In this study, Yagi et al. dissected the transcriptional and epigenetic dynamics of mouse fibroblasts undergoing reprogramming to either myotubes or iMPCs using a MyoD-inducible transgenic model. Their findings provide new insights into the role of MyoD in myogenic reprogramming and derive general principles by which transcription factors and signaling pathways cooperate to rewire cell identity.
Keywords: MyoD, transdifferentiation, dedifferentiation, induced myogenic progenitor cells (iMPCs), satellite cells, DNA methylation, epigenetic reprogramming
Abstract
The generation of myotubes from fibroblasts upon forced MyoD expression is a classic example of transcription factor-induced reprogramming. We recently discovered that additional modulation of signaling pathways with small molecules facilitates reprogramming to more primitive induced myogenic progenitor cells (iMPCs). Here, we dissected the transcriptional and epigenetic dynamics of mouse fibroblasts undergoing reprogramming to either myotubes or iMPCs using a MyoD-inducible transgenic model. Induction of MyoD in fibroblasts combined with small molecules generated Pax7+ iMPCs with high similarity to primary muscle stem cells. Analysis of intermediate stages of iMPC induction revealed that extinction of the fibroblast program preceded induction of the stem cell program. Moreover, key stem cell genes gained chromatin accessibility prior to their transcriptional activation, and these regions exhibited a marked loss of DNA methylation dependent on the Tet enzymes. In contrast, myotube generation was associated with few methylation changes, incomplete and unstable reprogramming, and an insensitivity to Tet depletion. Finally, we showed that MyoD's ability to bind to unique bHLH targets was crucial for generating iMPCs but dispensable for generating myotubes. Collectively, our analyses elucidate the role of MyoD in myogenic reprogramming and derive general principles by which transcription factors and signaling pathways cooperate to rewire cell identity.
Skeletal muscle is a tissue predominantly comprised of terminally differentiated, multinucleated myotubes responsible for motion (Yin et al. 2013; Comai and Tajbakhsh 2014; Almada and Wagers 2016). In addition, muscle contains rare mononucleated stem cells termed satellite cells, which are located in between the sarcolemma and basal lamina of myofibers. Satellite cells are maintained in a quiescent state under homeostatic conditions but undergo activation and cell division upon muscle damage or injury. Once activated, satellite cells produce proliferative progeny (myoblasts) that differentiate into myocytes and subsequently fuse with one another or with resident myofibers to restore tissue function. Satellite cells possess remarkable therapeutic potential. Studies in mice have shown that satellite cells can be isolated from healthy donors and transplanted into diseased or damaged recipient muscles, where they engraft, differentiate, and fuse to restore tissue function (Sherwood et al. 2004; Montarras et al. 2005; Cerletti et al. 2008; Sacco et al. 2008). While satellite cells can be propagated in vitro for short periods of time (Yin et al. 2013), they cannot be permanently maintained in culture, presenting a bottleneck for mechanistic studies and a hurdle for many possible therapeutic applications.
Muscle formation (myogenesis) from satellite cells is orchestrated by a cascade of transcription factors, including the paired homeodomain-containing protein Pax7 and the basic helix-loop-helix (bHLH)-containing myogenic regulatory factors (MRFs) (Brack and Rando 2012; Yin et al. 2013; Almada and Wagers 2016). Pax7 expression is restricted to satellite cells and is specifically required for their maintenance, while MRFs, including Myf5, MyoD, Myog, and Myf6 (MRF4), are activated at successive stages to drive myogenesis. Myf5, like Pax7, is expressed in quiescent and activated satellite cells, whereas MyoD protein is expressed in activated satellite cells and their progeny (i.e., myoblasts, myocytes, and myotubes). Accordingly, Myf5 and MyoD bind to shared sets of targets in myoblasts (Conerly et al. 2016), and they genetically compensate for one another during development (Rudnicki et al. 1993). Myog and Myf6 are expressed in myocytes committed toward terminal differentiation, with Myog being activated prior to Myf6 (Yin et al. 2013; Almada and Wagers 2016). Seminal experiments by Weintraub and others (Davis et al. 1987; Braun et al. 1989; Edmondson and Olson 1989; Rhodes and Konieczny 1989) elucidated the fate-instructive roles of MRFs following ectopic expression in heterologous cell types. Specifically, forced expression of MyoD and other MRFs was shown to be sufficient for the direct conversion of fibroblasts to myoblasts/myotubes, a process commonly referred to as “transdifferentiation.” While myogenic transdifferentiation has been a powerful tool to dissect the mechanisms by which MyoD rewires cell identity, the resultant myotubes are typically postmitotic and often incompletely reprogrammed (Manandhar et al. 2017; Cacchiarelli et al. 2018), consistent with observations in other transdifferentiation systems (Xu et al. 2015). In an effort to overcome these limitations, we recently showed that transient expression of MyoD, combined with exposure to particular small molecules generates induced myogenic progenitor cells (iMPCs) that share key hallmarks with satellite cells (Bar-Nur et al. 2018). Notably, the small molecules that facilitate the generation of iMPCs also facilitate the reprogramming of fibroblasts to iPSCs and include the cyclic AMP agonist forskolin (“F”), the TGF-β inhibitor RepSox (“R”), and the GSK3-β inhibitor CHIR99021 (“C”; collectively abbreviated as FRC) (Maherali and Hochedlinger 2009; Xu et al. 2013; Bar-Nur et al. 2018). Like satellite cells, iMPCs induced by MyoD + FRC express Pax7 in a subset of cells and require Pax7 for their maintenance; iMPCs spontaneously differentiate into contractile, multinucleated myotubes in vitro and regenerate myofibers in vivo when transplanted into dystrophic or wild-type mice; iMPCs also sustain myogenesis after repeated injury and populate the satellite cell niche in transplant recipients (Bar-Nur et al. 2018). In contrast to satellite cells, however, iMPCs can be extensively cultured in vitro without losing Pax7 expression or diminishing their myogenic potential. We refer to the particular lineage conversion process that generates iMPCs as “dedifferentiation” to indicate the gain in differentiation potential compared with the classical transdifferentiation paradigm.
Our recent discovery of iMPCs raises fundamental questions that are the basis for the present study. For example, it is unclear how the activity of a single transcription factor such as MyoD leads to a terminally differentiated cell type in one context (i.e., myocytes during transdifferentiation) and to an adult stem-like cell type in another context (i.e., iMPCs during dedifferentiation). Specifically, we lack insight into the molecular players and associated mechanisms that differentially rewire transcriptional and epigenetic patterns of fibroblasts toward either myocytes or iMPCs. Moreover, our understanding of the extent to which myotubes/iMPCs are stably reprogrammed and their resemblance to counterparts in vivo is incomplete. Addressing these questions is relevant not only for our basic understanding of transcription factor-dependent cell fate transitions but also for realizing the potential therapeutic utility of our reprogramming approach, as it may enable the capture of human muscle stem cell-like cells directly from fibroblasts in the future. By leveraging a novel MyoD-inducible transgenic model, we show here that MyoD and small molecules cooperatively reprogram cell identity toward a stable, self-renewing satellite cell-like state, whereas MyoD expression alone yields an unstable myogenic state. We also provide insights into the transcriptional, chromatin, and DNA methylation processes that underlie these profoundly different outcomes in cell fate, and propose strategies to produce stable, faithfully reprogrammed cell types in other transdifferentiation contexts.
Results
A versatile MyoD-inducible system for the study of muscle lineage reprogramming
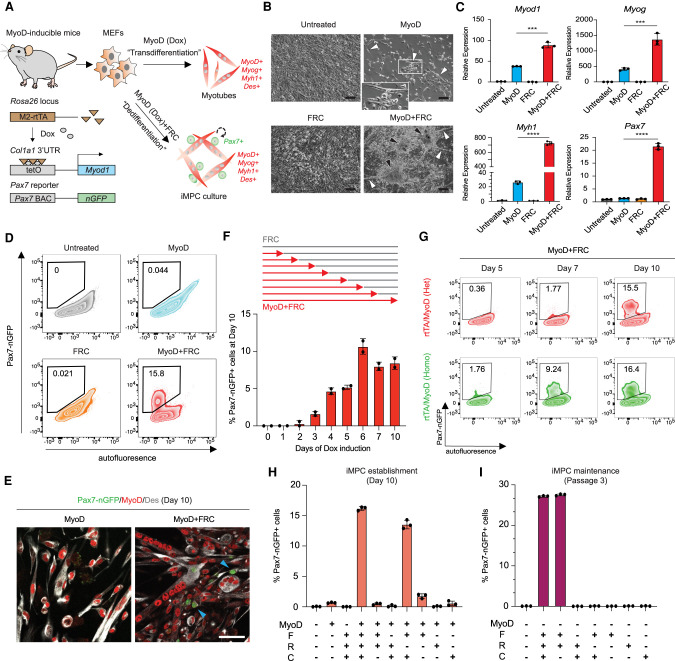
To dissect the mechanisms of myogenic reprogramming, we developed a novel Myod1-inducible transgenic model in embryonic stem cells (ESCs) and mice. Briefly, we introduced a single copy of Myod1 cDNA under the control of a doxycycline (Dox)-inducible promoter into the inert Col1a1 3′ UTR using site-specific recombination in KH2 ESCs (Hochedlinger et al. 2005). Correctly targeted ESCs were used to generate mice that were subsequently bred to animals harboring the Rosa26 promoter-driven M2rtTA Dox-dependent transactivator as well as a satellite cell-specific Pax7-nGFP reporter (Fig. 1A; Sambasivan et al. 2009). Offspring carrying Col1a1-tetO-Myod1, Rosa26-M2rtTA, and Pax7-nGFP were then used to derive murine embryonic fibroblasts (MEFs) and treated with Dox to induce transdifferentiation (referred to as the “MyoD” condition) or with Dox and FRC to induce dedifferentiation (referred to as the “MyoD + FRC” condition). MyoD protein was specifically and homogeneously induced in transgenic MEFs exposed to the MyoD condition (Supplemental Fig. S1A). We readily obtained multinucleated myotubes expressing Myog and Myh1 with the MyoD condition, while we obtained heterogeneous iMPC cultures expressing Myog, Myh1, and Pax7 with the MyoD + FRC condition (Fig. 1B,C; Supplemental Fig. S1B). Importantly, we also detected Pax7+ cells by immunofluorescence (IF) and flow cytometry in ∼10%–20% of cells exposed to the MyoD + FRC condition, but such cells were absent from the MyoD condition (Fig. 1D,E; Supplemental Fig. S1C). Exogenous MyoD expression was required for a minimum of 2 d to detect Pax7-nGFP+ cells by day 10, and the percentage of Pax7-nGFP+ cells further increased to ∼12% with 6 d of MyoD induction before plateauing (Fig. 1F). Intriguingly, Pax7-nGFP+ cells appeared earlier (around day 5) in cultures of MEFs carrying two copies of Col1a1-tetO-Myod1 and R26-M2rtTA compared with MEFs carrying only one copy (about day 7) (Fig. 1G; Supplemental Fig. S1D), indicating a dose-dependent effect of MyoD on dedifferentiation. Finally, we found that the cyclic AMP agonist forskolin (F) and the Gsk3 inhibitor CHIR99021 (C) were sufficient for the induction of iMPCs, whereas F and the Tgf-β inhibitor RepSox (R) were sufficient for the maintenance of iMPCs with our system, highlighting context-specific roles of Gsk3/Wnt and Tgf-β signaling (Fig. 1H,I). For consistency, we used FRC together with MEFs heterozygous for Col1a1-tetO-Myod1, Rosa26-M2rtTA, and Pax7-nGFP (referred to as MyoD-inducible MEFs) for experiments in the remainder of this study.
Figure 1.
