Abstract
The respiratory disease caused by the Coronavirus infectious disease 2019 (COVID19) has spread rapidly since December 2019 in Wuhan, China. This new strain of Coronavirus is similar to the SARS Corona virus and has been termed SARS-CoV-2. Both viruses have emerged from bats and adapted to humans. On March 11, 2020 COVID19 was declared Pandemic by the WHO and as of May 1, 2020 COVID19 disease continues to grow rapidly with 3,400,595 cases and 239,583 deaths world-wide. This review describes the biology of SARSCOV2, Detection, Macrophage-Mediated Pathogenesis and Potential Treatments.
Keywords: COVID-19, SARS-CoV-2, Endosomal, Polyproteins, Nucleocapsid, Pro-Inflammatory
Introduction
Coronaviruses (CoVs) are large, plus-stranded RNA, enveloped viruses that infect a large variety of avian and mammals. CoVs have displayed the ability to readily cross species barriers and this zoonotic transfer has caused three CoVs to recently emerge in the human population, including the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) that is responsible for the current pandemic. In humans, CoVs (HCoVs) are associated with upper respiratory tract infections (URTI) which can occasionally spread to the lungs. Four HCoVs (HCov OC43, NL63, 229E, and HKU1) are globally distributed and associated with 10 to 30% of all URTIs in adults [1]. Cryo-electron tomography has shown that CoV particles have a pleomorphic structure, a spherical shape and show a variation in size of 80–120 nm [2]. All CoVs are characterized by having three viral proteins anchored in their envelopes: the abundant triple-spanning membrane (M) protein, the envelope (E) protein (present in small amounts), and the spike (S) protein, which is critical for the viral entry process [2]. The CoV genome is a linear (+) RNA of approximately 30kb in length packed in a coiled nucleocapsid. Once inside the cell, the CoV genome employs two differentiated processes necessary for its replication. The continuous genome replication which results in multiple copies of genomic RNA strands and the discontinuous process of subgenomic mRNA transcription, which produces structural and accessory proteins necessary for viral assembly [3]. The (+)RNA is initially translated by host cell machinery to produce the RNA-dependent RNA polymerase (RdRp) which then initiates negative-strand synthesis. This polymerase is a multi-subunit protein with exonuclease and proofreading functions which helps the virus maintain viral progeny stability [3].
Severe Acute Respiratory Syndrome (SARS)-CoV-2 was first identified in Wuhan, China in December 2019 in clusters of patients with pneumonia of unknown etiology [4]. In January 2020 the causative agent of this pathology was determined to be a novel coronavirus placed in the betacoronavirus genus alongside the emerging coronaviruses of the century, Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) respectively [5]. As of March 2020 this novel coronavirus received the name of SARS-CoV-2 based on phylogeny, taxonomy and established practice, the Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, recognizes this virus as forming a sister clade to SARS-CoV [5]. According to the daily situation report assessed by the World Health Organization, the pandemic of SARS- CoV-2 has caused 1, 991, 562 confirmed cases and 130, 885 deaths worldwide by April 16th 2020.
SARS-CoV was first discovered and reported in November 2002 in Guangdong, China. Its pathogenesis is characterized by severe lung damage, an elevated innate immune response and upregulated number of proinflammatory cytokines and chemokines [6]. At the end of the epidemic, there were more than 8,000 reported cases worldwide and 774 deaths. The virus was finally contained in September 2003 and no new cases have been reported [7]. Nine years later, in 2012, MERS-CoV was the second coronavirus reported that resulted in a major threat to global health. Its pathogenesis is dependent on its cellular entry receptor dipeptidyl peptidase 4 (DPP4) which is widely expressed in the body’s organ tissues. Therefore, MERS-CoV tissue tropism is broader than that of any other coronavirus known [8]. This viral infection induces infected immune cells (like macrophages) to secrete a wide variety of proinflammatory cytokines that causes severe damage in the respiratory tract. A second MERS-CoV outbreak occurred 2 years later, in 2014, in the Arabian Peninsula, and cases are still being reported. Since 2012, there have been 2,494 confirmed cases and 858 confirmed deaths. Important to mention, MERS-CoV has a higher death rate than SARS-CoV and SARS-CoV-2. Nevertheless, what has maintained the virus contained and feasible to control is its low potential for transmission or (R0) (approximately 1) meaning that one infected person can infect only one more person [9]. However, SARS-CoV-1 has a R0 of 3 [7] and SARS-CoV-2 has a R0 of 3.28 [10].
Coronavirus in the Environment
Coronaviruses can be found in different birds and mammals. This includes domesticated and wild animals like dogs, cats, pigs, chicken, rabbits, ferrets, cattle, turkeys, camels, civets, rats, raccoon dogs, primates, pangolins, and bats [11]. The original animal reservoir source of SARS-CoV-2 is still unknown. Phylogenetic analyses revealed that the bat Rhinolophus genus coronavirus has a 96% homology to the current pandemic virus [12]. An intermediary host is suspected to be the Malaysian pangolin (Manis javanica) with amino acid sequences resemblance of 91%. Additional studies are required because these results come from an assembled genome draft of 73% coverage [13]. It must be acknowledged the pangolin as the potential intermediary because other preprint studies have shown protein gene resemblance of 100%, 98.2%, 96.7% and 90.4% to E, M, N and S genes, respectively [14]. Adding to this potential intermediary, both species of mammals are found in the markets of Wuhan where this pandemic was originated and they have share habitats in nature [15]. A recent study of animal models of SARS-CoV-2 found that dogs, pigs, chickens, ducks, ferrets and cats are permissive to the virus [16]. In experimental settings cats could transmit the infection to other cats while dogs were more resistant to the virus [16]. At present, studies to support that infection is spreading amongst populations of dogs and cats are lacking. At the current time there is no evidence of transmission from cats and dogs to humans. Additional research is needed to find evidence to determine if cats and dogs can transmit SARS-CoV-2 infection to humans.
