Abstract
Cells respond to contact with human cytomegalovirus (HCMV) virions by initiating intracellular signaling and gene expression characteristic of the interferon (IFN)-responsive pathway. Herein, we demonstrate that a principal mechanism of HCMV-induced signal transduction is via an interaction of the primary viral ligand, glycoprotein B (gB), with its cellular receptor. Cells incubated with a purified, soluble form of gB resulted in the transcriptional upregulation of IFN-responsive genes OAS and ISG54 (encoding 2′-5′ oligoadenylate synthetase and an IFN-stimulated gene product of 54 kDa) to a comparable level as virions or IFN. Gene induction was an immediate and direct response to gB which did not require de novo protein synthesis. Neither the initial virus attachment site, heparan sulfate proteoglycans, nor the IFN-α/β or IFN-γ receptors are involved in the response. Pleotropic protein phosphorylation was required for cellular gene induction, and the mitogen-activated protein kinases ERK1 and ERK2 were activated in response to the ligand. Together these data indicate that a principal means by which cytomegalovirus induces intracellular signaling and activation of the interferon-responsive pathway is via an interaction of gB with an as yet unidentified, likely novel cellular receptor that interfaces with the IFN signaling pathway.
Transmembrane receptors enable cells to respond to ligands by altering cellular metabolism. A consequence of binding of extracellular signaling proteins, such as growth factors, hormones, and cytokines, to their cognate receptors is transcriptional activation of previously quiescent genes. Interferons (IFNs) are cytokines that induce a well-characterized signal transduction pathway. The IFN-mediated signaling response begins with a ligand-receptor interaction and concludes with activated gene expression of family of genes known as the IFN-stimulated genes (ISGs) (reviewed in references 24 and 32). Type 1 IFN (IFN-α/β) activates gene expression by engaging a common cellular receptor complex, whereas type II IFN (IFN-γ) engages a distinct receptor (27, 29). Signaling requires tyrosine phosphorylation, but the receptor subunits themselves lack intrinsic tyrosine kinase activity. IFN receptors become rapidly phosphorylated by members of the Janus family of kinases (Jaks) which are associated with their cytoplasmic domains (24). Subsequently latent cytoplasmic transcription factors (STATs) are recruited, phosphorylated, and multimerized into functionally active transcriptional transactivators which traffic to the nucleus and induce gene expression by binding to particular cis-acting regulatory regions. In general, genes that respond to IFN-α contain a highly conserved upstream element termed the IFN-stimulated response element, whereas IFN-γ-responsive genes contain a IFN-γ activation site (24, 32). Unique and distinguishing complements of the Jak/STAT members along with a DNA binding protein known as p48 are responsible for the induction specificity (30, 42, 43). Although the IFN response pathway is the best characterized, it is now clear that a variety of membrane receptors respond to extracellular signaling molecules by activating combinations of Jak and STAT proteins, resulting in gene induction.
Infection of permissive cells with human cytomegalovirus (HCMV), a herpesvirus, results in the progression of a sequentially ordered set of physiological responses similar to growth factor- or cytokine-induced cellular activation. Cellular activation involves a rapid and measurable induction of a number of immediate-early signaling mediators that begins with membrane-associated events such as activation of phospholipase C and phospholipase A2 (2, 3, 72). Transient accumulations of activated cellular transcription factors such as NF-κB (9, 40, 58, 76), Sp1 (77), AP-1, and CRE/B (9, 58) are also detectable with rapid kinetics. Recently, differential display analysis was used to investigate the cellular changes that occur in response to HCMV infection (78). This study demonstrated that HCMV infection had a profound effect on host cell gene expression, as evidenced by the accumulation of a number of in HCMV-infected fibroblasts cytomegalovirus-inducible gene mRNAs compared to mock-infected cells. Interestingly, all of the cellular cytomegalovirus-inducible genes (15 independent mRNAs) were determined to be known or novel ISGs that were also induced by IFN-α. Activation of transcription factors and induction of the ISGs was independent of viral gene expression in that the response was as robust with UV-inactivated virus as with replication-competent virus. This fact indicates that a constituent of the virus particle mediates the induction. Virion components that could potentially mediate the gene induction are envelope proteins or tegument proteins that when delivered to cells after successful virus entry are known to activate transcription (44, 75).
