Abstract
Although many genes are regulated by estrogen, very few have been shown to directly bind the estrogen receptor complex. Therefore, transcriptional cascades probably occur in which the estrogen receptor directly binds to a target gene that encodes another transcription factor that subsequently regulates additional genes. Through the use of a differential display assay, a transcription factor has been identified that may be involved in estrogen transcriptional cascades. This report demonstrates that transcription factor δEF1 is induced eightfold by estrogen in the chick oviduct. Furthermore, the regulation by estrogen occurs at the transcriptional level and is likely to be a direct effect of the estrogen receptor complex, as it does not require concomitant protein synthesis. A putative binding site was identified in the 5′-flanking region of the chick ovalbumin gene identifying it as a possible target gene for regulation by δEF1. Characterization of this binding site revealed that δEF1 binds to and regulates the chick ovalbumin gene. Thus, a novel regulatory cascade that is triggered by estrogen has been defined.
Estrogen acting via the estrogen receptor complex is responsible for the transcriptional regulation of many target genes in both reproductive and nonreproductive tissues. In fact, estrogen is being implicated in a growing number of human disease states, including reproductive cancers (17), heart disease (2), osteoporosis (20), and Alzheimer’s disease (3). The mechanisms underlying these physiological manifestations have been the focus of a considerable amount of research in recent years. Mechanisms such as cross talk with other signaling pathways and the interaction of the estrogen receptor complex with both activating and inhibitory transcription complexes have emerged from this recent research. However, a large number of genes still exist that are known to be regulated by estrogen in an indirect manner but whose mechanism of regulation is not fully understood (6). A long-standing hypothesis for the mechanism of action of estrogen, as well as other steroid hormones, is the existence of a regulatory transcriptional hierarchy (1). The best evidence for the existence of this type of hierarchy is the control of molting in Drosophila melanogaster (33). This process is under the control of the steroid hormone ecdysone and proceeds through a transcriptional cascade. While this hypothesis has existed for many years and has been shown to exist in the Drosophila system, there has been little progress in elucidating this type of mechanism in vertebrate systems.
The ability to identify genes that are differentially regulated under various conditions was greatly aided by the description of differential display in 1992 by Liang et al. (13). This technique is ideally suited to the study of hormonal regulation, as it allows the comparison of gene expression in a given tissue plus and minus the hormone. Differential display was employed to identify genes that are induced in the chick oviduct following treatment with 17-β-estradiol. This line of investigation identified previously cloned chicken transcription factor δEF1 (δ-crystallin/E2-box factor) as being regulated by estrogen in the chick oviduct. This is the first report of the regulation of δEF1 by estrogen.
δEF1 was originally cloned in a Southwestern screen designed to identify proteins that bind to the intronic enhancer region of the lens-specific δ-crystallin gene (8). Since its cloning in 1993, homologs have also been identified in mice (9), hamsters (7), rats (9), and humans (34). Analysis of the amino acid sequence reveals intriguing structural features of this protein. δEF1 contains multiple DNA binding motifs with two clusters of zinc fingers, one at the N terminus and one at the C terminus, as well as a homeodomain in the middle of the protein (8). While DNA binding activity has been determined for the zinc finger clusters, the homeodomain failed to show an interaction with DNA (12). In fact, the δEF1 homeodomain has a striking amino acid sequence similarity to the POU homeodomains that have been suggested to play a role in protein-protein interaction rather than in DNA binding (8). Although δEF1 was originally identified in the chick lens, its expression is not limited to the lens. Expression of the δEF1 gene has been detected in mesodermal tissues, as well as the central nervous system (8), and many target genes have been identified (7, 10, 22–34). Some functions for this gene have been determined by investigation of genes that are regulated by δEF1, as well as by creation of transgenic animals. δEF1 is an essential gene, as demonstrated by knockout mice. Mice completely lacking δEF1 die in utero (11). Mutations that remove the N-terminal zinc finger cluster result in mice that have thymic abnormalities, including a deficiency in normal T cells (11). δEF1 mutations that lack the C-terminal zinc finger cluster result in bone abnormalities (32). The data presented herein add a role for δEF1 in the regulation of pathways triggered by estrogen in the chick oviduct and perhaps in other tissues. Estrogen is responsible for both the proliferation and differentiation of tubular gland cells in the chick oviduct. Tubular gland cells are responsible for the production of egg white proteins during egg development (26). Ovalbumin is the major egg white protein found in chicken eggs. Induction of the ovalbumin gene requires administration of estrogen, and maximal induction requires estrogen and a second steroid hormone, which is likely to be glucocorticoid in vivo (24). The proximal 900 bp of the ovalbumin 5′-flanking region are essential for induction by estrogen (27), but this region contains no canonical estrogen response elements (EREs). Furthermore, induction of the ovalbumin gene is sensitive to cycloheximide, a protein synthesis inhibitor (18). The lack of EREs and the cycloheximide-sensitive nature of the induction by estrogen place the ovalbumin gene in the class of secondary response genes (6). Therefore, the ovalbumin gene provides a good model system in which to study regulatory hierarchies that are triggered by estrogen. To this end, the role of δEF1 in a hierarchy of estrogen signaling was tested. Indeed, there exists a transcriptional cascade that is triggered by estrogen and culminates in induction of the ovalbumin gene. The role of δEF1 in the induction or repression of other genes following treatment with estrogen needs to be examined. The potential for understanding the regulatory mechanisms of many other secondary response genes is therefore greatly aided by the identification of the regulation of transcription factor δEF1 by estrogen.
