Abstract
This paper examines how ancient DNA data can enhance radiocarbon dating. Because there is a limit to the number of years that can separate the dates of death of related individuals, the ability to identify relatives through ancient DNA analysis can serve as a constraint on radiocarbon date range estimates. To determine the number of years that can separate related individuals, we modeled maximums derived from biological extremes of human reproduction and death ages and compiled data from historic and genealogical death records. We used these data to jointly study the date ranges of a global dataset of individuals that have been radiocarbon dated and for which ancient DNA analysis identified at least one relative. We found that many of these individuals could have their date uncertainties reduced by building in date of death separation constraints. We examined possible reasons for date discrepancies of related individuals, such as dating of different skeletal elements or wiggles in the radiocarbon curve. We also developed a program, refinedate, which researchers can download and use to help refine the radiocarbon date distributions of related individuals. Our research demonstrates that when combined, radiocarbon dating and ancient DNA analysis can provide a refined and richer view of the past.
Keywords: Ancient DNA, radiocarbon dating, genealogy, OxCal, Bayesian analysis
1. Introduction
This article examines how ancient DNA data can be used to help with a central aspect of archaeological research—chronology. Ancient DNA data are revolutionizing the field of archaeology. In slightly over a decade, ancient DNA analyses have discovered new hominins (Reich et al., 2010), elucidated the spread of farming through Europe (Lazaridis et al., 2016; Mathieson et al., 2015), shed light on the peopling of the Americas and Oceania (Lipson et al., 2018; Moreno-Mayar et al., 2018; Posth et al., 2018; Rasmussen et al., 2014; Skoglund et al., 2016), and more. While ancient DNA has helped provide insight on long-standing archaeological questions, exponentially increasing ancient DNA data have created unique opportunities for the examination of finer-grained issues, and even archaeological methods.
The basis of the work presented here is tied to the fact that there is a maximum number of years that can separate the dates of death (DOD) for two or more genetically related individuals (Mittnik et al., 2019). For example, it is exceedingly rare for a mother to die 100 or more years before her daughter, particularly in pre-industrial societies. Thus, if two or more individuals are identified as biological relatives through ancient DNA analysis and those individuals are radiocarbon dated, their relatedness can be used as a prior or constraint when analyzing their overlapping radiocarbon date ranges. Using these constraints, we examine how biological relatedness can be used to constrain radiocarbon date ranges and increase dating precision, if application of the methods to a large database of published ancient DNA data (https://reich.hms.harvard.edu/downloadable-genotypes-present-day-and-ancient-dna-data-compiled-published-papers) can reveal broad patterns, and present a software package, refinedate, which integrates prior radiocarbon date distributions with a prior distribution from DOD estimates to help refine the radiocarbon date range distributions of related individuals.
2. Materials and Methods
2.1. Identification of genetic relatives with ancient DNA
Identification of genetic relatives has become standard practice in ancient DNA analysis. Typically, individuals that are screened and produced working genomic data are compared against each other and previously analyzed individuals from similar geographic regions and time periods to identify unique genetic relationships. For each pair of individuals in this study, we computed the mean mismatch rate using all the autosomal single nucleotide polymorphisms (SNPs) with at least one sequencing read for both individuals in the comparison (this procedure to identify genetic relatives is described in Kennett et al. (2017:156) and van de Loosdrecht et al. (2018:15), and is similar to that in Kuhn et al. (2018:157)). In the cases with more than one sequencing read at a particular SNP for a given individual, we randomly sample one for analysis. We then estimate relatedness coefficients as in Kennett et al (2017:156): r = 1–((x-b)/b), with x being the mismatch rate and b the base mismatch rate expected for two genetically identical individuals from that population, which we estimate by computing intra-individual mismatch-rates. We also compute 95% confidence intervals using block jackknife standard errors (Olalde et al., 2019:S61). While such analysis can detect relationships up to the 5th degree, we limit relationships here to 3rd degree maximum, as DOD date separations become substantially less useful with decreasing genetic relatedness (e.g. great-grandparents and great-grandchildren). In some instances, we were able to determine specific relationships. Parent-offspring relationships must show r coefficient of 0.5 along all the autosomes because they always share one chromosome. Siblings will have 25% of the regions with r=0, 25% of the regions with r=1 and 50 % of the regions with r=0.5. Additionally, mtDNA lineages can add further specificity. Two male individuals identified as parent-offspring with different mtDNA must be father and son (or the other way around), and a male and a female with different mtDNA must be father-daughter.
2.2. Genetic relatives and DOD separation maximums
Below, two approaches—biological maximums and genealogically and historically derived estimates—are examined for determining the DOD separation of genetically related individuals. The biological maximums serve as theoretical extremes that are very unlikely to be exceeded, especially in pre-industrial archaeological cultures. Genealogically and historically (GH) derived DOD separations were created through the examination of genealogical records and historic data and reflect more realistic estimates of the number of years between the death of two related individuals.
2.3. Biological Maximum DOD Estimates
Biological maximum estimates use extremes of human reproduction and lifespan to produce maximum DOD separation estimates. Figures 1 and 2 (also see SM 1) are diagrams of how these estimates were modeled. To begin, a 15-year-old couple parent a male child. This child dies at birth, but both parents live to be 100 years old. Thus, the DOD separation between the child and parents would be 85 years. If instead the mother died during childbirth, but the child lived to 100 years old, the maximum DOD separation between parents-offspring would be 100 years. Siblings have an even greater potential maximum DOD separation, as Figure 2 demonstrates. In this model, the 15-year-old couple has a male child that dies at childbirth. That same couple has another child 30 years later (when they are 45 years old); that second child then dies 100 years later. So, the maximum separation between the siblings is 130 years.
Figure 1.
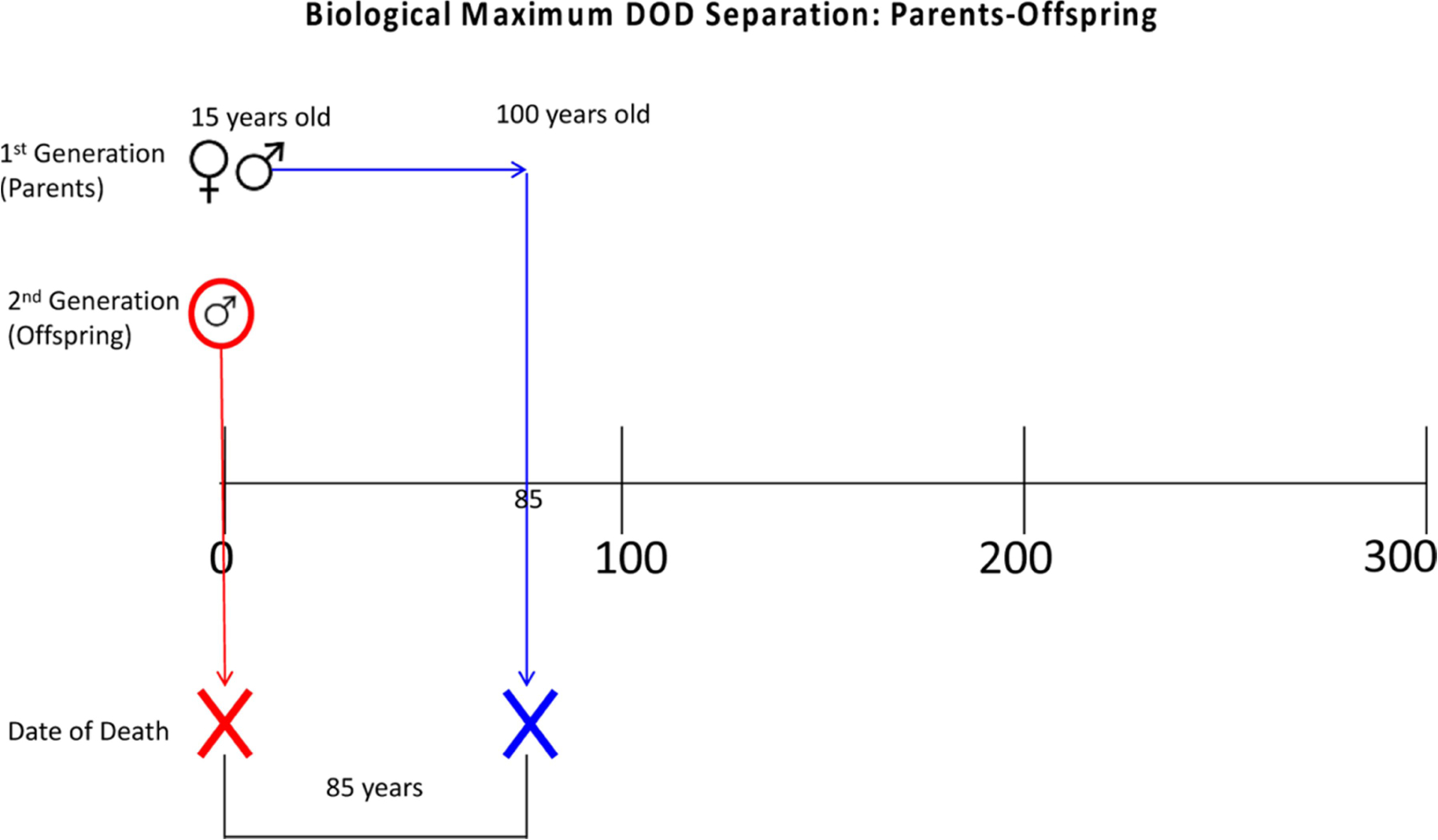
Model of biological maximum date of death separation for parents-offspring.
Figure 2.
