Abstract
Postoperative cognitive dysfunction (POCD) is a common complication in elderly patients. Circular RNAs (circRNAs) may contribute to neurodegenerative diseases. However, the role of circRNAs in POCD in aged mice has not yet been reported. This study aimed to explore the potential circRNAs in a POCD model. First, a circRNA microarray was used to analyze the expression profiles. Differentially expressed circRNAs were validated using quantitative real-time polymerase chain reaction. A bioinformatics analysis was then used to construct a competing endogenous RNA (ceRNA) network. The database for annotation, visualization, and integrated discovery was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of circRNA-related genes. Moreover, protein-protein interactions were analyzed to predict the circRNA-regulated hub genes using the STRING and molecular complex detection plug-in of Cytoscape. Microarray screen 124 predicted circRNAs in the POCD of aged mice. We found that the up/downregulated circRNAs were involved in multiple signaling pathways. Hub genes, including Egfr and Prkacb, were identified and may be regulated by ceRNA networks. These results suggest that circRNAs are dysexpressed in the hippocampus and may contribute to POCD in aged mice.
Keywords: circRNAs, expression profile, postoperative cognitive dysfunction, miRNAs, ceRNA network
Introduction
Postoperative cognitive dysfunction (POCD) causes significant harm to elderly patients (Deiner and Silverstein, 2009). It may prolong hospital stay, increase medical expenses, reduce patients’ daily living ability, and may even be associated with an increase in long-term mortality. In addition, it may exacerbate the decline in cognitive dysfunction and even increase the rate of dementia (Steinmetz et al., 2009). Age is considered one of the possible risk factors associated with the incidence of POCD, as older adults have reduced cognitive reserve in the fragile brain (Terrando et al., 2011). Aged people are expected to be the largest surgical population in the future. Researchers have suggested that neuroinflammation, neuronal apoptosis, mitochondrial dysfunction, and synaptic dysfunction may contribute to POCD progression (Miao et al., 2017, 2018, 2019; Chen et al., 2019; Qiu et al., 2020). However, the exact mechanisms of POCD remain unclear, and the biomarkers are also lacking (Vutskits and Xie, 2016). Therefore, an in-depth study of the molecular mechanism of POCD is of clinical significance.
Circular RNA (circRNA) is a newly discovered class of approximately 100 nucleotides (nt) in length non-coding RNAs (ncRNAs) as a closed-loop structure. Studies have suggested that circRNAs are potential candidates for clinical diagnostic biomarkers (Jeck and Sharpless, 2014) because they are more stable. A variety of functions and mechanisms have been discovered with circRNAs, such as acting as microRNA (miRNA) sponges, binding proteins, or deoxyribonucleic acid (DNA) sequences, thereby regulating gene expression (Hansen et al., 2013; Memczak et al., 2013). They are stable in tissue and developmental stages in different species (Szabo et al., 2015). New studies suggest that circRNAs may be involved in different physiological or pathological processes and play an important role (Cheng et al., 2020; Wang et al., 2020; Zhang et al., 2020). Evidence has also shown that circRNAs are involved in neurological diseases such as Alzheimer’s disease (AD; Zhou et al., 2018; Ma et al., 2019; Chu et al., 2020). However, the differently expressed circRNAs in the hippocampus of aged mice and the role of circRNA in POCD are largely unknown.
Genome analysis has been used to identify functional genes in POCD for several years (Li et al., 2015). For example, Liu et al. (2019) found that many miRNAs were differentially expressed between POCD and control-aged mice (Chen et al., 2019; Su et al., 2019). Furthermore, long non-coding RNAs (lncRNAs) were found to be differentially expressed in mRNAs and might contribute to POCD (Li et al., 2019). However, the role of circRNAs in POCD in aged mice remains unclear. Therefore, the current study aimed to illustrate the circRNA expression profile and the potential functions and mechanisms of POCD.
Materials and Methods
Animal Experiments
C57BL/6 male mice (weighing 30–35 g and aged 18 months) were purchased from Nanjing University’s Model Animal Research Center. Mice were housed under standardized conditions. Before the start of the study, all mice were acclimatized for at least 1 week. The animal protocol was approved by the Animal Care and Use Committee of Xuzhou Medical University.
Animal Model of POCD
Mice were separated into POCD (n = 10) and control groups (n = 10). The surgical operation of intramedullary fixation for open tibial fractures was performed in accordance with previous studies and with modifications (Feng et al., 2017; Chen et al., 2019). For the anesthesia induction period, 3.0% isoflurane was used, and 1.5% isoflurane was used for the maintenance period, which was supported by using 100% oxygen. First, we made an incision on the lateral side of the tibia, in order to expose the bone. Then, a minor hole was drilled into the tibial trochanter, and an intramedullary fixation needle was inserted. All mice were returned to their home cages after recovery from anesthesia. To manage postoperative pain, 2% lidocaine solution and 1% tetracaine hydrochloride glue were used locally twice daily.
Behavioral Tests
We used the open field test (OFT) to estimate motor activity 2 h ahead of the fear conditioning test (FCT) training. Mice were allowed free exploration in the OFT for 5 min. We used the system camera to track and record the mouse movement automatically, and selected the total distance to ascertain locomotor ability.
The test and training stages were included in the FCT. The mice were allowed to adapt to the chamber environment for 2 min, given six pairs stimulation, and then maintained for another 1 min. The cycle consisted of a conditional stimulus (70 dB tone, 20 s), a trace interval (contextual break, 25 s), and an unconditional stimulus (0.70 mA electrical footshock, 2 s). Every cycle was randomly divided into breaks of 45–60 s.
RNA Extraction
The hippocampal tissue of mice was preserved at −80°C after behavioral experimentation on day 3 postoperatively. TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA.
Microarray Detection
Kangchen Biotechnology (Shanghai, China) Arraystar circRNA microarray was used for RNA sample analysis and the obtained samples were labeled by Arraystar’s standard protocols. We used Limma for the original raw data processing.
Quantification With PCR
The cDNA samples were configured with the real-time PCR reaction system. 2X PCR Master Mix (Arraystar) was used. The primer sequences used for the predicted circRNAs are listed in Table 1.
Table 1.