MATERIALS AND METHODS
Differential display.
Two-week-old sexually immature chicks were subcutaneously implanted with two pellets containing diethylstilbesterol (DES; Hormone Pellet Press, Leawood, Kans.), a synthetic estrogen, for 2 weeks. This promotes the proliferation and differentiation of estrogen-responsive tubular gland cells. The pellets were then removed for 5 days to allow estrogen to return to basal levels. The chicks were injected in the wing vein with 17-β-estradiol (25 mg/kg of body weight) and sacrificed at 0.5, 1, 2, or 4 h. Total RNA was extracted from the magnum portion of the oviducts by using RNazol (TelTest, Friendswood, Tex.) and was used in reverse transcription reaction mixtures with a one-base anchored oligo(dT) primer (T11C) (16). PCR was performed on the reverse transcription reaction product by using the oligo(dT)-anchored primer and a nonspecific primer (5′-TGACGTACAC-3′). The products were electrophoresed on a 6% urea gel and analyzed for cDNAs that increased in abundance throughout all time points. The cDNAs identified were cut from the gel, eluted, and subcloned.
Northern blotting with chick oviduct RNA.
Sexually immature chicks were implanted with a pellet containing DES for 2 weeks. The pellets were then removed for 5 days prior to the wing vein injection of 17-β-estradiol for the indicated times. The oviducts were harvested, and total RNA was extracted by using RNazol (TelTest). Poly(A)+ RNA was obtained from the total RNA samples by using the Poly-ATRACT kit (Promega, Madison, Wis.). Northern blot analysis was performed with the indicated cDNA probes labeled by random priming using the Random Primer RmT kit (Stratagene, La Jolla, Calif.). Unless otherwise indicated, Northern blots were exposed to Hyperfilm (Amersham, Arlington Heights, Ill.) for 9 days.
Nuclear runon transcription assay.
Sexually immature chicks were subcutaneously implanted with two DES pellets for 2 weeks and then divided into three groups. (i) The pellets were withdrawn 5 days prior to the isolation of oviduct nuclei, (ii) They remained in place for the additional 5 days, or (iii) They were withdrawn and 5 days later, the chicks were injected intraperitoneally with cycloheximide 0.5 h prior to wing vein injection of 17-β-estradiol (25 mg/kg of body weight) for 2 h. Nuclei were obtained by Dounce homogenization followed by ultracentrifugation. The nuclei were then used in a nuclear runon transcription reaction as previously described (19). Radioactively labeled RNA was extracted from the nuclei and hybridized to filters containing either full-length δEF1 cDNA (courtesy of H. Kondoh) or a 1.4-kb fragment of the gene for osteoblast-specific factor 2 (OSF2), a gene that is not regulated by estrogen, for 36 h at 65°C. The filters were washed and exposed to Hyperfilm (Amersham) for 1 or 4 days as indicated. This was done twice; the blot shown is representative of both experiments.
Generation of a δEF1-specific antibody.
A peptide corresponding to amino acids 8 to 21 (KRRKQANPRRNNVTC) of the chick δEF1 gene was synthesized by the Microchemical Facility at the University of Minnesota. The peptide was conjugated to maleimide-activated keyhole limpet hemocyanin using the Imject activated-immunogen kit (Pierce, Rockford, Ill.). The conjugated peptide was sent to Bethyl Laboratories (Montgomery, Tex.) for generation of the antibody in rabbits. The antiserum was tested for antibody by immunoblotting in vitro-transcribed-translated δEF1 protein (TNT wheat germ kit, Promega). Once the titer was high, the antiserum was immunoglobulin G (IgG) fractionated by Bethyl Laboratories.
Western blotting.
Nuclear protein was extracted either from the oviducts of chicks that had DES pellets implanted for 19 days or from the oviducts of chicks that had DES pellets implanted for 14 days and then withdrawn for 5 days. One hundred micrograms of each of the protein extracts was concentrated by using a Microcon-50 spin column (Amicon, Beverly, Mass.). For both extracts, the resulting protein was split in half and loaded onto a 4 to 20% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (Bio-Rad, Hercules, Calif.). One half of the gel was stained with Coomassie to verify equal loading. The other half of the gel was electrotransferred onto nitrocellulose and used for Western blotting. The blots were blocked in 10% nonfat dry milk for 30 min. The δEF1 antibody was diluted 1:10,000, and the blots were incubated for 1 h. The secondary antibody, goat-anti-rabbit IgG coupled to alkaline phosphatase (Sigma, St. Louis, Mo.), was diluted 1:20,000, and the blots were incubated for 1 h. The antibody was visualized by development with a nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate solution (Pierce).
Gel mobility shift assay.
Synthetic oligonucleotides were generated by the Microchemical Facility at the University of Minnesota. The wild-type ovalbumin sequences (−159 to −141) are as follows, with the nucleotides critical for δEF1 binding underlined and a HindIII restriction site in small letters: agcttTTTGCTCTCCATTCAATCCa aAAACGAGAGGTAAGTTAGGttcga
The oligonucleotide was labeled by Klenow fill-in with [32P]dATP, followed by removal of free nucleotides by putting the reaction over a G-50 spin column (Worthington, Lakewood, N.J.). Nuclear protein extracts were isolated as previously described (19). Binding assays were performed essentially as described previously (29), using 8 μg of laying hen nuclear protein extracts or 2 μl of a 50-μl reaction mixture of in vitro-transcribed-translated protein and the Promega TNT wheat germ kit. When the δEF1 antibody was included, the protein and antibody were incubated together on ice for 15 min prior to their addition to the binding reaction mixture.