Envelope proteins mediate the early events in infection such as attachment and penetration. HCMV entry into host cells is a complex process involving sequential viral glycoprotein-host cell receptor interactions (21, 23). Virus attachment is initiated by binding to cell surface heparan sulfate proteoglycans (HSPGs) (23, 52). This initial binding is easily dissociable but rapidly converts to a more stable binding state thought to be mediated by engagement of a second receptor (21, 23). After virus-cell contact, fusion is initiated presumably involving rearrangements or conformational changes of fusogenic glycoproteins. The virion envelope fuses with the plasma membrane, resulting in localized deposits of envelope glycoproteins and delivery of virion contents including the tegument proteins, an amorphous, protein-dense material that includes transcriptional transactivators, as well as the DNA-containing capsid (22, 44). Glycoprotein B (gB) of HCMV is the most abundant component of the envelope, a target of neutralizing antibodies (14, 28, 35, 53) and an essential replication component (11a). Protein binding experiments with a purified, soluble form of gB (gB-S) revealed that gB serves as the primary viral ligand capable of interacting with two independent binding sites (11). A proportion of the binding activity was attributable to HSPGs, but the majority of gB’s binding was to an independent saturable binding site. Here we report that a consequence of the interaction of gB with the non-heparin receptor is the initiation of intracellular signaling and activation of the IFN-responsive pathway.
MATERIALS AND METHODS
Cell lines, virus, and reagents.
Human fibroblasts (HF cells) were cultured in Dulbecco’s minimal essential medium (DMEM; BioWhittaker, Walkersville, Md.) supplemented with 5% fetal bovine serum (FBS; HyClone, Logan, Utah), 1.0% penicillin-streptomycin-amphotericin B (Fungizone) (PSF; BioWhittaker), 0.3% l-glutamine (BioWhittaker), and 100 μg of Geneticin (Gibco BRL, Gaithersburg, Md.) per ml as described previously (20). Adherent cultures of Trichoplusia ni (TN-5) insect cells (15) were cultured in ExCell 401 medium (JRH Biosciences, Lenexa, Kans.) supplemented with 5% FBS and PSF. HCMV AD169 was grown and titered in HF cells as previously described (23). A recombinant strain of Autographa californica nuclear polyhedrosis virus encoding amino acids 1 to 692 of the gB homologue from the AD169 strain of HCMV was grown and titered as previously described (15). Recombinant gB-S was produced and purified as previously described (11). Recombinant human IFN-γ and IFN-α were purchased from Genzyme (Cambridge, Mass.).
Stimulation with signaling ligands.
Subconfluent monolayers of HF cells were incubated with the indicated stimuli for the time period specified for each experiment, generally 6 h. Cells were incubated with one of the following ligands: DMEM without FBS (mock treatment), gB-S (500 μg/ml), IFN-γ (100 U/ml), IFN-α (1,000 U/ml), CMV (10 PFU/ml), or bovine serum albumin (BSA; 500 μg/ml). Cellular RNA was isolated and processed for reverse transcriptase (RT)-mediated PCR (RT-PCR) analysis. To determine the conformational structure required to induce signaling, gB-S (200 μg/ml) was treated by one of the following methods: 70 or 94°C for 15 min, dithiothreitol (DTT; 100 μg/ml; Sigma, St. Louis, Mo.) for 30 min at 37°C, or trypsin (1 μg/ml; Sigma) for 60 min at 37°C. For some experiments, fibroblasts were treated with heparinase (2 U/ml; Sigma) for 60 min at 37°C, washed, stimulated for 60 min at 37°C, washed, and harvested 6 h poststimulation. In separate experiments, cycloheximide (100 μg/ml; Sigma) was added 2 h prior to stimulation and remained present during the subsequent 6-h stimulation regimen, whereas actinomycin D (20 μg/ml; Sigma) was added concurrently with the inducing ligand and incubated for 6 h prior to harvesting. To assay for mitogen-activated protein kinase (MAPK) activation, fibroblast monolayers were serum starved for 18 to 24 h and incubated with stimuli for 10 to 30 min, and the cellular cytoplasmic fraction was isolated as previously described (10).
RNA isolation and mRNA-dependent cDNA synthesis.
