Abstract
Transposable elements (TEs) account for more than 50% of the human genome and many have been co-opted throughout evolution to provide regulatory functions for gene expression networks. Several lines of evidence suggest that these networks are fine-tuned by the largest family of TE controllers, the KRAB-containing zinc finger proteins (KZFPs). One tissue permissive for TE transcriptional activation (termed “transposcription”) is the adult human brain, however comprehensive studies on the extent of this process and its potential contribution to human brain development are lacking. To elucidate the spatiotemporal transposcriptome of the developing human brain, we have analyzed two independent RNA-seq data sets encompassing 16 brain regions from eight weeks postconception into adulthood. We reveal a distinct KZFP:TE transcriptional profile defining the late prenatal to early postnatal transition, and the spatiotemporal and cell type–specific activation of TE-derived alternative promoters driving the expression of neurogenesis-associated genes. Long-read sequencing confirmed these TE-driven isoforms as significant contributors to neurogenic transcripts. We also show experimentally that a co-opted antisense L2 element drives temporal protein relocalization away from the endoplasmic reticulum, suggestive of novel TE dependent protein function in primate evolution. This work highlights the widespread dynamic nature of the spatiotemporal KZFP:TE transcriptome and its importance throughout TE mediated genome innovation and neurotypical human brain development. To facilitate interactive exploration of these spatiotemporal gene and TE expression dynamics, we provide the “Brain TExplorer” web application freely accessible for the community.
KZFPs constitute the largest family of transcription factors encoded by mammalian genomes. These proteins harbor an N-terminal Krüppel-associated box (KRAB) domain and a C-terminal zinc finger array, which for many mediates sequence-specific DNA recognition. The KRAB domain of a majority of KZFPs recruits the transcriptional corepressor TRIM28 (also known as KAP1), which acts as a scaffold for heterochromatin inducers such as the histone methyl-transferase SETDB1, the histone deacetylating NuRD complex, CBX5 (also known as HP1) and DNA methyltransferases (Ecco et al. 2017). Many KZFPs bind to and repress TEs, a finding that led to the “arms race” hypothesis, which states that waves of genomic invasion by TEs throughout evolution drove the selection of KZFP genes after they first emerged in the last common ancestor of tetrapods, lung fish and coelacanth some 420 million years ago (Jacobs et al. 2014; Imbeault et al. 2017). Although partly supportive of this proposal, functional and phylogenetic studies point to a more complex model, strongly suggesting that KZFPs have facilitated the co-option of TE-embedded regulatory sequences (TEeRS) into transcriptional networks throughout tetrapod evolution (Najafabadi et al. 2015; Imbeault et al. 2017; Helleboid et al. 2019). TEeRS indeed host an abundance of transcription factor (TF) binding sites (Bourque et al. 2008; Sundaram et al. 2014), and KZFPs and their TE targets influence a broad array of biological processes from early embryogenesis to adult life, conferring a high degree of species specificity (Chuong et al. 2013, 2016; Trono 2015; Pontis et al. 2019; Turelli et al. 2020). TEeRS can act as enhancers, repressors, promoters, terminators, insulators or via post-transcriptional mechanisms (Garcia-Perez et al. 2016; Chuong et al. 2017). Although these co-opted TE functions are key to human biology, their deregulation can also contribute to pathologies such as cancer and neurodegenerative diseases (Li et al. 2015; Chuong et al. 2016; Attig et al. 2019; Jang et al. 2019; Ito et al. 2020; Jönsson et al. 2020).
KZFPs and TEs are broadly expressed during human early development, playing key roles in embryonic genome activation and controlling transcription in pluripotent stem cells (Theunissen et al. 2016; Pontis et al. 2019; Turelli et al. 2020). However, how much TEeRS and their polydactyl controllers influence later developmental stages and the physiology of adult tissues is still poorly defined. KZFPs are collectively more highly expressed in the human brain than in other adult tissues, suggesting a prominent impact for these epigenetic regulators and their TEeRS targets in the function of this organ (Nowick et al. 2009; Imbeault et al. 2017; Farmiloe et al. 2020; Turelli et al. 2020). In line with this hypothesis, we recently described how ZNF417 and ZNF587, two primate specific KZFPs repressing HERVK (human endogenous retrovirus K) and SVA (SINE-VNTR-Alu) integrants in human embryonic stem cells (hESC), are expressed in specific regions of the human developing and adult brain (Turelli et al. 2020). Through the control of TEeRS, these KZFPs influence the differentiation and neurotransmission profile of neurons and prevent the induction of neurotoxic retroviral proteins and an interferon-like response (Turelli et al. 2020). Furthermore, activation of LINE-1, another class of TEs, has been noted in human neural progenitor cells (hNPCs) and in the adult human brain, occasionally leading to de novo retrotransposition events (Muotri et al. 2005, 2010; Coufal et al. 2009; Upton et al. 2015; Erwin et al. 2016; Guffanti et al. 2018). Finally, various patterns of TE derepression have been reported in several neurodevelopmental and neurodegenerative disorders, indicating that a deregulated “transposcriptome” may be detrimental to brain development or homeostasis (Tam et al. 2019; Jönsson et al. 2020).
A growing number of genomic studies relying on bulk RNA sequencing (RNA-seq), single-cell RNA sequencing (scRNA-seq), assay for transposase accessible chromatin using sequencing (ATAC-seq), and other types of epigenomic analyses are teasing apart the transcriptional landscape of the developing human brain, revealing its dynamism and the complexity of the underlying cellular make-up (Kang et al. 2011; Miller et al. 2014; Fullard et al. 2018; Keil et al. 2018; Li et al. 2018; Zhong et al. 2018; Cardoso-Moreira et al. 2019). The present work was undertaken to explore the contribution of TEs and their KZFP controllers to this process.
Results
Spatiotemporal patterns of KZFP gene expression during brain development
To determine the spatiotemporal patterns of KZFP and TE expression in human neurogenesis, we analyzed RNA-seq data from 507 samples corresponding to 16 different brain regions and 12 developmental stages (eight postconception weeks [pcw] to adulthood) available through the BrainSpan Atlas of the Human Brain (Miller et al. 2014) and Cardoso-Moreira et al. 2019 (Supplemental Fig. S1A,B). Although the latter data set comprises 114 samples exclusively from dorsolateral prefrontal cortex (DFC) and cerebellum (CB), transcriptomes for these regions were largely concordant with those documented in BrainSpan, justifying the two resources as suitable for reciprocal validation (Supplemental Fig. S1C,D; Supplemental Tables S1, S2). We first examined KZFP gene expression in these two brain regions, which are representative of the forebrain and the hindbrain, respectively. The large majority of KZFPs expressed in the DFC showed higher expression levels at early prenatal stages, which was reduced shortly before birth and remained low onward (Fig. 1A). When comparing early prenatal (2A–3B; 8–18 pcw) and adult (11; age 20–60+ yr) stages, about half (169/333) of KZFPs were more expressed in the former and only 1.5% (5/333) in the latter, the rest being stable (Fig. 1B). This temporal pattern was less evident in the CB (Supplemental Fig. S2A), with only 15.9% (53/333) and 2.1% (7/333) of KZFPs more strongly expressed in early prenatal and adult, respectively (Supplemental Fig. S2B). Thus, KZFP gene expression patterns are characterized by both temporal and regional specificity.
Figure 1.
KZFP genes show a global pre- to postnatal decrease in expression. (A) Heatmaps of KZFP expression across human neurogenesis in the DFC. Scale represents the row Z-score. See also Supplemental Table S2. (B) Dot plot of differential expression analysis of KZFP genes in the DFC comparing adult (stage 11) to early prenatal stages (stage 2A–3B) of neurogenesis. Only KZFPs differentially expressed in both data sets are shown. Up (orange) represents KZFPs significantly up-regulated in adult versus early prenatal (fold change ≥ 2, FDR ≤ 0.05). Down (blue) represents KZFPs significantly down-regulated in the adult (fold change ≤ –2, FDR ≤ 0.05). See also Supplemental Table S3. (C) Density plot depicting estimated age of KZFPs of each category in B (P ≤ 0.05, Wilcoxon test). (D) Heatmaps of TF expression across human neurogenesis in the DFC. Scale same as in A. (E) Dot plot of differential expression analysis of TFs (as defined by Lambert et al. 2018) in the DFC, excluding KZFP genes, comparing adult (stage 11) to early prenatal stages (stage 2A to 3B) of neurogenesis. Only TFs differentially expressed in both data sets are shown. Up (orange) represents TFs significantly up-regulated in the adult versus early prenatal (fold change ≥ 2, FDR ≤ 0.05). Down (blue) represents KZFPs significantly down-regulated in the adult (fold change ≤ –2, FDR ≤ 0.05). See also Supplemental Table S3. (F) Correlation plots representing the Pearson correlation coefficient of temporal KZFP expression (left) and TF expression (right) between all 16 regions. Size of spot and color both represent the correlation coefficient. (0) No correlation, (1) strong correlation. (G) Heatmaps depicting the log2 counts per million (CPM) for selected KZFPs and TFs over the 16 regions included. See also Supplemental Tables S1 and S2. All plots show expression data from BrainSpan.
KZFP genes have emerged continuously during higher vertebrate evolution, collectively undergoing a high turnover in individual lineages. Among some 360 human KZFPs, about half are primate restricted, whereas a few are highly conserved, with orthologous sequences present in species that diverged more than 300 million years ago (Huntley et al. 2006; Imbeault et al. 2017). To determine if the differentially expressed KZFPs arose at particular times in evolution, we determined their ages. We found KZFPs either significantly down-regulated or up-regulated from early prenatal to adult stages to be significantly younger than those displaying no differences between these developmental periods (Wilcoxon test P ≤ 0.01) (Fig. 1C). This delineates two subsets among KZFPs participating in brain development: one evolutionarily recent and more transcriptionally dynamic, the other more conserved and transcriptionally static.