The forward and reverse primers for qPCR.
| Gene | Primers | Sequence (5′ to 3′) |
|---|---|---|
| mmu_circRNA_28795 | Forward primer | AAGGAGGATACTAGCAGGTTGGA |
| Reverse primer | TGTAGTAGTAGAAGCGTGA | |
| TGTATTGG | ||
| mmu_circRNA_44122 | Forward primer | GACACACAGACAGACCCATACTGT |
| Reverse primer | CTTCTCTGCTCTGCAAGGTGG | |
| mmu_circRNA_22058 | Forward primer | AGGCAGCACTCAATGGACAAG |
| Reverse primer | ACCAGTCATGTCGTTTTCCTTTAC | |
| mmu_circRNA_44559 | Forward primer | CAAAGGTGTTGTGAAGGAAGTC |
| Reverse primer | TTTCTGTATCACACGTGCGTAA | |
| mmu_circRNA_45921 | Forward primer | ATGTGGGAGCGTGGAGATAA |
| Reverse primer | CATCACGGTCAGCATTCTTG | |
| mmu_circRNA_22673 | Forward primer | TCAAGTCCTGCACAAAGACAAC |
| Reverse primer | GCAAGGCTCCTAAATCAAAGTC | |
| Egfr | Forward primer | GCCATCTGGGCCAAAGATACC |
| Reverse primer | GTCTTCGCATGAATAGGCCAAT | |
| Prkacb | Forward primer | ATGGTTATGGAATACGTCCCTGG |
| Reverse primer | AATTAAGAGGTTTTCCGGCTTGA | |
| β-actin | Forward primer | GTACCACCATGTACCCAGGC |
| Reverse primer | AACGCAGCTCAGTAACAGTCC |
Annotation and Functional Prediction of Up/Downregulated circRNAs
CircRNA-microRNA interactions were anticipated using microRNA predicted estimate software based on TargetScan (Enright et al., 2003) and miRanda (Pasquinelli, 2012) manufactured by Arraystar Corporation. The software analysis procedure is as follows: First, the target estimate was obtained by employing the latest miRNA. Second, using Context +≤ −0.05 and Context ≤ −0.05, candidate miRNAs were concealed. Third, the demanded circRNA-microRNA interactions were acquired following miRNA-related MREs. We used MiRWalk 3.0 to predict the microRNA-related mRNA. The results of differentially expressed mRNAs were reported and overlapped in the gene expression omnibus (GEO) database (GSE 113738). Next, the acquired circRNAs possible target mRNAs’ protein-protein interactions were analyzed to predict the circRNA-regulated hub genes using Cytoscape STRING and molecular complex detection (MCODE) plug-ins.
Statistical Analysis
Based on our preliminary study, the sample size was chosen to obtain a 0.9 power and p < 0.05. Quantitative data are expressed as mean ± standard error (SEM). Data analysis was carried out using GraphPad Prism 6.0. The double-tailed student’s t-test was used to compare the statistical significance between groups.
Results
Cognitive Dysfunction of Aged Mice on Day 3 Postoperatively
The cognitive function of aged mice was assessed using the OFT and FCT 1 day before and 3 days after the orthopedic surgery under isoflurane anesthesia. The experimental schedule is shown in Figure 1A.
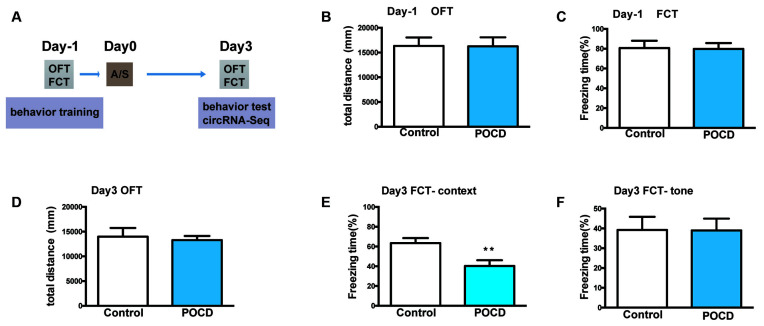
Figure 1.
Hippocampus-dependent learning and memory, but not hippocampus-independent learning and memory or motor activity was impaired by anesthesia/surgery in aged mice. (A) The experimental process’ schematic diagram. (B) The baseline of total distance the elderly mice traveled in open field test (OFT) during the training period 1 day before surgery. (C) The baseline of freezing time (%) in fear conditioning test (FCT) of aged mice during the training duration 1 day before surgery. (D) The total distance traveled in OFT 3 days after the operation. (E) Freezing time percentage in the FCT context test 3 days after surgery. (F) Freezing time percentage in the FCT tone test 3 days postoperatively. Data for each group are shown as the mean ± standard error (n = 10). **p < 0.01 compared with the control group.
First, the locomotor activity of mice tested by total distance in the OFT suggested that it did not change 1 day before (p > 0.05, Figure 1B) or 3 days (p > 0.05, Figure 1D) postoperatively. Second, FCT was used to evaluate the learning and memory functions. The context/tone test of the FCT was used to evaluate hippocampus-dependent/independent learning and memory. In the training session, the freezing time of FCT was not significantly changed (p > 0.05, Figure 1C), indicating that hippocampus-independent/dependent learning and memory were identical. For the context test, the freezing time was significantly reduced to 3 days postoperatively (p < 0.01, Figure 1E). Nevertheless, the freezing time for the tone test was not significantly different (p > 0.05, Figure 1F). In summary, the above results indicated that hippocampal-dependent cognition was impaired in aged mice postoperatively, which proves that the POCD mouse model has been successfully established.