Total cellular RNA was isolated by using RNA STAT-60 (Tel Test “B”; Friendswood, Tex.) as recommended by the manufacturer. Briefly, cells were lysed by addition of phenol-guanidinium thiocyanate and then chloroform extracted, and RNA was isopropanol precipitated. To eliminate DNA contamination, samples were treated with 75.4 U of DNase (Gibco BRL) for 1 h at 37°C in the presence of 3.75 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 100 U of Prime RNase inhibitor (5′-3′; Boulder, Colo.), after which time the samples were reextracted and reprecipitated. On average, a 100-mm-diameter tissue culture plate of fibroblast cells yielded 15 to 20 μg of total RNA. Ten micrograms of total RNA was reverse transcribed into mRNA-dependent cDNA by using Moloney murine leukemia virus RT (200 U/ml; Gibco BRL), 0.1 mg of oligo(dT) primer (Gibco BRL), 4 mM deoxynucleoside triphosphates (dNTPs; Pharmacia, Milwaukee, Wis.), Prime RNase inhibitor (1 U/ml), and 10 mM DTT. RNA was denatured at 65°C for 5 min and equilibrated at 37°C for 2 min, the RT mixture was added, and samples were allowed to incubate at 37°C for 1 h, after which time the reaction was terminated by heating at 95°C for 5 min followed by immediate cooling to 4°C. Equivalent amounts of cDNA were then subjected to PCR analysis.
PCR conditions and cycling parameters.
Each sample was analyzed by PCR in a final reaction volume of 50 μl under similar reaction conditions including 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 2.5 mM dNTPs, 500 mM primers, and 2.5 U of Taq polymerase (Perkin-Elmer, Branchburg, N.J.). Amplifications were performed in Perkin-Elmer 4800 thermocycler. The primers for 2′-5′ oligoadenylate synthetase and the ISG encoding a product of 54 kDa (OAS and ISG54) were designed with the Oligo 4.0 Macintosh program (National Biosciences, Inc., Plymouth, Minn.), β-Actin primers were purchased and used as instructed by the manufacturer (Stratagene, La Jolla, Calif.); the c-fos primers and reaction conditions have been previously described (57). The OAS primers (sense [5′ AAAGTGCCGGTAAAAGTCAT 3′] and antisense [5′ CTGTAGTGCAAGGGTTCTCA 3′]) amplified a 902-bp fragment in the presence of 2.5 mM MgCl2. The ISG54 primers (sense [5′ AGAAATCAAGGGAGAAAGAA 3′] and antisense [5′ AAGGTGACTAAGCAAATGGT 3′]) amplified a 506-bp fragment in the presence of 3.0 mM MgCl2. Identical amplification conditions were used for both sets of primers: 10-min denaturation step at 94°C followed by 30 cycles of 94°C for 1 min, 53°C for 1 min, and 72°C for 2 min, followed by 72°C for 10 min to allow for complete extension. PCR products were electrophoresed on 1% agarose (FMC, Rockland, Maine) stained with ethidium bromide for visualization.
Treatment of cells with protein kinase C (PKC) and protein tyrosine kinase (PTK) inhibitors.
Genistein, tyrphostin A25, calphostin C, H7 {[1-(5-isoquinolinylsulfonyl)-2-methylpiperazine]} and H8 {[(N-2-methylamino)ethyl]-5-isoquinolinesulfonamide} were purchased from Calbiochem (La Jolla, Calif.) and dissolved in dimethyl sulfoxide at concentrations of 74.0 mM (20.0 mg/ml), 49.4 mM (10.0 mg/ml), 50.0 μM (40.0 μg/ml), 27.5 mM (10.0 mg/ml), and 29.6 mM (10.0 mg/ml) respectively. Fibroblasts were incubated with dimethyl sulfoxide (solvent control), genistein (185.0 μM [50.0 μg/ml]), tyrphostin A25 (49.4 μM [10.0 μg/ml]), H7 (27.5 μM [10.0 μg/ml]), and H8 (29.6 μM [10.0 μg/ml]) for 2 h at 37°C; cells treated with calphostin C (50.0 nM [40.0 ng/ml]) were left for 2 h in the presence of light to activate the drug (33). Signaling ligands were added, and total cellular RNA was harvested 6 h posstimulation. The final concentrations of the inhibitors were chosen on the basis of the 50% inhibitory dose for enzyme inhibition and doses conventionally used in the literature (1, 31, 33, 39, 67). After 8 h of treatment, cell viability was determined by trypan blue exclusion to monitor toxicity.
Ligand binding assay.
Ligand binding assays were performed essentially as previously described (11). HF cells were chilled at 4°C and treated with ovalbumin (5 mg/ml) diluted in phosphate-buffered saline–1% FBS–0.1 mM CaCl2 (PBS-GC) for 30 min to block nonspecific binding. Purified, [35S]methionine-labeled gB-S protein was diluted in PBS-GC, added to the cell monolayers, and incubated for 90 min at 4°C in the presence or absence of IFN-γ or IFN-α. Unbound gB-S was removed; the cells were washed twice with PBS-GC and subsequently lysed in 1% sodium dodecyl sulfate (SDS)–1% Triton X-100. Both unbound and bound fractions were subjected to scintillation counting. All data points were performed in duplicate or triplicate.