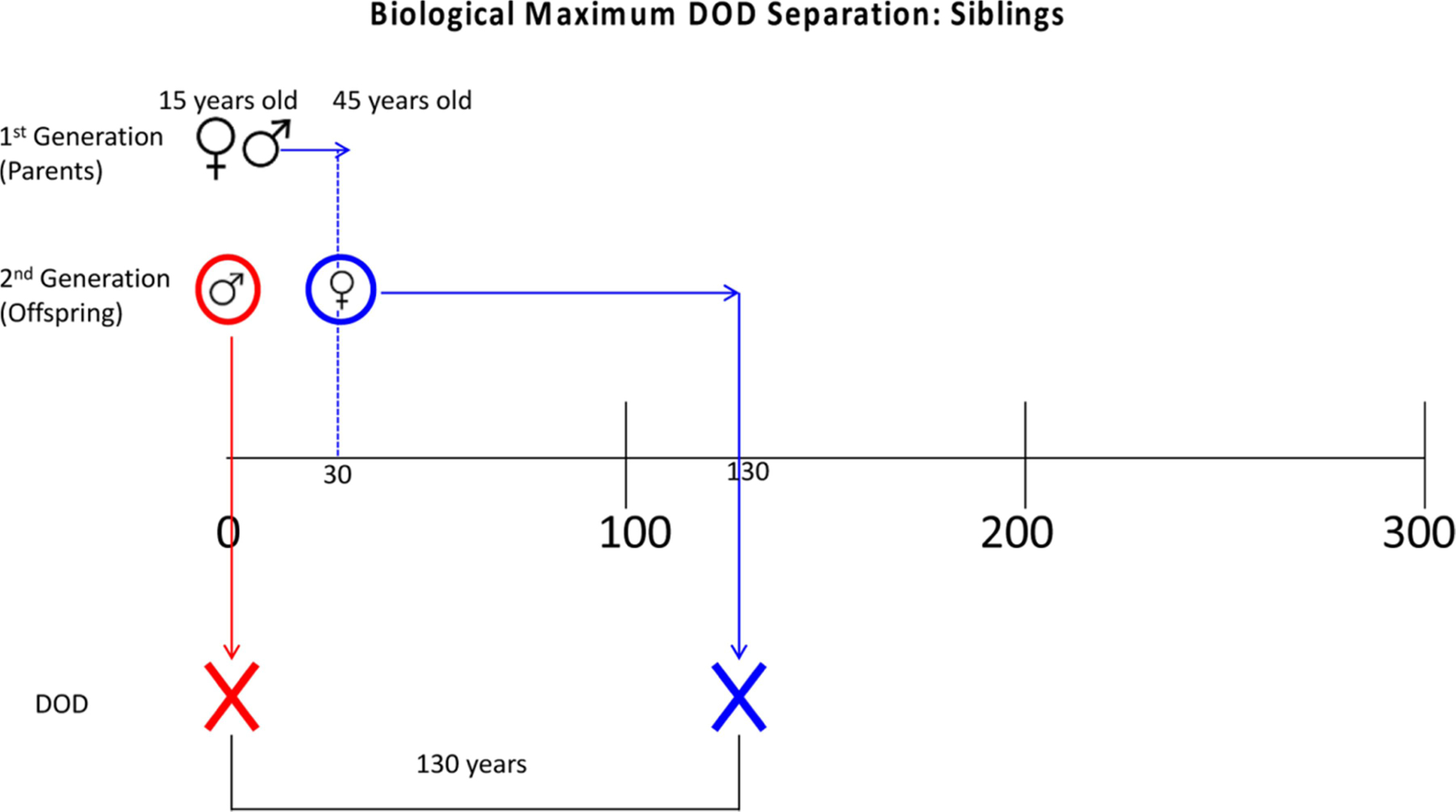
Model of biological maximum date of death separation for siblings.
Using these parameters, we modeled a number of potential biological maximums for various degrees of genetic relatedness (Table 1; note that we have only provided a few key examples here, for example cousins as 3rd degree relatives. While there are many more types of 3rd degree relationships, they typically would have DOD separations that exceed the 195-year cousin maximum). The biological DOD maximums presented above and in Table 1 are reliant on extremes—producing children at the biologically earliest and latest possible ages and living to extreme old age (see SM 1 for model diagrams). While possible, these DOD separations are not realistic and are largely ineffective as constraints on C14 date range distributions. Thus, to more effectively examine how DOD separations for related individuals can be applied to radiocarbon date ranges, we also compiled birth and death data from historical and genealogical records.
Table 1.
Theoretical DOD separation biological maximums (ordered by maximum year separation)
| Relation | Max Years Separation |
|---|---|
| 1st (Parents-Offspring) | 100 |
| 1st (Siblings) | 135 |
| 2nd (Grandparents-Grandchildren) | 180 |
| 3rd (Cousins) | 195 |
| 2nd (Aunts/Uncles-Nieces/Nephews) | 210 |
2.4. Genealogically and Historically Derived DOD Estimates
We began compiling data on the DOD separations for related individuals by consulting the plethora of genealogical and historical data that are publicly available online. Many of these databases contain data primarily from people of European ancestry who lived within the last two centuries. To create DOD estimates from more heterogenous data, we sought non-European focused databases for relatives’ death dates. Data were gathered from historic Anglo cemeteries, and online databases of birth and death dates for Cherokee, Tlingit, and other Native American groups (SM 2). Data were sorted by categories of relatedness: parent-offspring, sibling, grandparent-grandchild, and other 2nd–3rd degree.
The DOD separation for related individuals was compiled into a spreadsheet for each genealogical database (SM 2). DOD separations were calculated by identifying related individuals then subtracting the dates of death (i.e. if a mother and daughter were identified, and the mother died in 1800 CE and the daughter 1850 CE, the separation between the two entered in the database would be 50). For parent-child and grandparent-grandchild relationships the signed value of the DOD was recorded. As will be discussed later, knowing whether the child died before the parent, which would result in a negative value, is useful for building constraints of parent-child and grandparent-grandchild radiocarbon ranges. However, since in many instances ancient DNA cannot determine the relatedness direction of two individuals (e.g. which is the mother and which is the daughter due to shared mitochondrial haplogroups) the absolute value of DOD separation of each relative pair was also recorded for each relationship type and is primarily used for the analyses below.
A total of 5235 relative DOD separations were recorded: 800 parent-offspring, 813 sibling, 485 grandparent-grandchild, and 3137 other 2nd–3rd degree. The means, medians, and standard deviations of the absolute value for each relationship type were then calculated; the results are provided in Table 2 (see also SM 2).
Table 2.
Compiled genealogical and historical data for DOD absolute value separation
| Parent-Offspring | Sibling | Grandparent-Grandchild | Other 2nd–3rd relationships | |
|---|---|---|---|---|
| Mean | 28.84 | 26.33 | 35.00 | 34.94 |
| Median | 26 | 20 | 39 | 30 |
| Standard Dev. | 18.94 | 22.41 | 31.93 | 25.6 |
The data in Table 2 demonstrate that the biologically maximum DOD separation estimates in Table 1 are truly extremes. The largest mean separation in the GH dataset was 35.00 years between grandparents-grandchildren. The single greatest DOD separation in all the data was 117 years between Cherokee 2nd/3rd degree relatives—still 93 years short of the 2nd/3rd degree maximum theoretical estimate (210 years). The mean GH DOD separation estimates for parents-offspring, siblings, and grandparents-grandchildren are 71.16, 108.67, and 145.00 years less than the biological maximum separation estimates (Table 1), respectively.
The DOD separations above were produced by manual collection from online, publicly available resources. In a 2018 study, however, Kaplanis and colleagues developed software and an analysis pipeline to examine genealogies of millions of individuals downloaded from the online genealogical database geni.com. Kaplanis et al. (2018) used this data to construct family trees (sometimes containing millions of individuals); the anonymized data from this study were made available to download (https://familinx.org/). Significantly, the data contained information on which individuals had parent-offspring relationships, and the death date for each individual. We therefore downloaded these data and found the DOD separation for over 8 million parents and offspring (SM 3.1). We removed pairs with clear data errors (for example a death date of 3500) and used the biological DOD separations defined above for parent-offspring as constraints (i.e. 85 years for children dying before parents and 100 years for parents dying before children; SM 3.2). The mean absolute value DOD separation for these 8 million parent-offspring pairs was 31.43 years, slightly higher than the mean value we manually collected (28.84; SM 2); however, this should be expected as the geni.com data is heavily weighted toward individuals of European ancestry from industrialized societies who likely had longer life spans. Overall, the similarity between the Kaplanis et al. 2018 data and the genealogical and historical data we manually curated demonstrates that the DOD separations we obtained represent more realistic DOD separations for related individuals than the biological maximums.
Although the DOD separation estimates derived from GH data are more reflective of separations between genetic relatives than the biologically possible maximums, the GH data presented here should be viewed only as rough estimates. More precise estimates could be tailored for particular types of social organization, such as hunter-gatherers, pastoralists, agriculturalists, city-dwellers, nomads, etc., and if age of an individual can be determined from morphological assessment.
3. Application and analysis
3.1. Applications of relatedness data to radiocarbon dated individuals
We examined the Reich Laboratory ancient DNA database of published individuals from geographic locales across the globe spanning more than 30,000 years (although there is bias towards the last 10000 years in western Eurasia; see Marciniak and Perry, 2017; Reich, 2018) to test how DOD separation estimates can be applied to constrain radiocarbon data of related individuals and examine if any new insights can be revealed. As of March 2021, 5,699 published individuals were in the database, with 837 ancient individuals having at least one identified relative. Of those, 203 pairs (274 unique individuals, SM 4.1 and 4.2) had both individuals C14 dated (all dates generated with AMS and calibrated two-sigma, IntCal20; SM 5.1), allowing for analysis of DOD separations and constraints. Below, we examine the various ways knowledge of relatedness can be applied to radiocarbon dates and explore methods for constraining date ranges.