Coronavirus Entry into Susceptible Cells
The primary cell host attachment receptor used by SARS-CoV-2 is Angiotensin-converting enzyme 2 (ACE2) [12,17]. This protein is embedded in cell membranes of many organs and has two domains, an extracellular which is cleaved from the transmembrane domain. The extracellular domain is released to the blood and excreted through urine. The main function of the secreted ACE2 domain is to promote vasodilation by cleaving angiotensin I into angiotensin 1–9, and angiotensin into angiotensin 1–7 (National Center for Biotechnology Information (NCBI)). ACE2 can be found in lung type II alveolar cells, enterocytes of the small intestine, arterial and venous endothelial cells, and arterial smooth muscle cells in most organs [18]. By sequencing analyses, different groups have demonstrated the high similarity of SARS-CoV-2 to SARS-CoV, which leads to the hypothesis of ACE2 as the entry receptor of SARS-CoV-2, because SARS-CoV uses this receptor [12,19]. The expression of this receptor in different animal species that are permissive to the virus and its high expression in lung type II alveolar cells and the presence on type I pneumocytes, alveolar macrophages, ciliated and non-ciliated bronchiole epithelial cells present in the respiratory tract supported this hypothesis [12,20]. This hypothesis was confirmed when two independent research groups expressed the receptor in cells lacking the ACE2 that were not permissive to the virus and thereafter became ls susceptible to the virus [12,17]. One of these two groups confirmed the importance of the receptor for the susceptibility of the cell to the virus by using ACE2 inhibitors and blocking the viral entry [17].
After binding to its receptor, a fusion of the viral membrane with the endosomal membrane is required to complete the entry to host cells. The spike protein needs to be cleaved, which is known as priming 21]. This is performed by host endosomal cysteine proteases cathepsin B and L and the transmembrane protease, serine 2 (TMPRSS2) in susceptible cells [22]. The TMPRSS2 is indispensable to complete viral entry which led to the hypothesis that SARS-CoV-2 depends on this protease for entry. Using RNA sequencing a group of investigators determined that ACE2 and TMPRSS2 RNA were produced only in goblet cells of the nose, type II pneumocytes, and one type of enterocytes [23]. Hoffmann, et al. [17] performed various experiments inhibiting the function of TMPRSS2 with camostat mesylate and cathepsin B and L with E-64d, and found that using the combination of both compounds completely inhibited viral entry on the cell lines Caco-2 and Vero-TMPRSS2 [17]. In the cell lines 293T and Vero viral entry depended on cathepsin completely. They first, blocked viral entry using only E-64d and then rescued viral entry by using directed expression of TMPRSS2. Using an in-vitro model of human lung cells treated with camostat mesylate they concluded that SARS-CoV-2 entry depends on TMPRSS2 expression because the viral entry was significantly decreased with no cytotoxic effects.
Life Cycle
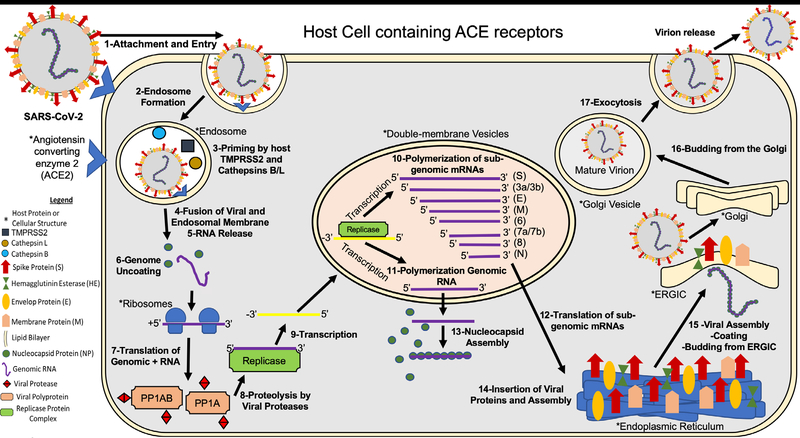
The SARSCoV2 life cycle is described in Figure 1.
Figure 1:
SARSCoV2 life cycle
Briefly, 1-SARS-CoV-2 attaches to the host cell surface receptors ACE2 and sialic acid receptor with its spike protein and hemagglutinin-esterase, respectively. 2-Following attachment of the spike an endosome is formed and transported to the cytoplasm of the cell through the endosomal pathway. 3- Inside the endosome the viral spike protein is primed by endosomal cysteine proteases cathepsin B and L and the serine protease TMPRSS2, the latter being essential for viral entry. The priming of the spike protein split the spike protein into subunits S1 and S2. The S1 contains the receptor-binding domain that directly binds to the peptidase domain (PD) of ACE2 causing 4- envelope fusion with the vesicle membrane 5- releasing the positive single-stranded viral genome to the host cell cytoplasm. 6-Nucleocapsid proteins detach from the genome uncoating for host ribosome binding. 7- The genome is translated by host ribosomes at the ORF1a and ORF1b synthesizing the viral replicase polyproteins. The replicase components are present in two translated polyproteins pp1a and 1ab 8- which are cleaved by viral proteases and assemble to form the replicase complex. 9- The replicase which is an RNA dependent RNA polymerase transcribes the positive-sense genomic RNA into a negative-sense RNA. 10- Using the negative-sense RNA it starts to polymerize a series of sub-genomic mRNAs by discontinuous transcription inside double-membrane vesicles. It is believed that these vesicles are derived from the endoplasmic reticulum and the Golgi apparatus. These sub-genomic RNAs are necessary to produce different viral polyproteins that are cleaved to generate structural proteins that will compose the new viral particles. 11- At the same time positive-sense genomic RNA is polymerized for the new viral particles. 12- The sub-genomic mRNAs are translated into viral proteins. All the structural proteins are exported to the endoplasmic reticulum (ER) except for the nucleocapsid proteins that are exported to the cytoplasm. 13- In the cytoplasm the nucleocapsid proteins link to the positive single- stranded viral RNA recently replicated to assemble a helical nucleocapsid. 14- At the same time the other viral structural proteins insert in the endoplasmic reticulum, finish their assembly, and are exported to ER-Golgi intermediate compartment (ERGIC). 15- In the ERGIC the nucleocapsid binds to the membrane proteins assembling the virion. 16-The viral particles bud from the Golgi and are transported by smooth-walled vesicles from the Golgi apparatus to the host cellular extracellular membrane where they are 17-released from the cell by exocytosis (Figure 1) [24–26].
Detection of Infection
Molecular Test
SARS-CoV-2 infection can be identified and diagnosed measuring presence of viral molecular compounds in patient secretions. A real time Reverse Transcriptase Polymerase Chain Reaction (rRT-PCR), is a nuclear method that detect the presence of viral RNA [27]. For this assay, a sample of viral infected cells or secretions from patients, TAQ polymerase (Thermus aquaticus), Reverse Transcriptase (RT) and Primers are needed in order to amplify RNA from the sample [28]. The viral RNA is commonly obtained by placing a mini-tip swab through the nostrils until the nasopharynx and left it for several seconds; swab is removed slowly while rotating to obtain infected cells. In Wyllie, et al. [29] study, they also tested if saliva sample is more sensitive to detect SARS-CoV-2 than nasopharyngeal swabs. In the study, it was asked to COVID-19 patients to self-collect saliva sample by repeatedly spit in a sterile urine cup until a third is full of liquid. Saliva shows more viral load than nasopharyngeal swabs, being more sensitive to detect respiratory pathogens including coronaviruses [29]. With further validation, widespread implementation of saliva sampling could transform public health efforts: saliva self-collection eliminates the need for direct healthcare worker-patient interaction, a major source of nosocomial infection risk [30].