Of note, KZFP expression appeared distinct from a random selection of genes or all other TFs (as defined by Lambert et al. 2018), as other members of this functional family showed far more diverse patterns of expression throughout development, whether in the DFC or in the CB (Fig. 1D,E; Supplemental Fig. S2C–F). Only about a quarter of TFs were indeed more highly expressed in early prenatal stages in either region, against ∼10% in the adult brain (Fig. 1E; Supplemental Fig. S2C,D). Furthermore, temporal expression patterns of KZFP genes were highly correlated across all 16 brain regions, albeit to a lesser extent in the CB (Fig. 1F). In contrast, other TFs displayed far more diverse behaviors, with the CB, mediodorsal nucleus of the thalamus (THA), and striatum (STR) showing reduced correlation values compared with other regions (Fig. 1F). Thus, KZFPs are collectively subjected to a remarkable degree of spatiotemporal coordination in spite of the diversity of their genomic targets and of cell types present in the various regions of the brain. The KZFP gene most differentially expressed in prenatal versus postnatal DFC was the hematopoietic differentiation–associated ZNF300 (Fig. 1B; Supplemental Table S3; Xu et al. 2010). This was true in all brain regions, although its transcripts persisted longer in the CB compared with other areas (Fig. 1G; Supplemental Tables S1, S2). ZNF445, which binds and controls imprinted loci in humans (Takahashi et al. 2019), similarly showed comparable patterns across all brain regions, but its expression was largely maintained in the CB all the way to adulthood (Fig. 1G; Supplemental Tables S1, S2).
We next examined TRIM28, which encodes a protein that serves as a corepressor for many KZFPs (Ecco et al. 2017). Its expression levels were globally higher than those of any KZFP, albeit also with a reduction from prenatal to postnatal stages except in the CB (Fig. 1G; Supplemental Tables S1, S2). We also probed DNMT1, which encodes the maintenance DNA methyltransferase important for TE repression in neural progenitor cells and other somatic tissues beyond the early embryonic period (Jönsson et al. 2019). Although displaying overall patterns comparable to those seen for KZFPs and TRIM28, DNMT1 expression progressively increased in the CB to reach its highest level in the adult (Fig. 1G; Supplemental Tables S1, S2). In sum, KZFPs and their main epigenetic cofactors show a largely homogenous, dynamic spatiotemporal reduction in expression during human brain development.
TE subfamilies are dynamically expressed throughout development
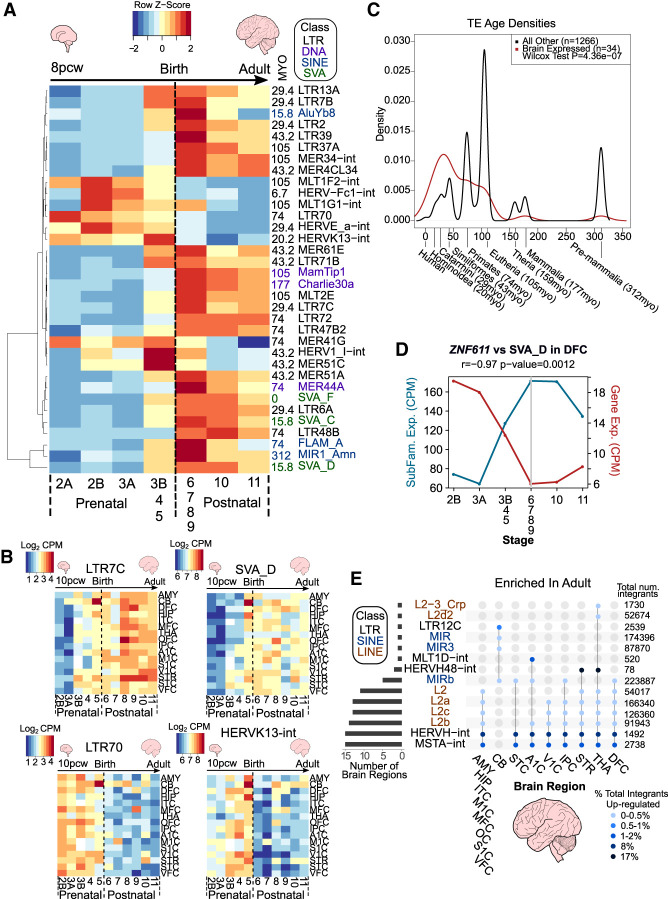
Having determined that the expression of most KZFPs is reduced at late stages of prenatal brain development, we examined the behavior of their TE targets. Young TEs are highly repetitive, which complicates the mapping of TE-derived RNA-seq reads to unique genomic loci, thus biasing against the scoring of their expression. We therefore first analyzed RNA-seq reads mapping to multiple TE loci within the same subfamily, regardless of positional information. In the DFC, discrete subfamilies, predominantly from the LTR class and to a lesser extent the SINE class, showed temporally distinct dynamics, concordant between data sets (Pearson correlation coefficient ≥ 0.7) (Fig. 2A; Supplemental Table S4). The same was true for the CB, but with moderately different subfamilies passing our threshold for concordance between data sets (Supplemental Fig. S3A; Supplemental Table S4). In the DFC, for example, the LTR7C and SVA-D subfamilies showed higher postnatal expression, whereas LTR70 and HERVK13-int behaved inversely, albeit without marked differences between brain regions (Fig. 2B; Supplemental Tables S4, S5). Similarly to KZFP genes, TEs have emerged continuously throughout evolution, with both young integrants and relics of ancient TEs reflective of different waves of genomic invasion. Using TE subfamily age estimates from Dfam (Hubley et al. 2016), we found that dynamically expressed TEs, concordant between both data sets, were significantly younger than nonconcordantly expressed subfamilies in the DFC and CB (Fig. 2C; Supplemental Fig. S3B).
Figure 2.
TE subfamilies and unique loci show spatiotemporal expression patterns. (A) Heatmap of TE subfamilies with concordant expression behaviors between both data sets (Pearson correlation coefficient ≥ 0.7) across human neurogenesis in the DFC. See also Supplemental Table S4. The mean expression values for stages 3B, 4, and 5 and for stages 6, 7, 8, and 9 were combined and averaged to reduce inherent variability owing to low numbers of samples for some stages (see Supplemental Fig. S1B). Scale represents the row Z-score. TE subfamily age in million years old (MYO) and class are shown to the right of the plot. (B) Heatmaps of TE subfamily expression across human neurogenesis in all 16 regions. See also Supplemental Tables S4 and S5. Scale represents log2 counts per million (CPM). Stage 2A was omitted owing to the lack of samples for some brain regions (see Supplemental Fig. S1B). (C) Density plot depicting estimated age of TEs in A (P ≤ 0.05, Wilcoxon test). Evolutionary stages and corresponding ages in MYO are shown beneath the plot. (D) Line plot showing expression in CPM of ZNF611 and its main TE target subfamily, SVA_D, and their Pearson correlation coefficient (−0.97, P-value = 0.0012). Gray line indicates birth at stage 6. (E) UpSet plot showing the significantly enriched differentially expressed subfamilies between adult and early prenatal stages per region from unique mapping analyses. Joined points represent combinations of significantly differentially expressed TE subfamilies. Points are colored with respect to the percentage of total integrants up-regulated. The total number of TE subfamily integrants in the genome is shown to the right of the plot. See also Supplemental Tables S6 and S7. All plots show expression data from BrainSpan.
Different TE subfamilies are bound by and harbor binding sites for a diverse array of TFs (Bourque et al. 2008; Sundaram et al. 2014). A combinatorial interplay between the reduced expression of transcriptional repressors and increased expression of activating TFs likely governs the transcription of individual TEs. Correspondingly, it was observed that depletion of TRIM28 or SETDB1, for instance, in embryonic stem cells, triggers the up-regulation of many but not all TE loci recruiting these corepressors (Matsui et al. 2010; Rowe et al. 2010; Turelli et al. 2014). To investigate the possible role of TFs in TE activation, we used a multimodal approach whereby we selected non-KZFP TF:TE pairs based on their significant positive (n = 3046) or negative correlations (n = 9003) to TE subfamilies from Figure 2A (Supplemental Fig. S4A). We next used the ENCODE data set of TF ChIP-seqs to determine if any TE subfamilies were significantly enriched in TF binding (The ENCODE Project Consortium 2012). Forty-five TF:TE pairs were highly significantly correlated ≥0.8 and were confirmed to be bona fide binding targets (Supplemental Fig. S4B). TF motif scanning of the consensus TE subfamily sequence with FIMO (Grant et al. 2011) revealed TF-specific motifs for 12 of these TF:TE pairs (Supplemental Fig. S4B,C). For example, LEF1, encoding a TF involved in specification and patterning of the mammalian cortex (Galceran et al. 2000), was significantly positively correlated in the DFC with, was bound to, and had a binding motif within LTR13A. ZNF845 was determined as a main binding KZFP of LTR13A and was significantly negatively correlated in expression (Supplemental Fig. S4D). TFE3, encoding a TF associated with neurological disorders (Lehalle et al. 2020), was also positively correlated with its binding target and motif containing LTR7C in the DFC, whereas the inverse was true of the significantly bound KZFP, ZNF468 (Supplemental Fig. S4D). Furthermore, ZNF611, encoding a previously characterized major regulator of SVA_D in early embryogenesis (Pontis et al. 2019), showed strongly anticorrelated expression with SVA_D throughout human brain development (Fig. 2D). Future work should investigate the interplay between KZFPs and other TFs in the regulation of TE transcription. To facilitate interactive exploration of these spatiotemporal gene and TE expression dynamics, we provide the “Brain TExplorer” web application freely accessible for the community at https://tronoapps.epfl.ch/BrainTExplorer/.