3.2. Outlier identification
The most basic example of how genetic relatedness can help refine radiocarbon dating is through the identification of anomalies. Archaeologists have long recognized that C14 sample contamination can occur and that other issues, such as the marine reservoir effect, can cause dates to be biased, in some cases by many hundreds of years (Taylor and Bar-Yosef, 2014). Genetic relatedness is a new independent measurement that can be used to test the validity of radiocarbon date ranges, particularly for dates that might not be obvious outliers. For example, if five individuals from the same stratigraphic layer in a cemetery were dated, and four of those individuals had calibrated ranges of approximately CE 1–150, while one had a calibrated date range of approximately 600–400 BCE, the early individual would seem suspicious and might be redated (Figure 3) even without knowledge of genetic relatedness. If the outlier individual instead had a range of approximately 150 BCE-CE 50 the individual might not be redated, as it is relatively close to the range of the other four individuals (Figure 3). But, if it was determined that the outlier was actually the father of individual 3, then it would be suspicious that individual 5 is inferred to potentially be hundreds of years older than individual 3, as the DOD separation between father and offspring cannot biologically be more than 100 years, and more realistically is around 29 years from GH DOD estimates (Table 1). Such an example was discovered in the database with individuals I2457 and I2600.
Figure 3.
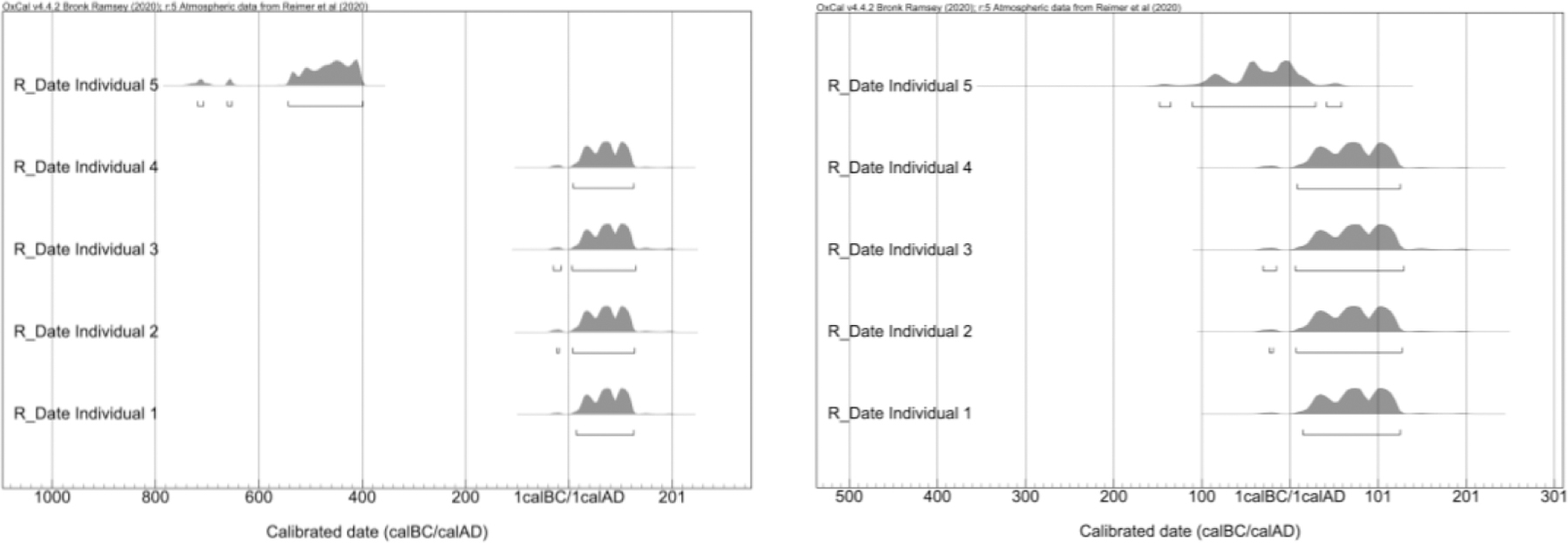
Hypothetical examples of clear outliers in radiocarbon dating (L) and an individual that is an outlier but does have overlap with other dates (R).
Individuals I2457 and I2600 (Olalde et al., 2018) were excavated from two Bell Beaker sites, Amesbury Down and Porton Down, Britain, separated by approximately 5km. The individuals had been radiocarbon dated prior to ancient DNA analysis, and the dates did not seem suspect based on the archaeological record (I2457= 3890±30; 2480–2280 calBCE, SUERC-36210; I2600=3646±27; 2140–1940 calBCE, SUERC-43374; Figure 4)1. Ancient DNA analysis of the individuals revealed that I2600 was the daughter of I2457 (due to them sharing 50% of their autosomal genomes and with differing mtDNA haplotypes), but there was no overlap in the calibrated distributions of the father-daughter pair. The minimum DOD separation between the father and daughter was 140 years, which exceeds even maximum biological estimates. Individual I2457 (the father) was therefore redated and the new date (3717±28; 2200–2031 calBCE; SUERC-69975) fit within the expected DOD range.
Figure 4.
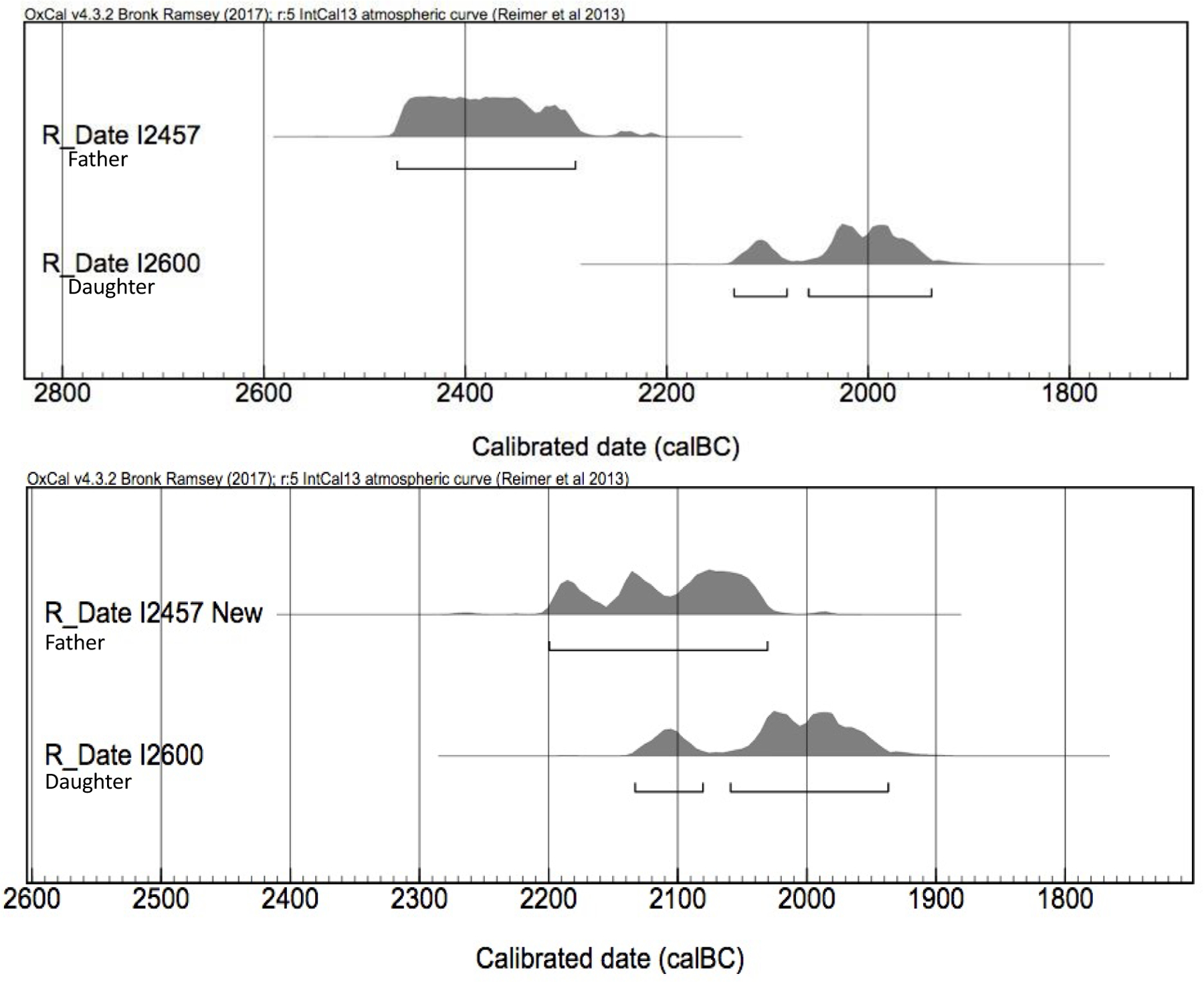
Original (top) and re-dated (bottom) 2sigma ranges for I2457 and I26001.
We identified five related pairs (10 individuals) that had non-overlapping ranges similar to the initial ranges of I2457 and I2600 (Figure 4). We removed these pairs and individuals from our dataset, leaving us with 198 pairs and 264 unique individuals.
3.3. Range tightening of radiocarbon date distributions
Along with detecting outliers, we examined the potential of using DOD separations to refine calibrated date ranges in instances where two (or more) related individuals with overlapping C14 date probability distribution ranges are identified. Using biological maximums as an example, consider a father whose AMS range is 1–500 calCE and a daughter whose range is 400–1000 calCE. Since the father cannot have died more than 100 years before or after the daughter, the father’s range can be constrained to approximately 300–500 CE; since the daughter cannot have died more than 100 years after the father, the daughter’s range can be constrained to 400–600 CE. Yet the maximum biological DOD separation is often too large and not applicable to most related and dated individuals in the dataset. We therefore examined how knowledge of relatedness and the GH DOD separations identified in Table 2 could be applied to related pairs.