A versatile MyoD-inducible system for the study of myogenic reprogramming. (A) Development of a doxycycline (Dox)-inducible MyoD transgenic system for studying myogenic reprogramming to myocytes and iMPCs. (B) Representative bright-field images of MEFs exposed to MyoD, FRC, or MyoD + FRC for 10 d. Scale bar, 100 µm. White arrowheads highlight myotubes; black arrowheads highlight iMPC colonies. (C) Quantitative RT-PCR analysis for myogenic markers after 10 d of exposure of MEFs to MyoD, FRC, or MyoD + FRC. Values normalized to untreated MEFs. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (***) P < 0.001, (****) P < 0.0001. (D) FACS analysis for Pax7-nGFP+ cells using MEFs exposed to MyoD, FRC, or MyoD + FRC for 10 d. (E) Immunofluorescence images showing expression of MyoD, Desmin (Des), and Pax7-nGFP in MEFs exposed for 10 d to MyoD or MyoD + FRC. Scale bar, 50 µm. Arrowheads indicate Pax7-nGFP+ cells. (F) Assay to determine temporal requirement of MyoD expression (Dox) to generate Pax7-nGFP+ iMPCs at day 10. Error bars indicate mean ± SD (n = 2). (G) Time-course FACS analysis for Pax7-nGFP+ cells using Rosa26-M2rtTA; Col1a1-tetO-MyoD (rtTA/MyoD) heterozygous (Het) and homozygous (Homo) MEFs exposed to Dox and FRC. (H,I) Requirement of small molecules for iMPC establishment (H) and maintenance (I). FACS analysis for Pax7-nGFP+ cells using transgenic MEFs exposed to the indicated small molecules for 10 d (H) or three passages (I). Error bars indicate mean ± SD (n = 3).
iMPCs share key transcriptional and epigenetic characteristics with satellite cells
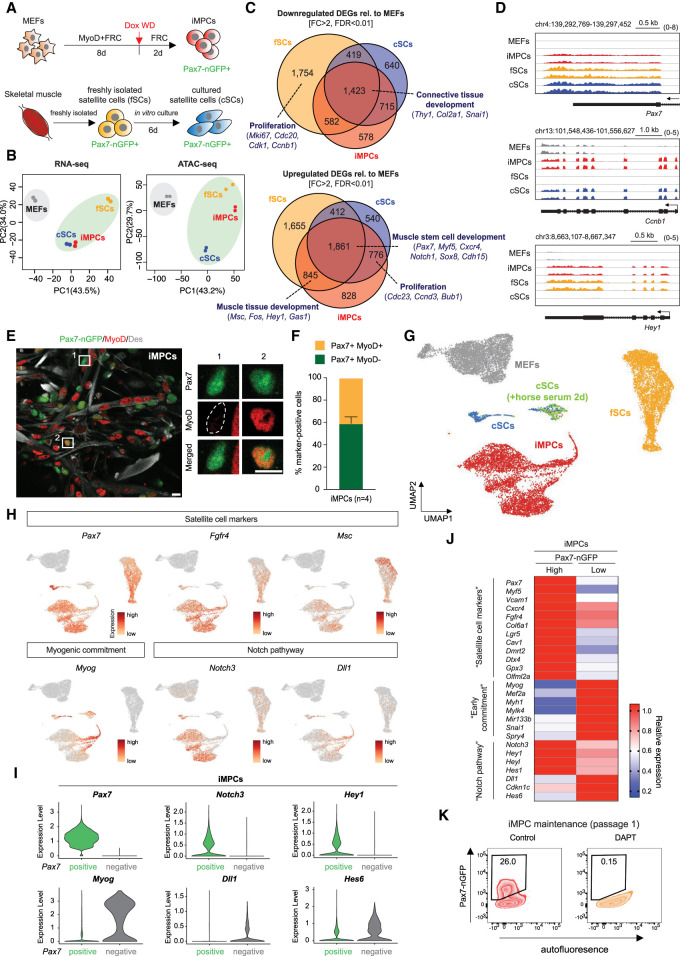
To determine the overall molecular similarity of iMPCs derived with our transgenic system compared with bona fide muscle stem cells, we performed RNA sequencing (RNA-seq) and ATAC sequencing (ATAC-seq) of Pax7-nGFP+ cells purified from established, Dox-independent iMPC cultures and primary muscle tissue (referred to as freshly isolated satellite cells [fSCs]) (Fig. 2A). Additionally, we cultured fSCs in vitro for 6 d before purifying Pax7-nGFP+ cells, which enabled activation and short-term expansion of satellite cells (referred to as cultured satellite cells [cSCs]) (Fig. 2A). Principal component analysis (PCA) revealed that Pax7-nGFP+ cells clustered away from MEFs, with iMPCs showing highest similarity to cSCs at the transcriptional level and to both fSCs and cSCs at the chromatin level (Fig. 2B). A comparison of differentially expressed genes (DEGs) between MEFs and either iMPCs, fSCs, or cSCs revealed a large group of commonly down-regulated genes (n = 1423) associated with fibroblast identity, including Thy1, Col2a1, and Snai1 as well as an equally large group of commonly up-regulated genes (n = 1861) associated with muscle stem cell identity, including Pax7, Myf5, and Myod1 (Fig. 2C,D; Supplemental Fig. S1E,F). Our ATAC-seq analysis of differentially accessible regions (DARs) between these samples yielded similar results as our RNA-seq/DEG analysis, with MEF-associated loci (e.g., Thy1, Runx1, and Col2a1) being closed and stem cell-associated loci (e.g., Pax7, Myf5, and Myod1) being opened in iMPCs, fSCs, and cSCs relative to MEFs (Supplemental Fig. S1G,H). Supporting the molecular similarity of iMPCs to fSCs and cSCs, we found that Pax7-nGFP+ iMPCs showed a forward/side scatter profile that overlapped with that of both fSCs and cSCs, and the Pax7-nGFP intensity of iMPCs was in between that of fSCs and cSCs (Supplemental Fig. S1I,J). Moreover, we showed by IF that ∼60% of mononucleated Pax7+ cells were MyoD−, which resembles phenotypic fSCs, while the remaining 40% were MyoD+, which resembles phenotypic cSCs (Fig. 2E,F). These results suggested that Pax7-nGFP+ iMPCs generated with our transgenic system acquired a transcriptional, epigenetic, and immunophenotypic state similar to muscle stem cells from adult mouse muscle.
Figure 2.
Pax7+ iMPCs share key characteristics with satellite cells. (A) Experimental outline to isolate Pax7-nGFP+ fresh SCs (fSCs), cultured SCs (cSCs), and iMPCs. (Dox WD) Dox withdrawal. (B) PCA of RNA-seq and ATAC-seq data for the indicated samples (n = 2 per sample). (C) Venn diagram showing the overlap of up-regulated and down-regulated differentially expressed genes (DEGs) when comparing MEFs with either iMPCs, fSCs, or cSCs (n = 2 per sample). (D) RNA-seq gene tracks of representative myogenic genes highlighted in C. (E) Immunofluorescence images showing expression of MyoD, Des, and Pax7-nGFP in established iMPCs maintained in FRC without exogenous MyoD (Dox). Scale bar, 10 µm. (F) Quantification of Pax7+/MyoD− and Pax7+/MyoD+ iMPC subsets shown in E. Error bars indicate mean ± SD (n = 4). (G) UMAP embedding of single-cell RNA-seq data for indicated samples. (H) Expression of representative genes using the same embedding as in G. (I) Expression levels of indicated genes in Pax7-nGFP+ iMPCs with (“positive”) or without (“negative”) endogenous Pax7 mRNA signal. (J) Heat map showing differentially expressed marker genes (RNA-seq) between Pax7-nGFPhigh (n = 2) and Pax7-nGFPlow (n = 2) iMPCs. (K) Effect of DAPT (Notch inhibitor) on iMPC maintenance.
To further define the heterogeneity of iMPCs relative to fSCs and cSCs, we performed single-cell RNA-seq analysis using the Chromium 10X platform. UMAP embedding of our samples suggested that iMPCs were overall closer to cSCs than to fSCs (Fig. 2G), consistent with our bulk RNA-seq data (Fig. 2B). Moreover, the majority of iMPCs expressed not only common fSC/cSC markers (e.g., Pax7, Fgfr4, and Cdh15) but also markers primarily associated with fSCs (e.g., Msc and Hey1) or cSCs (e.g., Ccnb1 and Cdc23), suggesting that iMPCs assume an intermediate state between fSCs and cSCs (Fig. 2H; Supplemental Fig. S1K). Of note, we detected small subpopulations of Pax7-nGFP+ iMPCs and cSCs that expressed the early commitment marker Myog, were closely aligned with differentiating cSCs (i.e., cSCs exposed to horse serum) in the UMAP embedding, and lacked endogenous expression of the satellite cell markers Pax7 and Msc. These observations indicated that Pax7-nGFP+ iMPCs, like Pax7-nGFP+ cSCs, contain primitive myogenic precursors as well as differentiating progeny, reflecting the various physiological stages of myogenesis (Fig. 2H). Intriguingly, we found that several components of the Notch signaling pathway previously implicated in satellite cell maintenance (Conboy et al. 2003; Mourikis et al. 2012) were differentially expressed between these two subsets of Pax7-nGFP+ iMPCs and cSCs. Specifically, Pax7-nGFP+ cells with endogenous Pax7 mRNA signal expressed the Notch receptor Notch3 and the canonical Notch target gene Hey1, while Pax7-nGFP+ cells lacking endogenous Pax7 mRNA signal expressed the Notch ligand Dll1 and the Notch inhibitor Hes6, which mirrored the expression pattern of Myog (Fig. 2H,I). We confirmed these results using RNA-seq analysis of FACS-purified Pax7-nGFPhigh and Pax7-nGFPlow cells from an established iMPC bulk culture (see Supplemental Fig. S1L for gating strategy). Indeed, Pax7-nGFPhigh cells expressed higher levels of muscle stem/progenitor cell-associated genes and Notch targets including Pax7, Myf5, Dmrt2 and Notch3, Hey1, Heyl, whereas Pax7-nGFPlow cells expressed higher levels of differentiation-associated genes and Notch ligands/Notch repressors including Myog, Mef2a, Mir-133b and Dll1, and Hes6 (Fig. 2J; Supplemental Fig. S1M). Accordingly, treatment of iMPCs with the pan-Notch inhibitor DAPT led to a complete loss of Pax7-nGFP+ cells, underscoring the functional relevance of our molecular observations (Fig. 2K). In addition to Notch components, we found that Lgr5, a potent mediator of Wnt/β-catenin signaling recently associated with injury-responsive satellite cells (Leung et al. 2020), was differentially regulated between Pax7-nGFPhigh and Pax7-nGFPlow cells (Fig. 2J). To further characterize Lgr5 expression in our reprogramming system, we derived iMPCs from MEFs carrying an Lgr5-GFP-DTR reporter (Supplemental Fig. S2A). Lgr5-GFP expression first became detectable at day 8 of dedifferentiation and persisted in 10%–15% of established iMPCs (Supplemental Fig. S2B,C). Strikingly, ablation of Lgr5-GFP+ cells in established iMPCs using diphtheria toxin (DT) treatment led to the loss of Pax7+ cells and a collapse of iMPC cultures (Supplemental Fig. S2D–G). Together, these results suggested that our iMPC cultures are maintained by a primitive group of Pax7+ cells that depend on Notch and Wnt/Lgr5 signaling.
Silencing of the MEF program precedes induction of the muscle stem cell program
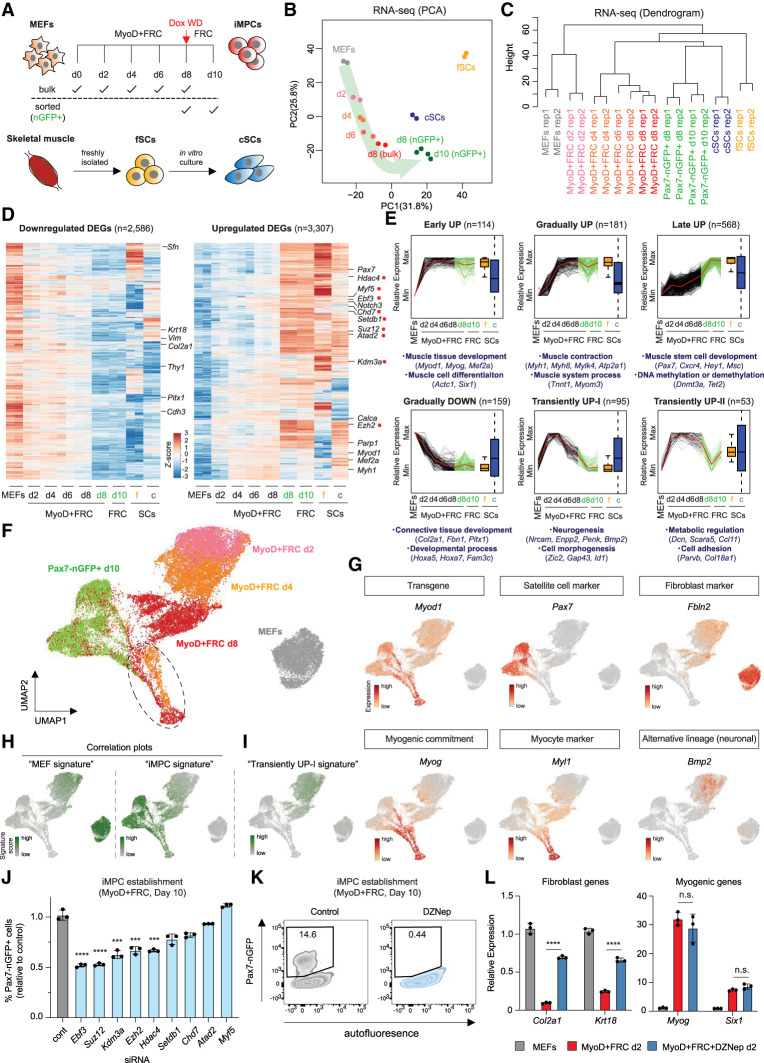
Having established fundamental similarities between iMPCs and native muscle stem and progenitor cells, we next sought to infer a molecular logic of dedifferentiation. We therefore treated our MyoD-inducible MEFs with Dox in the presence of FRC for 2, 4, 6, or 8 d before harvesting cells for RNA-seq (Fig. 3A). We isolated RNA from both bulk cultures as well as FACS-purified Pax7-nGFP+ cells at day 8. In addition, we isolated RNA from FACS-purified Pax7-nGFP+ cells following 8 d of Dox induction and 2 d of Dox withdrawal (day 10 time point) to assess the stability of transcriptional changes. PCA of these samples including fSCs and cSCs showed a progressive rewiring of transcriptional patterns from a MEF state toward a muscle stem cell-like state (Fig. 3B,C). Critically, transcriptional patterns of purified Pax7-nGFP+ cells at days 8 and 10 were closely aligned and most similar to cSCs, indicating that the induction of Pax7-nGFP signifies the acquisition of a stable muscle stem cell-like state regardless of the continuous presence of exogenous MyoD expression (Fig. 3B,C).