We next expanded our study by examining the expression of individual TE integrants, assigning RNA-seq reads to their genomic source loci and comparing early prenatal (stages 2A to 3B) and adult (stage 11) samples for the 16 available brain regions (Supplemental Fig. S1A,B). We found between 5000 and 7000 significant differentially expressed TE loci in each region, with 4000 loci common to both DFC and CB data sets (Supplemental Fig. S3C; Supplemental Tables S6, S7). Integrants belonging to 14 TE subfamilies from the LTR, LINE and SINE classes were significantly more expressed in adult samples, with HERVH-int, MSTA-int, and L2 elements significantly enriched in most brain regions (Fig. 2E). Eight percent and 17% of all HERVH-int and HERVH48-int integrants, respectively, were significantly increased in expression in adult, suggestive of their relatively widespread transcriptional derepression (Fig. 2E). The CB again showed distinct patterns, with significant enrichment of LTR12C and MIR elements instead (Fig. 2E). Conversely, integrants from 11 TE subfamilies were more expressed in the early prenatal period, largely in specific brain regions (Supplemental Fig. S3D). Together, these results highlight the spatiotemporal dynamic nature of the transposcriptome in the developing human brain.
Transpochimeric gene transcripts during human brain development
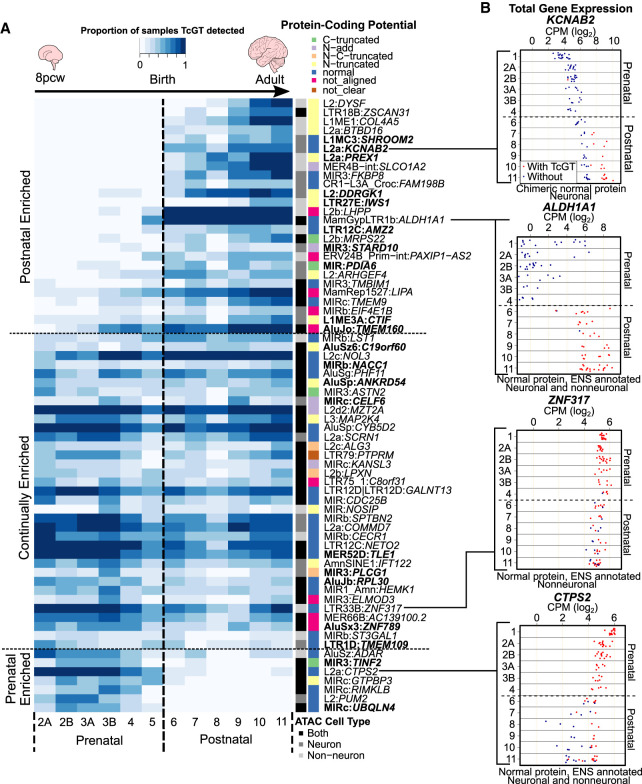
TE expression may be reflective of either “passive” cotranscription from genic transcripts or bona fide TE promoter activity (for review, see Lanciano and Cristofari 2020). Transpochimeric gene transcripts (TcGTs), that is, gene transcripts driven by TE-derived promoters, are the most easily interpretable and direct manifestation of the influence of TEeRS on gene expression. Some evidence for a role of TcGTs in the brain was provided by the recent observation that DNMT1 represses in hNPCs the expression of hominoid-restricted LINE-1 elements, which subsequently act as alternative promoters for genes involved in neuronal functions (Jönsson et al. 2019). To explore more broadly the potential role of TcGTs in human brain development and function, we performed de novo transcript assembly, searching for mature transcripts with a TE-derived sequence at their 5′-end and the coding sequence of a cellular gene downstream. Because of the distinct KZFP and global TE expression profiles between prenatal (stage 2A to stage 5) and postnatal stages (stage 6 to stage 11), we concentrated on these two periods, retaining only TcGTs present in >20% of either prenatal, postnatal, or both categories of samples and behaving in the same temporal manner in the BrainSpan and Cardoso data sets. If there was a twofold difference in the proportion of prenatal versus postnatal, the TcGT was annotated as either pre- or postnatal, whereas those below this threshold were deemed continual.
Our search yielded 480 high-confidence TcGTs, of which 9.8% (47/480) were prenatal, 12.3% (59/480) postnatal, and 72.3% (374/480) continual (Fig. 3A; Supplemental Table S8). Among pre- or postnatal TcGTs, developmental trajectories differed substantially, with some detected exclusively at either stage. For example, an L2a-driven isoform of CTP synthase 2 (CTPS2), whose product catalyzes CTP formation from UTP (van Kuilenburg et al. 2000), was found in 86% of all prenatal samples but only 12% of postnatal samples (Fig. 3A,B), whereas the inverse was observed for a MamGypLTR1b-driven isoform of the astrocyte-associated aldehyde dehydrogenase 1 family member A1 (ALDH1A1; 12% vs. 95%) (Adam et al. 2012) and an L2b-driven isoform of phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP; 0.9% vs. 97%) (Fig. 3A), the host of intronic single-nucleotide polymorphisms (SNPs) associated with major depressive disorder (Neff et al. 2009; Cui et al. 2016). The previously reported LTR12C-driven transcript of semaphorin 4D (SEMA4D), the product of which participates in axon guidance (Kumanogoh and Kikutani 2004; Cohen et al. 2009), was detected in 79% of postnatal and only 0.9% of prenatal samples in which it was instead expressed from a non-TE promoter, indicating a promoter switch during neurogenesis (Fig. 3A,B).
Figure 3.
TE co-option as genic promoters drives spatiotemporal gene expression in human neurogenesis. (A) Dot plot showing the proportion of pre- or postnatal samples TcGTs were detected in and behaving similarly in both data sets (prenatal, postnatal, or continual). A TcGT was classed as “detected” if one or more reads were spliced between a TE and a genic exon. (B) Sashimi browser plots from the Integrative Genomics Viewer (IGV; Robinson et al. 2011; Thorvaldsdóttir et al. 2013) showing the splicing events in representative samples for prenatal enriched TcGT L2a:CTPS2 and the postnatal enriched LTR12C:SEMA4D. (C) Heatmap indicating the proportion of samples per GTEx tissue in which each TcGT from A was detected. Each row represents an individual TcGT and each column a different tissue. (C, inset) Pie chart indicating the proportion of neurodevelopmental TcGTs detected in GTEx. (D) Stacked barplots indicating the proportion of TcGT TE TSS loci overlapping an ATAC-seq peak from BOCA (left) and CAGE-peak from FANTOM5 (right), and pie charts indicating their cell type distribution (bottom left); ATAC and CAGE peak overlaps (center) and highlighting 21 novel, non-Ensembl-annotated transcripts. (E) Stacked barplots indicating the TE subfamily, TE class, TE age, and the Ensembl overlap of each TcGT TE TSS loci. For all TcGT information, see also Supplemental Table S8.
We next examined the broader expression pattern of the 480 TcGTs detected during brain development. By applying our pipeline to the Genotype-Tissue Expression (GTEx) data set (Melé et al. 2015; The GTEx Consortium 2015), we detected around half of them in this collection of predominantly adult samples (Fig. 3C; Supplemental Table S8). Some were present in all available tissues, but the vast majority were brain restricted (Fig. 3C).
TcGTs show cell type–specific modes of expression with protein-coding potential
We next analyzed the state of the chromatin at the transcription start site (TSS) of the 480 TcGTs expressed during brain development by intersecting their proximal, TE-residing TSS (±200 bp) with ATAC-seq consensus peaks from neuronal (RBFOX3+) and nonneuronal (RBFOX3−) cells across 14 distinct adult brain regions from the Brain Open Chromatin Atlas (BOCA) (Fullard et al. 2018). About a quarter (111/480) of these TcGTs TSS overlapped with ATAC-seq peaks in the adult brain, indicating that their chromatin was opened in this setting (Fig. 3D). Of these, two-thirds showed cell type specificity, either to neurons (40.5%, 45/111) or to nonneuronal cells (22.5%, 25/111), whereas a third (41/111) were present in both cell types (Fig. 3D; Supplemental Table S8). These cell-restricted patterns were generally independent of the brain region considered, as illustrated by two postnatal enriched TcGTs; the nonneuronal L2-driven dysferlin (DYSF) (Supplemental Fig. S5A), a gene with protein accumulations in Alzheimer brains (Galvin et al. 2006) and mutations of which are associated with limb girdle muscular dystrophy 2B (Bashir et al. 1998; Liu et al. 1998); and the neuronal L2a-driven potassium voltage-gated channel subfamily A regulatory beta subunit 2 (KCNAB2) encoding a regulator of neuronal excitability (Supplemental Fig. S5B; McCormack et al. 2002).
To confirm that transcription of the TcGTs detected in the developing human brain was starting at the identified TE and to refine our highly sensitive “catch-all” RNA-seq-based approach, we intersected their TSS with cap analysis of gene expression (CAGE) peaks previously defined in around 1000 human cell lines and tissues (The FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014; Lizio et al. 2015). About a fifth of the TcGTs TSSs (19.5%, 94/480) overlapped with CAGE peaks, of which 68 also corresponded to ATAC-seq peaks, providing a subset of high-confidence TE-derived TSS loci driving gene transcription in the developing brain (Fig. 3D; Supplemental Table S8). The relatively small number of TcGTs intersecting a CAGE or ATAC peak may be owing to the high-sensitivity RNA-seq TcGT detection threshold of one spliced read spanning a TE and genic exon. The specific cell types, developmental stages, and tissues available in the CAGE and ATAC data sets may also not fully represent the in vivo cell types present in neurodevelopment; however, we cannot exclude the possibility of a small number of false-positive TcGTs. Of these, 21 were not annotated in Ensembl (Fig. 3D; Supplemental Table S8), indicating that co-opted TEs acting as promoter elements are contributing to a previously undetected TE-derived neurodevelopmental transcription network. Thirty-seven different TE subfamilies accounted for their promoters, but MIRs and L2s, belonging to the SINE and LINE families, respectively, contributed almost half, perhaps owing in part to their high prevalence in the genome (MiR3 and L2a: 87,870 and 166,340 integrants, respectively) (Fig. 3E), and LTRs about a fifth. A large range of evolutionary ages were represented, from the approximately 20-million-year-old (MYO) LTR12C to the approximately 177-MYO MIRs and L2s.