Differentially Expressed circRNAs in POCD of Aged Mice
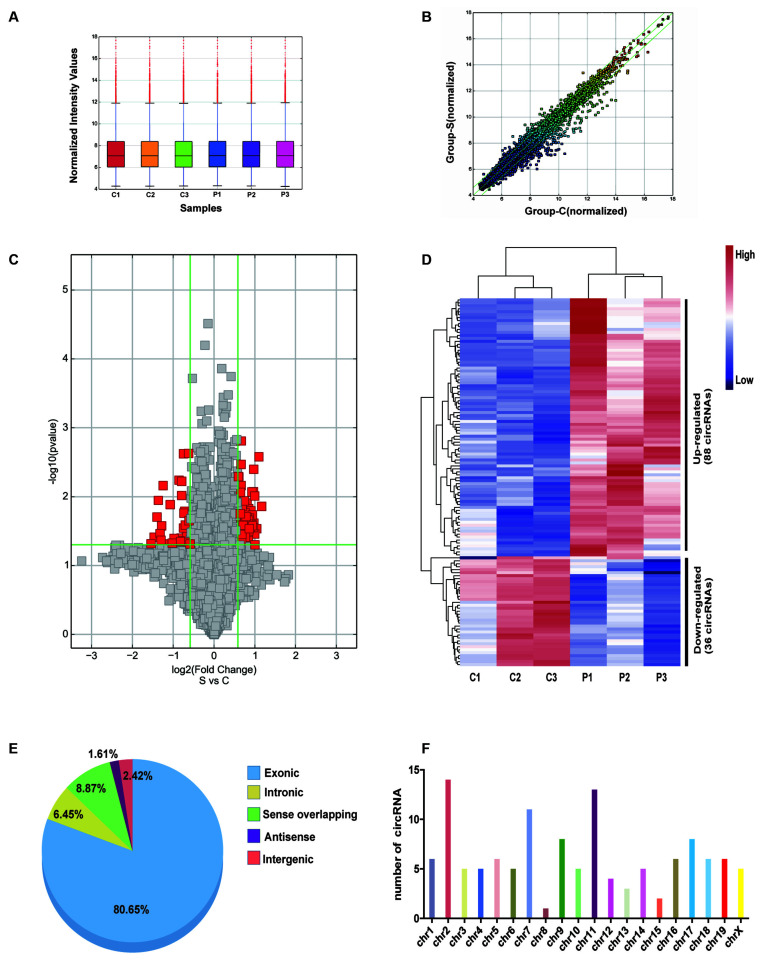
Differentially expressed circRNAs from the hippocampal tissues of control and POCD mice were screened by microarray-based profiling. The box plot shows a similar data distribution for both groups (Figure 2A). The scatter plot shows the original circRNA expression values between the two groups (Figure 2B). circRNA expression was observed in volcano plots and hierarchical clustering (Figures 2C,D). A total of 124 differentially expressed circRNAs in POCD hippocampal tissues compared with control tissues were identified, of which 88 were upregulated and 36 were downregulated when the fold change was ≥1.5, and the p-value was <0.05. The top 10 up/down-regulated circRNAs are listed in Table 2. Additionally, we found that 80.65% of circRNAs were exonic circRNAs, 8.87% were sense-overlapping circRNAs, 6.45% were intronic circRNAs, 2.42% were intergenic circRNAs, and 1.61% were antisense circRNAs (Figure 2E). Localization analysis revealed that these abnormally regulated circRNAs were mainly located in chr1-19 and chrX, but not chr20 and chrY (Figure 2F).
Figure 2.
Character of differentially expressed circRNA in the postoperative cognitive dysfunction (POCD) and control groups. (A) Distribution of circRNAs in the six samples (C1–3: control group; P1–3: POCD group). (B) The difference expression of circRNA is shown by a scatter plot between the control and POCD groups. (C) The differential circRNA expression is shown by volcano diagrams between the control and POCD groups. (D) Thirty-six downregulated and 88 upregulated circRNAs in POCD are shown in the heatmap. (E) The pie chart showed the transcriptional sources of the differentially expressed circRNAs. (F) Chromosome locations of the differentially expressed circRNAs.
Table 2.
Top 10 significantly up/downregulated circRNAs in POCD of aged mice.
| Significantly up/downregulated circRNAs | |||||
|---|---|---|---|---|---|
| probeID | P-value | FDR | FC(abs) | Regulation | circRNA |
| ASCRP4007024 | 0.013865474 | 0.529357952 | 2.2450955 | up | mmu_circRNA_22261 |
| ASCRP4007409 | 0.00262982 | 0.529357952 | 2.1437073 | up | mmu_circRNA_27408 |
| ASCRP4003389 | 0.029190777 | 0.529357952 | 2.0674603 | up | mmu_circRNA_42249 |
| ASCRP4013349 | 0.049867064 | 0.529357952 | 2.0025764 | up | mmu_circRNA_25474 |
| ASCRP4004614 | 0.034733688 | 0.529357952 | 1.9985147 | up | mmu_circRNA_44122 |
| ASCRP4010020 | 0.009783375 | 0.529357952 | 1.9958363 | up | mmu_circRNA_32165 |
| ASCRP4003327 | 0.029212904 | 0.529357952 | 1.9870759 | up | mmu_circRNA_41895 |
| ASCRP4000668 | 0.024166022 | 0.529357952 | 1.9686275 | up | mmu_circRNA_22058 |
| ASCRP4002143 | 0.030499114 | 0.529357952 | 1.9626064 | up | mmu_circRNA_39251 |
| ASCRP4006538 | 0.003998226 | 0.529357952 | 1.9615141 | up | mmu_circRNA_27407 |
| ASCRP4010600 | 0.048075886 | 0.529357952 | 2.9162194 | down | mmu_circRNA_33862 |
| ASCRP4001381 | 0.038744532 | 0.529357952 | 2.7699311 | down | mmu_circRNA_30227 |
| ASCRP4001879 | 0.019740651 | 0.529357952 | 2.6326581 | down | mmu_circRNA_38328 |
| ASCRP4001023 | 0.011304438 | 0.529357952 | 2.5770301 | down | mmu_circRNA_22673 |
| ASCRP4010800 | 0.034227559 | 0.529357952 | 2.5008209 | down | mmu_circRNA_31065 |
| ASCRP4011284 | 0.042921723 | 0.529357952 | 2.4593752 | down | mmu_circRNA_34414 |
| ASCRP4009404 | 0.02644451 | 0.529357952 | 2.4333386 | down | mmu_circRNA_45921 |
| ASCRP4011303 | 0.006882754 | 0.529357952 | 2.3732211 | down | mmu_circRNA_44559 |
| ASCRP4002514 | 0.048719463 | 0.529357952 | 2.200436 | down | mmu_circRNA_29699 |
| ASCRP4002584 | 0.013002798 | 0.529357952 | 2.1034439 | down | mmu_circRNA_29984 |
Validation of Dysregulated circRNAs Through Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
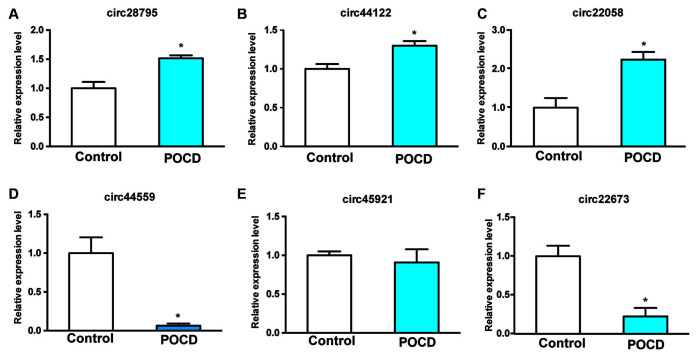
Six dysregulated circRNAs were chosen randomly, including upregulated (circRNA_28795, circRNA_44122, and circRNA_22058) and downregulated circRNAs (circRNA_44559, circRNA_45921, and circRNA_22673). qRT-PCR demonstrated that circRNA_28795, circRNA_44122, circRNA_22058, circRNA_44559, and circRNA_22673 expression in the control and POCD hippocampal tissues was verified with the microarray results. The expression of circRNA_28795, circRNA_44122, circRNA_22058, circRNA_44559, and circRNA_22673 expression were significantly different (Figures 3A–D,F, p < 0.05). The expression of circRNA_45921 was not significantly different (Figure 3E, p > 0.05).