In the first step for PCR expansion, Primers anneal to complementary zones of sample RNA, essential for Reverse transcriptase enzyme to synthesize and amplify complementary DNA (cDNA) genome sequence. RT activity creates cDNA from sample RNA template, providing start point for TAQ polymerase expansion. The polymerase chain reaction is a three steps process performed at specific required temperatures: Denaturation, Anneal and Elongation. Denaturation, first step, is required to expose genome of interest by increasing temperature to 94°C. Strands obtained anneal with Primers at 57°C, leading to the last step carried out by TAQ. TAQ polymerase work efficiently at 72°C, expand genome in at least 40 cycles in order to obtain enough amount of genetical compounds [31]. This amplification depends on the concentration and amount of initial RNA, factor that can affect the test result. Reasons for false negative rRT-PCR could be improper extraction of nucleic acid from clinical materials or insufficient molecular material for detection [32]. After performing the test, it can take from 24 to 72 hours to quantify and obtain the results, although they could be obtained faster in case of some clinical emergency like SARS.
In emergencies like these, the Emergency Use Authorization (EUA) allows the Food and Drug Administration (FDA) to make decisions about which medical products to use without waiting for any approval. The FDA has allowed companies to use medical products that are not approved to diagnose, treat, or prevent disease, when alternatives are not available or approved. They continue to approve test to detect SARS-CoV-2 and to date has approve forty-three molecular tests. One of the tests they approved is the ID NOW COVID-19 of the Abbott Company. In the emergency with Covid-19, the FDA has approved other PCR tests such as the STANDARD M nCoV Real-Time Detection kit and U-TOP COVID-19 Detection Kit. These detect nucleic acids from SARS-CoV-2 in oropharyngeal and nasopharyngeal swab specimens [33]. Abbott, et al. [34] received FDA authorization, due to this emergency, to use the ID NOW COVID-19 test only for the detection of nucleic acid from SARS-CoV-2, not for any other pathogens or viruses [34]. This statement will only last as long as circumstances may warrant authorization. This allows the results to be obtained faster, giving positive results in 5 minutes and negative results in 13 minutes after performing the test. This is possible, since having a small size allows testing in non-traditional places and obtaining the results faster.
Unlike this and the most common tests, the Aries SARS-CoV-2 Assay has the capacity to produce from 300 to 1,500 copies of each SARS-CoV-2 gene target, allowing results to be obtained in less time. ARIES Extraction Cassettes contain lysis and extraction reagents, which can extract nucleic acid, break cells, and remove inhibitors, something the aforementioned test cannot perform. The process of this test, sews in a short time thermal cycle. The first cycle consists of preheating to a temperature of 50 ° C for 7 minutes. The temperature is then increased to 95 ° C for 2 minutes for activation. Finally, the cycling process is carried out consisting of 45 cycles at a temperature of 95°C for 15 seconds and 60°C for 30 seconds [35]. At the end of the cycle, the test result can be obtained in approximately 10 minutes. When comparing both tests, two common factors can be observed: the ability to obtain the results faster due to knew mechanisms and their portability. These machines are accessible to be taken to unusual places where SARS-CoV-2 tests are required. However, it remains to be determined if their sensitivity and specificity are as good as the regular RT-PCR test.
Antibody Tests
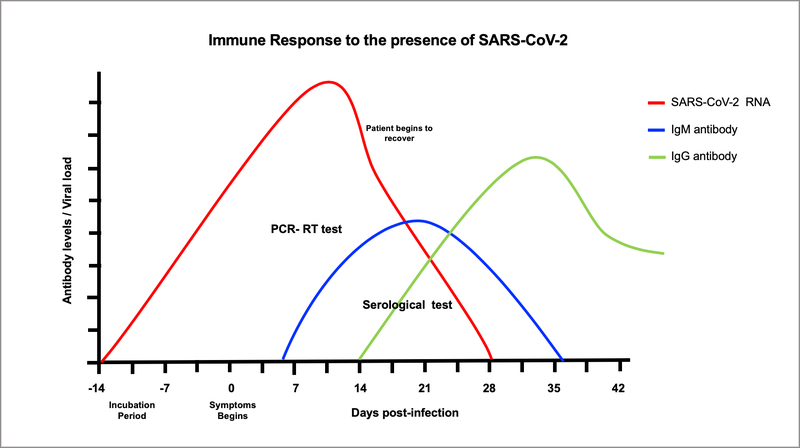
Unlike molecular tests (RT-PCR), serological tests detect COVID-19 infection by the presence of antibodies in the blood by the “Enzyme Immunosorbent Assay” (ELISA). The current ELISA technique is qualitative and allows detecting the presence of antibodies against an immobilized (SARS-CoV-2) antigen, through the use secondary antibodies. This test is useful for detecting a previous or active infection by the body’s immune response to the virus. The presence of viral proteins triggers the production of Immunoglobulin (Ig) M type antibodies at the beginning of the infection, about 6 days after the symptoms begin, followed by production of IgG, at the end of the infection and thereafter IgM disappear at the end of the infection while IgG start to be produced and remain high thereafter indicating immunity against SARSCoV2 (Figure 2). The presence of these antibodies begins approximately one week after the incubation period (2–14 days). The IgM test detects individuals responding to virus who are asymptomatic or with symptoms while the IgG test detect individuals that have recovered from COVID19 disease with or without symptoms. Immune individuals can donate plasma containing IgG antibodies to be used in the treatment of COVID-19 patients, called by the FDA as convalescent plasma. This plasma is used in the elaboration of a biological product called hyperimmune globulin that provides passive immunity against this virus. However, it is important to consider the fact that as yet, there are no studies indicating whether the use of these antibodies, are protective against reinfection. In most viral infections, the antibodies are protective against the specific virus that causes the disease. What is unknown if this SARSCoV2 does not change its antigens to respond to the original antibodies produced by infected patients.
Figure 2:
Once the patient gets infected with SARSCoV-2, viral replication in susceptible cells of the respiratory track increases and may remain active until 28 days of infections in symptomatic or asymptomatic patient (red line). One-week post infection IgM begins to be produced in response to the virus and remain for 2–3 weeks (blue line). After IgM declines, IgG begins to be produced and remain positive thereafter (green line).