Figure 3.
PRC2-dependent, gradual silencing of the fibroblast program precedes late induction of the stem cell program. (A) Experimental outline to study transcriptional dynamics of dedifferentiation. Pax7-nGFP+ fSC, cSC, and iMPC (d10) samples are identical to those in Figure 2. (B) PCA of RNA-seq data for the indicated samples (n = 2 per sample). (C) Hierarchical clustering dendrogram of RNA-seq data for the indicated samples. (D) Heat maps of gene expression (z-scores of log2 transformed RPKM relative to all samples) for genes differentially expressed (fold change [FC] > 2, FDR < 0.01) in iMPCs (GFP+) and at least fSCs or cSCs compared with MEFs (n = 2 for each). (E) k-means clustering of temporal bulk gene expression profiles with representative GO terms and genes shown below. Gene expression was normalized and scaled from minimum to maximum among MEFs, reprogramming intermediates (d2–d8) and iMPCs. (F) UMAP embedding of single-cell RNA-seq data of MEFs, Pax7-nGFP+ iMPCs, and intermediates (Dox + FRC d2, d4, and d8). Dotted oval indicates Myog+ population common to days 4 and 8 and iMPCs. (G) Expression of representative genes using the same UMAP embedding as in F. (H) Expression of MEF signature genes (MEF pseudobulk) and iMPC signature genes (iMPC pseudobulk) using the same UMAP embedding as shown in F. (I) Expression of “transient UP (I)” gene signature using the same UMAP embedding. transient UP (I) genes (n = 95) were extracted from bulk RNA-seq data shown in E. (J) Quantification of Pax7-nGFP+ cells using MEFs exposed to MyoD + FRC and indicated siRNAs for 10 d. Targeted genes are highlighted with red dots in D. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (***) P < 0.001, (****) P < 0.0001. (K) Effect of DZNep (Ezh2 inhibitor) on iMPC maintenance. (L) Quantitative RT-PCR analysis for indicated fibroblast and myogenic markers after 2 d of exposure to MyoD + FRC with or without DZNep. Values normalized to MEFs. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (****) P < 0.0001, (n.s.) not significant.
We next dissected the dynamics with which the fibroblast program is extinguished and the myogenic stem cell program induced during dedifferentiation by focusing on DEGs that distinguish MEFs from muscle stem/progenitor cells (i.e., Pax7-nGFP+ iMPCs, fSCs, and cSCs). This analysis revealed a gradual down-regulation of fibroblast genes (e.g., Col2a1, Thy1, and Vim) between days 2 and 8, which became further suppressed in Pax7-nGFP+ iMPCs (Fig. 3D; Supplemental Fig. S3A). In contrast, we observed a late induction of muscle stem cell-associated genes (e.g., Pax7, Myf5, and Notch3) between days 4 and 8, which was again most robust in cells that activated Pax7-nGFP (Fig. 3D; Supplemental Fig. S3A,B). We used k-means clustering of the temporal gene expression profiles to refine the dynamics of transcriptional changes in an unbiased manner and to infer functionally relevant gene categories beyond MEF/iMPC-enriched genes. We observed groups of genes that were up-regulated early, gradually, or late in MEFs exposed to MyoD + FRC, and these were enriched for the GO categories “muscle tissue development” (e.g., Myog and Mef2a), “muscle contraction” (e.g., Myh1 and Atp2a1), and “muscle stem cell function” (e.g., Pax7 and Cxcr4), respectively (Fig. 3E). This observation suggested that muscle genes associated with structural and developmental functions are reprogrammed more readily than are muscle genes associated with stem cell function. Additionally, we observed a group of genes that was induced transiently between days 2 and 4 (transiently UP-I) with association to alternative nonmuscle and nonfibroblast lineages, particularly “neurogenesis” and “cell morphogenesis” (e.g., Snap25, and Nrcam) (Fig. 3E; Supplemental Fig. S3A,C). This finding is consistent with the previous observation that myogenic and neurogenic bHLH transcription factors bind to shared E-box targets (Fong et al. 2015; Lee et al. 2020). Another group of genes with transient expression between days 2 and 8 (transiently UP-II) was associated with the GO categories “cell adhesion” and “metabolic regulation” (e.g., Dcn, Lum, Ccl11, and Col18a1), pointing to a possible remodeling of the extracellular matrix and reprogramming of the metabolic state during dedifferentiation (Fig. 3E; Supplemental Fig. S3A,C).
To determine whether up-regulated genes associated with mature myocytes, muscle stem/progenitor cells, and alternative lineages (Fig. 3E) were expressed in the same or different cells, we performed single-cell RNA-seq of MyoD-inducible MEFs exposed to Dox and FRC for 2, 4, or 8 d. Consistent with our bulk RNA-seq data, we observed a progressive transition of intermediates from MEFs to iMPCs using UMAP embedding (Fig. 3F). Myod1 was homogeneously expressed in intermediates and iMPCs but absent in MEFs, whereas Pax7 was exclusively expressed in iMPCs, as expected (Fig. 3G). Of note, we detected subpopulations of cells on days 4, 8, and 10 that appeared similar based on UMAP, clustered away from the main populations and expressed markers of mature myocytes such as Myog and Myl1 (Fig. 3G; cells highlighted with dotted oval in Fig. 3F). This result suggested that while the majority of dedifferentiation intermediates captured with single-cell analysis progressed as relatively homogenous cell populations, subsets of cells acquired a mature muscle state. We suspect these cells represent either directly transdifferentiating myocytes or differentiated progeny of nascent iMPCs.
To assess how global transcriptional programs associated with MEFs and iMPCs change within single cells over time, we calculated pseudobulk MEF/iMPC expression signatures for each single cell across time points/samples during dedifferentiation (Fig. 3H). We found that the fibroblast program was down-regulated between days 2 and 4, while the muscle stem cell program was up-regulated between days 4 and 10, supporting the existence of a hierarchy of events that is critical for successful reprogramming. Intriguingly, genes associated with alternative lineages (i.e., transient UP-I genes) were broadly induced in intermediates around day 4, coinciding with the silencing of the MEF program and the early induction of the iMPC program (Fig. 3I). We hypothesize that this transient expression pattern might therefore reflect a transcriptionally more promiscuous state as part of the dedifferentiation process rather than the parallel emergence of alternative (e.g., neuronal) cell types.
Perturbation of candidate genes reveals a role of PRC2 in MEF program silencing
We next determined whether genes that dynamically change during dedifferentiation are required for the generation of iMPCs. We selected three transcription factors (i.e., Ebf3, Myf5, and Atad2) and six chromatin regulators (i.e., Suz12, Ezh2, Setdb1, Hdac4, Kdm3a, and Chd7) for further analysis as they were progressively up-regulated during dedifferentiation (Fig. 3D; Supplemental Fig. S3D). Suppression of five of these nine regulators (Ebf3, Suz12, Ezh2, Hdac4, and Kdm3a) with siRNAs significantly impaired the formation of Pax7-nGFP+ iMPCs, indicating they play a functional role during dedifferentiation (Fig. 3J; Supplemental Fig. S3E). The fact that siRNAs targeting both Ezh2 and Suz12, which are key components of the Polycomb repressive complex 2 (PRC2), scored in our assay implied that dedifferentiation may require PRC2 activity. In support of this notion, we found that fibroblast-associated genes that became down-regulated during dedifferentiation were enriched for binding of the Polycomb complexes PRC1 (e.g., Rnf2) and PRC2 (e.g., Ezh2, Suz12, and Jarid2) (Supplemental Fig. S3F). Moreover, the treatment of MEFs undergoing dedifferentiation with the Ezh2 inhibitor DZNep failed to produce any Pax7-nGFP+ cells, suggesting that PRC2 is required for the generation of iMPCs (Fig. 3K). Last, we found that the MEF markers Col2a1 and Krt18 failed to be effectively silenced, whereas the muscle markers Myog and Six1 remained unaffected in early (day 2) dedifferentiation intermediates exposed to DZNep (Fig. 3L). These experiments validated the functional importance of dynamically expressed transcription/chromatin factors during dedifferentiation and uncovered a critical role of PRC2 in silencing the MEF program.
Muscle stem cell loci gain chromatin accessibility prior to transcriptional activation
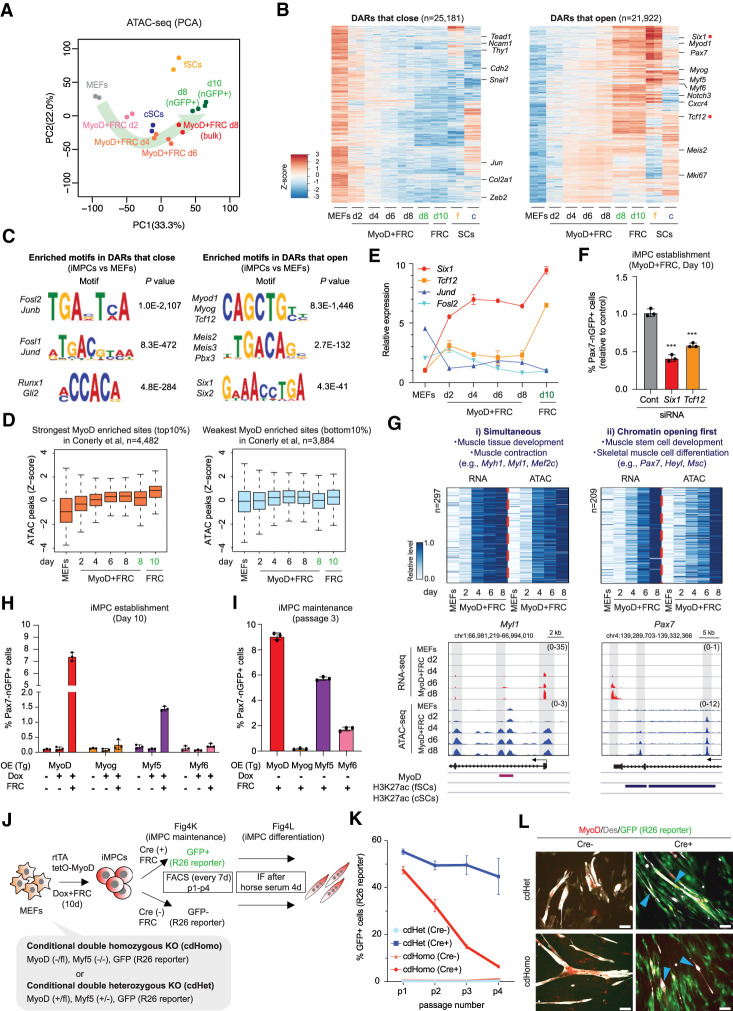
The dynamic changes in gene expression we observed during dedifferentiation implied a gradual remodeling of chromatin organization that is mediated by MyoD (Cao et al. 2010; Dall'Agnese et al. 2019) and possibly other transcription factors together with FRC. To explore this possibility, we performed chromatin accessibility and motif enrichment analyses during dedifferentiation using ATAC-seq analysis. Consistent with our transcriptional data, chromatin accessibility gradually changed from a fibroblast to an iMPC state with the most profound changes detected in Pax7-nGFP+ iMPCs (Fig. 4A,B; Supplemental Fig. S4A). Motifs enriched within differentially accessible regions (DARs) that closed in iMPCs relative to MEFs included transcription factor sites for AP-1 family members (Fosl1, Fosl2, JunB, and JunD) and Runx1 and Gli2 previously implicated in fibroblast identity (Fig. 4C; Chronis et al. 2017). In contrast, motifs enriched within DARs that opened in iMPCs relative to MEFs included the canonical myogenic regulatory factor (MRF) binding sequence (CAGCTG) as expected. Notably, MRF binding sites were also highly enriched in fSCs/cSCs based on ATAC-seq analysis, consistent with the reported ability of MRFs and Pax7 to open chromatin and bind to shared regulatory regions in myogenic cells (Fig. 4C; Supplemental Fig. S4B,C; Conerly et al. 2016; Lilja et al. 2017). When we compared our ATAC-seq data with a previously published ChIP-seq data set for MyoD (Conerly et al., 2016), we found that sites with the strongest MyoD enrichment (top 10%) led to a more significant increase in chromatin accessibility along our dedifferentiation time course than sites with the weakest MyoD enrichment (bottom 10%) (Fig. 4D; Supplemental Fig. S4D), confirming and extending the recent observation that MyoD binding strength directly correlates with the degree of chromatin opening (Lee et al. 2020). In addition to E-box motifs recognized by bHLH factors (i.e., MyoD, Myog, and Tcf12), we detected motifs recognized by the Pbx3/Meis2/Meis3 factors in iMPCs, which serve as cofactors for MyoD, as well as motifs for Six1/Six2 proteins previously implicated in myogenesis (Fig. 4C; Comai and Tajbakhsh 2014). Importantly, mRNAs for these muscle-associated transcription factors were up-regulated during dedifferentiation (Fig. 4E), and their knockdown with siRNAs impaired the formation of Pax7-nGFP+ iMPCs, suggesting functional relevance (Fig. 4F). These analyses confirm our hypothesis that dedifferentiation leads to a progressive reorganization of chromatin structure that appears to be largely driven by MRFs and related myogenic transcription factors such as Six1 and Tcf12.