Construction of CAT reporter genes and a δEF1 expression vector.
The 6-bp mutant vector pOvCAT-LS-δEF1 was generated by PCR using a primer homologous to nucleotides −177 to −152 of the ovalbumin promoter sequence that carries the mutation over the δEF1 binding site (−153 to −148) and the universal primer located in the vector. A second PCR used a primer homologous to ovalbumin promoter sequences from −147 to −126 harboring the mutant bases over the δEF1 binding site (−148 to −153) and a second primer homologous to chloramphenicol acetyltransferase (CAT) sequences. The resulting two PCR products were allowed to anneal and then subjected to first-round extension and then PCR. The resulting product was subcloned into the BglII/XhoI site of pOvCAT-.900, a vector carrying wild-type ovalbumin sequences from −900 to +9, by endogenous restriction sites. The LS-Z mutant vector was constructed in a similar manner. The δEF1 expression vector was generated by inserting the δEF1 cDNA into the NotI site of the pOPRSVI/MCS vector of the Stratagene Lac-switch system (Stratagene).
Transient transfections of primary tubular gland cells.
Tubular gland cells were isolated from sexually immature chicks from whom estrogen had been withdrawn and were transfected by CaPO4 precipitation as previously described (25). Following precipitation, cells were plated into serum-free medium containing either insulin (50 ng/ml) or insulin plus estrogen (10−7 M) and corticosterone (10−6 M) and cultured for 24 h. Cells were harvested and lysed in Promega Reporter Lysis Buffer. CAT assays were performed by a standard method, with normalization to protein concentration as measured by the Bradford assay, as previously described (28). The overexpression experiments were carried out by cotransfection of pOvCAT-.900 with a constant amount (68 pmol, per sample) of an empty expression vector plus the δEF1 expression vector.
RESULTS
Differential-display analysis identifies the gene for δEF1 as a gene that is regulated by estrogen.
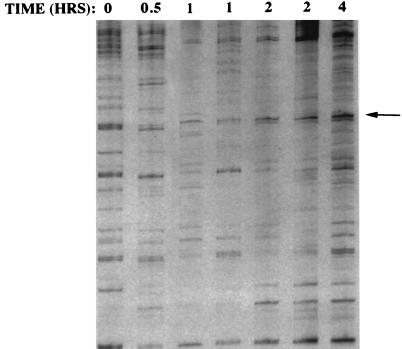
In order to identify genes that are regulated by estrogen in the chick oviduct, we conducted differential display analyses of samples taken from chicks treated with 17-β-estradiol for various times (15). The chick oviduct was chosen as a model system since treatment with 17-β-estradiol results in both the proliferation and differentiation of this tissue, making it a rich source of estrogen-responsive genes. In attempts to bias the system to the identification of transcription factors, only genes that were rapidly induced by estrogen were considered. A time course analysis with multiple points was done in order to limit the number of false positives. A 258-bp cDNA was identified as increasing throughout all time points after 0.5 h (Fig. 1). A BLAST search revealed that this cDNA was 98% identical at the nucleotide level and 100% identical at the amino acid level to previously cloned Gallus gallus transcription factor δEF1. While this transcription factor had been previously cloned, the observation of its regulation by estrogen is novel.
FIG. 1.
Differential display gel showing the time course of induction of oviduct cDNAs by estrogen. The arrow points to the δEF1 cDNA, which fits the criterion of increased abundance throughout the time points after 0.5 h. Each lane represents a sample from an individual chick. The values at the top are times of exposure to an intravenous injection of 17-β-estradiol.
δEF1 is rapidly induced by estrogen in the chick oviduct.
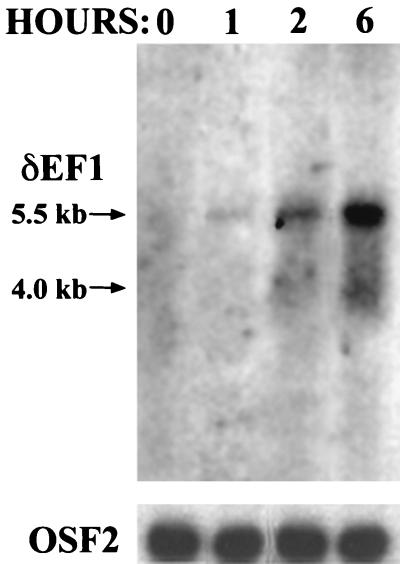
Differential display analysis can often yield false positives, partly due to unequal priming among samples in the reverse transcription reaction and the PCR (14). To verify that δEF1 is induced by estrogen and to determine the kinetics of the induction, a Northern blot analysis was preformed by using RNAs obtained from chicks that were injected in the wing vein with 17-β-estradiol for various times. δEF1 mRNA was induced within 1 h of treatment with estrogen (Fig. 2). The mRNA was induced approximately sevenfold at the 6-h time point, as determined by densitometry. A similar level of induction was seen in RNA isolated from laying hens, which contain physiological levels of estrogen (data not shown). Consistent with previously published results, two mRNA species were observed; the major species detected was 5.5 kb, and the minor species detected was 4.0 kb (8). These two transcripts arose from the use of different polyadenylation sites (8).