Of these 68 high-confidence TcGTs, 38.2% (26/68) were detected as postnatal specific, 51.5% (35/68) were continually detected, and 10.3% (7/68) were detected in prenatal stages only (Fig. 4A). Furthermore, the 5′-end of these TcGTs coincided with ATAC-seq peaks from neurons in 26.5% (18/68), from nonneuronal cells in 22% (15/68), and from both in 51.5% (35/68) of cases (Fig. 4A). Some TcGTs were present in all brain regions, whereas others showed regional specificity (Supplemental Fig. S6A). We next aimed to determine if the detected TcGTs had the capacity to code for protein. In silico prediction of the protein-coding potential of these TcGTs, found that about half (31/68) likely encoded the canonical protein sequence and a fifth (15/68) an N-truncated isoform, whereas other configurations (N-terminal addition, C or N and C truncation) were less frequent (Fig. 4A; Supplemental Table S8). The majority of TE-derived TSS loci (47/68) in each of the postnatal, continual, and prenatal categories retained binding sites for at least one KZFP, as determined by analysis of the library of over 300 KZFPs ChIP-exo peaks (Supplemental Fig. S6B; Imbeault et al. 2017).
Figure 4.
TcGTs are temporally expressed throughout neurogenesis in a cell type–specific manner, show protein-coding potential, and potentially drive transcript expression. (A) Heatmap showing the proportion of samples per developmental stage the 68 TcGTs (from Fig. 3D) were detected in the BrainSpan data set, regardless of region. Cell type–specific ATAC-seq overlaps and protein-coding potential determined via in silico translation are shown to the right of the plot. Bold indicates novel transcripts not annotated in Ensembl. See also Supplemental Table S8. (B) Dot plots showing the gene expression level per stage for the specified gene for samples in which the TcGT was detected (red) and in which it was not (blue) from the Cardoso data set in comparison to A. Dashed line represents birth at stage 6.
To determine the general expression of the 68 genes involved in high-confidence TcGTs, we compared their gene expression levels in samples in which the TcGT was or was not detected (Fig. 4B). In some cases, gene expression was higher in samples in which the TcGT was detected in a temporal manner such as the postnatally detected KCNAB2 (top) and ALDH1A1 (top mid) compared with samples in which the TcGT was not detected (Fig. 4B). Gene expression was higher throughout brain development for ZNF317 in samples in which the continually detected, nonneuronal TcGT was present (bottom mid), suggestive of a constitutive TE-derived promoter. Conversely, some genes such as CTPS2 showed higher prenatal gene expression in samples in which the TcGT was present (bottom), whereas for other genes, there were more moderate expression differences in samples with and without TcGT detection, as seen for DDRGK1, where the associated TcGT is postnatally detected in neurons (Supplemental Fig. S6C). A caveat with this analysis is that it does not directly address the relative contribution of the TcGT isoform versus canonical isoforms.
TcGTs are major contributors to neurodevelopmental transcript expression
To unequivocally determine the relative contributions of TcGT isoforms versus canonical isoforms to neurodevelopmental gene expression, we used a recent single-molecule real-time long-read Pacific Biosciences (PacBio) sequencing data set, which identified widespread isoform diversity in human neurodevelopment (Jeffries et al. 2020). Advances in long-read sequencing technologies now enable de novo full-transcript assembly and isoform expression quantification, analyses challenging to perform with short-read RNA-seq approaches (Oikonomopoulos et al. 2020). With this different technique, its lower sensitivity, analytical workflow from an independent laboratory, and far lower number of samples, long-read sequencing identified 41% (28/68) of TcGTs from Figure 4A with the identical TE TSS and largely identical predicted isoform structure (Fig. 5A). The TE TSS loci in hg19 were largely the same as in hg38, ensuring TE annotation did not alter between genome builds.
Figure 5.
TcGTs are major contributors to neurodevelopmental transcript expression. (A, inset) Pie chart showing the number of TcGTs from Figure 4A detected with the same TE-derived TSS and isoform structure in PacBio long-read sequencing in the adult from Jeffries et al. (2020) in hg38. Bar charts show the proportion of total transcripts that are TcGT derived. Numbers above each bar represent the TcGT isoform PacBio transcript counts (red numbers) and annotated non-TcGT isoform PacBio transcript counts (blue numbers) in adult samples as determined by Jeffries et al. (2020). If some non-TcGT Ensembl-annotated isoforms had appreciably higher counts than others, only these were used. 5′-Truncated incomplete splice matches (as defined by Jeffries et al. 2020) for non-TcGTs were omitted unless similar in number to nontruncated transcripts. Red bars indicate the TcGT isoform is the primary transcript; black bars, TcGT isoforms have “equivalent” expression to canonical isoforms; and blue bars, TcGTs are subsidiary transcript isoforms. (B) Genome browser images of TcGTs (red) detected in long-read sequencing in prenatal and adult samples in hg38. Only the non-TcGT isoforms (blue) with the most PacBio transcript counts are shown for clarity. Vertical orange bars highlight the TE-derived TSS of the TcGTs and are the same as detected in our short-read RNA-seq analyses in hg19.
We next directly assessed the transcriptional relevance of TcGTs relative to their canonical isoforms by using the provided transcript counts from Jeffries et al. (2020). Of the 28 TcGTs detected in the PacBio data set, 11 genes have the TE co-opted as their primary promoter in the adult brain (Fig. 5A, red bars). We determined four to be “equivalent” promoters, with similar transcript counts for TcGT and non-TcGT isoforms (Fig. 5A, black bars), and 13 as subsidiary promoters, where the transcript counts of non-TcGT isoforms were far greater than that of the TcGT isoforms (Fig. 5A, blue bars). This was further supported by quantifying the number of reads spanning the splice junction between the TE and genic exon from our short-read RNA-seq data set (Supplemental Fig. S6D). The median number of spliced reads for the PacBio-confirmed TcGTs and Ensembl canonical transcripts (6.5 and 6) was higher than for the TcGTs determined in Figure 3A and those with ATAC and CAGE peaks in Figure 4A (3 and 3.5), likely owing to the lower transcript detection sensitivity of single-molecule PacBio mRNA sequencing (Supplemental Fig. S7A). Using the detection criteria of one spliced read in our short-read RNA-seq analysis does not necessarily represent noise, as indicated by LTR27E:IWS1, a TcGT with a median of one spliced read detected only in postnatal samples despite high prenatal gene expression from the canonical IWS1 promoter, suggestive of a temporally regulated TE-derived promoter (Supplemental Fig. S7B,C). It was also detected in the PacBio data, represented by four long-read mRNA molecules (Fig. 5A) with the same splicing profile (Supplemental Fig. S7D). Only 14% of TcGTs from Figure 3A represented in Supplemental Figure S7A had a median of one spliced read. Together, these data show that the TE promoter co-option is indeed of relevance to transcriptional innovation in the developing brain and is indicative of a driving role of TEs in host gene expression throughout neurodevelopment.
We next focused our analyses on three antisense L2-driven, cell type–specific TcGTs predicted to encode for proteins involved in brain development and robustly detected in long-read sequencing: L2:DYSF, L2:DDRGK1 and L2a:KCNAB2; the first two as N-truncated isoforms and the last in its canonical protein isoform. For L2:DYSF, the TcGT isoform is the only transcript present and is only detected in the adult, in line with our short-read RNA-seq analyses (Fig. 5B, top). This is, therefore, the only DYSF promoter active in the brain. For L2:DDRGK1, the TcGT isoform was nearly equivalently expressed alongside the canonical isoform with similar transcript counts, again only in the adult as expected (Fig. 5B, mid). Indeed, L2:DDRGK1 had the most spliced reads between the TE and genic exon in short-read RNA-seq of any TcGT detected (Supplemental Fig. S6D). Our analysis of ATAC-seq data (Fig. 4A) indicates that L2:DDRGK1 is likely neuronal specific, and we hypothesize that the different isoforms may be transcribed simultaneously and/or in different cell types. L2a:KCNAB2 was detectable again only in the adult brain but with far lower transcript counts than its canonical “wild-type” (WT) counterpart; thus, it likely represents a subsidiary transcript isoform (Fig. 5B, bottom). We cannot rule out the relevance of this subsidiary isoform owing to potential cell type specificity. Indeed, these L2-driven TcGT isoforms represent a suite of TE transcriptional innovation, whereby the TE has been co-opted as the primary promoter, equivalent promoter, or subsidiary promoter throughout mammalian evolution.
