the C. elegans genome (Ensembl WBcel235/ce11 assembly) with concomitant trimming of the eight random bases was performed using Bowtie v.1.2.2 (http://bowtie-bio.sourceforge.net) with parameters “-v 1 -M 1 -y –best –strata –trim5 4 –trim3 4 -S,” and the SAM alignment files were converted into sorted BAM files using Samtools v.1.9 (http://www.htslib.org). C. elegans WBcel235/ce11 gene annotation in GTF format was downloaded from Ensembl release 96 (ftp://ftp.ensembl.org/pub). Aligned reads were assigned to small RNA loci and classes using Samtools, GNU Awk, and Subread featureCounts v.1.6.2 (http://bioinf.wehi.edu.au/featureCounts). Structural reads aligning sense to rRNA, tRNA, snRNA, and snoRNA loci were excluded from further analysis. 21U-RNAs were strictly defined as reads 21 bases long starting with T and fully overlapping with annotated piRNA (21ur) loci on the same DNA strand. For maximal specificity, excluded from analysis were a small number (3.6%) of ambiguous 21ur loci colocalizing on the same strand with miRNAs, snoRNAs, or any other RNA exons. The abundance of 21U-RNAs was normalized per million mapped nonstructural reads. Sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA743298 (prepublication reviewer link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA743298?reviewer=g2qqsvjnla2nn2h0r05bpjf4r3).
Accession numbers
The coordinates and the structure factors have been deposited in the Protein Data Bank with accession codes 7O6L, 7O6N, 7OCX, and 7OCZ. NMR backbone chemical shifts of free C. elegans ERH-2 were deposited at the Biological Magnetic Resonance Bank under accession number 50914. Sequencing data are available at the NCBI Sequence Read Archive (SRA) under BioProject PRJNA743298.
Supplementary Material
Acknowledgments
We thank all of the members of the Falk, Ketting, and Hennig laboratories for great help and discussion. We thank Svenja Hellmann for support in genome editing, and the Institute of Molecular Biology (IMB) Media Laboratory and the Microscopy Core Facilities for equipment and support. We thank the IMB Genomics Core Facilities for excellent support. We also thank the beamline scientists from the European Synchrotron Radiation Facility (ESRF) beamlines ID-23-1 and ID23-2 (Grenoble, France) and Swiss Light Source PXII (Villigen, Switzerland) for excellent support with data collection, as well as the European Molecular Biology Laboratory (EMBL) Biophysics Facility. The Hennig laboratory gratefully acknowledges support via an Emmy-Noether Fellowship of the Deutsche Forschungsgemeinschaft (DFG) (HE 7291). This study was supported by funding from the DFG via the priority program SPP1935 to J.H. (EP37/3-1 and EP37/3-2) and R.F.K. (KE1888/7-1) and laboratory startup funding from the University of Vienna to S.F. C.P.-B. thanks the EMBL Interdisciplinary Postdoc (EI3POD) Program fellowship under Marie Sklodowska-Curie Actions COFUND (cofunding of regional, national and international programmes; grant no. 664726) for support.
Author contributions: C.P.-B., K.H., N.P., R.L., J.B., J.H., and S.F. planned and executed experiments. K.H., R.L., and S.F. purified proteins and performed protein interaction analysis. K.H. and S.F. generated crystals. S.F. and J.B. collected crystallographic data. S.F. solved and modelled all crystal structures. N.P. generated and analyzed C. elegans strains and performed protein purifications. C.P.-B. and J.H. performed and analyzed ITC and NMR experiments. S.F., R.F.K., and J.H. supervised the projects and provided funds. C.P.-B., R.F.K., and S.F. wrote the initial draft. All authors edited and approved the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348648.121.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68: 352–367. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH-T. 2006. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J 25: 163–173. 10.1038/sj.emboj.7600918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78. 10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran T, Barroso C, Birkle TY, Stevens L, Schwartz HT, Sternberg PW, Fradin H, Gunsalus K, Piano F, Sharma G, et al. 2019. Comparative epigenomics reveals that RNA polymerase II pausing and chromatin domain organization control nematode piRNA biogenesis. Dev Cell 48: 793–810.e6. 10.1016/j.devcel.2018.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran T, Pahita E, Ghosh S, Lenhard B, Sarkies P. 2021. Integrator is recruited to promoter-proximally paused RNA Pol II to generate Caenorhabditis elegans piRNA precursors. EMBO J 40: e105564. 10.15252/embj.2020105564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkyurek AC, Furlan G, Lampersberger L, Beltran T, Weick E-M, Nischwitz E, Cunha Navarro I, Braukmann F, Akay A, Price J, et al. 2021. The RNA polymerase II subunit RPB-9 recruits the integrator complex to terminate Caenorhabditis elegans piRNA transcription. EMBO J 40: e105565. 10.15252/embj.2020105565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT. 2007. Version 1.2 of the crystallography and NMR system. Nat Protoc 2: 2728–2733. 10.1038/nprot.2007.406 [DOI] [PubMed] [Google Scholar]