Bayesian analysis to increase accuracy in a series of radiocarbon dates has become standard in archaeology (Bronk Ramsey, 2009; Taylor and Bar-Yosef, 2014). Although not designed explicitly to incorporate knowledge of genetic relatedness, the Bayesian functions built into OxCal can be used to constrain date ranges using genetic data on relatedness. We employed the “interval function” in OxCal to examine the relative pairs. The interval function allows users to constrain the duration between two or more dates. In archaeological context, for example, this would allow users to tell OxCal that a date must occur between certain years (e.g. CE 1200–1400). We used the GH DOD data we compiled as interval constraints in OxCal 4.4.2 (IntCal 20) for each relative pair in our dataset, applying the mean and standard deviation for each specific type of relationship identified in Table 2. If genetic analysis could not determine a specific type of relationship for each pair (for example it was determined that the individuals were first degree relatives, but it was unclear whether they were father-son or brothers), we used the largest mean and standard deviation for that degree relationship (e.g. 29 years with a 19 year SD for 1st degree relatives) Using I2457 and I2600 as examples (Figure 5), the interval function modelled 2-sigma ranges of 2200–2037 calBCE and 2137–1983 calBCE, respectively. This was a reduction of 9 years from the original unmodelled 2-sigma range for I2457 and 47 years for I2600.
Figure 5.
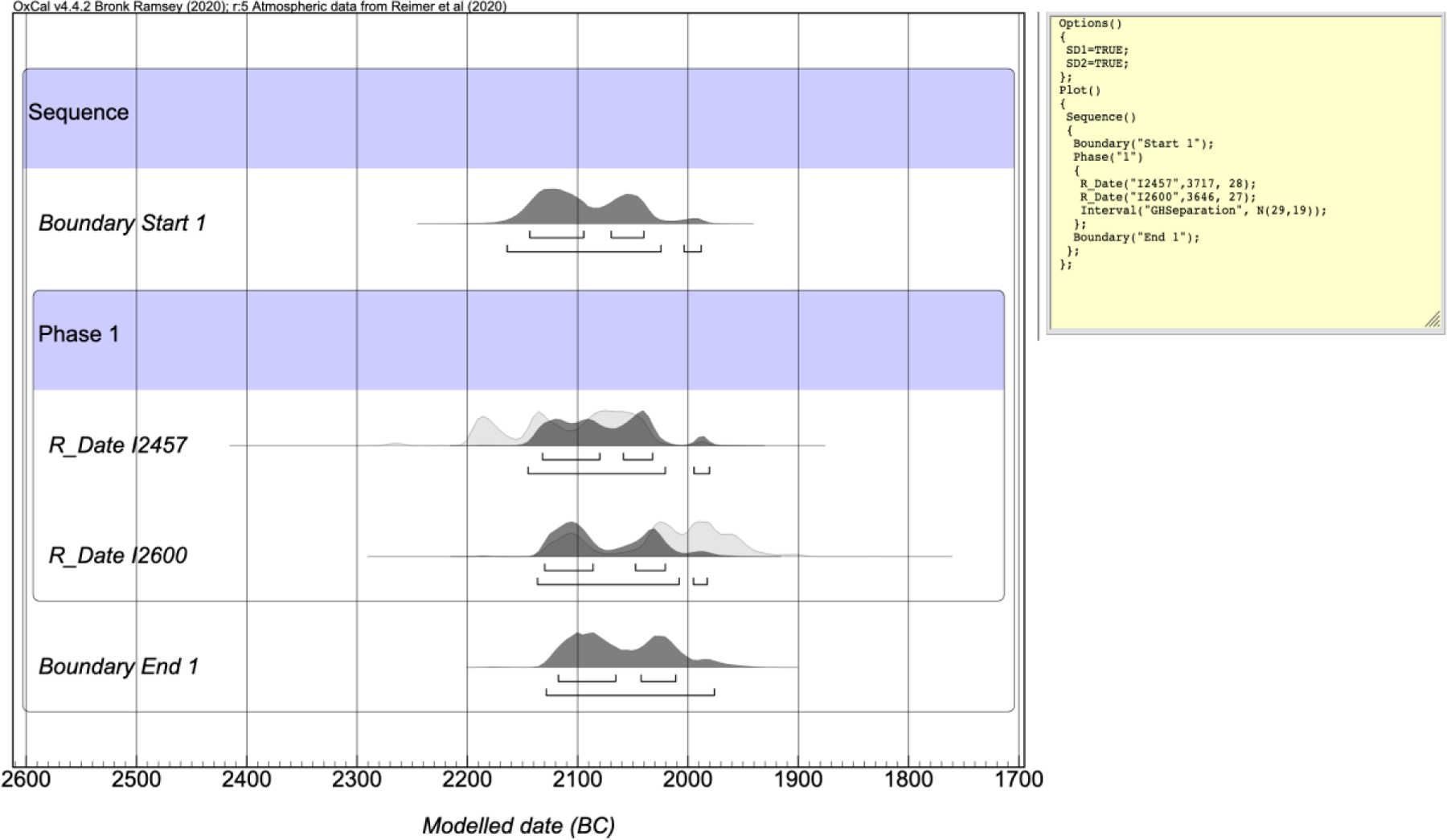
OxCal interval function analysis of father-daughter pair.
We also explored novel methods for constraining date ranges with relatedness data. We developed a new tool, which we call refinedate (described in SM 5.3; software available at https://github.com/DReichLab/refinedate01), that uses prior date distribution probabilities and the values compiled from GH data (SM 2). As mentioned above, the interval function analysis available in OxCal is only able to incorporate the mean GH DOD separation values (e.g. 26 years for siblings). Refinedate allows us to incorporate the more robust GH DOD data we compiled, along with prior date distributions for individuals from OxCal, to construct a joint posterior distribution. Refinedate requires three files; the prior date distribution file of relative a, the prior date distribution file for relative b, and a prior distribution file of the DOD difference for the specific type of relationship between the two individuals. Refinedate takes from the input prior distributions {a(i),b(j)} for the date of death in year i for sample A, year j for sample B. We take these priors to be independent. Then the joint distribution z(i,j) is given by:
where d is our difference distribution, and the constant of proportionality is chosen so that:
Refinedate outputs marginal posterior estimates a′(i),b′(j) and (optionally) z(i,j).
We again use I2600 and I2457 as examples. To obtain the date distributions for each individual we imported the raw calibrated date probability distributions for I2600 and I2457 from OxCal, which were generated in five-year intervals (SM 5.3; data provided in SM 6.1). We then ran these through gsmooth, a utility data smoother in refinedate that produces files in the program’s required one-year interval format. Refinedate then computed the posterior joint distributions for the individuals using the parent-offspring prior distribution derived from Kaplanis et al. (2018; SM 6.2). Figure 9 provides a plot of the distributions for I2457 and I2600 and Table 3 provides the mean calibrated date BCE and standard deviation for each individual.
Figure 9.
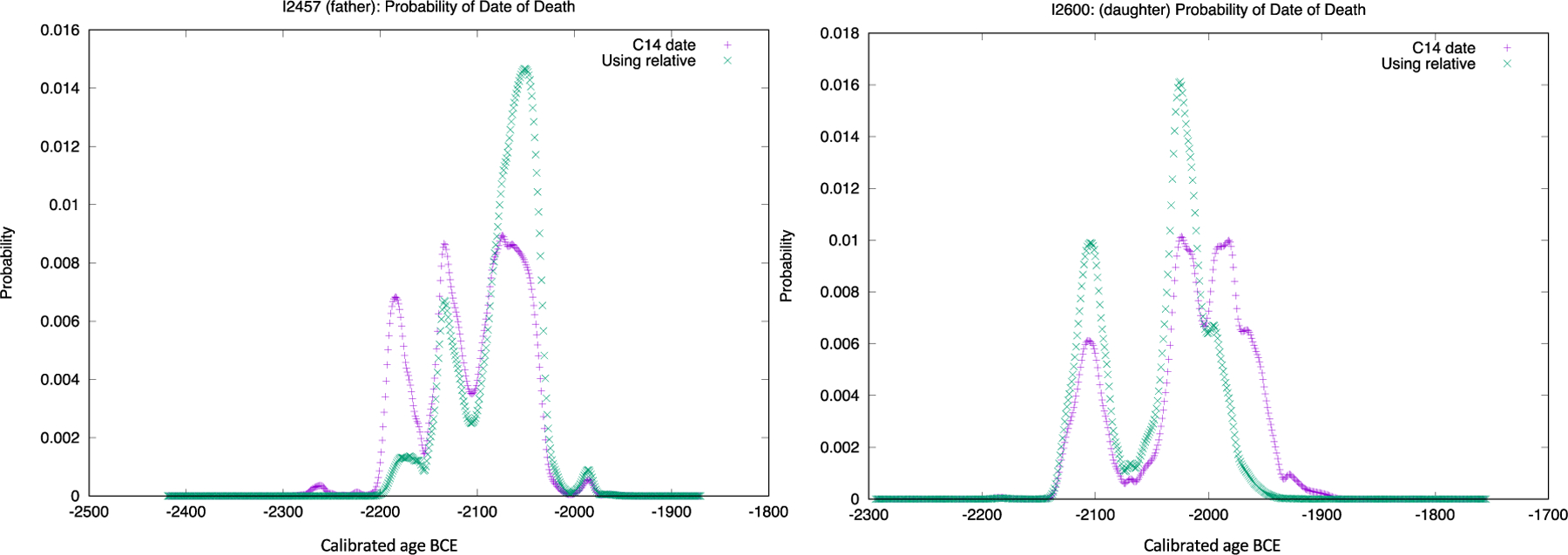
Prior and posterior probability distribution for I2457 (L) and I2600 (R). Original calibrated date probability distribution from OxCal in purple, new distribution plotted from results generated by refinedate (SM 5.1) in green.
Table 3.