Figure 4.
Chromatin opening at stem cell loci precedes their transcriptional activation. (A) PCA of ATAC-seq data for the indicated samples (n = 2 per sample). (B) Heat maps showing differentially accessible regions (DARs) that open or close (FC > 2, FDR < 0.01) in iMPCs (nGFP+) and at least fSCs or cSCs relative to MEFs (n = 2 for each). (C) Transcription factor motif analysis using DARs that open or close in iMPCs (Pax7-nGFP+) compared with MEFs. (D) Box plots of the levels of ATAC-seq signal (z-scores of log2 transformed RPKM relative to all samples) among subsets of MyoD-enriched sites with the strongest (top 10%, n = 4482) and the weakest (bottom 10%, n = 3884) MyoD occupancy calculated from public ChIP-seq data; two-tailed unpaired Student's t-test. (E) Expression levels of indicated genes during dedifferentiation (RNA-seq). Values for Six1 and Tcf12 are relative to MEFs, while values for Jund and Fosl2 are relative to iMPCs. Error bars indicate mean ± SD (n = 2). (F) FACS analysis of Pax7-nGFP+ cells using MyoD-inducible MEFs exposed to indicated siRNAs. Targeted genes are highlighted with red dots in B. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (***) P < 0.001. (G) Joint clustering analysis of RNA-seq and ATAC-seq dynamics for promoter regions (TSS ± 3 kb). RNA-seq expression (RPKM) and ATAC-seq signal (read density over TSS ± 3 kb) are normalized to the range between minimal and maximal levels for each gene. Representative gene tracks for each category are shown below. (H) Detection of Pax7-nGFP+ iMPCs using MEFs upon lentiviral overexpression of different MRFs. Error bars indicate mean ± SD (n = 3). (I) Detection of Pax7-nGFP+ cells in iMPCs (p3) derived from MRF-overexpressing MEFs. Error bars indicate mean ± SD (n = 3). (J) Experimental outline to determine the roles of Myf5 and Myod1 in iMPC establishment and maintenance. (R26 reporter) Cre-dependent Rosa26-GFP (R26NG) reporter. (K) Quantification of GFP+ cells in Cre-treated cdHomo and cdHet iMPCs maintained in FRC for three passages (p1–p4). Error bars indicate mean ± SD (n = 2). (L) Immunofluorescence images showing expression of MyoD, Des, and Rosa26-GFP after exposure of cdHomo and cdHet iMPCs (scale bar, 50 µm) to differentiation medium (DM; 2% horse serum) for 4 d following expansion in FRC for 6 d. Arrowheads for cdHomo iMPCs depict GFP−/Des+ myotubes. Arrowheads for cdHet iMPCs depict GFP+/Des+ myotubes.
To understand whether changes to chromatin accessibility are causal or consequential to transcriptional changes, we analyzed the relationship between the up-regulation of gene expression during dedifferentiation (see Fig. 3) and the gain of chromatin accessibility at the corresponding transcriptional start sites (TSSs) and distal “non-TSS” sites typically associated with enhancers. We defined genes that either increased expression and gained accessibility simultaneously or gained accessibility prior to increased expression (Fig. 4G; Supplemental Fig. S4E). Notably, canonical muscle regulators and structural muscle genes (e.g., GO term “muscle tissue development”) such as Mef2c, Myh1, and Myl1 increased expression simultaneously with changes to accessibility, pointing to immediate, direct effects of exogenous MyoD expression (Fig. 4G; Supplemental Fig. S4E). In contrast, muscle stem/progenitor cell-associated genes (e.g., GO term “muscle stem cell development”), including Pax7, Heyl, and Msc, gained accessibility prior to transcriptional activation, suggesting a gradual indirect process that is initiated by exogenous MyoD expression in combination with FRC (Fig. 4G; Supplemental Fig. S4E). Thus, our comparison of chromatin and transcriptional changes during dedifferentiation allowed us to classify responsive genes with different functional roles, with early responders being enriched for generic muscle function and late responders being enriched for stem cell function.
Distinct roles for MRFs in the induction and maintenance of iMPCs
Our observation that MRF recognition sequences were among the most enriched motifs in dedifferentiation intermediates and iMPCs using ATAC-seq analysis (Fig. 4C; Supplemental Fig. S4C) raised the question of whether MRFs other than MyoD are sufficient for the generation of iMPCs. Indeed, ectopic expression of each of the four MRFs (Myf5, MyoD, Myog, and Myf6) in MEFs gave rise to Myh1/Myog-expressing myotubes in the absence of FRC (Supplemental Fig. S4F,G), confirming and extending previous transdifferentiation results in immortalized fibroblasts (Davis et al. 1987; Braun et al. 1989; Edmondson and Olson 1989; Rhodes and Konieczny 1989). In the presence of FRC, expression of Myf5 and, to a lesser extent, expression of Myf6 also gave rise to transgene-independent Pax7-nGFP+ cells that could be propagated for at least three passages, although the fraction of Pax7-nGFP+ cells and the intensity of Pax7-nGFP signal remained lower than those of iMPCs generated with MyoD (Fig. 4H,I; Supplemental Fig. S4F,G; data not shown). Forced Myog expression was unable to generate Pax7-nGFP+ cells (Fig. 4H,I; Supplemental Fig. S4F,G). Unexpectedly, forced Pax7 expression also failed to give rise to either myocytes or iMPC-like colonies (Supplemental Fig. S4H). Thus, MRFs associated with both satellite cells and myoblasts (i.e., Myf5 and MyoD) are proficient in generating iMPCs, whereas MRFs associated with myogenic commitment and terminal differentiation (i.e., Myog and Myf6), as well as those exclusively associated with postnatal satellite cells (i.e., Pax7), are much less effective or deficient in generating phenotypic iMPCs.
To assess the requirement of MRFs for the induction and maintenance of iMPCs, we derived conditional double heterozygous (cdHet) Myf5+/−, Myod1+/fl control MEFs and conditional double-homozygous (cdHomo) Myf5−/−, Myod1−/fl experimental MEFs (Fig. 4J). To determine the functional role of Myf5/Myod1 in the establishment of iMPCs, we transduced cdHet and cdHomo MEFs with a Cre-expressing adenoviral vector as well as a Dox-inducible Myod1/rtTA-expressing vector to induce dedifferentiation. We detected normal levels of Pax7, Myh1, and Myog in cdHet + Cre and cdHomo + Cre cells after 10 d of dedifferentiation, indicating that endogenous Myf5 and Myod1 are not required for the acquisition of iMPCs (Supplemental Fig. S5A,B). To determine the functional role of Myf5/Myod1 in the maintenance of iMPCs, we treated established cdHet and cdHomo iMPCs with adenoviral Cre before measuring the fraction of Cre-recombined Rosa26-GFP reporter-positive (GFP+) cells over time (Fig. 4J). Notably, the fraction of GFP+ cdHomo + Cre experimental iMPCs (Fig. 4K, dark red line) progressively declined over three passages, while the fraction of GFP+ cdHet + Cre control iMPCs (Fig. 4K, dark blue line) remained stable over the same time period, indicating that both Myf5 and Myod1 are required for the maintenance of iMPCs (Fig. 4K). Indeed, reinduction of the lentiviral MyoD overexpression construct with Dox was sufficient to rescue the growth defect of cdHomo + Cre iMPCs (Supplemental Fig. S5C). Moreover, we found that GFP+ cdHomo + Cre iMPCs were enlarged compared with GFP+ cdHet + Cre iMPCs and lacked multinucleated myotubes and Desmin expression (Fig. 4L; Supplemental Fig. S5D–G), pointing to a severe differentiation defect that parallels Myf5/Myod1-deficient satellite cells (Yamamoto et al. 2018). In sum, our data defined distinct roles for endogenous MRFs in the establishment versus maintenance of iMPCs and uncovered additional functional similarities with satellite cells.
Active DNA demethylation by Tet enzymes is required for the generation of iMPCs
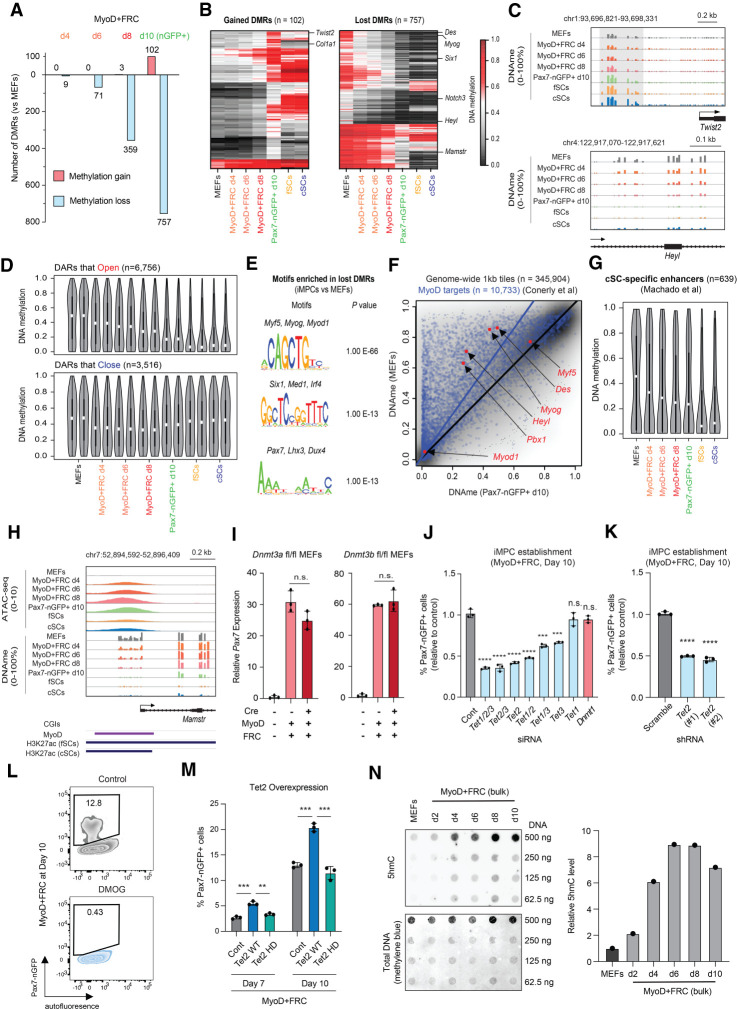
The regulatory regions of several MRFs, including Myf5 and Myog, undergo dynamic changes to DNA methylation during myogenesis, which correlates with gene expression (Carrió et al. 2015). These observations raised the question of whether MyoD-induced dedifferentiation involves changes to DNA methylation and whether DNA methylation itself is required for the generation of iMPCs. To measure methylation patterns during dedifferentiation, we performed reduced representation bisulfite sequencing (RRBS) of MEFs, Pax7-nGFP+ iMPCs, and intermediates (d4, d6, and d8) (Supplemental Fig. S6A). We observed a gradual and targeted loss of DNA methylation but extremely few gains in DNA methylation, which occurred almost exclusively late in dedifferentiation (Fig. 5A). Gained DMRs were nearby fibroblast-associated genes undergoing silencing such as Twist2 and Col1a1, while lost DMRs were nearby muscle-associated genes that underwent transcriptional activation such as Myog and Six1 as well as the Notch pathway components Notch3 and Heyl (Fig. 5B,C). Critically, these regions were also methylated and demethylated, respectively, in either fSCs or cSCs, suggesting they are physiologically relevant (Fig. 5B,C).
Figure 5.