The current antibody tests available in the market for SARS COV2 are five and provide results within 10 to 15 minutes. Two of these tests use the colloidal gold immunochromatography technique (rapid POC kit and VivaDiag COVID-19 IgG-IgM test), two others use the lateral flow immunoassay (COVID-19 IgM-IgG rapid test and COVID-19 rapid test cassette) and the last one uses the temporal resolution fluorescence immunoassay technique (GT-100 SARS-CoV-2 IgG/IgM kit). Although these are from different manufacturers and differ in the type of technique used, they are similar in that they mark the same types of antibodies (IgG/IgM). These tests are useful to preliminarily and quickly diagnose SARS-CoV-2 infection in an individual, due to its simplicity of procedure and equipment. Which unlike ELISAs, these are more practical and cost-effective for an emergency. However, their sensitivity and specificity are lower than the regular ELISAs and results must be confirmed by other tests.
Coronavirus and Macrophages
Macrophages: the main drivers of SARS-Cov pathogenesis?
Macrophages are one of the main phagocytic and antigen-presenting cells (APC) in the body that participate in the innate immunity and tissue homeostasis in response to different pathogenic stimuli [36]. Macrophages are present in most tissues -bone marrow, brain, liver, skin, gut, placenta, lung, heart, spleen, lymph nodes, etc- where they perform different functions [37,38]. After SARS-CoV-2 infection, ACE2-expressing cells in the lungs, liver and stomach could induce the migration of macrophages by releasing the macrophage migration inhibitory factor (MIF), a pro-inflammatory cytokine that binds to CD74 surface receptor on macrophages [39]. In SARS-CoV-1-infected mice, increased secretion of pro-inflammatory chemokines and cytokines correlate with the migration of macrophages to the lungs by day 2 post-infection [40]. Macrophages do not express ACE2, but the SARS-CoV-1 spike (S) antigen and the anti-S IgG antibody complexes can mediate entry of the virus through Fc-II receptors, a process known as antibody-dependent enhancement (ADE [18,41,42]. Since macrophages lack the ACE2 receptor, they do not get productively infected with SARS-CoV-1 [43]. Upon entry into macrophages by ADE, SARS-CoV-1 impairs the IFN-B response by suppressing the TRIM25-mediated and RIG-1 ubiquitination [44]. The coronavirus nonstructural protein (NSP) 15 evades intracellular dsRNA sensors and inhibits apoptosis of macrophages [45]. SARS-CoV-1 viroporin 3a activates the NLRP3 inflammasome leading to the release of IL-1B [46]. Release of IL-1B has the potential to induce pyroptosis [47]. Analysis of post-mortem tissues of SARS-CoV-2 patients has shown increased CD169+ macrophages in the marginal zones of the spleen and lymph nodes accompanied by lymphocyte apoptosis [48]. It is also known that SARS-CoV spike protein induces TNF-a and IL-16 release in macrophages by activation of NF-kB [48,49]. Increased NF-kB signaling was also observed in transcriptome analyses in lungs of patients with comorbidities associated to severe COVID-19 disease [50]. These studies suggest that macrophages could be the main driver of inflammation and lymphopenia associated to COVID-19 disease.
Increased inflammation could lead to the development of Acute Respiratory Distress Syndrome (ARDS) [51]. Monocyte-derived macrophages play an important role in this mechanism since previous studies in patients with ARDS demonstrated that inflammatory monocyte-derived FCN1+ macrophages, and not the alveolar macrophages, were predominant in the bronchoalveolar lavage fluid [52]. In addition, a recent study showed that COVID-19 patients with severe respiratory failure had macrophage activation syndrome (MAS) [53], a pathology seen in systemic inflammatory diseases that could lead to hyperferritinemia, pancytopenia, fibrinolytic coagulopathy, and liver dysfunction [54]. Also, autopsies revealed that among pathological changes in the lungs, there was increased infiltration of monocytes and macrophages containing inclusion bodies and SARS-CoV-2 antigen (National Health Commission of the People’s Republic of China, 2020) [55]. These changes were also observed in the spleen, where macrophage hyperplasia and phagocytosis were observed. Increased macrophage infiltration was also observed in the heart of SARS-CoV-1 infected patients [56]. A recent study determined that the accumulation of macrophages and complement C5b-9 deposition in the tubules of Kidneys play a role in the renal damage associated to SARS-CoV-2 [48]. Complement deposition could be contributing to the inflammatory process of SARS-CoV-2 infection through the activation of macrophages, and represents a potential target for treatment [57]. In an early clinical study, treatment with a compstatin-based complement C3 inhibitor, AMY-101, was safe and successful on a severely COVID-19 patient with ARDS [58]. Altogether, these studies suggest that macrophages play an essential role in the pathogenesis of SARS-CoV. Therefore, therapeutic interventions that target the altered immune responses of macrophages are urgently needed.
Potential Treatments:
In March 28th, 2020, the FDA issued an emergency-use authorization for chloroquine phosphate and hydroxychloroquine sulfate for the treatment of COVID-19 disease [59]. In April 29th, 2020, the National Institutes of Health (NIH) announced that the first clinical trial initiated in the United States using remdesivir for treatment against COVID-19 disease (NCT04280705) [60] obtained promising results [61]. In summary, it showed that those patients that received remdesivir, a broad-spectrum antiviral, had a 31% faster time to recovery, with a median of 11 days to recovery versus 15 days of those that received placebo. The mortality rate in those that received remdesivir was 8.0% compared to 11.6% in the placebo group. Therefore, it is expected that the FDA issues an emergency-use authorization for remdesivir against COVID-19 disease. Currently, there are hundreds of clinical trials related to COVID-19 in ClinicalTrials.gov. Among them, tens of studies are testing FDA-approved drugs in phase 4 of clinical trials. The FDA-approved drugs that are being tested in different experimental interventions related to COVID-19 in phase 4 of clinical trials are listed in Table 1: In these studies, the use of antivirals, immunomodulators, antihypertensive, anti-inflammatory, and antibiotics are the most common.