Date ranges, mean Dates and standard deviations for father-daughter pair from OxCal (prior) and refinedate (posterior).
| Prior | Posterior | |||||
|---|---|---|---|---|---|---|
| 2 Sigma Date Range | Mean Date | SD | 2 sigma Date Range | Mean Date | SD | |
|
| ||||||
| I2457 (father) | 2201–2029 calBCE | 2105.7 | 51.1 | 2173–2023 calBCE | 2077.7 | 39.3 |
|
| ||||||
| I2600 (daughter) | 2135–1945 calBCE | 2018.0 | 52.8 | 2126–1975 calBCE | 2045.4 | 45.5 |
Figure 9 and Table 3 demonstrate that refinedate can refine the date distributions of related individuals. The prior 2-sigma date ranges in OxCal for I2457 and I2600 were 172 and 201 years, respectively, while the posterior ranges from refinedate were reduced to 150 and 151 years, reductions of 22 and 50 years. Refinedate also reduced the mean date separation between the two related individuals, with the original being 87.7 years and the posterior 32.3 years. Additionally, the prior distributions for both were multimodal, whereas the posteriors were both bimodal, with an increased probability at one peak on the distributions (Figure 9). Thus, the most likely probability for both I2600 and I2457 is between 2100–2000 BCE.
4. Results and Applications
4.1. Comparing Methods
To explore the effectiveness of OxCal’s interval function and refinedate we applied them to each of the 198 relative pairs in our database. We then compiled the constrained ranges for each of the 264 unique individuals. Some individuals were in multiple relative pairs (e.g. I2600 is has a father-daughter relationship with I2457 but also a 2nd–3rd degree relationship with I2566). For those individuals we kept the largest range reduction that was generated from the two approaches. Summary data from applying the interval function and refinedate constraints are provided in Table 4 (SM 4.2 has data for each of the 264 individuals).
Table 4.
Results from applying range reduction methods to dataset
| Unmodified 2σ Range | OxCal Interval 2σ modeled range | Refinedate 2σ range | |
|---|---|---|---|
| N individuals w/constrained ranges | - | 239 | 250 |
| Mean | 207.51 | 156.33 | 157.72 |
| Standard Deviation | 84.23 | 69.24 | 63.81 |
| Reduction from unmodified 2σ range mean | - | 51.18 | 49.79 |
| Reduction from unmodified 2σ range standard deviation | - | 59.54 | 52.03 |
| N individuals w/largest reduction* | - | 134 | 118 |
There were 12 individuals for which the OxCal interval method and refinedate produced the same date range reductions.
The OxCal interval function method and refinedate performed very similarly at constraining the radiocarbon date ranges of related pairs. Refinedate was able to constrain the date ranges of slightly more individuals than the OxCal interval function (250 vs 239), though they had nearly identical mean ranges and mean reductions from an individual’s unmodified 2-sigma calibrated range. The OxCal interval function method produced the larger reduction for more individuals than refinedate (e.g. for AES12 the interval function reduced the range by 95 years and refinedate reduced the range by 89 years; SM 4.2). Overall, we find the similar performance of the two methods encouraging, though it is also clear that in some particular instances the two methods do generate different results (SM 4.2). This could be due to a variety of factors, perhaps most notably the prior DOD distribution used in refinedate. We therefore encourage researchers to consider the plethora behavioral and life history factors that could increase precision in DOD estimates and how those can be incorporated to further constrain date ranges.
4.2. Exploring results
After applying the constraints to the relative pairs, we explored if our results could provide further insight about the archaeological or radiocarbon record. In the discussion below, we use the data from the interval function method since it produced the smallest mean modelled ranges, largest mean range reductions, and largest reduction of the two methods for the most individuals.
We first considered why applying the constraints led some individuals to have range reductions that exceeded dozens or even hundreds of years. One possible reason for large date range reductions is if different skeletal elements were radiocarbon dated for each individual in a relative pair. Studies have demonstrated that different skeletal elements have different rates of remodeling and carbon uptake (Calcagnile et al., 2013; Cook et al., 2015; Hansen et al., 2017; Pinhasi et al., 2015). For example, a long bone (tibia, femur, etc.) remodels throughout an individual’s life and therefore regularly uptakes new carbon, whereas the petrous does not remodel during an individual’s lifetime. Thus, if a femur and petrous from the same individual are radiocarbon dated, two different dates may be generated, particularly in advanced-age individuals. This could potentially lead to discrepant date ranges for related individuals. If the petrous of an adult female who died giving childbirth was dated, while the femur of her daughter was used, there could in theory be a difference of more than 100 years. We therefore compiled data on which element of each individual was radiocarbon dated (SM 4.3). Unfortunately, in many instances no information on which element was dated was available in the published literature, or, if information was provided, it was sometimes imprecise or vague.
We hypothesized that there would be a higher proportion of individuals with OxCal interval functions reductions above the GH DOD separation values (Table 2) for related pair that had different skeletal elements radiocarbon dated. Table 5 provides counts of whether the skeletal element dated for each individual was different or the same (or if no information was available) as their relative. There was a higher percentage of individuals in relative pairs with reductions above the GH DOD separation values in instances where different elements were radiocarbon dated than individuals in pairs that had the same element dated (Table 5). A chi-square test confirmed that this was statistically significant (chi-square test; x2=26.74, p value= 1.562E-06, df=2; SM 4.4). While it is likely that some instances of large discrepancies and interval reductions can be explained by the C14 dating of different skeletal elements, we caution that this is a preliminary result as no information was available for many of the individuals. Dietary differences coupled with marine reservoir effects or freshwater reservoir effects could create even larger discrepancies between related individuals. We did not include any individuals that have been previously calibrated for marine reservoir effect (or diet) in our analysis, but hope that in the future archaeologists consider this as a possibility for related individuals with large date discrepancies (and correct for it as needed).
Table 5.
Counts and percentage of whether the same or different skeletal elements were used to date an individual in a relative pair.
| Comparison of skeletal element dated for each related pair | N individuals | N individuals above GH DOD Estimates | % with reductions above GH DOD separation per element category |
|---|---|---|---|
| Different elements | 32 | 18 | 56.25% |
| Same element | 178 | 94 | 52.81% |
| No information available | 54 | 31 | 57.41% |
| SUM | 264 | 143 |
We next binned the individuals in our dataset into 500-year intervals using the 2-sigma range mean date BP generated in OxCal to further explore if applying the OxCal interval function could reveal larger patterns in the dataset. We hypothesized that 500-year periods that had individuals with large range reductions may be a result of wiggles or plateaus on the radiocarbon curve during those 500-year periods. Figures 10a and 10b qualitatively demonstrate that the number of individuals with reductions over the GH DOD separation estimates (Table 2) generally corresponds with the total number of related individuals from a particular 500-year interval in the dataset. We performed a chi-squared test to test the null hypothesis that the number of individuals above the GH DOD separation estimates per 500-year interval correlates with the total number of related intervals per 500-year interval. The result was statistically significant (x2= 35.85, p value= 0.004805762, df=17, (SM 4.6)), suggesting that the number of individuals with reductions above the GH DOD per 500-year interval is not simply due to sampling and the overall number of related individuals per 500-year interval. The most notable intervals were 4500–4999BP, 4000–4499BP, 3500–3999BP, and 3499–3000BP (Figure 10.C), which had residual values of 7.81, 7.02, 9.07, and 5.69 respectively (SM 4.6). We also examined which 500-year periods had the largest mean range reduction (Figure 10.D). This produced a result that was somewhat different than number of reductions above the GH DOD separation estimates; the 500-year interval the greatest mean reduction was 3000–3499 BP, which also had 80% of individuals above the GH DOD values.
Figure 10.
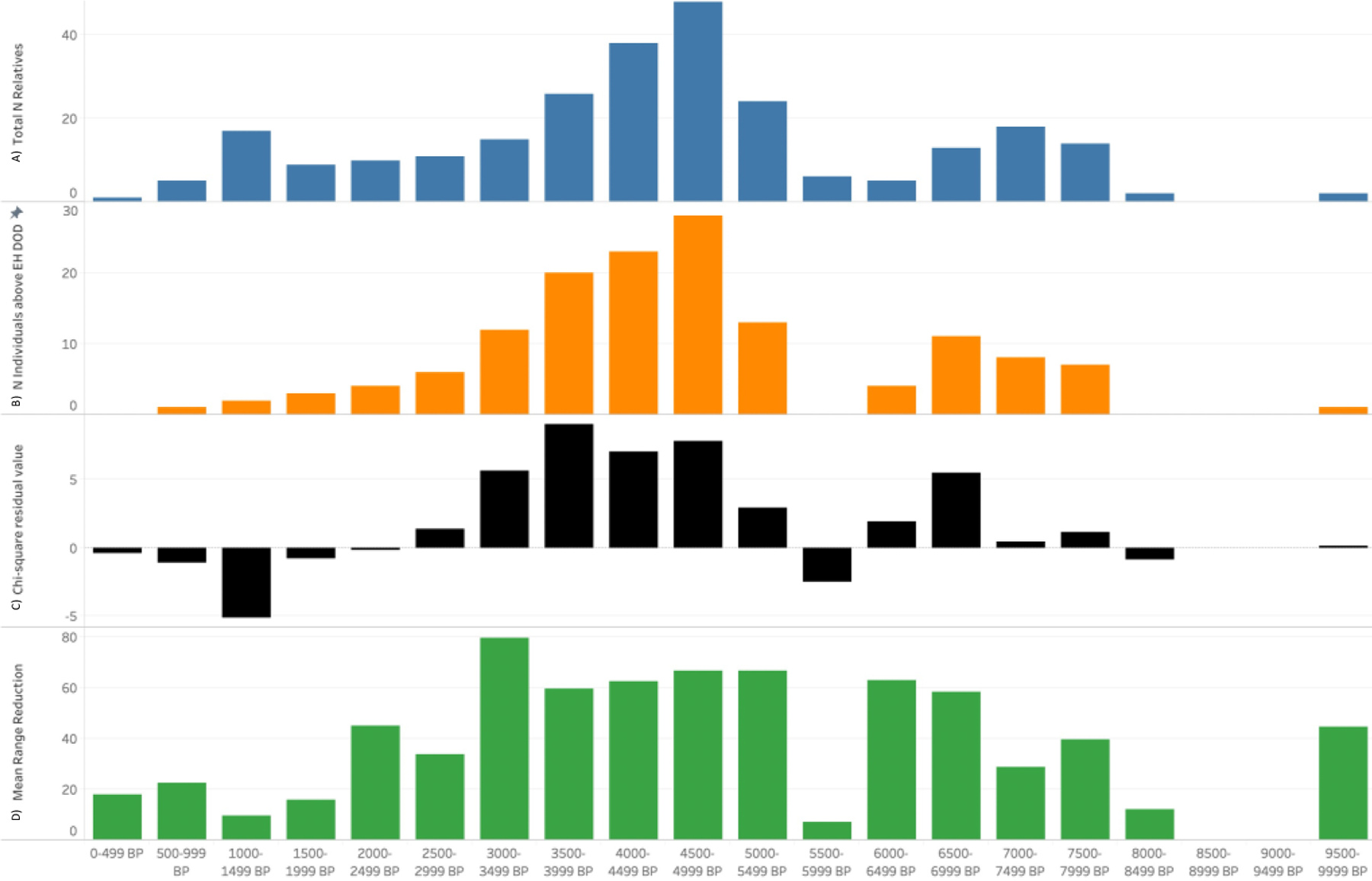
Data from the database and 264 individuals with relatives binned in 500-year intervals. A) #of individuals total in each 500-year interval. B) # of relatives per interval with reductions that exceed GH DOD estimates C) Residuals from chi-squared test of significance for individuals with reductions above GH DOD estimates D) Mean reduction of all relatives per 500 year-bin using OxCal’s interval function
To explore these results further, we examined the plotted radiocarbon date distributions for individuals in the four 500-year bins between 3000–5000BP. Figure 11 provides the results for each of these four intervals, separating the individuals in each bin with reductions below the GH DOD separations and those with reductions above the GH DOD separation estimates. As Figure 11 demonstrates, there is no apparent distinction between individuals with reductions above or below the GH DOD separation estimates; most individuals in these 500-year bins have radiocarbon ranges that fall on plateaus on the radiocarbon curve. Thus, the degree to which an individual in a related pair’s radiocarbon date range can be constrained does not seem to be tied to the location of that individual’s date range on the radiocarbon curve. The large reductions from these intervals also do not seem to be a result of a higher number of different skeletal elements dated; 8.7% (11/127) of individuals from 3000–5000BP had different skeletal elements dated than their relative, compared to 12.12% (32/264) overall.