Experimental validation of brain-detected TcGTs
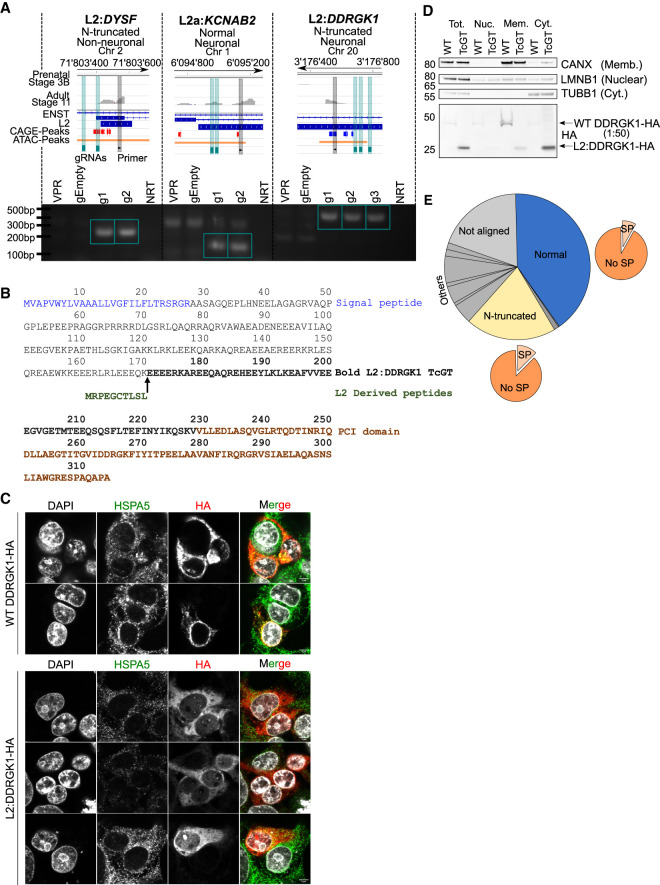
To verify that the TE and genic exon belonged to the same mRNA transcript, we next aimed to experimentally confirm TcGT candidates in the SH-SY5Y neuroblastoma cell line. Using qRT-PCR primers within the TE TSS and subsequent genic exon, we detected appreciable expression of TcGTs in this cell system (Supplemental Fig. S8A). However, this did not formally show that transcription was driven by the TE. To address this point, we targeted a CRISPR-based activation system (CRISPRa) to the TE-derived TSS region of L2:DYSF, L2a:KCNAB2, and L2:DDRGK1 TcGTs in HEK293T cells (Fig. 6A; Chavez et al. 2015). Activation of each of these three TcGTs could be induced, confirming that they were indeed driven by their respective TE promoters (Fig. 6A). Despite moderate gene expression in other tissues, the experimentally confirmed TcGTs L2:DYSF, L2a:KCNAB2, and L2:DDRGK1 are all highly brain specific and largely forebrain restricted, with lower detection levels in the THA, CB, and hippocampus (Supplemental Fig. S8B,C).
Figure 6.
Antisense L2 elements directly drive TcGTs and contribute to chimeric protein formation and cytosolic relocalization of the ER membrane–associated DDRGK1. (A) Schematic of TcGT TE TSS loci for indicated genes and representative prenatal (stage 3B) and adult (stage 11) RNA-seq tracks. Their associated protein-coding potential and cell type specificity are highlighted, and CAGE peak loci (red sense strand, blue antisense strand), CRISPRa gRNAs (green vertical bar), and TE-associated PCR primers are shown (black vertical bar; top). RT-PCR on cDNA generated from HEK293T cells transiently transfected with dCAS9-VPR plasmid and individual gRNA plasmids containing sequences targeting the TcGT TE TSS loci denoted in the schematic. dCAS9-VPR (VPR) or empty gRNA plasmids (gEmpty) alone were used as controls. Green box indicates bands of correct PCR product size absent in controls. (NRT) No reverse transcriptase. (B) Canonical WT DDRGK1- and TcGT L2:DDRGK1-derived protein sequence. (C) Overexpression of canonical WT DDRGK1-HA and L2:DDRGK1-HA in HEK293T cells followed by immunofluorescent staining for HSPA5 (an ER membrane–associated protein) and HA tag, followed by confocal imaging (scale bar, 5 µm). (D) Overexpression of canonical WT DDRGK1-HA and L2:DDRGK1-HA (TcGT) in HEK293T cells followed by cellular fractionation and western blot for the indicated marker proteins (right of western blot) and HA tag. For WT DDRGK1, 50× less protein lysate compared with L2:DDRGK1 was loaded for the HA blot owing to high levels of protein expressed. Image is representative of two independent experiments. (E) Pie charts showing the in silico protein-coding potential of the 480 TcGTs identified in Figure 3A with the proportion containing a signal peptide shown with the orange pie charts. See also Supplemental Table S8.
TcGT-encoded protein isoforms can display different subcellular localization
Having noted that 22% of high-confidence TcGTs were predicted to encode N-truncated proteins (Fig. 4A), we hypothesized that this could, in some cases, result in derivatives deprived of important subcellular localization domains, such as the endoplasmic reticulum (ER)–targeting N-terminal signal peptide. We focused on L2:DDRGK1 as the canonical DDRGK1 protein is anchored to the ER membrane by an N-terminal 27-amino-acid signal peptide (Fig. 6B) and plays a role in ER homeostasis and ER-phagy (Liu et al. 2017; Liang et al. 2020). GWAS studies have also identified a DDRGK1-associated risk locus for Parkinson's disease (Nalls et al. 2014; Chang et al. 2017). In the predicted translated product of the L2:DDRGK1 TcGT, the signal peptide is replaced by a 10-amino-acid L2-encoded sequence, conserved in New World primates but harboring nonsynonymous substitutions in Old World primates (Fig. 6B; Supplemental Figs. S8D, S9A). Of note, this L2 integrant is absent in mice (Supplemental Fig. S9A), but the L2:DDRGK1 TcGT is detected in the rhesus macaque developing brain with the same prenatal-to-postnatal expression dynamics as in humans (Supplemental Fig. S9B,C).
We therefore transfected HEK293T cells with plasmids expressing HA-tagged versions of either the canonical WT DDRGK1 transcript or its TcGT counterpart using cDNA from the bona fide L2:DDRGK1 transcript generated from the CRISPRa experiment (Fig. 6A), which was identical to the in silico predicted transcript. Confocal microscopy revealed that WT DDRGK1-HA largely colocalized with HSPA5, an ER membrane marker, whereas L2:DDRGK1-HA displayed a diffuse cytosolic pattern (Fig. 6C). Cellular fractionation further confirmed that the WT DDRGK1 isoform was sequestered in the membrane fraction, whereas the L2:DDRGK1 counterpart was enriched in cytosol (Fig. 6D).
As N-truncated isoforms made up the largest category of in silico predicted TcGT products besides full-length proteins, we next asked how widespread this type of TE-induced protein relocalization might be. For this, we intersected a database of signal peptide–containing proteins with our initial list of 480 TcGT-encoded protein products (Fig. 6E; Supplemental Table S8). Of 94 TcGT products predicted to be N-truncated, 12 contained a putative signal peptide in the canonical isoform. This prediction was supported in 11 cases in silico by signalP 5.0 (Almagro Armenteros et al. 2019), which predicted that in all of these instances, the TcGT isoforms lacked this putative signal peptide (Supplemental Fig. S10). Therefore, subcellular retargeting may be a frequent consequence of TE-driven protein innovation.
Discussion
An increasing number of studies are aimed at unravelling the transcriptional dynamics of human neurogenesis (Keil et al. 2018; Li et al. 2018; Cardoso-Moreira et al. 2019), yet so far, little attention has been paid to the participation of TEeRS in this process. Although retrotransposition of L1HS elements has been suggested to contribute to neuronal plasticity, experimental support for this model is lacking, and the vast majority of TEs hosted by the human genome have long lost the ability to spread (Brouha et al. 2003; Muotri et al. 2005). This prompted us to hypothesize that TEs might exert far greater influences on brain development through their ability to shape gene expression. As a first step toward testing this model, we analyzed two independent human neurogenesis RNA-seq data sets with a “TE-centric” approach. This led us to uncover that the transposcriptome and the global KZFP expression profile undergo profound changes at each stage of brain development. Correlative expression studies on genic KZFP targets suggest that KZFPs may directly regulate gene promoters during human neurogenesis independently from their TE binding ability (Farmiloe et al. 2020), a finding that may also extend to TEs. Increasing evidence also supports a regulatory role for KZFP-targeted TEs in this and other developmental contexts (Ecco et al. 2016, 2017; Chen et al. 2019; Pontis et al. 2019; Turelli et al. 2020). For example, we recently showed that two primate-restricted KZFPs, ZNF417 and ZNF587, control the expression of neuronal genes such as PRODH and AADAT via the regulation of HERVK-based TEeRS (Turelli et al. 2020). Furthermore, studies on the transcriptional corepressors TRIM28 and DNMT1 in hNPCs have highlighted their roles in the regulation of TEs and secondarily of cellular genes (Brattås et al. 2017; Jönsson et al. 2019). However, in vitro models do not recapitulate the global spatiotemporal complexity of gene and TE expression in the brain nor its diverse cell type milieu throughout development, hence the interest of performing large-scale “TE-centric” bioinformatics analyses on large postmortem brain RNA-seq data sets. A pertinent question from our analyses is whether the decline in KZFP expression per se is necessary and/or sufficient for the derepression of TE subfamilies in the in vivo human brain. We propose that there is likely a highly complex interplay between transcriptional repressor and activator TFs in the cell type–specific and developmental time point–specific control of diverse TE subfamilies.
Derepression of TEs, specifically of the LTR class, has been associated with various neurological disorders such as amyotrophic lateral sclerosis (ALS), Alzheimer's disease (AD), and multiple sclerosis (MS) (Tam et al. 2019; Jönsson et al. 2020). The up-regulation of LTR class elements in adult versus early prenatal brain suggests that LTR transposcription per se is a developmentally regulated feature of neurogenesis, which when deregulated is associated with a disease state. We propose that increased postnatal TE expression may possibly be reflective of the development of cell types not present in early prenatal stages, such as astrocytes, microglia, and oligodendrocytes, the developmental and transcriptional trajectories of which were identified by scRNA-seq analyses (Li et al. 2018). To determine the transposcriptome in scRNA-seq data remains technically challenging because many TE-derived transcripts are lowly abundant, a limitation that will hopefully be alleviated by progress in sequencing techniques and computational approaches (Linker et al. 2020; He et al. 2021). Of note, TEs heavily contribute to long noncoding RNAs (lncRNAs), which are abundant in the human brain (Derrien et al. 2012; Kelley and Rinn 2012; Zimmer-Bensch 2019). It is plausible that up-regulated TE transcripts play a role in this context, thereby exerting not cis- but trans-acting influences, the identification of which is far more challenging.