Table 1:
Drugs in Experimental Interventions related to COVID-19 in Phase 4 of Data from: ClinicalTrials.gov Searched as: “COVID-19”, “2019-nCoV”, “novel coronavirus 2019”, “SARS-CoV-2”, “coronavirus disease 2019.
| Clinical Trails | |||
|---|---|---|---|
| Drug(s) or Treatment(s) | ClinicalTrials.gov Identifier | Action (s) | PubChem Compound ID (CID) or Reference(s) |
| Hydroxychloroquine | NCT04359615 [62] | Immunosuppressive | CID=3652 [63] |
| NCT04359316 [64] | Anti-autophagy | ||
| NCT04343768 [65] | Anti-inflammatory | ||
| NCT04350671 [66] | Antirheumatologic | ||
| NCT04350684 [67] | Anti-malarial | ||
| NCT04355026 [68] | |||
| NCT04351919 [69] | |||
| NCT04351191 [70] | |||
| NCT04346667 [71] | |||
| NCT04334967 [72] | |||
| NCT04330495 [73] | |||
| NCT04341493 | |||
| NCT04338906 [74] | |||
| NCT04363866 [75] | |||
| Favipiravir | NCT04359615 [62] | Antiviral | CID=492405 [76] |
| Azithromycin |
NCT04359316[64] NCT04351919 [69] |
Antibiotic | CID=447043 [77] |
| Lopinavir / Ritonavir Or Ritonavir |
NCT04343768 [65] NCT04350671 [66] NCT04350684 [67] NCT04345276 [78] NCT04291729 [79] NCT04255017 [80] |
Antiviral | CID=11979606 [81] CID=392622 [82] |
| Interferon-βInterferon-β 1b, or PegIFN-α-2b |
NCT04343768 [65] NCT04350671 [66] NCT04350684 [67] NCT04291729 [79] NCT04254874 [83] |
Antiviral Antiproliferative Immunomodulator | Isaacs Lindenmann, et al. [84] |
| Lenalidomide | NCT04361643 [85] | Immunomodulator Antineoplastic | CID=216326 [86] |
| Arbidol (Umifenovir) |
NCT04350684 [67] NCT04255017 [80] NCT04254874 [83] NCT04260594 [87] |
Antiviral | CID=131411 [88] |
| Tetrandrine | NCT04308317 [89] | Immunosuppressive Antiproliferative Antioxidant | CID=73078 [90] |
| Chloroquine Sulfate | NCT04331600 [91] | Anti-malarial Anti-inflammatory | CID=91441 [92] |
| Carrimycin | NCT04286503 [93] | Antibiotic | Wang, et al. [49] |
| Ganovo (Danoprevir) |
NCT04345276 [78] NCT04291729 [79] |
Antiviral | CID=11285588 [94] |
| Bromhexine | NCT04355026 [68] | Expectorant Mucolytic | CID=2442 [95] |
| Thiazide or Thiazide-like diuretics | NCT04330300 [96] | Antihypertensive | Kjeldsen, et al. [97] |
| Calcium Channel Blockers | NCT04330300 [96] | Antihypertensive | Tocci, et al. [98] |
| Dexmedetomidine | NCT04350086 [99] | Analgesic Anxiolytic Sedative | CID=5311068 |
| Valsartan (Diovan) | NCT04335786 [100] | Antihypertensive | CID=60846 [101] |
| Sevoflurane | NCT04359862 [102] | Anesthetic | CID=5206 [103] |
| Oseltamivir | NCT04255017 [80] | Antiviral | CID=65028 [104] |
| Linagliptin | NCT04341935 | Dipeptidyl peptidase inhibitor | 4CID=10096344 [105] |
| Treatment for Diabetes Type 2 | |||
| Methylprednisolone | NCT04263402 [106] | Anti-inflammatory Immunosuppressive Neuroprotective Antiemetic Adrenergic | CID=6741 [107] |
| Ibuprofen | NCT04334629 [108] | Anti-inflammatory Analgesic Antipyretic | CID=3672 [109] |
| Nitazoxanide | NCT04341493 | Antiprotozoal Antiviral Antibiotic | CID=41684 [110] |
| Camostat Mesilate | NCT04338906 [74] | Anti-inflammatory | Gibo, et al. [111] |
| Budesonide | NCT04355637 [112] | Anti-inflammatory/ Bronchodilator | CID=5281004 [113] |
| Losartan | NCT04340557 | Antihypertensive | CID=3961 [114] |
| Oral nutrition supplement enriched in eicosapentaenoic acid, gamma-linolenic acid and antioxidants | NCT04323228 [115] | Anti-inflammatory Anti-oxidant | Gadek, et al., Singer, et al., Pontes-Arruda et al. [116–129] |
Acknowledgement
This work was supported by: 1 SC1 GM113691-01 (LMM), RCMI Translational Proteomics Center 8G12MD007600 (LMM), PR-INBRE-Institutional Developmental Award (IDEA) P20GM103475 (LMM), NIGMS-RISE R25 GM061838 (GBV and LRR)
Footnotes
Conflict of Interests
The authors declare no conflict of interests.