SDS-PAGE analysis and immunoblotting.
Cytoplasmic fractions from fibroblasts stimulated for MAPK activation were separated by SDS-polyacrylamide gel electrophoresis (PAGE) in the presence of reducing agents. Resolved proteins were transferred to nitrocellulose membranes (Millipore, Bedford, Mass.) and probed with antibodies directed toward the dually phosphorylated form of ERK1/2 (extracellular signal-regulated kinase 1/2) (0.5 μg anti-ACTIVE antibody; Promega, Madison, Wis.) or 5.0 μg of a pan anti-ERK1/2 (Upstate Biotechnology Inc., Lake Placid, N.Y.) polyclonal antibody. Primary antibodies were detected by incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibodies (Pierce, Rockford, Ill.) and the LumiGLO HRP substrate kit (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, Md.).
RESULTS
Upregulation of cellular gene expression by HCMV gB.
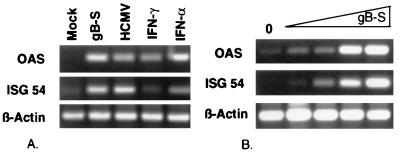
The HCMV gB protein is a major constituent of the virion envelope that possesses conventional ligand properties and is involved in virus-cell interactions (11, 49, 71). To examine the involvement of a defined virion component in the reported HCMV-induced cellular activation, fibroblast cells were stimulated with a recombinant soluble version of gB known to retain structural and functional features of the viral protein. Cells stimulated with gB, HCMV, or IFN were assayed for the upregulation of ISGs. OAS and ISG54, both of which were previously identified by differential display analysis as cellular mRNAs upregulated by HCMV infection (78), were selected as representative ISGs. As shown in Fig. 1A, gB was a potent inducer of cellular gene expression. Incubation of fibroblasts with gB or HCMV resulted in the upregulation of OAS and ISG54 mRNAs in a manner comparable to that for the IFNs. The gB-mediated signal transduction was dependent on the concentration of input ligand, since incubation with increasing amounts of gB resulted in a dose-dependent gene induction (Fig. 1B). These results are in agreement with previous data demonstrating that gB binding to fibroblasts was dose dependent (11). The protein concentration used for the remaining experiments (500 μg/ml, equivalent to 2.5 μM) was in the linear range of the binding curve and near the reported Kd (5 μM).
FIG. 1.
gB-mediated upregulation of cellular mRNA. (A) HF cells were incubated with medium alone (mock), gB-S (500 μg/ml), IFN-α (1,000 U/ml), IFN-γ (100 U/ml), or HCMV (10 PFU/ml). After 6 h of stimulation, total cellular RNA was isolated and RT-PCR analysis was performed with oligo(dT)-primed RNA and three pairs of PCR primers (OAS, ISG54, β-actin). PCR-amplified products were separated on a 1% agarose gel by electrophoresis and visualized by ethidium bromide staining. (B) HF cells were incubated with increasing concentrations of gB-S (0.05 to 500 μg/ml) for 6 h and subjected to RT-PCR analysis as for panel A.
Viral and recombinant gB protein assembles into a dimer shortly after translation and is proteolytically processed during intracellular transport into disulfide-linked fragments. The data in Fig. 2A demonstrate that the structural integrity of gB was important for signaling activity. Disruption of gB by proteolytic digestion or heat denaturation significantly reduced or abolished the response (Fig. 2A). The corresponding protein profiles show that the dimeric form of gB was lost upon heat treatment (Fig. 2B). Surprisingly, OAS and ISG54 mRNA levels were unaffected when the gB subunits were separated by disulfide bond reduction, suggesting that the oligomeric structure was not essential for signaling. Furthermore, mRNA upregulation did not occur with BSA treatment, suggesting that nonspecific protein-cell interactions were not responsible for signaling. We conclude that a defined component of the virus, gB, that can trigger intracellular signaling resulting and activation of the IFN-responsive genes OAS and ISG54.