One increasingly well-characterized aspect of TE co-option is the engagement of TEeRS as alternative promoters. A wide range of oncogene-encoding TE-driven TcGTs have been documented in recent surveys of cancer databases (Attig et al. 2019; Jang et al. 2019), but the role of these transcript variants in physiological conditions remains largely undefined. Tissue-specific TcGTs have also been detected in the mouse developing intestine, liver, lung, stomach, and kidney (Miao et al. 2020). Here, we show not only the spatially and temporally orchestrated expression of TcGTs in the developing human brain but also that these TcGTs are largely organ- and cell type–specific. Some of them appear to be solely responsible for the expression of the involved gene, whereas others were present alongside canonical non-TE-driven transcripts, indicating sophisticated levels of regulation. Future research efforts should be aimed at the detection of TcGTs in neurodegenerative conditions or aging and may yield previously undetected pathogenic associations.
By experimental activation of a selected subset of antisense L2-driven TcGTs with CRISPRa and functional analyses of the product of the L2:DDRGK1 transcript, we highlight the functional relevance of this phenomenon in vitro; however, the relevance “in vivo” in human neurogenesis remains an extensive avenue of future research. Indeed, L2 elements have cross-species promoter capacities, suggestive of potentially widespread evolutionary co-option (Roller et al. 2021). DDRGK1 is an ER membrane–associated protein with critical roles in UFMylation, an ubiquitin-like modification, and is involved in the unfolded protein response (UFP) and ER-phagy (Liu et al. 2017; Liang et al. 2020). DDRGK1 is essential to target interactors like UFL1, the UFMylation ligase, to the ER membrane. The novel cytosolic chimeric L2:DDRGK1 protein, where a short N-terminal sequence derived from the L2 integrant replaces the signal peptide characteristic of its canonical counterpart, may therefore exert novel functions in the cytosol of postnatal to adult neurons. We speculate that this may lead to UFL1 sequestration away from the ER membrane, perhaps leading to distinct UFMylation cascades in novel locations, the neurodevelopmental implications of which remain to be investigated. Of note, defects in ER transmembrane–associated proteins have severe implications for the UFP, and ER stress is a critical feature of neurodegenerative diseases (Martínez et al. 2016; Esk et al. 2020). As signal peptide excision seems to affect a number of other TcGT products, this example may illustrate a more general phenomenon, whereby TE-driven genome evolution generates novel protein isoforms, altering critical cell functions.
Our study indicates that the exaptation of TE-embedded regulatory sequences and its facilitation by TE-targeting KZFP controllers have significantly contributed to the complexity of transcriptional networks in the developing human brain. This warrants efforts aimed at delineating the evolutionary and functional impact of this phenomenon and at defining how its alterations, notably in the context of inter-individual differences at these genomic loci, translates into variations in brain development, function, and disease susceptibility.
Methods
Data sets
Raw RNA-seq FASTQ files for human and rhesus macaque brain development (Cardoso-Moreira et al. 2019) were downloaded from the European Nucleotide Archive (ENA: https://www.ebi.ac.uk/ena/browser/home) (data sets PRJEB26969 and PRJEB26956, respectively).
Raw RNA-seq FASTQ files for GTEx (phs000424.v7.p2, supported by the Common Fund of the Office of the Director of the National Institutes of Health) and BrainSpan (phs000755.v2.p1 provided by Dr. Nenad Sestan), were downloaded from the dbGaP-authorized access platform (Supplemental Acknowledgements). Long-read PacBio human neurodevelopmental transcript isoform hg38 GTFs were downloaded from http://genome.exeter.ac.uk/BrainIsoforms.html (Jeffries et al. 2020). Processed BED files containing regional neuronal or nonneuronal ATAC-seq peak loci from the BOCA (Fullard et al. 2018) were downloaded for hg19. To generate consensus neuronal and nonneuronal ATAC-peak BED files, BED coordinates from all regions were combined and overlapping peak coordinates merged using BEDTools merge (Quinlan and Hall 2010). Processed BED files for CAGE-seq peak loci from FANTOM5 (The FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014) were downloaded for hg19 (Lizio et al. 2015). TF binding data was downloaded from the ENCODE portal (The ENCODE Project Consortium 2012). Signal peptide–containing proteins in human were downloaded from http://signalpeptide.com/index.php. Processed BED files from KZFP ChIP-exo experiments were used from our previous study (Imbeault et al. 2017).
RNA-seq analysis
Reads were mapped to the human (hg19), or macaque (rheMac8) genome using HISAT2 (Kim et al. 2015) with parameters HISAT2 -k 5 ‐‐seed 42. Counts on genes and TEs were generated using featureCounts (Liao et al. 2014). To avoid read assignation ambiguity between genes and TEs, a GTF file containing both was provided to featureCounts. For repetitive sequences, an in-house curated version of the Repbase database was used (fragmented LTR and internal segments belonging to a single integrant were merged), generated as previously described (Turelli et al. 2020). Minor modifications to the repeat merging pipeline described by Turelli et al. (2020) were made for the macaque (RepeatMasker 4.0.5 20160202), with the distance between two LTR elements of the same orientation to an ERV-int fragment being <400 bp. For genes, the Ensembl release 75 annotation was used. Mapping and analyses were performed largely as by Turelli et al. (2020) with some data set–specific modifications (for details, see Supplemental Methods and Code).
TcGT detection pipeline
First, a per-sample transcriptome was computed from the RNA-seq BAM file using StringTie (Kovaka et al. 2019) with parameters –j 1 –c 1. Each transcriptome was then crossed using BEDTools (Quinlan and Hall 2010), to hg19 (or rheMac8) coding exons and curated RepeatMasker to extract TcGTs with one or more reads spliced between a TE and genic exon for each sample. Second, a custom Python program was used to annotate and aggregate the sample level TcGTs into counts per stages (defined in Supplemental Fig. S1B). In brief, for each data set, a GTF containing all annotated TcGTs was created, and TcGTs having their first exon overlapping an annotated gene, or TSS not overlapping a TE, were discarded. From this filtered file, TcGTs associated with the same gene and having a TSS within 100 bp of each other were aggregated. Finally, for each aggregate, its occurrence per group was computed, and a consensus transcript was generated for each TSS aggregate. For each exon of TcGT aggregate, its percentage of occurrence across the different samples was computed and integrated in the consensus if present in >30% of the samples the TcGT was detected in. All samples available in both data sets were used regardless of mapped read count.
From the resulting master file, additional criteria were applied to determine prenatal, postnatal, or continually expressed TcGTs. First, only TcGTs that were present in at least 20% of prenatal, postnatal, or 20% of both pre- and postnatal samples (continual) were kept for each data set. Second, to ensure TcGTs were robustly detectable in the different data sets, TcGT files were merged based on the same TSS TE and associated gene name. Third, TcGTs were required to show the same temporal transcriptional behavior in both data sets, that is, a twofold change in TcGT detection pre- versus postnatal and vice versa or a lower fold change in both data sets (continual). This resulted in the 480 robustly detectable temporal TcGTs in Figure 3A and Supplemental Table S8. These TcGTs were further filtered for strong promoter regions using BEDTools intersect (Quinlan and Hall 2010) of the 200 bp upstream of and downstream from the TcGT TSS with FANTOM5 CAGE-seq (The FANTOM Consortium and the RIKEN PMI and CLST (DGT) 2014) and BOCA neuronal and nonneuronal consensus ATAC-seq peak BED files (Fullard et al. 2018). TcGT TSS loci were also intersected with Ensembl (GRCh37.p13) transcriptional start sites to determine nonannotated transcripts. Reads spliced between the TE or canonical promoter region and first genic exon were quantified for samples in which the TcGT was detected using BEDTools and SAMtools. The promoter-containing exon of the canonical longest transcript from Ensembl was used with the caveat that this may not always reflect the most expressed genic transcript or may indeed reflect a previously annotated TcGT. The TcGT detection script is available in Supplemental Code.
Protein product prediction
DNA sequences were retrieved for each TcGTs consensus, and protein products were derived from the longest ORF in the three reading frames using Biopython (for details, see Supplemental Methods; Cock et al. 2009).
TE and KZFP age estimation
TE subfamily ages were downloaded from Dfam (Hubley et al. 2016). To compare KZFP ages, we developed a score we called complete alignment of zinc finger (CAZF) (for details, see Supplemental Methods; as described by Thorball et al. 2020).
Cell culture
Human embryonic kidney 293T (HEK293T) cells and SH-SY5Y neuroblastoma cells were cultured in DMEM supplemented with 10% fetal calf serum and 1% penicillin/streptomycin.
Transfection
Transient transfection of HEK293T cells was performed with FuGENE HD (Promega) as per the manufacturer's recommendation. Cells were harvested 48 h after transfection for either RNA extraction or immunofluorescence.
CRISPRa
The SP-dCas9-VPR (Addgene 63798) (Chavez et al. 2015) and the gRNA cloning vector (Addgene 41824) (Mali et al. 2013) were gifts from George Church. The CRISPRa experiment was performed and gRNAs were designed with CRISPOR (Supplemental Table S9; Concordet and Haeussler 2018) as described in the Supplemental Methods.
RT-PCR and qRT-PCR
Primers to detect TcGTs were designed with Primer3 (Untergasser et al. 2012) by inputting DNA sequences covering and flanking the splice junction between the TE and genic exon (Supplemental Table S9). Sanger sequencing confirmed the correct product was amplified (Supplemental Material). Further primer criteria and the standard RT-PCR protocol can be found in Supplemental Methods.