References
- 1.Paules CI, Marston HD, Fauci AS (2020) Coronavirus infections-more than just the common cold. Jama 323(8): 707–708. [DOI] [PubMed] [Google Scholar]
- 2.Barcena M, Oostergetel GT, Bartelink W, Faas FG, Verkleij A, et al. (2009) Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proceedings of the National Academy of Sciences 106(2): 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sola I, Almazan F, Zuniga S, Enjuanes L (2015) Continuous and discontinuous RNA synthesis in coronaviruses. Annual review of virology 2(1): 265–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, Li X, Yang B, et al. (2020) A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine 382(8): 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya AE (2020) Severe acute respiratory syndrome-related coronavirus-The species and its viruses, a statement of the Coronavirus Study Group. Bio Rxiv. [Google Scholar]
- 6.Nicholls J, Dong XP, Jiang G, Peiris M (2003) SARS: clinical virology and pathogenesis. Respirology 8(S1): 6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson RM, Fraser C, Ghani AC, Donnelly CA, Riley S, et al. (2004) Epidemiology, transmission dynamics and control of SARS: the 2002–2003 epidemic. Philosophical Transactions of the Royal Society of London Series B Biological Sciences 359(1447): 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widagdo W, Raj VS, Schipper D, Kolijn K, van Leenders GJ, (2016) Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. Journal of virology 90(9): 4838–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarner J (2020) Three emerging coronaviruses in two Decades. National Library of Medicine 153(4): 420–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Gayle AA, Wilder Smith A, Rocklöv J (2020) The reproductive number of COVID-19 is higher compared to SARS coronavirus. Journal of travel medicine 27(2): 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: Genome structure, replication, and pathogenesis. Journal of Medical Virology 92(4): 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798): 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Zheng W, Huang X, Bell EW, Zhou X, et al. (2020) Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. Journal of Proteome Research 19(4): 1351–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, et al. (2020) Isolation and Characterization of 2019-nCoV-like Coronavirus from Malayan Pangolins. BioRxiv. [Google Scholar]
- 15.Sun Z, Thilakavathy K, Kumar SS, He G, Liu SV (2020) Potential factors influencing repeated SARS outbreaks in China. International Journal of Environmental Research and Public Health 17(5): 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Wen Z, Zhong G, Yang H, Wang C, et al. (2020) Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS- coronavirus 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M, Kleine Weber H, Schroeder S, Kruger N, Herrler T, et al. (2020) SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181(2): 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology 203(2): 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu F, Lan Z, Xin Z, He C, Guo Z, et al. (2020) Emerging insights into molecular mechanisms underlying pyroptosis and functions of inflammasomes in diseases. Journal of Cellular Physiology 235(4): 3207–3221. [DOI] [PubMed] [Google Scholar]
- 20.Van Den Brand JMA, Lhaagmans B, Leijten L, Van Riel D, Emartina BE, et al. (2008) Pathology of experimental SARS coronavirus infection in cats and ferrets. Veterinary Pathology 45(4): 551–562. [DOI] [PubMed] [Google Scholar]
- 21.Belouzard S, Chu VC, Whittaker GR (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences of the United States of America 106(14): 5871–5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwata Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, et al. (2019) TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. Journal of Virology 93(6): 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, et al. (2020) HCA Lung Biological Network, SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 8674(20): 30500–30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cascella M, Rajnik M, Cuomo A (2020) Figure, Covid 19, Corona Replication. National Center for Biotechnology Information, U.S. National Library of Medicine. Contributed by Singh Rohan Bir. [Google Scholar]
- 25.Song Z, Xu Y, Bao L, Zhang L, Yu P, et al. (2019) From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 11(1): 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knipe DM, Fields BN, Knipe DM, Howley PM, Cohen JI (2013) Fields Virology. 6th (Edn.), Volume 1. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health. [Google Scholar]
- 27.Mo J, Angelichio M, Gow L, Leathers V, Jackwood MW (2020) Validation of specific quantitative real-time RT-PCR assay panel for Infectious Bronchitis using synthetic DNA standards and clinical specimens. Journal of virological methods 276: 113773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green K, Graziadio S, Turner P, Fanshawe T, Allen J (2020) Molecular and antibody point-of-care tests to support the screening, diagnosis and monitoring of COVID-19. Oxford Covid-19 Evidence Service. [Google Scholar]
- 29.Wyllie AL, Fournier J, Casanovas Massana A, Campbell M, Tokuyama M, et al. (2020) Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv. [Google Scholar]
- 30.Wang D, Hu Bo, Hu Chang, Fangfang Z, Xing L, et al. (2020) Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 323(11): 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rydzak P, Corona FMO, Whitfield AE, Wayadande AC (2020) Combining multiplex PCR and high-resolution melting for the detection and discrimination of arthropod transmitted viruses of cereals. Journal of Virological Methods 278: 113823. [DOI] [PubMed] [Google Scholar]
- 32.Xie X, Zhong Z, Zhao W, Zheng C, Wang F, (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(2020a) Emergency Use Authorization (EUA) information and list of all current EUAs. U.S. Food Drug Administration (FDA). [Google Scholar]
- 34.Abbott (2020) Detect COVID-19 in as little as 5 minutes.
- 35.Rao A, Goldstein DY, Wolk DM, Wolf LA (2020) Development and Evaluation of Two SARS CoV-2 RT-PCR Laboratory Developed Tests on the ARIES® Automated, Sample-to-Answer, Real-Time PCR System.
- 36.Hirayama D, Iida T, Nakase H (2017) The Phagocytic Function of Macrophage- Enforcing Innate Immunity and Tissue Homeostasis. International Journal of Molecular Sciences 19(1): 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon S, Pluddemann A (2017) Tissue macrophages: heterogeneity and functions. BMC Biology 15(1): 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zulu MZ, Martinez FO, Gordon S, Gray CM (2019) The Elusive Role of Placental Macrophages: The Hofbauer Cell. Journal of Innate Immunity 11(6): 447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi F, Qian S, Zhang S, Zhang Z (2020) Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochemical and Biophysical Research Communications. 526(1): 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Lau YF, Lamirande EW, Paddock CD, Bartlett JH, et al. (2009) Cellular immune responses to severe acute respiratory syndrome coronavirus (SARS-CoV) infection in senescent BALB/c mice: CD4+ T cells are important in control of SARS-CoV infection. Journal of Virology 84(3): 1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harmer D, Gilbert M, Borman R, Clark KL (2002) Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Letters. 532(1–2): 107–110. [DOI] [PubMed] [Google Scholar]
- 42.Yip MS, Leung NH, Cheung CY, Li PH, Lee HH, et al. (2014) Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virology Journal 11: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tseng CT, Perrone LA, Zhu H, Makino S, Peters CJ (2005) Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. The Journal of Immunology 174(12): 7977–7985. [DOI] [PubMed] [Google Scholar]
- 44.Hu Y, Li W, Gao T, Cui Y, Jin Y, et al. (2017) The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. Journal of Virology 91(8): 2143–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng X, Hackbart M, Mettelman RC, OBrien A, Mielech AM, et al. (2017) Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proceedings of the National Academy of Sciences USA 114(21): 4251–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen IY, Moriyama M, Chang MF, Ichinohe T (2019) Severe Acute Respiratory Syndrome Coronavirus Viroporin 3a Activates the NLRP3 Inflammasome. Frontiers in Microbiology 10: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu R, Zhao X, Li J, Niu P, Yang B, et al. (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 395(10224): 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Y, Feng Z, Diao B, Wang R, Wang G, et al. (2020) The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Directly Decimates Human Spleens and Lymph Nodes. medRxiv pp: 1–18. [Google Scholar]
- 49.Wang W, Ye L, Ye L, Li B, Gao B, et al. (2007) Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Research 128(1–2): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinto BGG, Oliveira AER, Singh Y, Jimenez L, Goncalves ANA, et al. (2020) ACE2 Expression is increased in the Lungs of Patients with Comorbidities Associated with Severe COVID- 19. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang C, Wang Y, Li X, Ren L, Zhao J, et al. (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395(10223): 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao M, Liu Y, Yuan J, Wen Y, Xu G, et al. (2020) The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. medRxiv. [Google Scholar]
- 53.Giamarellos Bourboulis EJ, Netea MG, Rovina N, Akinosoglou K, Antoniadou A, et al. (2020) ComplexImmune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe 3128(20): 30235–30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crayne CB, Albeituni S, Nichols KE, Cron RQ (2019) The Immunology of Macrophage Activation Syndrome. Frontiers in Immunology 10: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.National Health Commission of the People’s Republic of China and National Health Commission of the People’s Republic of China. (2020) Guidelines for the diagnosis and treatment of coronavirus disease 2019. [Google Scholar]
- 56.Oudit GY, Kassiri Z, Jiang C, Liu PP, Poutanen SM, et al. (2009) SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. European Journal of Clinical Investigation 39(7): 618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Risitano AM, Mastellos DC, Huber Lang M, Yancopoulou D, Garlanda C, et al. (2020) Complement as a target in COVID-19? Nature Reviews Immunology 23: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mastaglio S, Ruggeri A, Risitano AM, Angelillo P, Yancopoulou D, et al. (2020) The first case of COVID-19 treated with the complement C3 inhibitor AMY-101. Clinical Immunology 215: 108450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.(2020) Chloroquine phosphate and hydroxychloroquine sulfate for treatment of COVID-19. U.S. Food & Drug Administration (FDA). [Google Scholar]
- 60.(2020) Adaptive COVID-19 Treatment Trial (ACTT) ClinicalTrials.gov National Library of Medicine NCT04280705.