- Cecere G, Zheng GXY, Mansisidor AR, Klymko KE, Grishok A. 2012. Promoters recognized by forkhead proteins exist for individual 21U-RNAs. Mol Cell 47: 734–745. 10.1016/j.molcel.2012.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro Rodrigues RJ, de Jesus Domingues AM, Hellmann S, Dietz S, de Albuquerque BFM, Renz C, Ulrich HD, Sarkies P, Butter F, Ketting RF. 2019. PETISCO is a novel protein complex required for 21U RNA biogenesis and embryonic viability. Genes Dev 33: 857–870. 10.1101/gad.322446.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62: 1002–1011. 10.1107/S0907444906022116 [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 41: 324–337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31: 79–90. 10.1016/j.molcel.2008.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Albuquerque BFM, Luteijn MJ, Cordeiro Rodrigues RJ, van Bergeijk P, Waaijers S, Kaaij LJT, Klein H, Boxem M, Ketting RF. 2014. PID-1 is a novel factor that operates during 21U-RNA biogenesis in Caenorhabditis elegans. Genes Dev 28: 683–688. 10.1101/gad.238220.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293. 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- Drakouli S, Lyberopoulou A, Papathanassiou M, Mylonis I, Georgatsou E. 2017. Enhancer of rudimentary homologue interacts with scaffold attachment factor B at the nuclear matrix to regulate SR protein phosphorylation. FEBS J 284: 2482–2500. 10.1111/febs.14141 [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller M, Matsunaga M, Yin S, Loo JA, Guo F. 2007. Heme is involved in microRNA processing. Nat Struct Mol Biol 14: 23–29. 10.1038/nsmb1182 [DOI] [PubMed] [Google Scholar]
- Fang W, Bartel DP. 2020. MicroRNA clustering assists processing of suboptimal microRNA hairpins through the action of the ERH protein. Mol Cell 78: 289–302.e6. 10.1016/j.molcel.2020.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF chimerax: meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh WSS, Seah JWE, Harrison EJ, Chen C, Hammell CM, Hannon GJ. 2014. A genome-wide RNAi screen identifies factors required for distinct stages of C. elegans piRNA biogenesis. Genes Dev 28: 797–807. 10.1101/gad.235622.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Lee H-C, Chaves D, Youngman EM, Pazour GJ, Conte D Jr, Mello CC. 2012. CapSeq and CIP-TAP identify Pol II start sites and reveal capped small RNAs as C. elegans piRNA precursors. Cell 151: 1488–1500. 10.1016/j.cell.2012.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra D, Andrić V, Palancade B, Rougemaille M, Graille M. 2020. Formation of S. pombe Erh1 homodimer mediates gametogenic gene silencing and meiosis progression. Sci Rep 10: 1034. 10.1038/s41598-020-57872-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocine S, Singer RH, Grünwald D. 2010. RNA processing and export. Cold Spring Harb Perspect Biol 2: a000752. 10.1101/cshperspect.a000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K, Lohmüller M, Jukic A, Eichin F, Avci S, Labi V, Szabo TG, Hoser SM, Hüttenhofer A, Villunger A, et al. 2020. SAFB2 enables the processing of suboptimal stem-loop structures in clustered primary miRNA transcripts. Mol Cell 78: 876–889.e6. 10.1016/j.molcel.2020.05.011 [DOI] [PubMed] [Google Scholar]
- Joosten RP, Joosten K, Murshudov GN, Perrakis A. 2012. PDB_REDO: constructive validation, more than just looking for errors. Acta Crystallogr D Biol Crystallogr 68: 484–496. 10.1107/S0907444911054515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper DM, Wang G, Gardner KE, Johnstone TG, Reinke V. 2014. The C. elegans SNAPc component SNPC-4 coats piRNA domains and is globally required for piRNA abundance. Dev Cell 31: 145–158. 10.1016/j.devcel.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper BD, Lamphear BJ, Deshpande AM, Jankowska-Anyszka M, Aamodt EJ, Blumenthal T, Rhoads RE. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J Biol Chem 275: 10590–10596. 10.1074/jbc.275.14.10590 [DOI] [PubMed] [Google Scholar]