Figure 3.
Conformation of the six prospected circRNAs with qRT-PCR. RNAs from the POCD and control groups were involved (A–F). *p < 0.05 compared with the control group.
MicroRNA Response Elements (MREs) Analysis
circRNAs may regulate target gene expression through competitive binding with their target miRNAs (Han et al., 2017; Qiu et al., 2018). The circRNA-microRNA networks were predicted using Arraystar’s self-made miRNA target prediction software. A 2D structure was established through sequence analysis of MREs (Supplementary Figures 1A–E). For circRNA_28795, the predicted miRNAs included miR-151-5p, miR-669b-5p, miR-453, miR-138-5p, and miR-298-5p, circRNA_44122, the predicted miRNAs included miR-7033-5p, miR-6914-3p, miR-6974-3p, miR-7010-3p, and miR-361-3p; for circRNA_22058, the predicted miRNAs included miR-196a-5p, miR-7048-3p, miR-322-5p, miR-6964-3p, and miR-670-3p; for circRNA_44559, the predicted miRNAs included miR-1903, miR-207, miR-6946-3p, miR-6896-3p, and miR-6976-3p; for circRNA_22673, the predicted miRNAs included miR-6952-5p, miR-6982-5p, miR-7092-3p, miR-6957-3p, and miR-6986-5p (Table 3).
Table 3.
Predicted miRNA response elements of the five confirmed circRNAs.
| circRNA ID | Predicted miRNA response elements (MREs) | ||||
|---|---|---|---|---|---|
| MRE1 | MRE2 | MRE3 | MRE4 | MRE5 | |
| circRNA_28795 | miR-151-5p | miR-669b-5p | miR-453 | miR-138-5p | miR-298-5p |
| circRNA_44122 | miR-7033-5p | miR-6914-3p | miR-6974-3p | miR-7010-3p | miR-361-3p |
| circRNA_22058 | miR-196a-5p | miR-7048-3p | miR-322-5p | miR-6964-3p | miR-670-3p |
| circRNA_44559 | miR-1903 | miR-207 | miR-6946-3p | miR-6896-3p | miR-6976-3p |
| circRNA_22673 | miR-6952-5p | miR-6982-5p | miR-7092-3p | miR-6957-3p | miR-6986-5p |
circRNA-miRNA-mRNA Interaction Analysis of Validated circRNAs
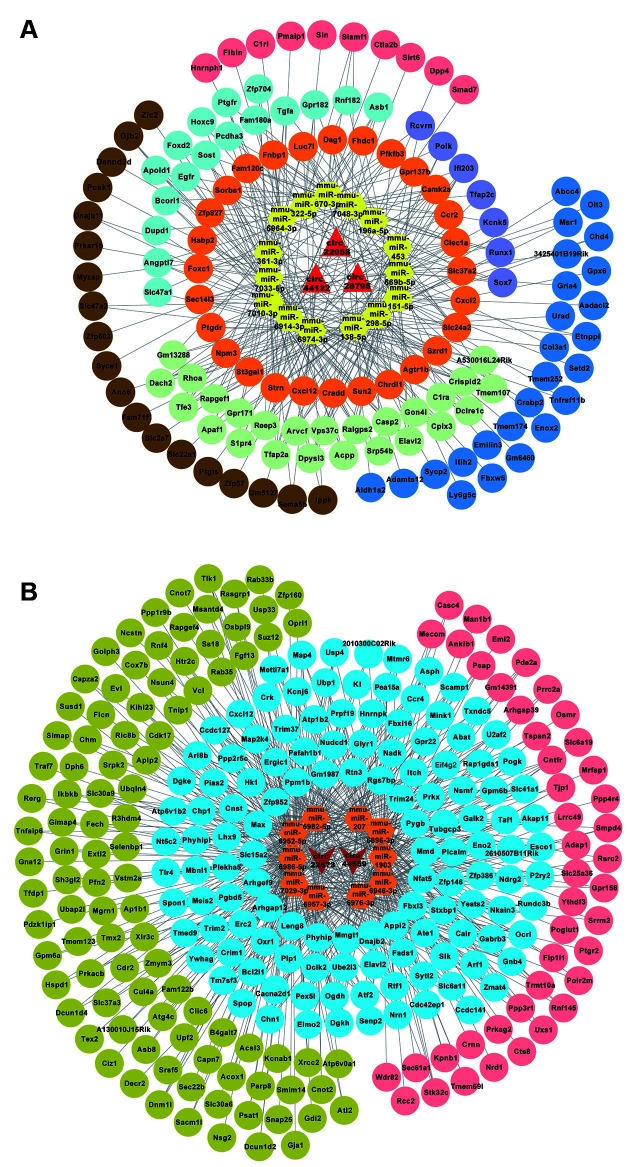
MiRWalk 3.0 was used to predict target genes of the above-mentioned circRNA-targeted miRNAs. Then, the predicted genes of the three upregulated circRNAs were overlapped by the reported differentially upregulated mRNAs from the GEO database (GSE 113738, from POCD model of aged mice), 131 upregulated mRNAs were screened to build the ceRNA networks, and 268 downregulated mRNAs were screened in the same way. The circRNA-miRNA-mRNA regulatory networks for circRNA_28795, circRNA_44122, and circRNA_22058 included 15 miRNAs and 131 mRNAs (Figure 4A), and for circRNA_44559 and circRNA_22673 included 10 miRNAs and 268 mRNAs (Figure 4B). These results indicate that the ceRNA network might be involved in the POCD mechanism.