Active DNA demethylation is required, while de novo methylation is dispensable for reprogramming to a Pax7+ state. (A) Number of differentially methylated regions (DMRs) that are gained or lost when comparing MEFs with the indicated time points using RRBS analysis (FDR < 0.01, CpGs > 8, and difference > 0.2). (B) Heat maps showing DMRs that are gained or lost between MEFs and iMPCs, with inclusion of fSC and cSC samples. (C) Genome browser tracks of representative DMRs shown in B. (D) Violin plots showing DNA methylation (DNAme) levels for CpGs located within DARs (differentially accessible regions) that open (n = 6756) or close (n = 3516) between MEFs and iMPCs. (E) Transcription factor motif enrichment for regions that gain accessibility and lose methylation (Pax7-nGFP+ d10 iMPCs vs. MEFs). (F) Scatter plot showing correlation between global DNAme (1-kb tiles, n = 345,904) and known MyoD targets (n = 10,733) when comparing MEFs (n = 2) with Pax7-nGFP+ iMPCs d10 (n = 2). (G) Violin plots for DNAme at CpGs located within cSC-specific enhancers (n = 639). (H) Genome browser track showing chromatin accessibility and DNAme changes at the Mamstr locus during dedifferentiation and in established iMPCs, fSCs, and cSCs. (I) Quantitative RT-PCR analysis for Pax7 in conditional Dnmt3a knockout (Dnmt3a fl/fl) or Dnmt3b knockout (Dnmt3b fl/fl) MEFs exposed to Cre and MyoD + FRC for 10 d. Values normalized to untreated control (no Cre, MyoD, or FRC). Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (n.s.) not significant. (J) Detection of Pax7-nGFP+ cells using MyoD-inducible MEFs treated with the indicated siRNAs. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (***) P < 0.001, (****) P < 0.0001, (n.s.) not significant. (K) Detection of Pax7-nGFP+ cells using MyoD-inducible MEFs transduced with two different shRNA vectors targeting Tet2 or a scrambled control. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (****) P < 0.0001. (L) Effect of DMOG (Tet inhibitor) on iMPC maintenance. (M) Detection of Pax7-nGFP+ cells using MyoD-inducible MEFs overexpressing a Tet2 WT or Tet2 HD (catalytic mutant) lentiviral construct. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (**) P < 0.01, (***) P < 0.001. (N, top) Dot blot analysis for global 5hmC levels in MEFs and dedifferentiation intermediates. (N, bottom) Values are normalized to methylene blue and MEFs (n = 1).
DMRs with reduced methylation in iMPCs relative to MEFs were associated with regions that gained chromatin accessibility but not with regions that lost chromatin accessibility (Fig. 5D; Supplemental Fig. S6B) and showed a parallel pattern in fSCs and cSCs, suggesting biological significance (Fig. 5D). Moreover, these regions were enriched for transcription factor motifs associated with myogenic stem and progenitor cells, including MyoD, Six1, and Pax7 (Fig. 5E). Indeed, we observed a significant enrichment for ChIP-seq-validated MyoD binding sites among DMRs with reduced methylation compared with all detected CpGs, including at the key myogenic genes Myf5, Heyl, and Pbx1 (Fig. 5F). Of note, DMRs that lost methylation and gained accessibility were also enriched for enhancers previously characterized in satellite cells (Fig. 5G; Supplemental Fig. S6C,D), as exemplified by the gene Mamstr (Fig. 5H). Thus, demethylation is enriched at open chromatin sites associated with MyoD binding and satellite cell-specific regulatory elements.
To determine whether methylation and demethylation are required for dedifferentiation, we assessed the expression and functional roles of enzymes responsible for de novo methylation (Dnmt3a and Dnmt3b), maintenance methylation (Dnmt1), and demethylation (Tet1, Tet2, and Tet3). Dnmt3a/Dnmt3b as well as Tet1/Tet2 and, to a lesser extent, Tet3 were transcriptionally up-regulated during dedifferentiation as well as in established iMPCs relative to MEFs (Supplemental Fig. S6E). However, neither Cre-mediated deletion of Dnmt3a or Dnmt3b nor siRNA-mediated knockdown of Dnmt1 had a discernable effect on the formation of iMPCs (Fig. 5I,J; Supplemental Fig. S6F–H). In contrast, knockdown of the Tet enzymes reduced the formation of Pax7-nGFP+ iMPCs by ∼50%–60% (Fig. 5J,K; Supplemental Fig. S6I), and treatment with the pan-Tet inhibitor DMOG (Amouroux et al. 2016) completely abrogated the formation of Pax7-nGFP+ iMPCs (Fig. 5L). Conversely, retroviral overexpression of catalytically active Tet2 significantly enhanced the generation of Pax7-nGFP+ iMPCs, while forced expression of a catalytically inactive Tet2 mutant had no effect (Fig. 5M; Supplemental Fig. S6J,K). Supporting the role of Tet enzymes in DNA demethylation, we observed a marked increase of global 5hmC levels between days 4 and 6 of dedifferentiation (Fig. 5N), which preceded the loss of 5mC levels between days 6 and 10 (Fig. 5A). These results showed that Tet-mediated demethylation is required and limiting for cellular dedifferentiation to iMPCs, while de novo methylation via Dnmt3a or Dnmt3b is dispensable for dedifferentiation.
Dedifferentiation and transdifferentiation follow different trajectories
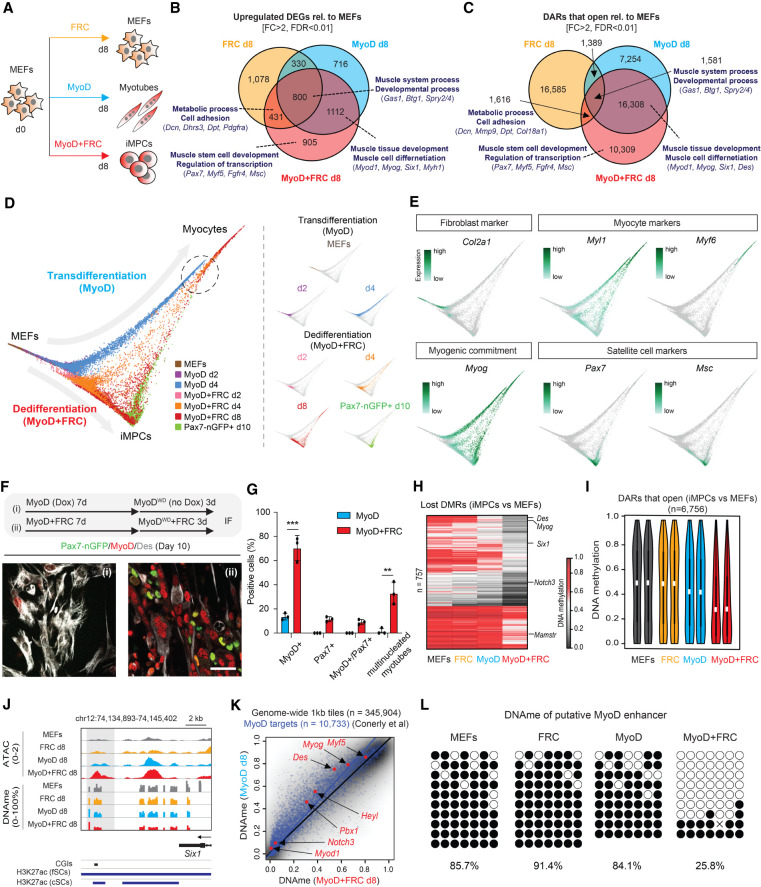
Key stem/progenitor cell markers such as Pax7 and Lgr5 were up-regulated in the MyoD + FRC condition but not in the MyoD or FRC conditions, suggesting that MyoD and FRC cooperatively rewire transcriptional and epigenetic patterns (Fig. 1; Supplemental Fig. S2). To further investigate this observation, we assessed the individual and shared contributions of MyoD, FRC, and MyoD + FRC to transcription, chromatin accessibility, and DNA methylation (Fig. 6A). Analysis of up-regulated DEGs and gained DARs between untreated MEFs and MEFs exposed to either MyoD, FRC, or MyoD + FRC suggested that MyoD's individual role is the induction of a myogenic program as predicted (e.g., GO terms “muscle tissue development” and “muscle cell differentiation”), while FRC's individual role appears to be the rewiring of cell adhesion and metabolism-related programs (e.g., GO terms “metabolic process” and “cell adhesion”). Importantly, muscle stem cell-specific genes such as Pax7, Myf5, and Fgfr4 were up-regulated and gained accessibility exclusively in the MyoD + FRC condition (Fig. 6B,C; Supplemental Fig. S7A), supporting cooperative effects between MyoD and FRC. Analogous to the exclusive up-regulation of muscle stem cell genes during dedifferentiation, we observed the exclusive down-regulation of certain fibroblast-associated genes including Tgfb1 and Junb in the MyoD + FRC condition (Supplemental Fig. S7A–C). Together, these results suggested that MyoD and FRC act in concert to more effectively silence the fibroblast program and specifically induce the muscle stem cell-like program.
Figure 6.
Dedifferentiation and transdifferentiation follow different trajectories due to cooperative effects of MyoD and FRC. (A) Experimental outline. (B) Venn diagram depicting the overlap of DEGs up-regulated in MEFs exposed to FRC, MyoD, or MyoD + FRC (n = 2 for each) for 8 d relative to untreated MEFs. (C) Venn diagram depicting the overlap of DARs that open in MEFs exposed to FRC, MyoD, or MyoD + FRC (n = 2 for each) for 8 d relative to untreated MEFs. Enriched GO terms (bold) and selected genes are highlighted. (D) Diffusion pseudotime (DPT) representation of single-cell RNA-seq data, comparing transdifferentiating (MyoD) and dedifferentiating (MyoD + FRC) cells. Data from MyoD + FRC cells are equivalent to those in Figure 3. (E) Expression of representative genes in the DPT representation shown in D. (F) Immunofluorescence analysis of MyoD, Des, and Pax7-nGFP expression in MyoD-inducible MEFs exposed to Dox ± FRC for 7 d before withdrawing Dox for another 3 d. Scale bar, 50 µm. (G) Quantification of MyoD+, Pax7+, and MyoD+/Pax7+ cells and multinucleated myotubes shown in F. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (**) P < 0.01, (***) P < 0.001. (H) Heat map showing DNAme levels for DMRs that are lost in iMPCs compared with MEFs (n = 757) with selected genes highlighted. (I) Violin plots showing DNAme levels at CpGs located within DARs that open in iMPCs relative to MEFs (n = 6756). (J) Gene tracks of the Six1 locus in MyoD, FRC, and MyoD + FRC cells with annotation of fSC/cSC enhancers below. (K) Analysis of DNAme and previously validated MyoD targets between MyoD (n = 2) and MyoD + FRC cells at day 8 (n = 2). (L) Bisulfite sequencing of putative MyoD enhancer in MEFs exposed to FRC, MyoD, or MyoD + FRC for 10 d.
To understand whether transdifferentiation and dedifferentiation are distinct processes or whether they share a common molecular trajectory, we performed single-cell RNA-seq of MEFs expressing MyoD and compared results with our previous single-cell data of MEFs expressing MyoD + FRC. When we compared all samples using either UMAP embedding (Supplemental Fig. S7D) or diffusion pseudotime (DPT) analysis (Fig. 6D), transdifferentiating and dedifferentiating cells formed a continuum of related cells within each condition that reflected the progression from MEFs toward either myocytes or iMPCs. Importantly, we observed no obvious overlap between these conditions except for a major fraction of d4 cells from the MyoD condition that aligned with minor fractions of d4 and d8 cells from the MyoD + FRC condition in the UMAP and DPT embeddings (Fig. 6D; Supplemental Fig. S7D, see dotted ovals). Inspection of myogenic markers revealed that these related cell populations were Myog+ myocytes present in both conditions (Fig. 6E; Supplemental Fig. S7E). However, Myog+ cells derived in the MyoD condition lacked expression of certain differentiation markers compared with Myog+ cells derived in the MyoD + FRC condition (e.g., Myf6, Myoz1, and Myh4), suggesting that dedifferentiation generates more mature muscle cells (Fig. 6E; Supplemental Fig. S7E). We also failed to detect expression of satellite cell markers among the transdifferentiating cells, thus ruling out that FRC selects for rare undifferentiated cells transiently generated by MyoD expression. Moreover, we found that perturbation of transcription, chromatin, and signaling molecules impacting dedifferentiation (i.e., Suz12, Ezh2, Ebf3, Kdm3a, Hdac4, and Tet2/3) (Fig. 3J) did not significantly impair transdifferentiation (Supplemental Fig. S7F,G). These findings support the conclusion that transdifferentiation and dedifferentiation represent largely distinct processes owing to the cooperative effects of MyoD and FRC.
iMPCs are stably reprogrammed compared with transdifferentiated myotubes
We reasoned that the different transcriptional and chromatin patterns of transdifferentiating and dedifferentiating intermediates might impact the stability of the final cell states (i.e., myocytes and iMPCs). We therefore induced MyoD with Dox for 7 d in the presence or absence of FRC and then withdrew Dox for another 3 d (referred to as MyoDWD and MyoDWD+FRC conditions) before scoring stably reprogrammed myogenic cells expressing endogenous MyoD, Desmin, and Pax7-nGFP. Unexpectedly, MyoDWD cultures failed to maintain endogenous MyoD expression in the vast majority of cells and reverted toward a fibroblast-like morphology despite continuous expression of Desmin (Fig. 6F,G). We obtained similar results when forcing MyoD cultures into terminal differentiation with horse serum prior to Dox withdrawal, thus excluding that the failure to efficiently activate endogenous Myod1 is due to our specific culture conditions (Supplemental Fig. S8A). In contrast, MyoDWD + FRC cultures contained multinucleated myotubes with robust expression of endogenous Myod1 and Desmin, and we consistently observed nearby Pax7+ cells (Fig. 6F,G; Supplemental Fig. S8A). Similarly, we detected endogenous MyoD expression in Myf5WD + FRC and Myf6WD + FRC cultures but not in MyogWD + FRC cultures (Supplemental Fig. S8B), mirroring the presence of Pax7-GFP+ cells in these conditions (Fig. 4H,I). Thus, MyoD + FRC-mediated dedifferentiation achieves a stable muscle stem cell-like state, while MyoD-mediated transdifferentiation generates an unstable state that depends on the continuous expression of exogenous MyoD.