FIG. 2.
δEF1 mRNA is induced quickly upon treatment with 17-β-estradiol. Poly(A)+ RNA (2 μg) was extracted from chick oviducts at the indicated times after injection with 17-β-estradiol and subjected to Northern blot analysis using a 0.25-kb cDNA fragment corresponding to nucleotides 2199 to 2449 of the δEF1 gene. This time course agrees well with the induction seen in Fig. 1. The lower panel is the same Northern blot probed with a 1.4-kb fragment of OSF2 cDNA, a gene that is not regulated by estrogen, to show equal loading among the lanes.
The regulation of δEF1 by estrogen occurs at the transcriptional level and does not require concomitant protein synthesis.
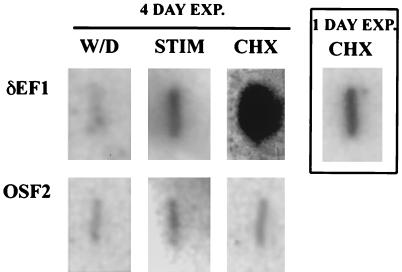
A change in the mRNA level of a gene may be the result of a change in the transcriptional rate of that gene, a change in the stability of that message, or a combination of both. In order to determine whether estrogen causes an increase in the rate of transcription of the δEF1 gene, a nuclear runon experiment was performed. The nuclear runon experiment showed an eightfold increase in the amount of δEF1 mRNA in nuclei isolated from DES-stimulated chick oviducts over nuclei isolated from the oviducts of chicks from whom DES had been withdrawn (Fig. 3). The level of induction seen in the nuclear runon assay was in strict agreement with the level of induction seen in Northern blots, indicating that most, if not all, of the regulation of δEF1 by estrogen occurs at the transcriptional level. To determine whether or not this was likely due to a direct effect of the estrogen receptor complex, the nuclear runon assay was performed with nuclei isolated from chicks that had been treated with both 17-β-estradiol and cycloheximide, an inhibitor of protein synthesis. Inclusion of cycloheximide did not prevent the induction of δEF1 and, in fact, resulted in superinduction of the gene. Superinduction in the presence of cycloheximide has been seen for other transcription factors and is most likely due to the loss of a labile repressor of the gene (4, 21). δEF1 induction is not dependent on concomitant protein synthesis, suggesting that the induction of this gene may well be a direct effect of the estrogen receptor complex.
FIG. 3.
Regulation of δEF1 gene expression by estrogen is at the transcriptional level and does not require concomitant protein synthesis. Nuclei isolated from the oviducts of chicks that were either withdrawn from DES for 5 days (W/D), DES stimulated (STIM), or withdrawn from DES for 5 days and then injected with cycloheximide for 2.5 h and estrogen for 2 h (CHX) were used in a nuclear runon transcriptional assay. The blot reveals that δEF1 is regulated at the transcriptional level and does not require concomitant protein synthesis for expression. The main group of six panels shows the nuclear runon after 4 days of exposure (EXP.) to Hyperfilm (Amersham). The boxed panel on the right shows the effect of cycloheximide and estrogen after 1 day of exposure for clarity. OSF2 cDNA was used as a control.
There is a corresponding increase in δEF1 protein levels following treatment with estrogen.
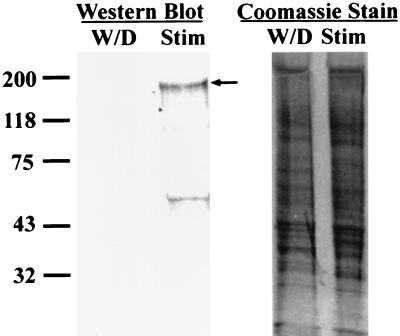
To determine whether protein levels showed a corresponding increase in δEF1 mRNA upon treatment with estrogen, an antibody specific for δEF1 was generated. The antibody was directed against a peptide corresponding to amino acids 8 to 21 of the chick δEF1 protein. Oviduct nuclear protein extracts were prepared from chicks that had been either stimulated with DES or withdrawn from DES treatment for 5 days. Coomassie staining of the gel showed equal loading between lanes (Fig. 4, right panel). Immunoblots with these nuclear extracts showed that the δEF1 protein levels were also induced by estrogen treatment (Fig. 4, left panel, an arrow marks the full-length δEF1). The smaller band of approximately 60 kDa most likely represents a proteolytic fragment of δEF1 generated in the preparation of the extracts, since inclusion of a protease cocktail in the extraction medium significantly reduced the intensity of this band (data not shown). The levels of δEF1 protein in stimulated chick nuclear protein extracts and laying hen nuclear protein extracts were similar (data not shown).
FIG. 4.
δEF1 protein levels are also induced by estrogen. Chick nuclear protein was extracted from DES-stimulated chicks (Stim) or chicks from which DES had been withdrawn for 5 days (W/D). Fifty micrograms of nuclear protein was blotted with an antibody specific for δEF1. The arrow indicates the full-length δEF1 protein. The smaller band likely represents a proteolytic fragment, as inclusion of a protease inhibitor cocktail greatly reduced the intensity of this band (data not shown). The panel on the right shows Coomassie staining to demonstrate approximate equal loading of samples. The values on the left are molecular sizes in kilodaltons.