- Kovalevskiy O, Nicholls RA, Long F, Carlon A, Murshudov GN. 2018. Overview of refinement procedures within REFMAC5: utilizing data from different sources. Acta Crystallogr D Struct Biol 74: 215–227. 10.1107/S2059798318000979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Jang H, Shen S, Baek SC, Kim K, Yang J, Kim J, Kim J-S, Wang S, Shi Y, et al. 2020. ERH facilitates microRNA maturation through the interaction with the N-terminus of DGCR8. Nucleic Acids Res 48: 11097–11112. 10.1093/nar/gkaa827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tonelli M, Markley JL. 2015. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31: 1325–1327. 10.1093/bioinformatics/btu830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linge JP, Habeck M, Rieping W, Nilges M. 2003. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics 19: 315–316. 10.1093/bioinformatics/19.2.315 [DOI] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. 2013. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet 14: 523–534. 10.1038/nrg3495 [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH-T. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131. 10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, Zamore PD. 2019. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 20: 89–108. 10.1038/s41576-018-0073-3 [DOI] [PubMed] [Google Scholar]
- Pabis M, Popowicz GM, Stehle R, Fernández-Ramos D, Asami S, Warner L, García-Mauriño SM, Schlundt A, Martínez-Chantar ML, Díaz-Moreno I, et al. 2019. Hur biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs. Nucleic Acids Res 47: 1011–1029. 10.1093/nar/gky1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36: 2295–2300. 10.1093/nar/gkn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wüthrich K. 1997. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci 94: 12366–12371. 10.1073/pnas.94.23.12366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter D, Weber R, Köne C, Chung M-Y, Ebertsch L, Truffault V, Weichenrieder O, Igreja C, Izaurralde E. 2015. Mextli proteins use both canonical bipartite and novel tripartite binding modes to form eIF4E complexes that display differential sensitivity to 4E-BP regulation. Genes Dev 29: 1835–1849. 10.1101/gad.269068.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton L, Agirre J, Ballard C, Cowtan K, Dodson E, Evans PR, Jenkins HT, Keegan R, Krissinel E, Stevenson K, et al. 2018. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr D Struct Biol 74: 68–84. 10.1107/S2059798317016035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pühringer T, Hohmann U, Fin L, Pacheco-Fiallos B, Schellhaas U, Brennecke J, Plaschka C. 2020. Structure of the human core transcription-export complex reveals a hub for multivalent interactions. Elife 9: e61503. 10.7554/eLife.61503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripin N, Boudet J, Duszczyk MM, Hinniger A, Faller M, Krepl M, Gadi A, Schneider RJ, Šponer J, Meisner-Kober NC, et al. 2019. Molecular basis for AU-rich element recognition and dimerization by the HuR C-terminal RRM. Proc Natl Acad Sci 116: 2935–2944. 10.1073/pnas.1808696116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, He C. 2017. Dynamic RNA modifications in gene expression regulation. Cell 169: 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. 2006. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127: 1193–1207. 10.1016/j.cell.2006.10.040 [DOI] [PubMed] [Google Scholar]
- Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K. 1998. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci 95: 13585–13590. 10.1073/pnas.95.23.13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl J, Wang J, Hohmann U, Gehre M, Batki J, Andreev VI, Purkhauser K, Fasching N, Duchek P, Novatchkova M, et al. 2021. Molecular principles of Piwi-mediated cotranscriptional silencing through the dimeric SFiNX complex. Genes Dev 35: 392–409. 10.1101/gad.347989.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller SK, Schuller JM, Prabu JR, Baumgärtner M, Bonneau F, Basquin J, Conti E. 2020. Structural insights into the nucleic acid remodeling mechanisms of the yeast THO-Sub2 complex. Elife 9: e61467. 10.7554/eLife.61467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. 2012. Identification of helix capping and β-turn motifs from NMR chemical shifts. J Biomol NMR 52: 211–232. 