Figure 11.
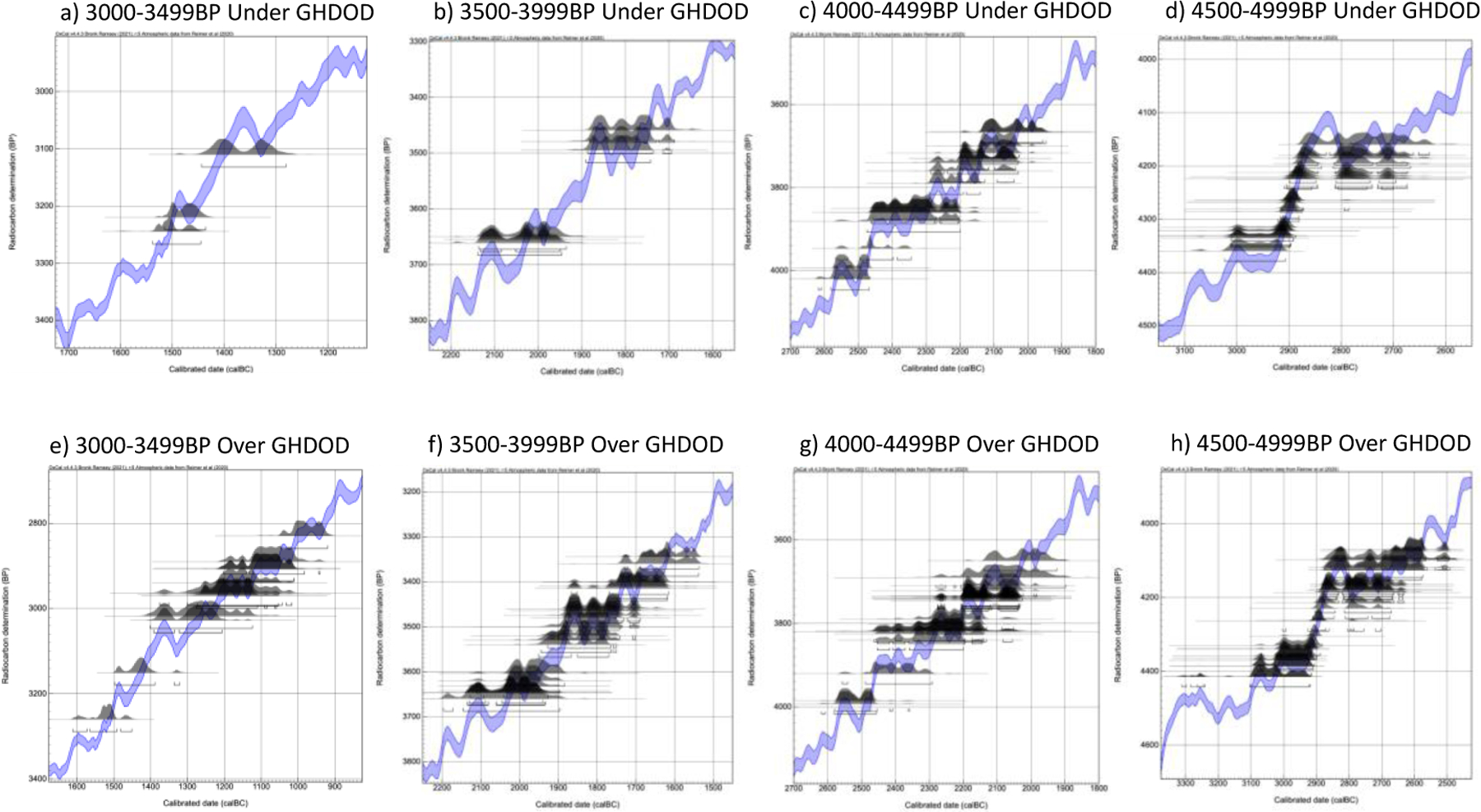
Radiocarbon distributions plotted on radiocarbon curve for individuals with reductions below the GH DOD separation estimates (top) and individuals with reductions above GH DOD separation estimates (bottom) for the 500-year intervals with the most and largest reductions (Figure 10) using OxCal’s interval function.
5. Discussion
Through the application of OxCal’s interval function and refinedate, it is evident that knowledge of genetic relatedness can be used to constrain radiocarbon date distributions. These refinements are not universally applicable; related pairs often have date distributions that overlap, sometimes almost entirely, limiting the extent to which DOD estimates can refine date ranges. Yet, overlap is what should be expected; related individuals should not have large date separations. Date distributions of related individuals that do not overlap could reveal an error in radiocarbon dating (such as I2600 and I2457) or genetic analysis, or other issues, such as an uncorrected marine reservoir effect. In other words, the more substantially DOD separation estimates can constrain C14 date ranges, the more likely a significant issue exists in dating for any of a variety of reasons (unaccounted marine reservoir effect, curve plateau, etc).
Combining previously independent lines of data—knowledge of genetic relatedness derived from ancient DNA; biological and estimated DOD separations for relatives; and radiocarbon dates—creates potential benefits for researchers examining the ancient past. Perhaps the most apparent is evaluation of data generated through disparate methods. Relatedness often confirms radiocarbon dates (and vice-versa, Saag et al., 2019:5). Using genetic relatedness and DOD separation estimates to evaluate radiocarbon dates can also help attend to some of the pitfalls in radiocarbon dating. According to Taylor and Bar-Yosef (2014:132), “the most common reason why C14 dating evidence is considered to be anomalous can be traced to failures to clearly establish and document the physical relationship between a C14 dated sample and a specific targeted event or cultural expression.” Somewhat counterintuitively, incorporating genetic relationship DOD-separations addresses Taylor and Bar-Yosef’s concerns by circumventing taphonomic processes. Instead of focusing on potential confounding factors of when individuals were buried, removed, reburied, etc., date ranges are examined with an independent line of evidence that is not prone to issues associated with taphonomic processes and archaeological context.
This research is a first step in combining two discrete analytical methods to add refinement to interpretation of the archaeological record and is meant to demonstrate that knowledge of genetic relatedness can be used to augment radiocarbon dating. As ancient DNA databases continue to grow, and more relatives are identified and radiocarbon dated, researchers will likely feel compelled to refine GH and DOD estimates as they see fit, as some have already done (Kennett et al., 2017; Saag et al., 2019). Levels of social organization (e.g. hunter-gatherer vs. agriculturalist), age of skeletons (adult vs. juvenile), and lifespan estimates could also be incorporated into estimates. Additionally, the prior GH DOD constraints we determined and employed were derived primarily from modern industrialized populations; archaeologists could develop their own particular separation estimates and prior distributions to be used in OxCal and refinedate. Once enough related individuals are identified and dated, specific regions, sub-regions, or even sites can be examined for anomalies in the associated radiocarbon records. The approaches outlined above represent only a small number of applications for how knowledge of genetic relatedness can help with radiocarbon dating. The potential for further applicability also needs to be explored; one promising application could be the use of extended families for radiocarbon curve “wiggle matching” with multi-generational lineages.
Ancient DNA innovations are providing archaeologists with unprecedented insight into the past. As ancient DNA becomes increasingly integral to archaeological studies, researchers should explore novel applications of genetic data to archaeological studies. This paper used ancient DNA to identify radiocarbon outliers, examine methods for constraining date distribution ranges for related pairs—including the creation of a program (refinedate) that researchers can download and use in analyses of relative pairs—and delineate potential issues unaccounted for in the radiocarbon record of particular eras and locales. We hope this study helps move ancient DNA and archaeology forward together into the next era of research on the human past.