To define possible mechanisms underlying these different cellular outcomes, we compared chromatin accessibility and DNA methylation patterns between the MyoD and MyoD + FRC conditions. Motif enrichment analysis of DARs detected by ATAC-seq in MyoD + FRC cells relative to MyoD cells revealed an overrepresentation of binding sites for the classical MRFs as well as for Msc, Tcf12, and Ascl2 previously implicated in muscle stem and progenitor cells (Supplemental Fig. S8C,D). Strikingly, we did not observe substantial changes in DNA methylation in MyoD-only and FRC-only cells, respectively, which is in contrast to the robust demethylation we detected in the MyoD + FRC cells and underscores the synergistic effects of MyoD and FRC on the epigenome (Fig. 6H). DMRs that distinguished MyoD + FRC cells from MyoD cells were enriched for demethylated sites with open chromatin as well as validated MyoD targets such as Myog, Des, and Myf5 (Fig. 6I–K; Supplemental Fig. S8E). Some of the regions exclusively demethylated in MyoD + FRC cells overlapped with previously mapped satellite cell enhancer elements for myogenic regulators including Six1 and the Myod1 gene itself, specifically the core enhancer (CE) ∼20 kb upstream of the TSS (Fig. 6J; Supplemental Fig. S8F; Goldhamer et al. 1995). Intriguingly, we identified another DMR ∼37 kb upstream of the Myod1 TSS that overlapped with the enhancer mark H3K27ac in satellite cells and was demethylated with open chromatin in Pax7-nGFP+ iMPCs, fSCs, and cSCs, suggesting it plays a physiological role (Supplemental Fig. S8F). We used bisulfite sequencing to confirm demethylation of this enhancer in the MyoD + FRC condition (Fig. 6L; Supplemental Fig. S8G). This enhancer was also demethylated in the Myf5 + FRC condition and to a lesser extent in the Myf6 + FRC and Myog + FRC conditions (Supplemental Fig. S8G), which correlated with the ability of these MRFs to generate Pax7-nGFP+ iMPCs (Fig. 4H,I). Moreover, FC treatment was minimally required for Myod1 enhancer demethylation and for genomic 5hmC up-regulation, highlighting a critical role of cAMP and GSK3/Wnt signaling in the demethylation process (Supplemental Fig. S8H,I). Indeed, motif enrichment analysis of the putative Myod1 enhancer revealed binding sites for the Wnt effector Tcf3 (Tcf7l1) in addition to canonical muscle stem/progenitor factors such as Pax7, Msc, and Sox8 (Supplemental Fig. S8J). Thus, our molecular comparison of MyoD + FRC and MyoD cells revealed potential effectors of dedifferentiation and provided a plausible epigenetic mechanism underlying the differential stability of transdifferentiated versus dedifferentiated cell states.
Genetic uncoupling of MyoD's potential for dedifferentiation vs. transdifferentiation
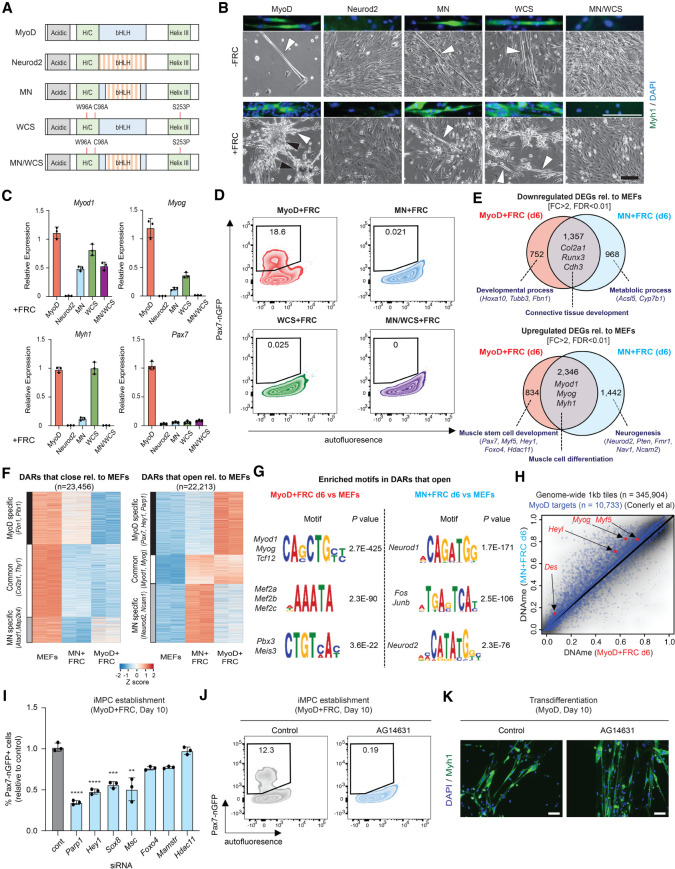
To determine which domains of MyoD are required for the induction of a stem cell program in one context but for the induction of a differentiated program in another context, we used MyoD mutants with perturbed DNA binding and/or transactivation potential (Fig. 7A; Fong et al. 2015). Briefly, we used the “MN” mutant in which MyoD's bHLH DNA-binding domain was replaced with that of the related neurogenic bHLH factor Neurod2; MN was previously shown to associate with shared MyoD/NeuroD2 targets but no longer with unique MyoD targets in P19 cells. The “WCS” mutant contains point mutations in MyoD's transactivation domains, which abrogate binding to the Pbx/Meis group of cofactors and impairs myogenic gene induction. “MN/WCS” is a combination of MN and WCS mutants and entirely fails to induce muscle genes in P19 cells. We lentivirally expressed each of these mutants in MEFs in the presence or absence of FRC (Supplemental Fig. S9A,B). We found that both the MN and WCS mutants supported the transdifferentiation of MEFs to myotubes expressing Myog and Myh1, although with lower efficiency than wild-type MyoD, whereas MN/WCS entirely failed to support transdifferentiation (Fig. 7B; Supplemental Fig. S9C,D). Critically, neither the MN nor the WCS mutant supported the dedifferentiation of MEFs to iMPCs as determined by flow cytometry for Pax7-nGFP and qPCR for endogenous Pax7 expression (Fig. 7C,D; Supplemental Fig. S9E). These results implied that MyoD's potential to associate with exclusive targets, specified by the MyoD-specific DNA and cofactor binding domains, is required for dedifferentiation to iMPCs but dispensable for transdifferentiation to myocytes/myotubes, thus uncoupling these two processes genetically.
Figure 7.
DNA and cofactor binding activity of MyoD uncouple its potential for dedifferentiation versus transdifferentiation. (A) Depiction of MyoD/Neurod2 mutants used in this experiment. (B) Representative bright-field images of MEFs expressing either MyoD, Neurod2, MN, WCS, or MNWCS for 10 d with or without FRC. Scale bar, 100 µm. Myotubes are highlighted by white arrowheads, and iMPC colonies are highlighted by black arrowheads. Immunofluorescence images (top of each bright-field image) show expression of Myh1. Scale bar, 100 µm. (C) Quantitative RT-PCR analysis for indicated genes in MEFs expressing MyoD, Neurod2, MN, WCS, or MNWCS for 10 d in the presence of FRC. Values are normalized to the MyoD + FRC condition. Error bars indicate mean ± SD (n = 3). (D) Detection of Pax7-nGFP+ cells using MEFs expressing MyoD, MN, WCS, or MNWCS for 10 d in the presence of FRC. (E) Venn diagram showing the overlap of down-regulated and up-regulated DEGs (FC > 2, FDR < 0.01) between MyoD + FRC and MN + FRC cells (day 6) relative to MEFs (n = 2 per sample). Enriched GO terms (bold) and selected genes are highlighted. (F) Heat maps showing DARs that close or open (FC > 2, FDR < 0.01) relative to MEFs in MyoD + FRC and MN + FRC cells at day 6 (n = 2 per sample). (G) Transcription factor motif enrichment analysis based on DARs that open in MyoD + FRC and MN + FRC cells relative to MEFs at day 6. (H) Analysis of DNAme and published MyoD targets (n = 10,733) between MyoD + FRC (n = 2) and MN + FRC (n = 2) cells at day 6. (I) Quantification of Pax7-nGFP+ cells using MyoD-inducible MEFs exposed to the indicated siRNAs. Error bars indicate mean ± SD (n = 3). Two-tailed unpaired Student's t-test: (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001. (J) Effect of AG14631 (Parp1 inhibitor) on iMPC induction. (K) Effect of AG14631 on transdifferentiation. Scale bars, 50 µm.
To define the molecular consequences and possible effectors of these MyoD variants, we compared the patterns of gene expression, chromatin accessibility, and DNA methylation between MEFs expressing MyoD versus MN in the presence of FRC. We detected a large fraction of DEGs that were commonly regulated between the two conditions relative to MEFs (Fig. 7E). These genes were associated with muscle cell identity for up-regulated DEGs and fibroblast identity for down-regulated DEGs, respectively (Fig. 7E). In contrast, genes specifically up-regulated in the MyoD + FRC condition were enriched for stem cell-associated GO terms and included the satellite cell regulators Pax7, Myf5, and Foxo4. Genes specifically up-regulated in the MN + FRC condition were enriched for neurogenesis-associated GO terms and included the neuronal regulators Ncam2, Neurod2, and Nav1 (Fig. 7E; Supplemental Fig. S9F). While chromatin accessibility patterns were overall similar to gene expression patterns, we found that as many as 48% of lost DARs were shared between the MyoD + FRC and MN + FRC conditions, while only 19% of gained DARs were shared (Fig. 7F; Supplemental Fig. S9G). This result suggested that fibroblast loci were efficiently closed regardless of the DNA binding specificity of MyoD, whereas the opening of muscle and neuronal loci, respectively, was more reliant on DNA binding specificity. Indeed, motifs enriched in MyoD + FRC cells were strongly associated with members of the MRF and Mef2 family (Fig. 7G; Supplemental Fig. S9H). In contrast, motifs enriched in MN + FRC cells were strongly associated with neuronal functions, including Neurod1/2 and AP-1 (Fos/Jun) (Fig. 7G). Accordingly, myogenic loci targeted by MyoD were more hypomethylated in the MyoD + FRC condition than in the MN + FRC condition (Fig. 7H). Together, these results showed that chromatin remodeling, DNA demethylation, and transcriptional activation of muscle stem cell-associated genes require the potential of MyoD to associate with its unique targets in the presence of FRC.
Finally, we compared our expression data of MyoD + FRC and MN + FRC cells to identify possible transcriptional and chromatin regulators critical for dedifferentiation. We identified seven candidates (Parp1, Hey1, Sox8, Msc, Foxo4, Mamstr, and Hdac11) that were differentially expressed between MyoD + FRC and MN + FRC cells (Supplemental Fig. S9I) and then performed siRNA-mediated knockdown during dedifferentiation. Suppression of three out of seven candidates (Parp1, Hey1, and Sox8) significantly impaired the formation of Pax7-nGFP+ iMPCs (Fig. 7I). While Hey1 and Sox8 were previously implicated in muscle stem or progenitor cells (Schmidt et al. 2003; Mourikis et al. 2012), the role of Parp1 in acquiring a muscle stem cell-like identity remains less well understood. Indeed, we observed that treatment with the Parp1 inhibitor AG-14631 completely blunted the generation of Pax7-nGFP+ cells, confirming that Parp1 is essential for dedifferentiation (Fig. 7J). Importantly, the suppression of Parp1, Hey1, or Sox8 did not have a discernible effect on transdifferentiation (Fig. 7K; Supplemental Fig. S9J). Thus, by comparing the transcriptional effects of MyoD and MN on the dedifferentiation process, we defined key downstream regulators required for dedifferentiation but dispensable for transdifferentiation.