Figure 4.
circRNA-miRNA-mRNA network analysis. (A) The ceRNA network for the three upregulated circRNAs. (B) The ceRNA network for the three downregulated circRNAs.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of the Predicted Network Genes
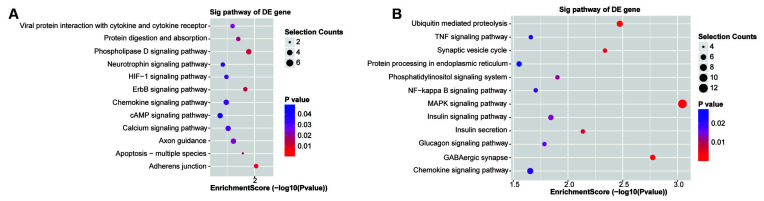
The functions of the 131 upregulated and 268 downregulated genes were analyzed using GO and KEGG analyses. The top highly enriched GO terms of biological process, cellular component, and molecular function for upregulated and downregulated genes are shown in Supplementary Figures 2A,B. The top terms of upregulated genes were identical protein binding (GO:0042802), extracellular region (GO:0005576), and tube morphogenesis (GO:0035239). For downregulated genes, the top terms were protein binding (GO:0005515), cell part (GO:0044464), and localization (GO:0051179). The KEGG pathway analysis revealed that the upregulated genes were involved in the adherens junction, phospholipase D signaling pathway, and ErbB signaling pathway (Figure 5A). Accordingly, the downregulated genes were significantly enriched in the MAPK signaling pathway, GABAergic synapse, and ubiquitin-mediated proteolysis (Figure 5B).
Figure 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. (A,B) The KEGG enrichment analysis of the up/downregulated circRNA’s predicted genes.
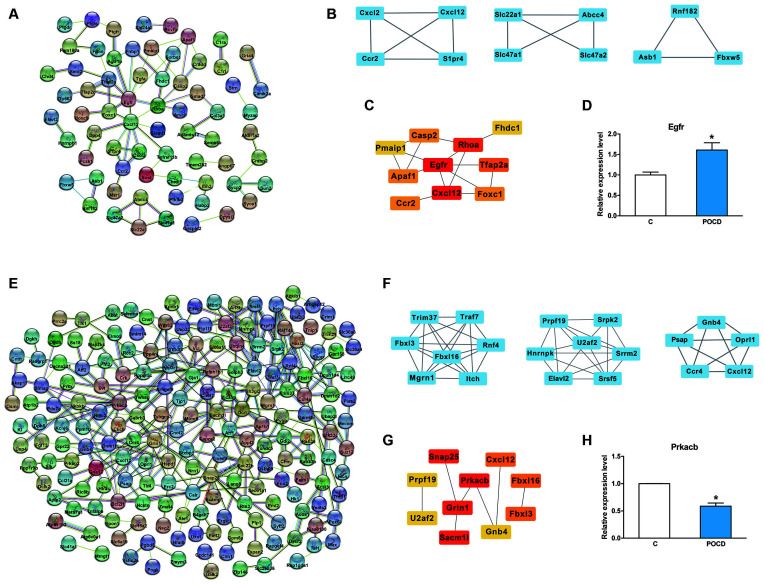
Protein-Protein Analysis of circRNAs Regulated Genes
Further analysis was performed to identify the most promising related genes using the proven circRNAs. Analysis of the protein-protein interaction network for these up/downregulated genes was performed using STRING (11.0 version) under default parameters (Figures 6A,E). Using the Cytoscape plug-in MCODE, the top three sub-network modules were aggregated and extracted from the protein-protein interaction network (Figures 6B,F). For the upregulated network, module A contained four nodes, including cxcl2 and cxcl12. For the downregulated network, module A contained seven nodes, including fbxl3 and fbxl16. Moreover, based on the protein-protein interaction results, the hub genes were screened by degree using CytohHubba of the Cytoscape software. As shown in Figures 6C,G, the hub genes Egfr/Prkacb were in the up/downregulated networks, and the mRNA expression levels of Egfr and Prkacb were also measured by qRT-PCR (Figures 6D,H).
Figure 6.
Protein-protein analysis of circRNAs regulated genes. (A,E) The protein-protein interaction network analysis for the up/downregulated genes. (B,F) Top three sub-network modules that are predicted by MCODE in the Cytoscape software. (C,G) Top 10 hub genes screened by CytohHubba in the Cytoscape software. (D,H) The mRNA expression level of Egfr and Prkacb validation. *p < 0.05 compared with the control group.
Discussion
POCD has a high incidence among elderly patients, especially those undergoing major surgeries. The pathogenesis of POCD is complex and lacks effective diagnosis and treatment. Therefore, several recent studies have focused on the epigenetic regulation of the pathogenesis of POCD to discover potential therapies (Li et al., 2015; Liu et al., 2019; Zhong and Xu, 2019). miRNAs and lncRNAs in the brain have been reported to contribute to POCD (Li et al., 2019; Su et al., 2019). However, the roles of circRNAs in POCD remain largely unknown.
CircRNAs are evolutionarily conserved at varying species sequence levels (Jeck et al., 2013). For example, 28% of the mouse circRNA molecules are conserved in humans (Rybak-Wolf et al., 2015). CircRNAs are extremely rich, conserved, and dynamically present in the mammalian brain (Westholm et al., 2014). Because circRNAs are preferentially expressed in nervous genes and nervous tissues (Floris et al., 2017), they are associated with neurological diseases, such as AD, one of the most common neurodegenerative diseases (Burns and Iliffe, 2009). Recent studies have shown that several circRNAs are present in the nervous tissues of patients with sporadic AD and AD mice (Huang et al., 2018; Sekar et al., 2018). Additionally, Cao et al. (2020) reported that dexmedetomidine alleviates POCD through circRNA in aged rats. This study compared the differentially expressed circRNAs between dexmedetomidine and POCD groups; however, our study is the first to examine circRNA profiles between POCD and control groups in the hippocampus of aged mice. Wang et al. (2019) found that circRNA_089763 expression in the plasma exosomes of POCD patients after coronary artery bypass graft surgeries, indicating that circRNA may be a potential biomarker for POCD.
CircRNA has been reported to carry out various kinds of modulating roles, including the interaction with RNA-binding proteins, acting as miRNA sponges, and regulation of paternal gene transcription, mainly at the posttranscriptional and transcriptional levels. In the human genome, a total of 519 canonical miRNA genes have been identified (Denzler et al., 2014; Bartel, 2018). Approximately 70% of the discovered miRNAs are present in the temporary brain and neurons (Cao et al., 2006), implying that miRNAs play a significant regulatory function during the development of the nervous system. As in psychiatric dysfunctions such as schizophrenia (Xu et al., 2013), deregulation of miRNAs is involved in neurodegenerative abnormalities such as Parkinson’s disease (PD) and AD as well (Esteller, 2011). Previous studies have reported a significant correlation that supports the role of miRNAs in POCD (Yu et al., 2015; Wei et al., 2017; Chen et al., 2019; Yazit et al., 2020).