Supplementary Material
Highlights.
Ancient DNA allows archaeologists to identify individuals that are biologically related
There are biological limitations to the number of years that can separate the death dates of related individuals
A number of specialized analyses, including a program we developed, (refinedate) can use knowledge of biological relatedness as a prior to constrain radiocarbon date ranges
Applying constraints to a large dataset can reveal patterns in the archaeological record.
Acknowledgements
This work was funded by NIH grant GM100233, the Paul Allen Foundation, John Templeton Foundation (grant number 6122). David Reich is an Investigator of the Howard Hughes Medical Institute. We thank Melissa Gymrek for providing assistance with familinx data from the Kaplanis et al. 2018 study, Greg Hodgins for reviewing an early draft of the paper, Iain Mathieson and Vagheesh Narasimhan for statistical assistance, Matthew Mah for help with loading refinedate to github, and members of our laboratory for feedback on the study during its development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interests
None
I2600 and I2457 were published in 2018 and originally calibrated in OxCal v4.3.2 with IntCal13; those original calibrations are used in Figure 4 since the discrepancy was discovered in that version of OxCal. Recalibrating the original dates with IntCal20 still results in a discrepancy that exceeds expected parent-offspring date of death separation.
References Cited
- Bronk Ramsey C, 2009. Bayesian Analysis of Radiocarbon Dates. Radiocarbon 51, 337–360. 10.1017/S0033822200033865 [DOI] [Google Scholar]
- Calcagnile L, Quarta G, Cattaneo C, D’Elia M, 2013. Determining 14 C Content in Different Human Tissues: Implications for Application of 14 C Bomb-Spike Dating in Forensic Medicine. Radiocarbon 55, 1845–1849. 10.1017/S003382220004875X [DOI] [Google Scholar]
- Cook GT, Ainscough LAN, Dunbar E, 2015. Radiocarbon Analysis of Modern Skeletal Remains to Determine Year of Birth and Death—A Case Study. Radiocarbon 57, 327–336. 10.2458/azu_rc.57.18394 [DOI] [Google Scholar]
- Hansen HB, Damgaard PB, Margaryan A, Stenderup J, Lynnerup N, Willerslev E, Allentoft ME, 2017. Comparing Ancient DNA Preservation in Petrous Bone and Tooth Cementum. PLOS ONE 12, e0170940. 10.1371/journal.pone.0170940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplanis J, Gordon A, Shor T, Weissbrod O, Geiger D, Wahl M, Gershovits M, Markus B, Sheikh M, Gymrek M, Bhatia G, MacArthur DG, Price AL, Erlich Y, 2018. Quantitative analysis of population-scale family trees with millions of relatives. Science 360, 171–175. 10.1126/science.aam9309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennett DJ, Plog S, George RJ, Culleton BJ, Watson AS, Skoglund P, Rohland N, Mallick S, Stewardson K, Kistler L, LeBlanc SA, Whiteley PM, Reich D, Perry GH, 2017. Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nature Communications 8, 14115. 10.1038/ncomms14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JMM, Jakobsson M, Günther T, 2018. Estimating genetic kin relationships in prehistoric populations. PLOS ONE 13, e0195491. 10.1371/journal.pone.0195491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, Connell S, Stewardson K, Harney E, Fu Q, Gonzalez-Fortes G, Jones ER, Roodenberg SA, Lengyel G, Bocquentin F, Gasparian B, Monge JM, Gregg M, Eshed V, Mizrahi A-S, Meiklejohn C, Gerritsen F, Bejenaru L, Blüher M, Campbell A, Cavalleri G, Comas D, Froguel P, Gilbert E, Kerr SM, Kovacs P, Krause J, McGettigan D, Merrigan M, Merriwether DA, O’Reilly S, Richards MB, Semino O, Shamoon-Pour M, Stefanescu G, Stumvoll M, Tönjes A, Torroni A, Wilson JF, Yengo L, Hovhannisyan NA, Patterson N, Pinhasi R, Reich D, 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424. 10.1038/nature19310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M, Skoglund P, Spriggs M, Valentin F, Bedford S, Shing R, Buckley H, Phillip I, Ward GK, Mallick S, Rohland N, Broomandkhoshbacht N, Cheronet O, Ferry M, Harper TK, Michel M, Oppenheimer J, Sirak K, Stewardson K, Auckland K, Hill AVS, Maitland K, Oppenheimer SJ, Parks T, Robson K, Williams TN, Kennett DJ, Mentzer AJ, Pinhasi R, Reich D, 2018. Population Turnover in Remote Oceania Shortly after Initial Settlement. Current Biology 28, 1157–1165.e7. 10.1016/j.cub.2018.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak S, Perry GH, 2017. Harnessing ancient genomes to study the history of human adaptation. Nature Reviews Genetics 18, 659–674. 10.1038/nrg.2017.65 [DOI] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, Sirak K, Gamba C, Jones ER, Llamas B, Dryomov S, Pickrell J, Arsuaga JL, de Castro JMB, Carbonell E, Gerritsen F, Khokhlov A, Kuznetsov P, Lozano M, Meller H, Mochalov O, Moiseyev V, Guerra MAR, Roodenberg J, Vergès JM, Krause J, Cooper A, Alt KW, Brown D, Anthony D, Lalueza-Fox C, Haak W, Pinhasi R, Reich D, 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503. 10.1038/nature16152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittnik A, Massy K, Knipper C, Wittenborn F, Friedrich R, Pfrengle S, Burri M, Carlichi-Witjes N, Deeg H, Furtwängler A, Harbeck M, von Heyking K, Kociumaka C, Kucukkalipci I, Lindauer S, Metz S, Staskiewicz A, Thiel A, Wahl J, Haak W, Pernicka E, Schiffels S, Stockhammer PW, Krause J, 2019. Kinship-based social inequality in Bronze Age Europe. Science eaax6219. 10.1126/science.aax6219 [DOI] [PubMed] [Google Scholar]
- Moreno-Mayar JV, Vinner L, de Barros Damgaard P, de la Fuente C, Chan J, Spence JP, Allentoft ME, Vimala T, Racimo F, Pinotti T, Rasmussen S, Margaryan A, Iraeta Orbegozo M, Mylopotamitaki D, Wooller M, Bataille C, Becerra-Valdivia L, Chivall D, Comeskey D, Devièse T, Grayson DK, George L, Harry H, Alexandersen V, Primeau C, Erlandson J, Rodrigues-Carvalho C, Reis S, Bastos MQR, Cybulski J, Vullo C, Morello F, Vilar M, Wells S, Gregersen K, Hansen KL, Lynnerup N, Mirazón Lahr M, Kjær K, Strauss A, Alfonso-Durruty M, Salas A, Schroeder H, Higham T, Malhi RS, Rasic JT, Souza L, Santos FR, Malaspinas A-S, Sikora M, Nielsen R, Song YS, Meltzer DJ, Willerslev E, 2018. Early human dispersals within the Americas. Science 362, eaav2621. 10.1126/science.aav2621 [DOI] [PubMed] [Google Scholar]
- Narasimhan VM, Patterson N, Moorjani P, Rohland N, Bernardos R, Mallick S, Lazaridis I, Nakatsuka N, Olalde I, Lipson M, Kim AM, Olivieri LM, Coppa A, Vidale M, Mallory J, Moiseyev V, Kitov E, Monge J, Adamski N, Alex N, Broomandkhoshbacht N, Candilio F, Callan K, Cheronet O, Culleton BJ, Ferry M, Fernandes D, Freilich S, Gamarra B, Gaudio D, Hajdinjak M, Harney É, Harper TK, Keating D, Lawson AM, Mah M, Mandl K, Michel M, Novak M, Oppenheimer J, Rai N, Sirak K, Slon V, Stewardson K, Zalzala F, Zhang Z, Akhatov G, Bagashev AN, Bagnera A, Baitanayev B, Bendezu-Sarmiento J, Bissembaev AA, Bonora GL, Chargynov TT, Chikisheva T, Dashkovskiy PK, Derevianko A, Dobeš M, Douka K, Dubova N, Duisengali MN, Enshin D, Epimakhov A, Fribus AV, Fuller D, Goryachev A, Gromov A, Grushin SP, Hanks B, Judd M, Kazizov E, Khokhlov A, Krygin AP, Kupriyanova E, Kuznetsov P, Luiselli D, Maksudov F, Mamedov AM, Mamirov TB, Meiklejohn C, Merrett DC, Micheli R, Mochalov O, Mustafokulov S, Nayak A, Pettener D, Potts R, Razhev D, Rykun M, Sarno S, Savenkova TM, Sikhymbaeva K, Slepchenko SM, Soltobaev OA, Stepanova N, Svyatko S, Tabaldiev K, Teschler-Nicola M, Tishkin AA, Tkachev VV, Vasilyev S, Velemínský P, Voyakin D, Yermolayeva A, Zahir M, Zubkov VS, Zubova A, Shinde VS, Lalueza-Fox C, Meyer M, Anthony D, Boivin N, Thangaraj K, Kennett DJ, Frachetti M, Pinhasi R, Reich D, 2019. The formation of human populations in South and Central Asia. Science 365, eaat7487. 