Discussion
Our study provides fundamental insight into the transcriptional, epigenetic, and signaling events required for the conversion of a differentiated cell into a stable, self-renewing adult stem cell. Specifically, by dissecting the dedifferentiation of fibroblasts to iMPCs with a versatile new transgenic system, we have shown that (1) iMPCs share key molecular and functional characteristics with muscle stem/progenitor cells, including a requirement for Myf5/Myod1 and Notch signaling; (2) MyoD and FRC cooperate to gradually silence the fibroblast program before inducing the muscle stem cell-like program; (3) key stem cell-associated loci gain chromatin accessibility prior to transcriptional activation and preferentially undergo DNA demethylation; (4) Tet-dependent DNA demethylation of myogenic loci is required, while de novo methylation is dispensable for dedifferentiation; (5) dedifferentiation leads to a stable muscle stem cell-like state, whereas transdifferentiation leads to an unstable fibroblast/muscle hybrid state; and (6) DNA and cofactor binding activity of MyoD uncouples its potential for dedifferentiation versus transdifferentiation (Supplemental Fig. S10). Collectively, our data illuminate the instructive role MyoD plays in the induction of two distinct developmental states and provides a valuable resource for studying myogenesis in vitro.
An intriguing theme emerging from our analyses is that key mechanisms associated with the dedifferentiation of MEFs to iMPCs are shared with mechanisms associated with the reprogramming of somatic cells to iPSCs (Doege et al. 2012; Polo et al. 2012; Sardina et al. 2018; Schwarz et al. 2018). Specifically, our observation that dedifferentiation involves cell proliferation and requires 7–10 d until stem cell-associated genes are robustly activated parallels observation in iPSC reprogramming. Moreover, our finding that altered chromatin accessibility precedes transcriptional activation of crucial stem cell-associated loci and correlates with local DNA demethylation is another notable analogy to reprogramming and highlights the importance of epigenetic remodeling in both processes (Schwarz et al. 2018). Indeed, DNA demethylation via Tet enzymes is requiredwhile de novo methylation by Dnmt3a or Dnmt3b is dispensable for iMPC generation, as was previously shown for iPSC generation (Pawlak and Jaenisch 2011; Sardina et al. 2018). Last, we detected transient expression of adhesion, metabolic, and alternative lineage genes, particularly neuronal genes, during dedifferentiation, which is typically observed during iPSC reprogramming and may signify the acquisition of an epigenetically more plastic state (Polo et al. 2012; Schiebinger et al. 2019).
Our data suggest that MyoD endows fibroblasts with a myogenic fate that is poised to undergo terminal differentiation, while the presence of an appropriate signaling environment (i.e., FRC) diverts cell fate toward a more primitive myogenic state that undergoes continuous self-renewal and differentiation. While the precise contributions of FRC to the dedifferentiation process remain to be elucidated and are likely complex, it is noteworthy that R was dispensable for the induction of iMPCs, whereas C was dispensable for the maintenance of iMPCs. This observation implies that GSK3/Wnt signaling via C is more critical for the establishment of iMPCs, while Tgf-β signal inhibition via R is more critical for the maintenance of iMPCs, with cAMP signaling via F being important for both processes. Interestingly, studies of myogenic induction from human pluripotent cells likewise point to activation of both Wnt and cAMP signaling as key for the emergence of Pax7+ muscle progenitors (Xu et al. 2013; Chal et al. 2015). Thus, insights gained through our analyses will facilitate the future dissection of the role each pathway plays in the establishment and maintenance of mouse iMPCs and of ESC/iPSC-derived muscle precursors, and it may aid in the derivation of human iMPCs. Surprisingly, and in contrast to dedifferentiation, transdifferentiating MEFs failed to effectively maintain endogenous Myod1 expression and instead reverted to a hybrid fibroblast/muscle cell state upon discontinuation of exogenous MyoD expression. We ascribe these differences to the inability of exogenous MyoD to enable sufficient chromatin remodeling and demethylation of myogenic genes, including at the Myod1 locus itself. Thus, we hypothesize that effective remodeling of chromatin and DNA methylation patterns, as is the case during iPSC reprogramming (Apostolou and Hochedlinger 2013), is important for the induction of faithfully reprogrammed iMPCs with closer molecular and functional resemblance to primary satellite cells in vivo. This notion is consistent with our finding that iMPC-derived myotubes are more mature than transdifferentiated myotubes in terms of structure/function (i.e., contractile myotubes with more myonuclei) and expression of mature muscle markers. Finally, our MyoD domain swapping experiments allowed us to genetically uncouple MyoD's potential for transdifferentiation from its potential for dedifferentiation and define relevant downstream effectors. Our discovery that Parp1 was exclusively required for dedifferentiation is particularly intriguing in light of the fact that Parp1 collaborates with Tet2 during the demethylation of the Nanog promoter in iPSC reprogramming (Doege et al. 2012). We therefore speculate that, analogous to iPSC reprogramming, Parp1 may interact with Tet2 to rewire satellite cell-associated genes and facilitate the acquisition of a stable iMPC state. It will be important to test whether MyoD itself targets Tets and/or Parp1 to relevant targets specifically during dedifferentiation.
Our study not only illuminates the mechanisms underlying myogenic transdifferentiation and dedifferentiation but also may inform other direct lineage conversion approaches that are currently hampered by incomplete reprogramming. Specifically, ineffective silencing of the starting cell transcriptome and incomplete induction of the target cell transcriptome have been observed across alternative transdifferentiation paradigms such as MEF/hepatocyte-to-neuron (Marro et al. 2011; Treutlein et al. 2016), fibroblast-to-hepatocyte, and B cell or MEF-to-macrophage conversion (Feng et al. 2008; Morris et al. 2014). Thus, we propose that the modulation of pathways previously shown to facilitate the generation of iPSCs from somatic cells, including FRC (Maherali and Hochedlinger 2009; Bar-Nur et al. 2018), will facilitate the generation of more faithfully reprogrammed, stable cell types using conventional transdifferentiation approaches.
Materials and methods
Plasmid generation and cloning
The coding sequence for Myod1 was amplified using Phusion polymerase (Thermo Fisher Scientific). The resulting cDNA fragment was subjected to A-tailing with Taq polymerase and inserted into pCR2.1 (Thermo Fisher Scientific) via TOPO-TA cloning. The plasmid was sequence verified, and the coding sequence for Myod1 was subcloned into pBS31 using EcoRI to generate pBS31-Myod1 (New England Biolabs). Plasmids of pRRLSIN-Myod1, Neurod2, MN, and MNWCS, as well as pCS-WCS, were gifts from Dr. Stephen Tapscott (Fong et al. 2015). To generate pRRLSIN-WCS, the pRRLSIN-MyoD and pCS-WCS vectors were digested with PmI1 and Sbf1-HF (New England Biolabs) and fragments ligated using T4 ligase (New England Biolabs). Retroviral plasmids of pMMLV-Tet2 WT or Tet2 HD were gifts from Dr. Jose Luis Sardina and Dr. Thomas Graf (Di Stefano et al. 2014; Sardina et al. 2018).
ES cell culture and gene targeting
KH2 mouse embryonic stem cells were cultured on irradiated mouse embryonic fibroblasts in growth media (KO-DMEM [Life Technologies], 15% fetal bovine serum [FBS; Hyclone], 2 mM L-glutamine [Life Technologies], 100 U/mL penicillin, 100 µg/mL streptomycin [Life Technologies], 1× MEM nonessential amino acid solution [Life Technologies], 50 µM β-mercaptoethanol [Life Technologies]). For targeting, 40 µg of pCAGGS-flpE-puro and 40 µg of pBS31-Myod1 were added to the resuspended ES cell pellet and subjected to electroporation. The cells were then seeded at clonal density onto DR4 irradiated mouse embryonic fibroblasts (GlobalStem) before initiating selection with 200 µg/mL hygromycin (Gibco). Individual colonies were selected, expanded, and genotyped via PCR.
Blastocyst injection and generation of transgenic mice
To generate transgenic mice, MyoD-inducible KH2 ES cells were injected into E3.5 blastocysts as previously described (Nagy et al. 1993; Eggan et al. 2001). High-grade chimeras were crossed to B6/C57 wild-type mice, and agouti offspring were genotyped to confirm germline transmission. These mice were then bred to M2-rtTA mice (B6/C57 background) and subsequently to Pax7-nGFP mice (Sambasivan et al. 2009). Lgr5-GFP-DTR mice (Tian et al. 2011) were obtained from Dr. Frederic J. de Sauvage. Mice used in this study were housed and bred in specific pathogen-free (SPF) rooms located in the AAALAC-accredited Center for Comparative Medicine vivarium at Massachusetts General Hospital. All procedures involving mice adhered to the guidelines of the approved Massachusetts General Hospital Institutional Animal Care and Use Committee (IACUC) protocol number 2006N000104.
Mouse embryonic fibroblasts (MEFs)
Embryos carrying the tetO-MyoD, R26-rtTA, and Pax7-nGFP alleles were harvested at E13.5; the heads and internal organs were removed; and the remaining tissue was chopped and dissociated with trypsin to isolate MEFs. MEFs were cultured in DMEM supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 1× MEM nonessential amino acid solution, 50 µM β-mercaptoethanol, and 10% FBS. MEFs were expanded to passages 1–2 prior to transdifferentiation or dedifferentiation.
Transdifferentiation (MEFs to myotubes)
MyoD-inducible MEFs were seeded on plates at different densities (six-well plates: 1 × 105 cells/well; 12-well plates: 4 × 104 cells/well; 24-well plates: 2 × 104 cells/well; µ-Slide eight-well [Ibidi]: 1 × 104 cells/well). Transdifferentiation experiments were performed in KO-DMEM supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 1× MEM nonessential amino acid solution, 50 µM β-mercaptoethanol, 10% FBS, 10% knockout serum replacement (KSR; Life Technologies), 10 ng/mL bFGF (Peprotech), and 2 µg/mL doxycycline (Dox; Sigma).
Dedifferentiation (MEFs to iMPCs)
Dedifferentiation experiments were performed in KO-DMEM supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 1× MEM nonessential amino acid solution, 50 µM β-mercaptoethanol, 10% FBS, 10% KSR, 10 ng/mL bFGF, 2 µg/mL Dox, 5 µM forskolin (Sigma), 5 µM Repsox (Sigma), and 3 µM CHIR99021 (Tocris).
Isolation of freshly isolated satellite cells (fSCs) and cultured satellite cells (cSCs)
fSCs were isolated from 8–10 wk age of mice as previously described (Maesner et al. 2016). For cSCs, fSCs were seeded on 96-well plated coated with 1 µg/mL collagen type I from rat tail (Sigma) and 10 µg/mL mouse Laminin (Invitrogen) and were cultured for 6 d in F10 (Gibco) supplemented with 20% horse serum (Gibco), 100 U/mL penicillin, 100 µg/mL streptomycin, and GlutaMAX (Thermo). bFGF (5 ng/mL; Sigma) was added daily.
Differentiation of cSCs to mature myotubes
To induce differentiation of cSCs into mature myotubes, cells were cultured in DMEM supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, 1× MEM nonessential amino acid solution, and 2% horse serum.
RNA-seq
RNA-seq libraries were constructed from polyadenosine (polyA)-selected RNA using NEBNext Ultra directional RNA library prep kit for Illumina (New England BioLabs). Libraries were amplified for 14 cycles and validated using a 2200 TapeStation system and high sensitivity D1000 ScreenTape kit. Libraries were quantified using the Kapa Biosystems library quantification kit (KK4828) and the Bio-Rad CFX96 instrument. These pools were then denatured to 16 pM with 1% PhiX and sequenced on a Illumina HiSeq2000 instrument.
Reduced representation bisulfite sequencing (RRBS)
Genomic DNA was quantified using a Qubit 3.0 fluorometer. RRBS was performed on 100 ng of each sample using the NuGEN Ovation RRBS methyl-seq system following the manufacturer's recommendations. Bisulfite conversion of DNA was conducted using the Qiagen EpiTect fast bisulfite conversion kit. Libraries were purified with Agencourt RNAclean XP beads and quality-assessed using an Agilent 4150 TapeStation D1000 ScreenTape. Libraries were then sequenced on the Illumina NovaSeq6000, generating 100-bp single-end reads.
ATAC-seq
ATAC-seq was performed as previously described (Buenrostro et al. 2013). Sixty-thousand cells were washed once with 100 mL of PBS and resuspended in 50 mL of lysis buffer (10 mM Tris-HCl at pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.2% IGEPAL CA-630). The suspension of nuclei was then centrifuged at 500g for 10 min at 4°C, followed by the addition of 50 mL of transposition reaction mix (25 mL of TD buffer, 2.5 mL of Tn5 transposase, 22.5 mL of nuclease-free H2O) and incubation for 30 min at 37°C. DNA was isolated using MiniElute kit (Qiagen). Libraries were amplified by PCR (13 cycles). After the PCR reaction, the library was selected for fragments between 100 and 1000 bp with AMPure XP beads (Beckman Coulter). Libraries were purified with Qiaquick PCR (Qiagen) and integrity-checked on a Bioanalyzer before sequencing.