- 61.NIH (2020) NIH clinical trial shows Remdesivir accelerates recovery from advanced COVID-19.
- 62.(2020) Favipiravir in Hospitalized COVID-19 Patients. ClinicalTrials.gov National Library of Medicine; Identifier NCT04359615. [Google Scholar]
- 63.National Center for Biotechnology Information (2020) PubChem Database. Hydroxychloroquine, CID=3652.
- 64.(2020) Azithromycin in Hospitalized COVID-19 Patients (AIC). ClinicalTrials.gov National Library of Medicine; Identifier NCT04359316. [Google Scholar]
- 65.(2020) An Investigation Into Beneficial Effects of Interferon Beta 1a, Compared to Interferon Beta 1b And The Base Therapeutic Regiment in Moderate to Severe COVID-19: ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04343768. [Google Scholar]
- 66.(2020) Interferon Beta 1a in Hospitalized COVID-19 Patients. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04350671. [Google Scholar]
- 67.(2020) Umifenovir in Hospitalized COVID-19 Patients. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US). Identifier NCT04350684. [Google Scholar]
- 68.(2020) Use of Bromhexine and Hydroxychloroquine for Treatment of COVID-19 Pneumonia. ClinicalTrials. gov, Bethesda (MD): National Library of Medicine (US). Identifier NCT04355026. [Google Scholar]
- 69.(2020) Assessment of Efficacy and Safety of HCQ and Antibiotics Administrated to Patients COVID19 (+). ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US). Identifier NCT04351919. [Google Scholar]
- 70.(2020) PRophylaxis of Exposed COVID-19 Individuals with Mild Symptoms Using choloroquinE Compounds. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US). Identifier NCT04351191. [Google Scholar]
- 71.(2020) Post-Exposure Prophylaxis for Asymptomatic SARS-CoV-2 COVID-19 Patients With choloroquinE Compounds. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US). Identifier NCT04346667. [Google Scholar]
- 72.(2020) Hydroxychloroquine in Patients With Newly Diagnosed COVID-19 Compared to Standard of Care.” ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US). Identifier NCT04334967. [Google Scholar]
- 73.(2020) Randomized, Controlled, Double-blind Clinical Trial Comparing the Efficacy and Safety of Chemoprophylaxis With Hydroxychloroquine in Patients Under Biological Treatment and or JAK Inhibitors in the Prevention of SARS-CoV-2 Infection. ClinicalTrials. gov, Bethesda (MD): National Library of Medicine (US). Identifier NCT04330495. [Google Scholar]
- 74.(2020) Combination Therapy with Camostat Mesilate + Hydroxychloroquine for COVID-19. ClinicalTrials. gov. Bethesda (MD): National Library of Medicine (US). Identifier NCT04338906. [Google Scholar]
- 75.(2020) A Pilot Study to Assess Hydroxychloroquine in Patients with SARS-CoV-2(COVID-19). ClinicalTrials. gov, Bethesda (MD): National Library of Medicine (US). Identifier NCT04363866. [Google Scholar]
- 76.National Center for Biotechnology Information (2020) PubChem Database. Favipiravir, CID=492405.
- 77.National Center for Biotechnology Information (2020) PubChem Database. Azithromycin, CID=447043.
- 78.(2020) Efficacy and Safety of Ganovo (Danoprevir) Combined With Ritonavir in the Treatment of SARS-CoV-2 Infection Identifier NCT04345276.
- 79.(2020) Evaluation of Ganovo (Danoprevir) Combined With Ritonavir in the Treatment of SARS-CoV-2 Infection. >ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US). Identifier NCT04291729. [Google Scholar]
- 80.(2020) A Prospective/Retrospective, Randomized Controlled Clinical Study of Antiviral Therapy in the 2019-nCoV Pneumonia. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US). Identifier NCT04255017. [Google Scholar]
- 81.National Center for Biotechnology Information (2020) PubChem Database. Lopinavir/Ritonavir, CID=11979606.
- 82.National Center for Biotechnology Information (2020) PubChem Database. Ritonavir, CID=392622.
- 83.(2020) A Prospective/Retrospective, Randomized controlled Clinical Study of Interferon Atomization in the 2019-nCoV Pneumonia. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04254874. [Google Scholar]
- 84.Isaacs A, Lindenmann J (1957) Virus interference. I. the interferon. Proceedings of the Royal Society of London. Series B, Biological Sciences 147(927): 258–267. [DOI] [PubMed] [Google Scholar]
- 85.(2020) Low-dose Lenalidomide for Non-severe COVID-19 Treatment Trial. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04361643. [Google Scholar]
- 86.National Center for Biotechnology Information (2020) PubChem Database. Lenalidomide, CID=216326.
- 87.(2020) Clinical Study of Arbidol Hydrochloride Tablets in the Treatment of Pneumonia Caused by Novel Coronavirus. Identifier NCT04260594.
- 88.National Center for Biotechnology Information (2020) PubChem Database. Arbidol, CID=131411.
- 89.(2020) Tetrandrine Tablets Used in the Treatment of COVID-19. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US), Identifier NCT04308317. [Google Scholar]
- 90.National Center for Biotechnology Information (2020) PubChem Database. Tetrandrine, CID=73078.
- 91.(2020) ChloroQUine As antiviRal treAtmeNT In coroNavirus infection 2020. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04331600. [Google Scholar]
- 92.National Center for Biotechnology Information (2020) PubChem Database. Chloroquine sulfate, CID=91441.