δEF1 binds to a site in the ovalbumin promoter.
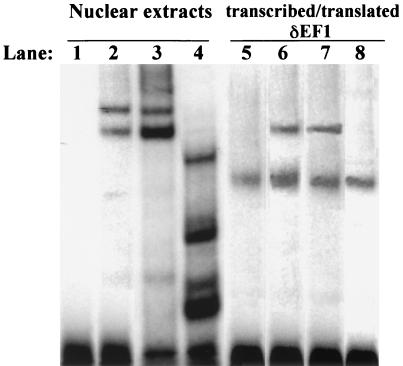
A consensus binding site for δEF1 and its homologs has been established by comparison of sites found in the promoters of genes regulated by δEF1, as well as by the PCR-assisted CASTing technique (12). The binding sites identified all have a C/TACCT core, in either orientation, in common (12, 29). Examination of the ovalbumin 5′-flanking region identified a consensus binding site for δEF1. The gene for ovalbumin is an egg white gene that is induced by estrogen in the chick oviduct (24). The regulation of ovalbumin by estrogen is known to be a secondary response of the estrogen receptor complex (5). To determine whether δEF1 was capable of binding to the putative site in the ovalbumin promoter, a gel mobility shift assay was performed. An oligonucleotide corresponding to the putative δEF1 site in the ovalbumin 5′-flanking region was capable of shifting a complex (Fig. 5, lane 2) from stimulated chick nuclear extracts. This complex was completely abolished by addition of the δEF1 antibody (Fig. 5, lane 4) but not by addition of preimmune serum (Fig. 5, lane 3). While the δEF1 band was abolished by the addition of antibody, several smaller complexes were seen. This could be due to other proteins with weaker binding affinities binding to the site left available by the removal of δEF1 binding activity. There are several proteins which could bind to this site, as it is an E-box binding site recognized by many basic helix-loop-helix proteins (23, 30). Furthermore, the shifted complex seen with chick oviduct nuclear extracts was identical in size to a complex formed by addition of in vitro-transcribed-translated δEF1 protein (Fig. 5, lane 6). The lower band present when the δEF1 in vitro-transcribed-translated protein was shifted was due to a binding activity in the wheat germ extract, as a mock transcription-translation reaction resulted in the formation of the same complex (Fig. 5, lane 5). The complex formed by the addition of the δEF1 in vitro-transcribed-translated protein was also abolished by addition of the δEF1 antibody (Fig. 5, lane 8), whereas addition of preimmune serum did not affect binding (Fig. 5, lane 7). These results demonstrate that δEF1 is capable of binding to the ovalbumin gene and that it is the major component present in oviduct nuclear protein extracts that binds to this element.
FIG. 5.
δEF1 is the component of oviduct nuclear extracts that binds to the ovalbumin gene. A gel mobility shift assay was performed by using the δEF1 binding site from the ovalbumin gene as a probe. Lane 1 shows the probe (−159 to −141) alone. The ovalbumin probe was incubated with 8 μg of oviduct nuclear protein extract (lanes 2 to 4) or with in vitro-transcribed-translated δEF1 protein (lanes 6 to 8) or a mock in vitro transcription-translation reaction mixture (lane 5). Addition of preimmune sera (lane 3) did not affect binding to the ovalbumin probe. Addition of 1 μl of δEF1 antibody (lane 4) abolished the complexes seen with the probe and oviduct nuclear protein (lane 2). Several smaller complexes were seen when δEF1 antibody was included in the binding reaction mixture and were likely due to other proteins binding to the probe in the absence of δEF1 binding. Incubation of the probe with 2 μl of a 50-μl δEF1 in vitro transcription-translation reaction mixture resulted in the formation of a complex (lane 6) that was the same size as complexes formed with nuclear protein extracts (compare lanes 2 and 6). Addition of the δEF1 antibody to the in vitro-transcribed-translated δEF1 abolished all binding (lane 8), while preimmune serum did not affect binding (lane 7). Lane 5 shows the ovalbumin probe with 2 μl of a mock in vitro transcription-translation reaction mixture added.
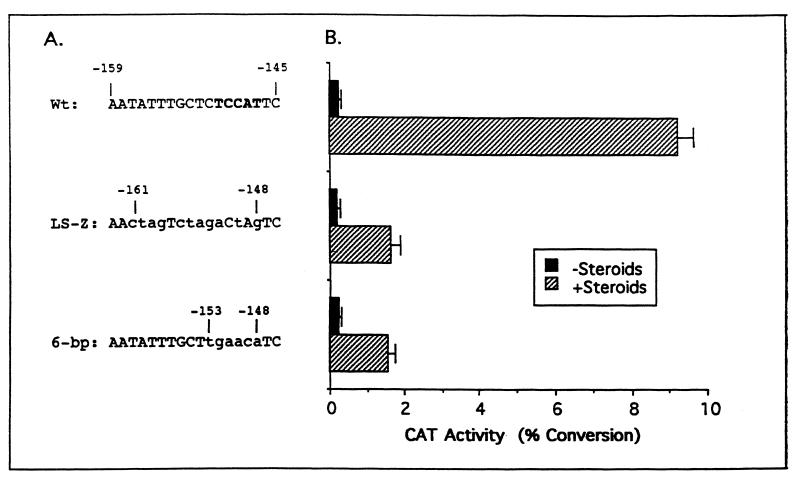
Mutation of the putative δEF1 site in the ovalbumin promoter greatly attenuates transcriptional activity.