Cloning of WT DDRGK1 and L2:DDRGK1
The WT DDRGK1 cDNA clone (NCBI GenBank database [https://www.ncbi.nlm.nih.gov/genbank/] accession number HQ448262 ImageID:100071664) was obtained from the ORFeome Collaboration (http://www.orfeomecollaboration.org/) in the pENTR223 vector without a stop codon. The WT DDRGK1 and L2:DDRGK1 sequences were cloned into pTRE-3HA, which produces proteins with three C-terminal HA tags in a doxycycline-dependent manner using standard cloning procedures (Supplemental Methods).
Cellular fractionation
Approximately 400,000 HEK293T cells in different wells of a six-well plate were transfected with either pTRE-WT:DDRGK1-HA or pTRE-L2:DDRGK1-HA, whose expression was induced for 48 h by adding 1 µg/mL doxycycline to the media. After 48 h, wells were washed with 1 mL ice-cold PBS, and cells were scraped and transferred to Eppendorf tubes on the second wash. After centrifugation at 300rcf for 5 min at 4°C, PBS was aspirated, and cells were resuspended in 400 µL ice-cold cytoplasmic isolation buffer (10 mM KOAc, 2 mM MgOAC, 20 mM HEPES at pH 7.5, 0.5 mM DTT, 0.015% digitonin) and centrifuged at 900rcf for 5 min at 4°C. Supernatant was collected as the cytoplasmic fraction, and the remaining pellet was resuspended in 400 µL of membrane isolation buffer (10 mM HEPES, 10 mM KCl, 0.1 mM EDTA at pH 8, 1 mM DTT, 0.5% Triton X-100, 100 mM NaF) and then centrifuged for 10 min at 900rcf at 4°C to pellet nuclei with the supernatant collected as the membrane fraction. Pelleted nuclei were resuspended in 400 µL of lysis buffer (1% NP-40, 500 mM Tris-HCL at pH 8, 0.05% SDS, 20 mM EDTA, 10 mM NaF, 20 mM benzamidine) for 10 min on ice and centrifuged for 10 min at 900rcf at 4°C, and the supernatant was collected as the nuclear fraction. One hundred microliters of 4× NuPAGE LDS sample buffer (Thermo Fisher Scientific) was added to the 400 µL cellular fractions and samples boiled for 5 min at 95°C followed by western blot (Supplemental Methods).
Immunofluorescence
Immunofluorescence was performed as previously described (Supplemental Methods; Helleboid et al. 2019).
Data access
The interactive Brain TExplorer can be accessed at https://tronoapps.epfl.ch/BrainTExplorer/. Custom analysis code can be found in the Supplemental Code.
Supplementary Material
Acknowledgments
We thank all members of the Trono laboratory and Retha Ritter for helpful and insightful discussions, along with Samuel Corless and Nezha Benabdallah for critical reading of the manuscript. This study was supported by grants from the Personalized Health and Related Technologies (École Polytechnique Fédérale de Lausanne, PHRT-508), the European Research Council (KRABnKAP, #268721; Transpos-X, #694658), and the Swiss National Science Foundation (310030_152879 and 310030B_173337) to D.T.
Author contributions: C.J.P. and D.T. conceived the study, interpreted the data, and wrote the manuscript. C.J.P. performed bioinformatics analyses and all experiments. J.D. and S.S. developed key code and performed bioinformatics analyses. S.D. performed the GTEx TcGT analysis and in silico translation of TcGTs. A.C. performed the KZFP aging analysis and determined KZFP TE subfamily targets. E.P. contributed to bioinformatics tools and code. All authors reviewed the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.275133.120.
Freely available online through the Genome Research Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Adam SA, Schnell O, Pöschl J, Eigenbrod S, Kretzschmar HA, Tonn J-C, Schüller U. 2012. ALDH1A1 is a marker of astrocytic differentiation during brain development and correlates with better survival in glioblastoma patients. Brain Pathol 22: 788–797. 10.1111/j.1750-3639.2012.00592.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37: 420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- Attig J, Young GR, Hosie L, Perkins D, Encheva-Yokoya V, Stoye JP, Snijders AP, Ternette N, Kassiotis G. 2019. LTR retroelement expansion of the human cancer transcriptome and immunopeptidome revealed by de novo transcript assembly. Genome Res 29: 1578–1590. 10.1101/gr.248922.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir R, Britton S, Strachan T, Keers S, Vafiadaki E, Lako M, Richard I, Marchand S, Bourg N, Argov Z, et al. 1998. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat Genet 20: 37–42. 10.1038/1689 [DOI] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew J-L, Ruan Y, Wei C-L, Ng HH, et al. 2008. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res 18: 1752–1762. 10.1101/gr.080663.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brattås PL, Jönsson ME, Fasching L, Nelander Wahlestedt J, Shahsavani M, Falk R, Falk A, Jern P, Parmar M, Jakobsson J. 2017. TRIM28 controls a gene regulatory network based on endogenous retroviruses in human neural progenitor cells. Cell Rep 18: 1–11. 10.1016/j.celrep.2016.12.010 [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285. 10.1073/pnas.0831042100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, Shao Y, Liechti A, Ascenção K, Rummel C, Ovchinnikova S, et al. 2019. Gene expression across mammalian organ development. Nature 571: 505–509. 10.1038/s41586-019-1338-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, Kerchner GA, Ayalon G, Bingol B, Sheng M, et al. 2017. A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet 49: 1511–1516. 10.1038/ng.3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, Iyer EPR, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. 2015. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 12: 326–328. 10.1038/nmeth.3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Schwalie PC, Pankevich EV, Gubelmann C, Raghav SK, Dainese R, Cassano M, Imbeault M, Jang SM, Russeil J, et al. 2019. ZFP30 promotes adipogenesis through the KAP1-mediated activation of a retrotransposon-derived Pparg2 enhancer. Nat Commun 10: 1809. 10.1038/s41467-019-09803-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet 45: 325–329. 10.1038/ng.2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet 18: 71–86. 10.1038/nrg.2016.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. 2009. Biopython: freely available python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114. 10.1016/j.gene.2009.06.020 [DOI] [PubMed] [Google Scholar]
- Concordet J-P, Haeussler M. 2018. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46: W242–W245. 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. 2009. L1 retrotransposition in human neural progenitor cells. Nature 460: 1127–1131. 10.1038/nature08248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Gong X, Tang Y, Kong L, Chang M, Geng H, Xu K, Wang F. 2016. Relationship between the LHPP gene polymorphism and resting-state brain activity in major depressive disorder. Neural Plast 2016: 9162590. 10.1155/2016/9162590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. 2016. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev Cell 36: 611–623. 10.1016/j.devcel.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Imbeault M, Trono D. 2017. KRAB zinc finger proteins. Development 144: 2719–2729. 10.1242/dev.132605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Paquola ACM, Singer T, Gallina I, Novotny M, Quayle C, Bedrosian TA, Alves FIA, Butcher CR, Herdy JR, et al. 2016. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat Neurosci 19: 1583–1591. 10.1038/nn.4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esk C, Lindenhofer D, Haendeler S, Wester RA, Pflug F, Schroeder B, Bagley JA, Elling U, Zuber J, von Haeseler A, et al. 2020. A human tissue screen identifies a regulator of ER secretion as a brain-size determinant. Science 370: 935–941. 10.1126/science.abb5390 [DOI] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN PMI and CLST (DGT). 2014. A promoter-level mammalian expression atlas. Nature 507: 462–470. 10.1038/nature13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmiloe G, Lodewijk GA, Robben SF, van Bree EJ, Jacobs FMJ. 2020. Widespread correlation of KRAB zinc finger protein binding with brain-developmental gene expression patterns. Philos Trans R Soc B Biol Sci 375: 20190333. 10.1098/rstb.2019.0333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullard JF, Hauberg ME, Bendl J, Egervari G, Cirnaru M-D, Reach SM, Motl J, Ehrlich ME, Hurd YL, Roussos P. 2018. An atlas of chromatin accessibility in the adult human brain. Genome Res 28: 1243–1252. 10.1101/gr.232488.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JLR, Grosschedl R. 2000. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 127: 469–482. 10.1242/dev.127.3.469 [DOI] [PubMed] [Google Scholar]
- Galvin JE, Palamand D, Strider J, Milone M, Pestronk A. 2006. The muscle protein dysferlin accumulates in the Alzheimer brain. Acta Neuropathol 112: 665–671. 10.1007/s00401-006-0147-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez JL, Widmann TJ, Adams IR. 2016. The impact of transposable elements on mammalian development. Development 143: 4101–4114. 10.1242/dev.132639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti G, Bartlett A, Klengel T, Klengel C, Hunter R, Glinsky G, Macciardi F. 2018. Novel bioinformatics approach identifies transcriptional profiles of lineage-specific transposable elements at distinct loci in the human dorsolateral prefrontal cortex. Mol Biol Evol 35: 2435–2453. 10.1093/molbev/msy143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Babarinde IA, Sun L, Xu S, Chen R, Shi J, Wei Y, Li Y, Ma G, Zhuang Q, et al. 2021. Identifying transposable element expression dynamics and heterogeneity during development at the single-cell level with a processing pipeline scTE. Nat Commun 12: 1456. 10.1038/s41467-021-21808-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleboid P, Heusel M, Duc J, Piot C, Thorball CW, Coluccio A, Pontis J, Imbeault M, Turelli P, Aebersold R, et al. 2019. The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification. EMBO J 38: e101220. 10.15252/embj.2018101220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubley R, Finn RD, Clements J, Eddy SR, Jones TA, Bao W, Smit AFA, Wheeler TJ. 2016. The Dfam database of repetitive DNA families. Nucleic Acids Res 44: D81–D89. 10.1093/nar/gkv1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L. 2006. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 16: 669–677. 10.1101/gr.4842106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbeault M, Helleboid P-Y, Trono D. 2017. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543: 550–554. 10.1038/nature21683 [DOI] [PubMed] [Google Scholar]