Salmena et al. (2011) proposed the ceRNA hypothesis, suggesting that circRNAs interact with miRNAs to modulate gene expression at the transcriptional or posttranscriptional level. Over the past few years, the ceRNA assumption has been confirmed by many experiments. For instance, circRNA_2837 targets miR-34 family members to regulate LC3-II/p62 and protect neurons by autophagy in the sciatic nerve injury model (Zhou et al., 2018). In neural stem cells, circRNA TTC3 sponges miR-372-3p to regulate TLR4 expression and prevent cerebral ischemia reperfusion injury (Yang et al., 2021). With a new understanding of the ceRNA networks in different diseases, therapies targeting ceRNAs rather than miRNAs alone may become more significant and valuable (Bak and Mikkelsen, 2014). Therefore, GO and KEGG enrichment analyses were carried out for the up/downregulated genes in the ceRNA network. In the GO analyses, the top three terms of the upregulated genes were identical protein binding, extracellular region, and tube morphogenesis; for downregulated genes, the top three terms were protein binding, cell part, and localization. Next, KEGG pathway analysis showed that the upregulated genes included adherens junction, phospholipase D, ErbB signaling pathway, and the downregulated genes involved in the MAPK signaling pathway and mediated proteolysis, which were revealed to be associated with POCD (Li et al., 2014, 2016; Vutskits and Xie, 2016; Zhang et al., 2016; Ding et al., 2017; Lu et al., 2017; Wang D. S. et al., 2018; Wang W. X. et al., 2018; Orser and Wang, 2019; Zheng et al., 2019; Zhong and Xu, 2019). Thus, these bioinformatics analyses of possible pathways also suggested the potential functions of the identified circRNAs in POCD of aged mice.
Meanwhile, based on the protein-protein interaction analysis, Egfr and Prkacb were predicted in the interaction network. In addition, Egfr may be modified by circRNA_22058 and circRNA_44122. Prkacb may be modified by circRNA_22673. Egfr has also been proposed as a fundamental disease-associated protein in AD pathogenesis (Quan et al., 2020; Yuen et al., 2020). They have therapeutic abilities through autophagy induction and the attenuation of reactive astrocytes (Tavassoly et al., 2020). Prkacb might be involved in the MAPK signaling pathway, GABAergic synapse, insulin secretion, insulin signaling pathway, and chemokine signaling pathway. PKA has also been reported to contribute to neurodegenerative diseases, such as PD and AD (Dagda and Das Banerjee, 2015; Myeku et al., 2016; Kumar and Singh, 2017; Sanders and Rajagopal, 2020; Zhang et al., 2020). PKA has also been reported to be involved in POCD mechanism (Wang W. X. et al., 2018; Zhu et al., 2019). Therefore, the mmu-circRNA_22058 and circRNA_44122/Egfr ceRNA network or circRNA_22673/Prkacb ceRNA network may contribute to the pathological process of POCD, which will be validated in future studies.
The present study had several limitations. First, we only screened differentially expressed circRNAs in POCD mice 3 days after anesthesia/surgery. Consequently, we did not know whether the differentially expressed circRNAs were time-dependent. Second, although we found that some circRNAs might be associated with POCD, the mechanism needs to be elucidated in further studies.
In summary, we found that circRNAs were differentially expressed in the POCD of aged mice hippocampus. Using bioinformatics analysis of predicted circRNAs, we evaluated the roles and relevant pathways of relative circRNA-target genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165798.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Xuzhou Medical University.
Author Contributions
Y-QW and H-HM designed the research. QL, H-BW, CC, HH, and Y-MS contributed to behavioral test and sample collection. L-HM, JW, and Y-YS performed the molecular experiments. Y-QW and H-HM analyzed the bioinformatics data and wrote the article. Y-QW, QL, and H-HM revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Kangchen Bio-tech (Shanghai, China) for the excellent technical assistance.
Footnotes
Funding. This work was supported by the Natural Science Foundation of Beijing (7212023) and the National Natural Science Foundation of China (82071180) awarded to H-HM; by the Natural Science Foundation of Jiangsu Province (BK20191464), awarded to Y-QW.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnagi.2021.716383/full#supplementary-material.
References
- Bak R. O., Mikkelsen J. G. (2014). miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip. Rev. RNA 5, 317–333. 10.1002/wrna.1213 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2018). Metazoan MicroRNAs. Cell 173, 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns A., Iliffe S. (2009). Alzheimer’s disease. BMJ 338:b158. 10.1136/bmj.b158 [DOI] [PubMed] [Google Scholar]
- Cao C., Deng F., Hu Y. (2020). Dexmedetomidine alleviates postoperative cognitive dysfunction through circular RNA in aged rats. 3 Biotech 10:176. 10.1007/s13205-020-2163-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Yeo G., Muotri A. R., Kuwabara T., Gage F. H. (2006). Noncoding RNAs in the mammalian central nervous system. Ann. Rev. Neurosci. 29, 77–103. 10.1146/annurev.neuro.29.051605.112839 [DOI] [PubMed] [Google Scholar]
- Chen L., Dong R., Lu Y., Zhou Y., Li K., Zhang Z., et al. (2019). MicroRNA-146a protects against cognitive decline induced by surgical trauma by suppressing hippocampal neuroinflammation in mice. Brain Behav. Immun. 78, 188–201. 10.1016/j.bbi.2019.01.020 [DOI] [PubMed] [Google Scholar]
- Cheng J., Meng J., Zhu L., Peng Y. (2020). Exosomal noncoding RNAs in glioma: biological functions and potential clinical applications. Mol. Cancer 19:66. 10.1186/s12943-020-01189-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu K. J., Ma Y. S., Jiang X. H., Wu T. M., Wu Z. J., Li Z. Z., et al. (2020). Whole-transcriptome sequencing identifies key differentially expressed mRNAs, miRNAs, lncRNAs and circRNAs associated with CHOL. Mol. Ther. Nucleic. Acids 21, 592–603. 10.1016/j.omtn.2020.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda R. K., Das Banerjee T. (2015). Role of protein kinase a in regulating mitochondrial function and neuronal development: implications to neurodegenerative diseases. Rev. Neurosci. 26, 359–370. 10.1515/revneuro-2014-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiner S., Silverstein J. H. (2009). Postoperative delirium and cognitive dysfunction. Br. J. Anaesth. 103, i41–46. 10.1093/bja/aep291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R., Agarwal V., Stefano J., Bartel D. P., Stoffel M. (2014). Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 54, 766–776. 