- 93.(2020) The Clinical Study of Carrimycin on Treatment Patients With COVID-19. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04286503. [Google Scholar]
- 94.National Center for Biotechnology Information (2020) PubChem Database. Danoprevir, CID=11285588.
- 95.National Center for Biotechnology Information (2020) PubChem Database. Bromhexine, CID=2442.
- 96.(2020) Coronavirus (COVID-19) ACEi/ARB Investigation. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04330300. [Google Scholar]
- 97.Kjeldsen SE, Os I (2020) Assessing hypertension therapies: randomization or confounding by indication? Nature Reviews Cardiology 17(2): 73–74. [DOI] [PubMed] [Google Scholar]
- 98.Tocci G, Battistoni A, Passerini J, Musumeci MB, Francia P, et al. (2015) Calcium channel blockers and hypertension. Journal of Cardiovascular Pharmacology and Therapeutics 20(2): 121–130. [DOI] [PubMed] [Google Scholar]
- 99.(2020) Use of Dexmedetomidine in Light to Moderate Sedation in the Patient in the Palliative Situation of a Sars-cov-2 COVID-19 Infection, Identifier NCT04350086.
- 100.(2020) Valsartan for Prevention of Acute Respiratory Distress Syndrome in Hospitalized Patients with SARS-COV-2 (COVID-19) Infection Disease. ClinicalTrials. gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04335786. [Google Scholar]
- 101.National Center for Biotechnology Information (2020) PubChem Database. Valsartan, CID=60846.
- 102.(2020) Sedation With Sevoflurane Versus Propofol in Patients With Acute Respiratory Distress Syndrome Caused by COVID19 Infection. ClinicalTrials.gov. Bethesda (MD): National Library of Medicine (US), Identifier NCT04359862. [Google Scholar]
- 103.National Center for Biotechnology Information (2020) PubChem Database. Sevoflurane, CID=5206.
- 104.National Center for Biotechnology Information (2020) PubChem Database. Oseltamivir, CID=65028.
- 105.National Center for Biotechnology Information (2020) PubChem Database. Linagliptin, CID=10096344.
- 106.(2020) The Efficacy of Different Hormone Doses in 2019-nCoV Severe Pneumonia. ClinicalTrials.gov, Bethesda (MD: ): National Library of Medicine (US), Identifier NCT04263402. [Google Scholar]
- 107.National Center for Biotechnology Information (2020) PubChem Database. Methylprednisolone, CID=6741.
- 108.(2020) LIBA Trial in COVID-19. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US), Identifier NCT04334629. [Google Scholar]
- 109.National Center for Biotechnology Information (2020) PubChem Database. Ibuprofen, CID=3672.
- 110.National Center for Biotechnology Information (2020) PubChem Database. Nitazoxanide, CID=41684.
- 111.Gibo J, Ito T, Kawabe K, Hisano T, Inoue M, et al. (2005) Camostat mesilate attenuates pancreatic fibrosis via inhibition of monocytes and pancreatic stellate cells activity. Laboratory Investigation 85(1): 75–89. [DOI] [PubMed] [Google Scholar]
- 112.(2020) Inhaled Corticosteroid Treatment of COVID19 Patients with Pneumonia. ClinicalTrials.gov, Bethesda (MD): National Library of Medicine (US). Identifier NCT04355637. [Google Scholar]
- 113.National Center for Biotechnology Information (2020) PubChem Database. Budesonide, CID=5281004.
- 114.National Center for Biotechnology Information (2020) PubChem Database. Losartan, CID=3961.
- 115.(2020) Anti-inflammatory/Antioxidant Oral Nutrition Supplementation in COVID-19. ClinicalTrials. gov Bethesda (MD): National Library of Medicine (US), Identifier NCT04323228. [Google Scholar]
- 116.Gadek JE, DeMichele SJ, Karlstad MD, Pacht ER, Donahoe M, et al. (1999) Effect of enteral feeding with eicosapentaenoic acid, gamma-linolenic acid and antioxidants in patients with acute respiratory distress syndrome. Enteral Nutrition in ARDS Study Group. Critical Care Medicine 27(8): 1409–1420. [DOI] [PubMed] [Google Scholar]
- 117.Singer P, Theilla M, Fisher H, Gibstein L, Grozovski E, et al. (2006) Benefit of an enteral diet enriched with eicosapentaenoic acid and gamma-linolenic acid in ventilated patients with acute lung injury. Critical Care Medicine 34(4): 1033–1038. [DOI] [PubMed] [Google Scholar]
- 118.Pontes Arruda A, Aragao AM, Albuquerque JD (2006) Effects of enteral feeding with eicosapentaenoic acid, gamma-linolenic acid, and antioxidants in mechanically ventilated patients with severe sepsis and septic shock. Critical Care Medicine 34(9): 2325–2333. [DOI] [PubMed] [Google Scholar]
- 119.Do Angiotensin Receptor Blockers Mitigate Progression to Acute Respiratory Distress Syndrome With SARS-CoV-2 Infection? Clinical Trails Gov.
- 120.Hugo Mendieta Zeron (2020) Hydroxychloroquine vs Nitazoxanide in Patients with COVID-19. Clinical Trails Gov.
- 121.Effects of DPP4 Inhibition on COVID-19. ClinicalTrials.gov.
- 122.Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, et al. (2020) HCA Lung Biological Network, SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell (2020) 8674(20): 30500–30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li Z, Yi Y, Luo X, Xiong N, Liu Y, et al. (2020) Development and Clinical Application of Rapid IgM-IgG Combined Antibody Test for SARS-CoV-2 Infection Diagnosis. Journal of Medical Virology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Loeffelholz M, Tang Y (2020) Laboratory diagnosis of emerging human coronavirus infections- the state of the art. Emerging Microbes & Infections 9(1): 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.National Center for Biotechnology Information (2020) PubChem Database. Spironolactone, CID=5833.
- 126.(2020) ACE2 angiotensin I converting enzyme 2 [Homo sapiens (human)] National Library of Medicine, Gene, pp: 100. [Google Scholar]
- 127.Petherick A (2020) Developing antibody tests for SARS-CoV-2. World Report 395(10230): 1101–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R (2020) COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. Journal of Advanced Research 24: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Xiao S, Wu Y, Liu H (2020) Evolving status of the 2019 novel coronavirus infection: Proposal of conventional serologic assays for disease diagnosis and infection monitoring. Journal of Medical Virology 92(5): 464–467. [DOI] [PMC free article] [PubMed] [Google Scholar]