10.1126/science.aat7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalde I, Brace S, Allentoft ME, Armit I, Kristiansen K, Booth T, Rohland N, Mallick S, Szécsényi-Nagy A, Mittnik A, Altena E, Lipson M, Lazaridis I, Harper TK, Patterson N, Broomandkhoshbacht N, Diekmann Y, Faltyskova Z, Fernandes D, Ferry M, Harney E, de Knijff P, Michel M, Oppenheimer J, Stewardson K, Barclay A, Alt KW, Liesau C, Ríos P, Blasco C, Miguel JV, García RM, Fernández AA, Bánffy E, Bernabò-Brea M, Billoin D, Bonsall C, Bonsall L, Allen T, Büster L, Carver S, Navarro LC, Craig OE, Cook GT, Cunliffe B, Denaire A, Dinwiddy KE, Dodwell N, Ernée M, Evans C, Kuchařík M, Farré JF, Fowler C, Gazenbeek M, Pena RG, Haber-Uriarte M, Haduch E, Hey G, Jowett N, Knowles T, Massy K, Pfrengle S, Lefranc P, Lemercier O, Lefebvre A, Martínez CH, Olmo VG, Ramírez AB, Maurandi JL, Majó T, McKinley JI, McSweeney K, Mende BG, Mod A, Kulcsár G, Kiss V, Czene A, Patay R, Endrődi A, Köhler K, Hajdu T, Szeniczey T, Dani J, Bernert Z, Hoole M, Cheronet O, Keating D, Velemínský P, Dobeš M, Candilio F, Brown F, Fernández RF, Herrero-Corral A-M, Tusa S, Carnieri E, Lentini L, Valenti A, Zanini A, Waddington C, Delibes G, Guerra-Doce E, Neil B, Brittain M, Luke M, Mortimer R, Desideri J, Besse M, Brücken G, Furmanek M, Hałuszko A, Mackiewicz M, Rapiński A, Leach S, Soriano I, Lillios KT, Cardoso JL, Pearson MP, Włodarczak P, Price TD, Prieto P, Rey P-J, Risch R, Rojo Guerra MA, Schmitt A, Serralongue J, Silva AM, Smrčka V, Vergnaud L, Zilhão J, Caramelli D, Higham T, Thomas MG, Kennett DJ, Fokkens H, Heyd V, Sheridan A, Sjögren K-G, Stockhammer PW, Krause J, Pinhasi R, Haak W, Barnes I, Lalueza-Fox C, Reich D, 2018. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555, 190–196. 10.1038/nature25738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalde I, Mallick S, Patterson N, Rohland N, Villalba-Mouco V, Silva M, Dulias K, Edwards CJ, Gandini F, Pala M, Soares P, Ferrando-Bernal M, Adamski N, Broomandkhoshbacht N, Cheronet O, Culleton BJ, Fernandes D, Lawson AM, Mah M, Oppenheimer J, Stewardson K, Zhang Z, Jiménez Arenas JM, Toro Moyano IJ, Salazar-García DC, Castanyer P, Santos M, Tremoleda J, Lozano M, García Borja P, Fernández-Eraso J, Mujika-Alustiza JA, Barroso C, Bermúdez FJ, Viguera Mínguez E, Burch J, Coromina N, Vivó D, Cebrià A, Fullola JM, García-Puchol O, Morales JI, Oms FX, Majó T, Vergès JM, Díaz-Carvajal A, Ollich-Castanyer I, López-Cachero FJ, Silva AM, Alonso-Fernández C, Delibes de Castro G, Jiménez Echevarría J, Moreno-Márquez A, Pascual Berlanga G, Ramos-García P, Ramos-Muñoz J, Vijande Vila E, Aguilella Arzo G, Esparza Arroyo Á, Lillios KT, Mack J, Velasco-Vázquez J, Waterman A, Benítez de Lugo Enrich L, Benito Sánchez M, Agustí B, Codina F, de Prado G, Estalrrich A, Fernández Flores Á, Finlayson C, Finlayson G, Finlayson S, Giles-Guzmán F, Rosas A, Barciela González V, García Atiénzar G, Hernández Pérez MS, Llanos A, Carrión Marco Y, Collado Beneyto I, López-Serrano D, Sanz Tormo M, Valera AC, Blasco C, Liesau C, Ríos P, Daura J, de Pedro Michó MJ, Diez-Castillo AA, Flores Fernández R, Francès Farré J, Garrido-Pena R, Gonçalves VS, Guerra-Doce E, Herrero-Corral AM, Juan-Cabanilles J, López-Reyes D, McClure SB, Merino Pérez M, Oliver Foix A, Sanz Borràs M, Sousa AC, Vidal Encinas JM, Kennett DJ, Richards MB, Werner Alt K, Haak W, Pinhasi R, Lalueza-Fox C, Reich D, 2019. The genomic history of the Iberian Peninsula over the past 8000 years. Science 363, 1230–1234. 10.1126/science.aav4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Fernandes D, Sirak K, Novak M, Connell S, Alpaslan-Roodenberg S, Gerritsen F, Moiseyev V, Gromov A, Raczky P, Anders A, Pietrusewsky M, Rollefson G, Jovanovic M, Trinhhoang H, Bar-Oz G, Oxenham M, Matsumura H, Hofreiter M, 2015. Optimal Ancient DNA Yields from the Inner Ear Part of the Human Petrous Bone. PLOS ONE 10, e0129102. 10.1371/journal.pone.0129102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posth C, Nakatsuka N, Lazaridis I, Skoglund P, Mallick S, Lamnidis TC, Rohland N, Nägele K, Adamski N, Bertolini E, Broomandkhoshbacht N, Cooper A, Culleton BJ, Ferraz T, Ferry M, Furtwängler A, Haak W, Harkins K, Harper TK, Hünemeier T, Lawson AM, Llamas B, Michel M, Nelson E, Oppenheimer J, Patterson N, Schiffels S, Sedig J, Stewardson K, Talamo S, Wang C-C, Hublin J-J, Hubbe M, Harvati K, Nuevo Delaunay A, Beier J, Francken M, Kaulicke P, Reyes-Centeno H, Rademaker K, Trask WR, Robinson M, Gutierrez SM, Prufer KM, Salazar-García DC, Chim EN, Müller Plumm Gomes L, Alves ML, Liryo A, Inglez M, Oliveira RE, Bernardo DV, Barioni A, Wesolowski V, Scheifler NA, Rivera MA, Plens CR, Messineo PG, Figuti L, Corach D, Scabuzzo C, Eggers S, DeBlasis P, Reindel M, Méndez C, Politis G, Tomasto-Cagigao E, Kennett DJ, Strauss A, Fehren-Schmitz L, Krause J, Reich D, 2018. Reconstructing the Deep Population History of Central and South America. Cell 175, 1185–1197.e22. 10.1016/j.cell.2018.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TW, Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, Poznik GD, Gudmundsdottir V, Yadav R, Malaspinas A-S, V, S.S.W., Allentoft ME, Cornejo OE, Tambets K, Eriksson A, Heintzman PD, Karmin M, Korneliussen TS, Meltzer DJ, Pierre TL, Stenderup J, Saag L, Warmuth VM, Lopes MC, Malhi RS, Brunak S, Sicheritz-Ponten T, Barnes I, Collins M, Orlando L, Balloux F, Manica A, Gupta R, Metspalu M, Bustamante CD, Jakobsson M, Nielsen R, Willerslev E, 2014. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature 506, 225–229. 10.1038/nature13025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, 2018. Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past. Pantheon, New York. [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, Maricic T, Good JM, Marques-Bonet T, Alkan C, Fu Q, Mallick S, Li H, Meyer M, Eichler EE, Stoneking M, Richards M, Talamo S, Shunkov MV, Derevianko AP, Hublin J-J, Kelso J, Slatkin M, Pääbo S, 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060. 10.1038/nature09710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer PJ, Austin WEN, Bard E, Bayliss A, Blackwell PG, Ramsey CB, Butzin M, Cheng H, Edwards RL, Friedrich M, Grootes PM, Guilderson TP, Hajdas I, Heaton TJ, Hogg AG, Hughen KA, Kromer B, Manning SW, Muscheler R, Palmer JG, Pearson C, Plicht J.van der, Reimer RW, Richards DA, Scott EM, Southon JR, Turney CSM, Wacker L, Adolphi F, Büntgen U, Capano M, Fahrni SM, Fogtmann-Schulz A, Friedrich R, Köhler P, Kudsk S, Miyake F, Olsen J, Reinig F, Sakamoto M, Sookdeo A, Talamo S, 2020. The IntCal20 Northern Hemisphere Radiocarbon Age Calibration Curve (0–55 cal kBP). Radiocarbon 62, 725–757. 10.1017/RDC.2020.41 [DOI] [Google Scholar]
- Saag Lehti, Laneman M, Varul L, Malve M, Valk H, Razzak MA, Shirobokov IG, Khartanovich VI, Mikhaylova ER, Kushniarevich A, Scheib CL, Solnik A, Reisberg T, Parik J, Saag Lauri, Metspalu E, Rootsi S, Montinaro F, Remm M, Mägi R, D’Atanasio E, Crema ER, Díez-del-Molino D, Thomas MG, Kriiska A, Kivisild T, Villems R, Lang V, Metspalu M, Tambets K, 2019. The Arrival of Siberian Ancestry Connecting the Eastern Baltic to Uralic Speakers further East. Current Biology S0960982219304245 10.1016/j.cub.2019.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Posth C, Sirak K, Spriggs M, Valentin F, Bedford S, Clark GR, Reepmeyer C, Petchey F, Fernandes D, Fu Q, Harney E, Lipson M, Mallick S, Novak M, Rohland N, Stewardson K, Abdullah S, Cox MP, Friedlaender FR, Friedlaender JS, Kivisild T, Koki G, Kusuma P, Merriwether DA, Ricaut F-X, Wee JTS, Patterson N, Krause J, Pinhasi R, Reich D, 2016. Genomic insights into the peopling of the Southwest Pacific. Nature 538, 510–513. 10.1038/nature19844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RE, Bar-Yosef O, 2014. Radiocarbon dating: an archaeological perspective, Second edition. ed. Left Coast Press, Inc, Walnut Creek, California. [Google Scholar]
- van de Loosdrecht M, Bouzouggar A, Humphrey L, Posth C, Barton N, Aximu-Petri A, Nickel B, Nagel S, Talbi EH, El Hajraoui MA, Amzazi S, Hublin J-J, Pääbo S, Schiffels S, Meyer M, Haak W, Jeong C, Krause J, 2018. Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548–552. 10.1126/science.aar8380 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