10.1016/j.molcel.2014.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Shi C., Chen L., Ma P., Li K., Jin J., et al. (2017). Effects of andrographolide on postoperative cognitive dysfunction and the association with NF-κB/MAPK pathway. Oncol. Lett. 14, 7367–7373. 10.3892/ol.2017.7088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., John B., Gaul U., Tuschl T., Sander C., Marks D. S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5:R1. 10.1186/gb-2003-5-1-r1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- Feng X., Valdearcos M., Uchida Y., Lutrin D., Maze M., Koliwad S. K. (2017). Microglia mediate postoperative hippocampal inflammation and cognitive decline in mice. JCI Insight 2:e91229. 10.1172/jci.insight.91229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floris G., Zhang L., Follesa P., Sun T. (2017). Regulatory role of circular RNAs and neurological disorders. Mol. Neurobiol. 54, 5156–5165. 10.1007/s12035-016-0055-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D., Li J., Wang H., Su X., Hou J., Gu Y., et al. (2017). Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 66, 1151–1164. 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- Huang J. L., Qin M. C., Zhou Y., Xu Z. H., Yang S. M., Zhang F., et al. (2018). Comprehensive analysis of differentially expressed profiles of Alzheimer’s disease associated circular RNAs in an Alzheimer’s disease mouse model. Aging (Albany NY) 10, 253–265. 10.18632/aging.101387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sharpless N. E. (2014). Detecting and characterizing circular RNAs. Nat. Biotechnol. 32, 453–461. 10.1038/nbt.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck W. R., Sorrentino J. A., Wang K., Slevin M. K., Burd C. E., Liu J., et al. (2013). Circular RNAs are abundant, conserved and associated with ALU repeats. RNA 19, 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Singh N. (2017). Pharmacological activation of protein kinase a improves memory loss and neuropathological changes in a mouse model of dementia of Alzheimer’s type. Behav. Pharmacol. 28, 187–198. 10.1097/FBP.0000000000000294 [DOI] [PubMed] [Google Scholar]
- Li M., Chen C., Zhang W., Gao R., Wang Q., Chen H., et al. (2019). Identification of the potential key long non-coding RNAs in aged mice with postoperative cognitive dysfunction. Front. Aging Neurosci. 11:181. 10.3389/fnagi.2019.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. M., Su F., Ji M. H., Zhang G. F., Qiu L. L., Jia M., et al. (2014). Disruption of hippocampal neuregulin 1-ErbB4 signaling contributes to the hippocampus-dependent cognitive impairment induced by isoflurane in aged mice. Anesthesiology 121, 79–88. 10.1097/ALN.0000000000000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Pan K., Chen L., Ning J. L., Li X., Yang T., et al. (2016). Deferoxamine regulates neuroinflammation and iron homeostasis in a mouse model of postoperative cognitive dysfunction. J. Neuroinflammation 13:268. 10.1186/s12974-016-0740-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wang S., Ran K., Hu Z., Liu Z., Duan K., et al. (2015). Differential hippocampal protein expression between normal aged rats and aged rats with postoperative cognitive dysfunction: a proteomic analysis. Mol. Med. Rep. 12, 2953–2960. 10.3892/mmr.2015.3697 [DOI] [PubMed] [Google Scholar]
- Liu Q., Hou A., Zhang Y., Guo Y., Li J., Yao Y., et al. (2019). MiR-190a potentially ameliorates postoperative cognitive dysfunction by regulating tiam1. BMC Genomics 20:670. 10.1186/s12864-019-6035-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Liufu N., Dong Y., Xu G., Zhang Y., Shu L., et al. (2017). Sevoflurane acts on ubiquitination-proteasome pathway to reduce postsynaptic density 95 protein levels in young mice. Anesthesiology 127, 961–975. 10.1097/ALN.0000000000001889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Pan J., Ye X., Yu B., Zhang W., Wan J., et al. (2019). Whole-transcriptome analysis of APP/PS1 mouse brain and identification of circRNA-miRNA-mRNA networks to investigate AD pathogenesis. Mol. Ther. Nucleic. Acids 18, 1049–1062. 10.1016/j.omtn.2019.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- Miao H. H., Wang M., Wang H. X., Tian M., Xue F. S. (2019). Ginsenoside Rg1 attenuates isoflurane/surgery-induced cognitive disorders and sirtuin 3 dysfunction. Biosci. Rep. 39:BSR20190069. 10.1042/BSR20190069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H. H., Zhang Y., Ding G. N., Hong F. X., Dong P., Tian M., et al. (2017). Ginsenoside Rb1 attenuates isoflurane/surgery-induced cognitive dysfunction via inhibiting neuroinflammation and oxidative stress. Biomed. Environ. Sci. 30, 363–372. 10.3967/bes2017.047 [DOI] [PubMed] [Google Scholar]
- Miao H., Dong Y., Zhang Y., Zheng H., Shen Y., Crosby G., et al. (2018). Anesthetic isoflurane or desflurane plus surgery differently affects cognitive function in Alzheimer’s disease transgenic mice. Mol. Neurobiol. 55, 5623–5638. 10.1007/s12035-017-0787-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeku N., Clelland C. L., Emrani S., Kukushkin N. V., Yu W. H., Goldberg A. L., et al. (2016). Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat. Med. 22, 46–53. 10.1038/nm.4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orser B. A., Wang D. S. (2019). GABAA receptor theory of perioperative neurocognitive disorders. Anesthesiology 130, 618–619. 10.1097/ALN.0000000000002562 [DOI] [PubMed] [Google Scholar]
- Pasquinelli A. E. (2012). MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282. 10.1038/nrg3162 [DOI] [PubMed] [Google Scholar]
- Qiu L. L., Pan W., Luo D., Zhang G. F., Zhou Z. Q., Sun X. Y., et al. (2020). Dysregulation of BDNF/TrkB signaling mediated by NMDAR/Ca(2+)/calpain might contribute to postoperative cognitive dysfunction in aging mice. J. Neuroinflammation 17:23. 10.1186/s12974-019-1695-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z., et al. (2018). The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 78, 2839–2851. 10.1158/0008-5472.CAN-17-2808 [DOI] [PubMed] [Google Scholar]