10.1007/s10858-012-9602-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon B, Köstler H. 2019. Improving the sensitivity of FT-NMR spectroscopy by apodization weighted sampling. J Biomol NMR 73: 155–165. 10.1007/s10858-019-00243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon B, Madl T, Mackereth CD, Nilges M, Sattler M. 2010. An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angew Chem Int Ed Engl 49: 1967–1970. 10.1002/anie.200906147 [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Thillainadesan G, Chalamcharla VR, Meng Z, Balachandran V, Dhakshnamoorthy J, Zhou M, Grewal SIS. 2016. Enhancer of rudimentary cooperates with conserved RNA-processing factors to promote meiotic mRNA decay and facultative heterochromatin assembly. Mol Cell 61: 747–759. 10.1016/j.molcel.2016.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Tu S, Lee HC, Weng Z, Mello CC. 2016. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell 164: 974–984. 10.1016/j.cell.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Farazi TA, Tuschl T, Patel DJ. 2016. Structural basis underlying CAC RNA recognition by the RRM domain of dimeric RNA-binding protein RBPMS. Q Rev Biophys 49: e1. 10.1017/S0033583515000207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64: 61–69. 10.1107/S090744490705024X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. 2009. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr 65: 582–601. 10.1107/S0907444909012098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay SK, Mackereth CD. 2020. Structural basis of UCUU RNA motif recognition by splicing factor RBM20. Nucleic Acids Res 48: 4538–4550. 10.1093/nar/gkaa168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Tempel W, Liu Z-J, Wang B-C, Rose RB. 2005. Structure of the conserved transcriptional repressor enhancer of rudimentary homolog. Biochemistry 44: 5017–5023. 10.1021/bi047785w [DOI] [PubMed] [Google Scholar]
- Wang G, Reinke V. 2008. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol 18: 861–867. 10.1016/j.cub.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weick E-M, Miska EA. 2014. piRNAs: from biogenesis to function. Development 141: 3458–3471. 10.1242/dev.094037 [DOI] [PubMed] [Google Scholar]
- Weick E-M, Sarkies P, Silva N, Chen RA, Moss SMM, Cording AC, Ahringer J, Martinez-Perez E, Miska EA. 2014. PRDE-1 is a nuclear factor essential for the biogenesis of Ruby motif-dependent piRNAs in C. elegans. Genes Dev 28: 783–796. 10.1101/gad.238105.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng MT, Luo J. 2013. The enigmatic ERH protein: its role in cell cycle, RNA splicing and cancer. Protein Cell 4: 807–812. 10.1007/s13238-013-3056-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng C, Kosalka J, Berkyurek AC, Stempor P, Feng X, Mao H, Zeng C, Li W-J, Yan Y-H, Dong M-Q, et al. 2019. The USTC co-opts an ancient machinery to drive piRNA transcription in C. elegans. Genes Dev 33: 90–102. 10.1101/gad.319293.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CJ, Headd JJ, Moriarty NW, Prisant MG, Videau LL, Deis LN, Verma V, Keedy DA, Hintze BJ, Chen VB, et al. 2018. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci 27: 293–315. 10.1002/pro.3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson MP. 2013. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc 73: 1–16. 10.1016/j.pnmrs.2013.02.001 [DOI] [PubMed] [Google Scholar]
- Winter G, Lobley CMC, Prince SM. 2013. Decision making in xia2. Acta Crystallogr D Biol Crystallogr 69: 1260–1273. 10.1107/S0907444913015308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie G, Vo TV, Thillainadesan G, Holla S, Zhang B, Jiang Y, Lv M, Xu Z, Wang C, Balachandran V, et al. 2019. A conserved dimer interface connects ERH and YTH family proteins to promote gene silencing. Nat Commun 10: 251. 10.1038/s41467-018-08273-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER suite: protein structure and function prediction. Nat Methods 12: 7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Kim VN. 2020. A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing. Nat Rev Mol Cell Biol 21: 542–556. 10.1038/s41580-020-0246-8 [DOI] [PubMed] [Google Scholar]
- Zeng C, Weng C, Wang X, Yan YH, Li WJ, Xu D, Hong M, Liao S, Dong MQ, Feng X, et al. 2019. Functional proteomics identifies a PICS complex required for piRNA maturation and chromosome segregation. Cell Rep 27: 3561–3572.e3. 10.1016/j.celrep.2019.05.076 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.