Single-cell RNA-seq (scRNA-seq)
Cells (1000 cells/µL) were resuspended with PBS and were loaded into a Chromium 10× system. scRNA-seq libraries were generated from approximately 5000 cells per sample using a Chromium 10× instrument with version 3.0 kit.
Method details are provided in the Supplemental Material.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank Michael Rundnicki for sharing Pax7-nGFP mice and the FUW-tetO-Pax7 plasmid; Stephen J Tapscott for sharing the pRRLSIN-MyoD, Neurod2, MN, MNWCS, and pCS-WCS vectors; Jose L. Sardina and Thomas Graf for sharing Tet2 WT and HD retroviral plasmids; the Massachusetts General Hospital next-generation sequencing facility and the Harvard Stem Cell Institute flow cytometry core for technical assistance; and Jiho Choi for initial help with the project. M. Yagi was supported by a Uehara Memorial Foundation Research Fellowship, a Mochida Memorial Foundation Research Fellowship, and a Japan Society for the Promotion of Science Overseas Research Fellowship. D.J.G. received support from the National Institutes of Health (NIH; R01AR076394). N.H. was supported by an NIH training grant (T32 DK007260). A.E.A. was supported by an NIH diversity supplement award (3R01AG048917-02S1) and a postdoctoral enrichment award from the Burroughs Wellcome Fund. A.J.W. was supported by funds from the Glenn Foundation for Medical Research and the NIH (R01 AG048917 and DP1 AG063419). Support from the Dana-Farber Cancer Institute's Center for Cancer Evolution and Physical Sciences-Oncology Center (U54CA193461 to F.M.) is gratefully acknowledged. R.I.S. was supported by a P30 DK040561 grant from the NIH. K.H. was supported by funds from Massachusetts General Hospital, the NIH (R01 HD058013, R01AR077695, and P01 GM099134), and the Gerald and Darlene Jordan Chair in Regenerative Medicine.
Author contributions: M. Yagi and K.H. conceived the study and wrote the manuscript. M. Yagi, B.D.S., N.T., A.J.H., O.B.-N., and M.S.H. performed experiments and analyzed data. F.J. and R.I.S. analyzed the RNA-seq and ATAC-seq data. J.C. and A.M. performed and analyzed the RRBS experiments. S.C. and F.M. analyzed scRNA-seq data. K.M., N.H, A.E.A., and A.J.W. provided advice on experimental design and assisted with the sorting of satellite cells. M. Yamamoto, A.P., and D.J.G. provided Myod1/Myf5 conditional knockout (cdHet and cdHomo) MEFs.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348678.121.
Freely available online through the Genes & Development Open Access option.
References
- Almada AE, Wagers AJ. 2016. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat Rev Mol Cell Biol 17: 267–279. 10.1038/nrm.2016.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, et al. 2016. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol 18: 225–233. 10.1038/ncb3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. 2013. Chromatin dynamics during cellular reprogramming. Nature 502: 462–471. 10.1038/nature12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nur O, Gerli MFM, Di Stefano B, Almada AE, Galvin A, Coffey A, Huebner AJ, Feige P, Verheul C, Cheung P, et al. 2018. Direct reprogramming of mouse fibroblasts into functional skeletal muscle progenitors. Stem Cell Reports 10: 1505–1521. 10.1016/j.stemcr.2018.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brack AS, Rando TA. 2012. Tissue-specific stem cells: lessons from the skeletal muscle satellite cell. Cell Stem Cell 10: 504–514. 10.1016/j.stem.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Buschhausen-Denker G, Bober E, Tannich E, Arnold HH. 1989. A novel human muscle factor related to but distinct from MyoD1 induces myogenic conversion in 10T1/2 fibroblasts. EMBO J 8: 701–709. 10.1002/j.1460-2075.1989.tb03429.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacchiarelli D, Qiu X, Srivatsan S, Manfredi A, Ziller M, Overbey E, Grimaldi A, Grimsby J, Pokharel P, Livak KJ, et al. 2018. Aligning single-cell developmental and reprogramming trajectories identifies molecular determinants of myogenic reprogramming outcome. Cell Syst 7: 258–268.e3. 10.1016/j.cels.2018.07.006 [DOI] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18: 662–674. 10.1016/j.devcel.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrió E, Díez-Villanueva A, Lois S, Mallona I, Cases I, Forn M, Peinado MA, Suelves M. 2015. Deconstruction of DNA methylation patterns during myogenesis reveals specific epigenetic events in the establishment of the skeletal muscle lineage. Stem Cells 33: 2025–2036. 10.1002/stem.1998 [DOI] [PubMed] [Google Scholar]
- Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, Wagers AJ. 2008. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell 134: 37–47. 10.1016/j.cell.2008.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. 2015. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol 33: 962–969. 10.1038/nbt.3297 [DOI] [PubMed] [Google Scholar]
- Chronis C, Fiziev P, Papp B, Butz S, Bonora G, Sabri S, Ernst J, Plath K. 2017. Cooperative binding of transcription factors orchestrates reprogramming. Cell 168: 442–459.e20. 10.1016/j.cell.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai G, Tajbakhsh S. 2014. Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol 110: 1–73. 10.1016/B978-0-12-405943-6.00001-4 [DOI] [PubMed] [Google Scholar]
- Conboy IM, Conboy MJ, Smythe GM, Rando TA. 2003. Notch-mediated restoration of regenerative potential to aged muscle. Science 302: 1575–1577. 10.1126/science.1087573 [DOI] [PubMed] [Google Scholar]
- Conerly ML, Yao Z, Zhong JW, Groudine M, Tapscott SJ. 2016. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev Cell 36: 375–385. 10.1016/j.devcel.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall'Agnese A, Caputo L, Nicoletti C, di Iulio J, Schmitt A, Gatto S, Diao Y, Ye Z, Forcato M, Perera R, et al. 2019. Transcription factor-directed re-wiring of chromatin architecture for somatic cell nuclear reprogramming toward trans-differentiation. Mol Cell 76: 453–472.e8. 10.1016/j.molcel.2019.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000. 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- Di Stefano B, Sardina JL, van Oevelen C, Collombet S, Kallin EM, Vicent GP, Lu J, Thieffry D, Beato M, Graf T. 2014. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 506: 235–239. 10.1038/nature12885 [DOI] [PubMed] [Google Scholar]
- Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. 2012. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488: 652–655. 10.1038/nature11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Olson EN. 1989. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev 3: 628–640. 10.1101/gad.3.5.628 [DOI] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM 3rd, Yanagimachi R, Jaenisch R. 2001. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci 98: 6209–6214. 10.1073/pnas.101118898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. 2008. PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci 105: 6057–6062. 10.1073/pnas.0711961105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong AP, Yao Z, Zhong JW, Johnson NM, Farr GH 3rd, Maves L, Tapscott SJ. 2015. Conversion of MyoD to a neurogenic factor: binding site specificity determines lineage. Cell Rep 10: 1937–1946. 10.1016/j.celrep.2015.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP Jr . 1995. Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development 121: 637–649. 10.1242/dev.121.3.637 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. 2005. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121: 465–477. 10.1016/j.cell.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Lee QY, Mall M, Chanda S, Zhou B, Sharma KS, Schaukowitch K, Adrian-Segarra JM, Grieder SD, Kareta MS, Wapinski OL, et al. 2020. Pro-neuronal activity of Myod1 due to promiscuous binding to neuronal genes. Nat Cell Biol 22: 401–411. 10.1038/s41556-020-0490-3 [DOI] [PubMed] [Google Scholar]
- Leung C, Murad KBA, Tan ALT, Yada S, Sagiraju S, Bode PK, Barker N. 2020. Lgr5 marks adult progenitor cells contributing to skeletal muscle regeneration and sarcoma formation. Cell Rep 33: 108535. 10.1016/j.celrep.2020.108535 [DOI] [PubMed] [Google Scholar]
- Lilja KC, Zhang N, Magli A, Gunduz V, Bowman CJ, Arpke RW, Darabi R, Kyba M, Perlingeiro R, Dynlacht BD. 2017. Pax7 remodels the chromatin landscape in skeletal muscle stem cells. PLoS One 12: e0176190. 10.1371/journal.pone.0176190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maesner CC, Almada AE, Wagers AJ. 2016. Established cell surface markers efficiently isolate highly overlapping populations of skeletal muscle satellite cells by fluorescence-activated cell sorting. Skelet Muscle 6: 35. 10.1186/s13395-016-0106-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Hochedlinger K. 2009. Tgfβ signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol 19: 1718–1723. 10.1016/j.cub.2009.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar D, Song L, Kabadi A, Kwon JB, Edsall LE, Ehrlich M, Tsumagari K, Gersbach CA, Crawford GE, Gordân R. 2017. Incomplete MyoD-induced transdifferentiation is associated with chromatin remodeling deficiencies. Nucleic Acids Res 45: 11684–11699. 10.1093/nar/gkx773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Südhof TC, Wernig M. 2011. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9: 374–382. 10.1016/j.stem.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montarras D, Morgan J, Collins C, Relaix F, Zaffran S, Cumano A, Partridge T, Buckingham M. 2005. Direct isolation of satellite cells for skeletal muscle regeneration. Science 309: 2064–2067. 10.1126/science.1114758 [DOI] [PubMed] [Google Scholar]
- Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. 2014. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 158: 889–902. 10.1016/j.cell.2014.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, Tajbakhsh S. 2012. A critical requirement for Notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30: 243–252. 10.1002/stem.775 [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. 1993. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci 90: 8424–8428. 10.1073/pnas.90.18.8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak M, Jaenisch R. 2011. De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state. Genes Dev 25: 1035–1040. 10.1101/gad.2039011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. 2012. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151: 1617–1632. 10.1016/j.cell.2012.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes SJ, Konieczny SF. 1989. Identification of MRF4: a new member of the muscle regulatory factor gene family. Genes Dev 3: 2050–2061. 10.1101/gad.3.12b.2050 [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75: 1351–1359. 10.1016/0092-8674(93)90621-V [DOI] [PubMed] [Google Scholar]
- Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. 2008. Self-renewal and expansion of single transplanted muscle stem cells. Nature 456: 502–506. 10.1038/nature07384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivan R, Gayraud-Morel B, Dumas G, Cimper C, Paisant S, Kelly RG, Tajbakhsh S. 2009. Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev Cell 16: 810–821. 10.1016/j.devcel.2009.05.008 [DOI] [PubMed] [Google Scholar]
- Sardina JL, Collombet S, Tian TV, Gómez A, Di Stefano B, Berenguer C, Brumbaugh J, Stadhouders R, Segura-Morales C, Gut M, et al. 2018. Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell 23: 727–741.e9. 10.1016/j.stem.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Schiebinger G, Shu J, Tabaka M, Cleary B, Subramanian V, Solomon A, Gould J, Liu S, Lin S, Berube P, et al. 2019. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176: 928–943.e22. 10.1016/j.cell.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Glaser G, Wernig A, Wegner M, Rosorius O. 2003. Sox8 is a specific marker for muscle satellite cells and inhibits myogenesis. J Biol Chem 278: 29769–29775. 10.1074/jbc.M301539200 [DOI] [PubMed] [Google Scholar]
- Schwarz BA, Cetinbas M, Clement K, Walsh RM, Cheloufi S, Gu H, Langkabel J, Kamiya A, Schorle H, Meissner A, et al. 2018. Prospective isolation of poised iPSC intermediates reveals principles of cellular reprogramming. Cell Stem Cell 23: 289–305.e5. 10.1016/j.stem.2018.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ. 2004. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell 119: 543–554. 10.1016/j.cell.2004.10.021 [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. 2011. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478: 255–259. 10.1038/nature10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M, et al. 2016. Dissecting direct reprogramming from fibroblast to neuron using single-cell RNA-seq. Nature 534: 391–395. 10.1038/nature18323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Tabebordbar M, Iovino S, Ciarlo C, Liu J, Castiglioni A, Price E, Liu M, Barton ER, Kahn CR, et al. 2013. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell 155: 909–921. 10.1016/j.cell.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Du Y, Deng H. 2015. Direct lineage reprogramming: strategies, mechanisms, and applications. Cell Stem Cell 16: 119–134. 10.1016/j.stem.2015.01.013 [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Legendre NP, Biswas AA, Lawton A, Yamamoto S, Tajbakhsh S, Kardon G, Goldhamer DJ. 2018. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Reports 10: 956–969. 10.1016/j.stemcr.2018.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Price F, Rudnicki MA. 2013. Satellite cells and the muscle stem cell niche. Physiol Rev 93: 23–67. 10.1152/physrev.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.