In order to determine whether there is a functional consequence of δEF1 binding to the ovalbumin gene, transient transfections were performed. A construct containing ovalbumin 5′-flanking sequences from −900 to +9 (pOvCAT-.900) placed upstream of cat was used as a positive control, as this is the minimal promoter that confers a full response to estrogen (24). The putative δEF1 site lies at −152 to −148 of the ovalbumin gene. Initial experiments conducted in this area of the ovalbumin gene were carried out by using 10-bp linker scanning mutant constructs. The linker scanning mutant construct that covers the putative δEF1 site, termed LS-Z, mutated bases −161 to −147 and resulted in a sixfold reduction in promoter activity (Fig. 6). This mutation only mutated half of the nucleotides that comprise the δEF1 site and mutated other nucleotides that have been shown to be unnecessary for δEF1 binding and activity (8). In order to determine whether the δEF1 site contributed to the ovalbumin activity in this region, a new mutation was made that mutated 6 bp over the δEF1 binding site (Fig. 6A, 6-bp). Mutation of just the 6 bp of the δEF1 binding site resulted in the same reduction of activity as the LS-Z mutation (Fig. 6B), implying that δEF1 is the only factor acting in this region of the ovalbumin promoter. These results indicate that the δEF1 site is essential for maximal induction of the ovalbumin gene by steroids.
FIG. 6.
Mutation of the δEF1 site in the ovalbumin gene causes sixfold attenuation of activity. (A) Sequences of the ovalbumin gene in which mutations were made. Nucleotide positions relative to the transcription start site are indicated. The δEF1 site is indicated by boldface type and lowercase letters indicate mutant bases. (B) Transient transfection of constructs into chick primary tubular gland cells. Following transfection, the cells were cultured in the absence (black bars) or the presence (cross-hatched bars) of estrogen and glucocorticoid. Glucocorticoid is needed to achieve maximal activation of the ovalbumin promoter. Standard errors are indicated by bars. The graph depicted here is a composite of four experiments with each construct tested in duplicate for each treatment. Wt, wild type.
Overexpression of δEF1 results in activation of the ovalbumin promoter in the absence of steroid hormones.
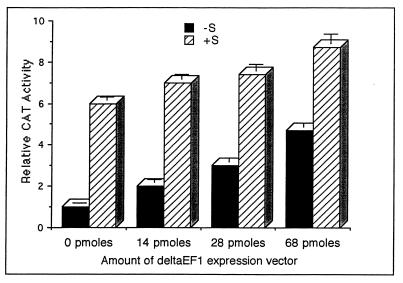
To directly test the hypothesis that steroid activation of the ovalbumin promoter involves δEF1, it was overexpressed in the absence and presence of steroid hormones (Fig. 7). Overexpression of δEF1 caused an increase in the expression of the wild-type ovalbumin promoter in the absence of steroid hormones. The increase in pOvCAT-.900 expression was dose dependent. The largest amount of the δEF1 expression vector used (68 pmol) gave rise to nearly the same level of activation as seen with pOvCAT-.900 plus steroid hormones. Inclusion of steroid hormones in the media of cells overexpressing δEF1 did induce a slight but significant increase in the pOvCAT-.900 response, which may have been due to higher levels of δEF1. These results indicate that δEF1 is involved in the activation of the ovalbumin promoter and may be a key regulator in the steroid hormone response of the ovalbumin promoter.
FIG. 7.
Overexpression of δEF1 in the absence of steroid hormones activates the ovalbumin gene. The δEF1 expression construct was transiently cotransfected into chick primary oviduct cells along with the wild-type ovalbumin reporter construct pOvCAT-.900. The level of induction was normalized to that seen in oviduct cells cotransfected with an empty expression vector in the absence of steroid hormones (−S). The amount of δEF1 expression plasmid transfected is shown with the total amount of DNA being held constant by the addition of an empty expression vector. Transfection was done in both the absence (−S) and the presence (+S) of estrogen and glucocorticoid. The graph shown is for a representative experiment, one of three such experiments, with each construct done in duplicate per treatment. The error bars represent the ranges of duplicate samples.
DISCUSSION
Regulation of δEF1 transcription by estrogen.
δEF1 was isolated in a differential display analysis designed to identify chick oviduct genes that are rapidly induced following treatment with estrogen. We found that estrogen causes an eightfold induction of δEF1 mRNA. Moreover, the induction occurs quickly, with an increase in mRNA being detected within 1 h following estrogen treatment (Fig. 2). The regulation of δEF1 occurs at the transcriptional level, as determined by nuclear runon analysis (Fig. 3). There is also a corresponding induction of δEF1 protein following estrogen stimulation (Fig. 4). This is the first report of the regulation of δEF1 by estrogen or any other specific stimulus. BZP, the hamster homolog of δEF1, is regulated by serum, although the serum component responsible for the regulation is unidentified (7). The proximal 1,100-bp of the δEF1 5′-flanking region have been isolated and do not contain a canonical ERE, although there are two half EREs located at −550 to −546 and −447 to −443 (31).
δEF1 is a positive regulator of the chick ovalbumin gene.