- Quan X., Liang H., Chen Y., Qin Q., Wei Y., Liang Z., et al. (2020). Related network and differential expression analyses identify nuclear genes and pathways in the hippocampus of Alzheimer disease. Med. Sci. Monit. 26:e919311. 10.12659/MSM.919311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved and dynamically expressed. Mol. Cell 58, 870–885. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders O., Rajagopal L. (2020). Phosphodiesterase inhibitors for Alzheimer’s Disease: a systematic review of clinical trials and epidemiology with a mechanistic rationale. J. Alzheimers Dis. Rep. 4, 185–215. 10.3233/ADR-200191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar S., Cuyugan L., Adkins J., Geiger P., Liang W. S. (2018). Circular RNA expression and regulatory network prediction in posterior cingulate astrocytes in elderly subjects. BMC Genomics 19:340. 10.1186/s12864-018-4670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz J., Christensen K. B., Lund T., Lohse N., Rasmussen L. S. (2009). Long-term consequences of postoperative cognitive dysfunction. Anesthesiology 110, 548–555. 10.1097/ALN.0b013e318195b569 [DOI] [PubMed] [Google Scholar]
- Su R., Sun P., Zhang D., Xiao W., Feng C., Zhong L., et al. (2019). Neuroprotective effect of miR-410–3p against sevoflurane anesthesia-induced cognitive dysfunction in rats through PI3K/Akt signaling pathway via targeting C-X-C motif chemokine receptor 5. Genes Genomics 41, 1223–1231. 10.1007/s13258-019-00851-5 [DOI] [PubMed] [Google Scholar]
- Szabo L., Morey R., Palpant N. J., Wang P. L., Afari N., Jiang C., et al. (2015). Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 16:126. 10.1186/s13059-015-0690-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoly O., Sato T., Tavassoly I. (2020). Inhibition of brain epidermal growth factor receptor activation: a novel target in neurodegenerative diseases and brain injuries. Mol. Pharmacol. 98, 13–22. 10.1124/mol.120.119909 [DOI] [PubMed] [Google Scholar]
- Terrando N., Brzezinski M., Degos V., Eriksson L. I., Kramer J. H., Leung J. M., et al. (2011). Perioperative cognitive decline in the aging population. Mayo Clin. Proc. 86, 885–893. 10.4065/mcp.2011.0332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vutskits L., Xie Z. (2016). Lasting impact of general anaesthesia on the brain: mechanisms and relevance. Nat Rev. Neurosci. 17, 705–717. 10.1038/nrn.2016.128 [DOI] [PubMed] [Google Scholar]
- Wang D. S., Kaneshwaran K., Lei G., Mostafa F., Wang J., Lecker I., et al. (2018). Dexmedetomidine prevents excessive gamma-aminobutyric acid type a receptor function after anesthesia. Anesthesiology 129, 477–489. 10.1097/ALN.0000000000002311 [DOI] [PubMed] [Google Scholar]
- Wang M., Su P., Liu Y., Zhang X., Yan J., An X., et al. (2019). Abnormal expression of circRNA_089763 in the plasma exosomes of patients with post-operative cognitive dysfunction after coronary artery bypass grafting. Mol. Med. Rep. 20, 2549–2562. 10.3892/mmr.2019.10521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Yu F., Li P., Wang K. (2020). Emerging function and clinical significance of exosomal circRNAs in cancer. Mol. Ther. Nucleic Acids 21, 367–383. 10.1016/j.omtn.2020.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. X., Wu Q., Liang S. S., Zhang X. K., Hu Q., Chen Q. H., et al. (2018). Dexmedetomidine promotes the recovery of neurogenesis in aged mouse with postoperative cognitive dysfunction. Neurosci. Lett. 677, 110–116. 10.1016/j.neulet.2018.03.043 [DOI] [PubMed] [Google Scholar]
- Wei C., Luo T., Zou S., Zhou X., Shen W., Ji X., et al. (2017). Differentially expressed lncRNAs and miRNAs with associated ceRNA networks in aged mice with postoperative cognitive dysfunction. Oncotarget 8, 55901–55914. 10.18632/oncotarget.18362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm J. O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., et al. (2014). Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 1966–1980. 10.1016/j.celrep.2014.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Hsu P. K., Stark K. L., Karayiorgou M., Gogos J. A. (2013). Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell 152, 262–275. 10.1016/j.cell.2012.11.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Zang L., Cui J., Wei L. (2021). Circular RNA TTC3 regulates cerebral ischemia-reperfusion injury and neural stem cells by miR-372–3p/TLR4 axis in cerebral infarction. Stem Cell Res. Ther. 12:125. 10.1186/s13287-021-02187-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazit N. A. A., Juliana N., Das S., Teng N., Effendy N., Azmani S., et al. (2020). Association of micro RNA and post-operative cognitive dysfunction: a review. Mini Rev. Med. Chem. 20, 1781–1790. 10.2174/1389557520666200621182717 [DOI] [PubMed] [Google Scholar]
- Yu X., Liu S., Li J., Fan X., Chen Y., Bi X., et al. (2015). MicroRNA-572 improves early post-operative cognitive dysfunction by down-regulating neural cell adhesion molecule 1. PLoS One 10:e0118511. 10.1371/journal.pone.0118511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen S. C., Zhu H., Leung S. W. (2020). A systematic bioinformatics workflow with meta-analytics identified potential pathogenic factors of Alzheimer’s disease. Front. Neurosci. 14:209. 10.3389/fnins.2020.00209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Huo S. T., Wu Z., Chen L., Wen C., Chen H., et al. (2020). Rapid development of targeting circRNAs in cardiovascular diseases. Mol. Ther. Nucleic Acids 21, 568–576. 10.1016/j.omtn.2020.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dong H., Li N., Zhang S., Sun J., Zhang S., et al. (2016). Activated brain mast cells contribute to postoperative cognitive dysfunction by evoking microglia activation and neuronal apoptosis. J. Neuroinflammation 13:127. 10.1186/s12974-016-0592-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Shen Q., Wu X., Zhang D., Xing D. (2020). Activation of PKA/SIRT1 signaling pathway by photobiomodulation therapy reduces Aβ levels in Alzheimer’s disease models. Aging Cell 19:e13054. 10.1111/acel.13054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Min S., Hu B., Liu Q., Wan Y. (2019). Nrdp1 is involved in hippocampus apoptosis in cardiopulmonary bypass-induced cognitive dysfunction via the regulation of ErbB3 protein levels. Int. J. Mol. Med. 43, 1747–1757. 10.3892/ijmm.2019.4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Xu W. (2019). Characterization of DNA hydroxymethylation in the hypothalamus of elderly mice with post-operative cognitive dysfunction. Exp. Ther. Med. 18, 4002–4010. 10.3892/etm.2019.8056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z. B., Niu Y. L., Huang G. X., Lu J. J., Chen A., Zhu L., et al. (2018). Silencing of circRNA.2837 plays a protective role in sciatic nerve injury by sponging the miR-34 family via regulating neuronal autophagy. Mol. Ther. Nucleic Acids 12, 718–729. 10.1016/j.omtn.2018.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. S., Xiong Y. F., Luo F. Q., Min J. (2019). Dexmedetomidine protects rats from postoperative cognitive dysfunction via regulating the GABA(B) R-mediated cAMP-PKA-CREB signaling pathway. Neuropathology 39, 30–38. 10.1111/neup.12530 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE165798.