- Ito J, Kimura I, Soper A, Coudray A, Koyanagi Y, Nakaoka H, Inoue I, Turelli P, Trono D, Sato K. 2020. Endogenous retroviruses drive KRAB zinc-finger protein family expression for tumor suppression. Sci Adv 6: eabc3020. 10.1126/sciadv.abc3020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516: 242–245. 10.1038/nature13760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, Zhang D, Li D, Xing X, Kim S, et al. 2019. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet 51: 611–617. 10.1038/s41588-019-0373-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries AR, Leung SK, Castanho I, Moore K, Davies JP, Dempster EL, Bray NJ, O'Neill P, Tseng E, Ahmed Z, et al. 2020. Full-length transcript sequencing of human and mouse identifies widespread isoform diversity and alternative splicing in the cerebral cortex. bioRxiv 10.1101/2020.10.14.339200 [DOI] [PMC free article] [PubMed]
- Jönsson ME, Ludvik Brattås P, Gustafsson C, Petri R, Yudovich D, Pircs K, Verschuere S, Madsen S, Hansson J, Larsson J, et al. 2019. Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat Commun 10: 3182. 10.1038/s41467-019-11150-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson ME, Garza R, Johansson PA, Jakobsson J. 2020. Transposable elements: a common feature of neurodevelopmental and neurodegenerative disorders. Trends Genet 36: 610–623. 10.1016/j.tig.2020.05.004 [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature 478: 483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil JM, Qalieh A, Kwan KY. 2018. Brain transcriptome databases: a user's guide. J Neurosci 38: 2399–2412. 10.1523/JNEUROSCI.1930-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DR, Rinn JL. 2012. Transposable elements reveal a stem cell specific class of long noncoding RNAs. Genome Biol 13: R107. 10.1186/gb-2012-13-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol 20: 278. 10.1186/s13059-019-1910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumanogoh A, Kikutani H. 2004. Biological functions and signaling of a transmembrane semaphorin, CD100/Sema4D. Cell Mol Life Sci 61: 292–300. 10.1007/s00018-003-3257-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, Weirauch MT. 2018. The human transcription factors. Cell 172: 650–665. 10.1016/j.cell.2018.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciano S, Cristofari G. 2020. Measuring and interpreting transposable element expression. Nat Rev Genet 21: 721–736. 10.1038/s41576-020-0251-y [DOI] [PubMed] [Google Scholar]
- Lehalle D, Vabres P, Sorlin A, Bierhals T, Avila M, Carmignac V, Chevarin M, Torti E, Abe Y, Bartolomaeus T, et al. 2020. De novo mutations in the X-linked TFE3 gene cause intellectual disability with pigmentary mosaicism and storage disorder-like features. J Med Genet 57: 808–819. 10.1136/jmedgenet-2019-106508 [DOI] [PubMed] [Google Scholar]
- Li W, Lee M-H, Henderson L, Tyagi R, Bachani M, Steiner J, Campanac E, Hoffman DA, von Geldern G, Johnson K, et al. 2015. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med 7: 307ra153. 10.1126/scitranslmed.aac8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, Sunkin SM, Li Z, Shin Y, Zhu Y, et al. 2018. Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362: eaat7615. 10.1126/science.aat7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Luong T, Ahmed S, Muhar M, Nguyen T, Olzmann JA, Corn JE. 2020. A genome-wide ER-phagy screen highlights key roles of mitochondrial metabolism and ER-resident UFMylation. Cell 180: 1160–1177.e20. 10.1016/j.cell.2020.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Linker SB, Randolph-Moore L, Kottilil K, Qiu F, Jaeger BN, Barron J, Gage FH. 2020. Identification of bona fide B2 SINE retrotransposon transcription through single-nucleus RNA-seq of the mouse hippocampus. Genome Res 30: 1643–1654. 10.1101/gr.262196.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Aoki M, Illa I, Wu C, Fardeau M, Angelini C, Serrano C, Urtizberea JA, Hentati F, Hamida MB, et al. 1998. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat Genet 20: 31–36. 10.1038/1682 [DOI] [PubMed] [Google Scholar]
- Liu J, Wang Y, Song L, Zeng L, Yi W, Liu T, Chen H, Wang M, Ju Z, Cong Y-S. 2017. A critical role of DDRGK1 in endoplasmic reticulum homoeostasis via regulation of IRE1α stability. Nat Commun 8: 14186. 10.1038/ncomms14186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, et al. 2015. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol 16: 22. 10.1186/s13059-014-0560-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez G, Vidal RL, Mardones P, Serrano FG, Ardiles AO, Wirth C, Valdés P, Thielen P, Schneider BL, Kerr B, et al. 2016. Regulation of memory formation by the transcription factor XBP1. Cell Rep 14: 1382–1394. 10.1016/j.celrep.2016.01.028 [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova Ia, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- McCormack K, Connor JX, Zhou L, Ho LL, Ganetzky B, Chiu S-Y, Messing A. 2002. Genetic analysis of the mammalian K+ channel β subunit Kvβ2 (Kcnab2). J Biol Chem 277: 13219–13228. 10.1074/jbc.M111465200 [DOI] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. The human transcriptome across tissues and individuals. Science 348: 660–665. 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Fu S, Lyu C, Gontarz P, Wang T, Zhang B. 2020. Tissue-specific usage of transposable element-derived promoters in mouse development. Genome Biol 21: 255. 10.1186/s13059-020-02164-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, et al. 2014. Transcriptional landscape of the prenatal human brain. Nature 508: 199–206. 10.1038/nature13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910. 10.1038/nature03663 [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. 2010. L1 retrotransposition in neurons is modulated by MeCP2. Nature 468: 443–446. 10.1038/nature09544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. 2015. C2h2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol 33: 555–562. 10.1038/nbt.3128 [DOI] [PubMed] [Google Scholar]
- Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, DeStefano AL, Kara E, Bras J, Sharma M, et al. 2014. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet 46: 989–993. 10.1038/ng.3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff CD, Abkevich V, Packer JCL, Chen Y, Potter J, Riley R, Davenport C, DeGrado Warren J, Jammulapati S, Bhathena A, et al. 2009. Evidence for HTR1A and LHPP as interacting genetic risk factors in major depression. Mol Psychiatry 14: 621–630. 10.1038/mp.2008.8 [DOI] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L. 2009. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci 106: 22358–22363. 10.1073/pnas.0911376106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomopoulos S, Bayega A, Fahiminiya S, Djambazian H, Berube P, Ragoussis J. 2020. Methodologies for transcript profiling using long-read technologies. Front Genet 11: 606. 10.3389/fgene.2020.00606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, Theunissen TW, Jaenisch R, Trono D. 2019. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell 24: 724–735.e5. 10.1016/j.stem.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller M, Stamper E, Villar D, Izuogu O, Martin F, Redmond AM, Ramachanderan R, Harewood L, Odom DT, Flicek P. 2021. LINE retrotransposons characterize mammalian tissue-specific and evolutionarily dynamic regulatory regions. Genome Biol 22: 62. 10.1186/s13059-021-02260-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res 24: 1963–1976. 10.1101/gr.168872.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Coluccio A, Thorball CW, Planet E, Shi H, Offner S, Turelli P, Imbeault M, Ferguson-Smith AC, Trono D. 2019. ZNF445 is a primary regulator of genomic imprinting. Genes Dev 33: 49–54. 10.1101/gad.320069.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Ostrow LW, Gale Hammell M. 2019. Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob DNA 10: 32. 10.1186/s13100-019-0176-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Friedli M, He Y, Planet E, O'Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, et al. 2016. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell 19: 502–515. 10.1016/j.stem.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorball CW, Planet E, de Tribolet-Hardy J, Coudray A, Fellay J, Turelli P, Trono D. 2020. Ongoing evolution of KRAB zinc finger protein-coding genes in modern humans. bioRxiv 10.1101/2020.09.01.277178 [DOI]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trono D. 2015. Transposable elements, polydactyl proteins, and the genesis of human-specific transcription networks. Cold Spring Harb Symp Quant Biol 80: 281–288. 10.1101/sqb.2015.80.027573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. 2014. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res 24: 1260–1270. 10.1101/gr.172833.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli P, Playfoot C, Grun D, Raclot C, Pontis J, Coudray A, Thorball C, Duc J, Pankevich EV, Deplancke B, et al. 2020. Primate-restricted KRAB zinc finger proteins and target retrotransposons control gene expression in human neurons. Sci Adv 6: eaba3200. 10.1126/sciadv.aba3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3: new capabilities and interfaces. Nucleic Acids Res 40: e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sánchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. 2015. Ubiquitous L1 mosaicism in hippocampal neurons. Cell 161: 228–239. 10.1016/j.cell.2015.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. 2000. Identification of a cDNA encoding an isoform of human CTP synthetase. Biochim Biophys Acta 1492: 548–552. 10.1016/S0167-4781(00)00141-X [DOI] [PubMed] [Google Scholar]
- Xu J-H, Wang T, Wang X-G, Wu X-P, Zhao Z-Z, Zhu C-G, Qiu H-L, Xue L, Shao H-J, Guo M-X, et al. 2010. PU.1 can regulate the ZNF300 promoter in APL-derived promyelocytes HL-60. Leuk Res 34: 1636–1646. 10.1016/j.leukres.2010.04.009 [DOI] [PubMed] [Google Scholar]
- Zhong S, Zhang S, Fan X, Wu Q, Yan L, Dong J, Zhang H, Li L, Sun L, Pan N, et al. 2018. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555: 524–528. 10.1038/nature25980 [DOI] [PubMed] [Google Scholar]
- Zimmer-Bensch G. 2019. Emerging roles of long non-coding RNAs as drivers of brain evolution. Cells 8: 1399. 10.3390/cells8111399 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.