A putative binding site for δEF1 was identified in the chick ovalbumin gene from −148 to −152. δEF1 is capable of binding to this site, as shown by gel mobility shift assay (Fig. 5). Furthermore, mutation of these base pairs attenuates ovalbumin promoter activity, suggesting that δEF1 acts upon this gene (Fig. 6). δEF1 was shown to be directly involved in the activation of this gene by overexpression experiments (Fig. 7). Overexpression of δEF1 in oviduct transient cotransfections shows that δEF1 is capable of inducing the ovalbumin promoter in the absence of steroid hormones. Furthermore, the level of induction seen with the largest amount of δEF1 expression vector used in the absence of steroid hormones was nearly the same as that seen in the presence of steroid hormones without overexpression of δEF1. This indicates that the δEF1 protein is a key regulator of ovalbumin transcription. Although it is known that multiple other sites are required for induction of the ovalbumin gene (25), it is clear from these experiments that δEF1 is necessary to achieve activation in response to estrogen.
Role of δEF1 as a transcriptional regulator.
Transcription factor δEF1 was originally identified as a negative regulator of the lens-specific gene for δ-crystallin (8). Now it is known that δEF1 and its homologs regulate a wide variety of genes, including those for δ-crystallin (8), alpha4integrin (22), MyoD (29), interleukin-2 (35), IgH (9), and the Na,K-ATPase (34). While δEF1 has been shown to act as a repressor of transcription in most instances, a role for δEF1 as an activator has been suggested for two reasons. First, analysis of the amino acid sequence reveals proline-rich and acidic-residue-rich domains, as is common in other transcriptional activators (8). Second, δEF1 can activate a construct that contains a consensus δEF1 binding site upstream of the hsp70 promoter in rat neuroblastoma or HeLa cells (34). The hypothesis has been proposed that δEF1 can exert opposite effects on genes based upon what zinc finger cluster is binding to DNA (12). The data presented herein demonstrate that δEF1 is capable of positively regulating the ovalbumin gene.
Role of δEF1 in estrogen transcription cascades.
Regulation of transcription factor δEF1 by estrogen and subsequent identification of the ovalbumin gene as a target for this regulation complete a regulatory cascade in the chick oviduct. Despite the long-standing hypothesis of the existence of transcriptional regulatory cascades, little evidence, until now, has supported this hypothesis in vertebrate systems. It is interesting to speculate that δEF1 may be involved in the regulation of many other genes that are regulated by estrogen in an indirect manner. The ability of δEF1 to act as either an activator or a repressor of target gene transcription further expands the regulatory potential of this single protein. The observation that δEF1 is expressed in tissues such as the brain, heart, breast, and oviduct, where estrogen is known to play an important physiological role, supports an attractive model in which δEF1 is involved in mediating the effects of estrogen in these tissues, as well as others.
ACKNOWLEDGMENTS
We thank Hisato Kondoh for his generous gift of the full-length δEF1 cDNA. We also thank Karl Sensenbaugh, Dave Monroe, and Steve Hagen for their technical assistance.
This research was supported by NIH grant RO1 DK40082 to M.M.S.
REFERENCES
- 1.Ashbruner M, Chihara C, Meltzer C, Richard G. Temporal control of puffing in polytene chromosomes. Cold Spring Harbor Symp Quant Biol. 1974;38:655–662. doi: 10.1101/sqb.1974.038.01.070. [DOI] [PubMed] [Google Scholar]
- 2.Barrett C E. Postmenopausal estrogen and heart disease. Atherosclerosis. 1995;118:S7–S10. [PubMed] [Google Scholar]
- 3.Behl C, Widmann M, Trapp T, Holsboer F. 17-Beta estradiol protects neurons from oxidative stress-induced cell death in vitro. Biochem Biophys Res Commun. 1995;216:473–482. doi: 10.1006/bbrc.1995.2647. [DOI] [PubMed] [Google Scholar]
- 4.Chiou S T, Chang W C. Insulin-like growth factor I stimulates transcription of the c-jun proto-oncogene in Balb/C 3T3 cells. Biochem Biophys Res Commun. 1992;183:524–531. doi: 10.1016/0006-291x(92)90513-k. [DOI] [PubMed] [Google Scholar]
- 5.Dean D M, Jones P S, Sanders M M. Regulation of the chick ovalbumin gene by estrogen and corticosterone requires a novel DNA element that binds a labile protein, Chirp-1. Mol Cell Biol. 1996;16:2015–2024. doi: 10.1128/mcb.16.5.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean D M, Sanders M M. Ten years after—reclassification of steroid-responsive genes. Mol Endocrinol. 1996;10:1489–1495. doi: 10.1210/mend.10.12.8961259. [DOI] [PubMed] [Google Scholar]
- 7.Franklin A J, Jetton T L, Shelton K D, Magnuson M A. BZP, a novel serum-responsive zinc finger protein that inhibits gene transcription. Mol Cell Biol. 1994;14:6773–6788. doi: 10.1128/mcb.14.10.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 9.Genetta T, Kadesch T. Cloning of a cDNA encoding a mouse transcriptional repressor displaying striking sequence conservation across vertebrates. Gene. 1996;169:289–290. doi: 10.1016/0378-1119(95)00824-1. [DOI] [PubMed] [Google Scholar]
- 10.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda K, Kawakami K. DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur J Biochem. 1995;233:73–82. doi: 10.1111/j.1432-1033.1995.073_1.x. [DOI] [PubMed] [Google Scholar]
- 13.Liang P, Averboukh L, Keyomarsi K, Sager R, Pardee A B. Differential display and cloning of messenger RNAs from human breast cancer versus mammary epithelial cells. Cancer Res. 1992;52:6966–6968. [PubMed] [Google Scholar]
- 14.Liang P, Averboukh L, Pardee A B. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993;21:3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang P, Pardee A B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 16.Liang P, Zhu W, Zhang X, Guo Z, O’Connell R P, Averboukh L, Wang F, Pardee A B. Differential display using one-base anchored oligo-dT primers. Nucleic Acids Res. 1994;22:5763–5764. doi: 10.1093/nar/22.25.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobo R A. Benefits and risks of estrogen replacement therapy. Am J Obstet Gynecol. 1995;173:982–989. doi: 10.1016/0002-9378(95)90247-3. [DOI] [PubMed] [Google Scholar]
- 18.McKnight G S, Palmiter R D. Transcriptional regulation of the ovalbumin and conalbumin genes by steroid hormones in chick oviduct. J Biol Chem. 1979;254:9050–9058. [PubMed] [Google Scholar]
- 19.Nordstrom L A, Dean D M, Sanders M M. A complex array of double-stranded and single-stranded DNA-binding proteins mediates induction of the ovalbumin gene by steroid hormones. J Biol Chem. 1993;268:13193–13202. [PubMed] [Google Scholar]
- 20.Pacifici R. Estrogen, cytokines, and pathogenesis of postmenopausal osteoporosis. J Bone Mineral Res. 1996;11:1043–1051. doi: 10.1002/jbmr.5650110802. [DOI] [PubMed] [Google Scholar]
- 21.Paquette R L, Minosa M R, Verma M C, Nimer S D, Koeffler H P. An interferon-gamma activation sequence mediates the transcriptional regulation of the IgG Fc receptor type IC gene by interferon-gamma. Mol Immunol. 1995;32:841–851. doi: 10.1016/0161-5890(95)00056-k. [DOI] [PubMed] [Google Scholar]
- 22.Postigo A A, Dean D C. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postigo A A, Sheppard A M, Mucenski M L, Dean D C. c-Myb and Ets proteins synergize to overcome transcriptional repression by ZEB. EMBO J. 1997;16:3924–3934. doi: 10.1093/emboj/16.13.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders M M, McKnight G S. Chicken egg white genes: multihormone regulation in a primary cell culture system. Endocrinology. 1985;116:398–405. doi: 10.1210/endo-116-1-398. [DOI] [PubMed] [Google Scholar]
- 25.Sanders M M, McKnight G S. Positive and negative regulatory elements control the steroid-responsive ovalbumin promoter. Biochemistry. 1988;27:6550–6557. doi: 10.1021/bi00417a053. [DOI] [PubMed] [Google Scholar]
- 26.Schutz G, Nguyen-Huu M C, Giesecke K, Hynes N E, Groner B, Wurtz T, Sippel A E. Hormonal control of egg white protein messenger RNA synthesis in the chicken oviduct. Cold Spring Harbor Symp Quant Biol. 1978;42:617–624. doi: 10.1101/sqb.1978.042.01.064. [DOI] [PubMed] [Google Scholar]
- 27.Schweers L A, Frank D E, Weigel N L, Sanders M M. The steroid-dependent regulatory element in the ovalbumin gene does not function as a typical steroid response element. J Biol Chem. 1990;265:7590–7595. [PubMed] [Google Scholar]
- 28.Schweers L A, Sanders M M. A protein with a binding specificity similar to NF-kappa B binds to a steroid-dependent regulatory element in the ovalbumin gene. J Biol Chem. 1991;266:10490–10497. [PubMed] [Google Scholar]
- 29.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa S A, Nabeshima Y, Kondoh H. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekido R, Murai K, Kamachi Y, Kondoh H. Two mechanisms in the action of repressor deltaEF1: binding site competition with an activator and active repression. Genes Cells. 1997;2:771–783. doi: 10.1046/j.1365-2443.1997.1570355.x. [DOI] [PubMed] [Google Scholar]
- 31.Sekido R, Takagi T, Okanami M, Moribe H, Yamamura M, Higashi Y, Kondoh H. Organization of the gene encoding transcriptional repressor delta EF1 and cross-species conservation of its domains. Gene. 1996;173:227–232. doi: 10.1016/0378-1119(96)00185-0. [DOI] [PubMed] [Google Scholar]
- 32.Takagi T, Moribe H, Kondoh H, Higashi Y. DeltaEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 33.Thummel C S. From embryogenesis to metamorphosis: the regulation and function of Drosophila nuclear receptor superfamily members. Cell. 1995;83:871–877. doi: 10.1016/0092-8674(95)90203-1. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe Y, Kawakami K, Hirayama Y, Nagano K. Transcription factors positively and negatively regulating the Na,K-ATPase alpha 1 subunit gene. J Biochem. 1993;114:849–855. doi: 10.1093/oxfordjournals.jbchem.a124267. [DOI] [PubMed] [Google Scholar]
- 35.Yasui D H, Genetta T, Kadesch T, Williams T M, Swain S L, Tsui L V, Huber B T. Transcriptional repression of the IL-2 gene in Th cells by ZEB. J Immunol. 1998;160:4433–4440. [PubMed] [Google Scholar]