FIG. 2.
gB-mediated signaling requires native structure. Prior to cell stimulation, gB (200 μg/ml) was denatured by heating to 70 or 94°C for 15 min, reduced by the presence of DTT (100 μg/ml) for 30 min at 37°C, or proteolytically digested with trypsin for 60 min at 37°C. As a control, cells were stimulated with BSA (500 μg/ml) under identical conditions. (B) The protein profiles for the heat-denatured and DTT-treated samples were determined by SDS-PAGE and immunoblotting with an antibody specific for an epitope on the carboxy-terminal domain. The positions of the dimeric (gB-D), monomeric (gB-M), carboxy-terminal (gB-C) fragments and the heat-induced aggregate (heat agg.) are indicated.
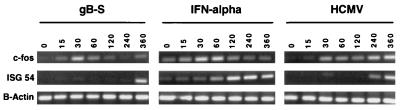
To investigate the kinetic regulation of the IFN-responsive gene induction by the viral ligands, two IFN-responsive genes with different induction kinetics were examined. c-fos, a member of the AP-1 transcription factor family, exhibits rapid and transient activation upon stimulation (8), whereas ISG54 requires a prolonged period prior to gene induction (78). Time course analysis demonstrated that stimulation of cells with either gB or IFN-α resulted in rapid induction of c-fos mRNA levels (Fig. 3). The gB-treated cells exhibited with maximum levels after 30 min of stimulation which declined over time. The activation response to IFN-α was very similar except that the induction lingered slightly longer, to 60 min. HCMV treatment appeared to induce c-fos gene expression in a biphasic manner; the first wave of induction occurred approximately 30 min poststimulation and the second occurred at 4 to 6 h postinfection, correlating with viral gene expression. For ISG54, gB and HCMV-induced mRNA upregulation was delayed by ca. 2 h relative to the natural cytokine. OAS gene expression by HCMV and gB proceeded at similar times as ISG54 induction (data not shown). These findings suggest that the signaling pathway initiated by HCMV via gB may not be identical to the IFNs themselves.
FIG. 3.
Kinetic analysis of gene expression. HF cells were incubated with gB-S, HCMV, or IFN-α and harvested for mRNA-dependent RT-PCR analysis at increasing time intervals as indicated in minutes. PCR primer pairs for c-fos and ISG54 were used to assess upregulation of IFN-responsive genes; the β-actin gene was used as a housekeeping control gene.
gB mediates signaling through its cellular receptor.
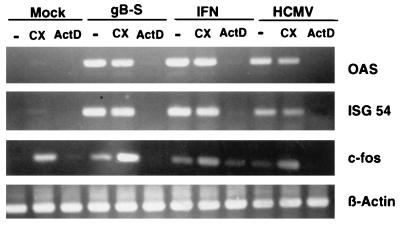
We next questioned whether the gB-mediated signaling was initiated via an interaction with its cellular receptors. This hypothesis posits that protein synthesis is not required for the signal transduction response. To test this hypothesis, the signaling assay was conducted in the presence of cycloheximide. As shown in Fig. 4, fibroblasts incubated with gB, HCMV, or IFN in combination with cycloheximide treatment demonstrated no appreciable decrease in gene expression. The superinduction of c-fos mRNA levels by cycloheximide treatment was not unexpected, as this effect has been reported for serum- or growth factor-stimulated cells (34). These data confirm that neither viral replication nor IFN synthesis in infected cells was required for HCMV-induced gene expression. Most significantly, the result indicates that gB-mediated signaling was a direct response to the ligand which activated existing pools of cellular transcription factors. The cycloheximide conditions were determined to be effective since parallel experiments using identical concentrations of cycloheximide inhibited protein synthesis by 96%, as measured by the incorporation of [35S]methionine into trichloroacetic acid-precipitable material, and HCMV IE gene expression was completely blocked, as assayed by Western blot analysis (data not shown). To confirm that the induction of gene expression was at the level of transcription, cells were treated during stimulation with actinomycin D, a potent inhibitor of DNA-dependent RNA synthesis and elongation. Actinomycin D treatment had a profound effect on gene induction by all ligands such that levels were reduced to an undetectable basal level (Fig. 4). Thus, the data suggest that signal transduction triggered by gB was a direct response regulated at the level of receptor engagement.
FIG. 4.
gB-mediated signaling is a direct response requiring transcription. Cycloheximide (CX; 100 μg/ml) was added to HF cells for 2 h before and during the 6-h stimulation period, while actinomycin D (ActD; 20 μg/ml) was present during the 6-h stimulation period. RNA was harvested at 6 h poststimulation and subjected to RT-PCR as for Fig. 1 and 3.
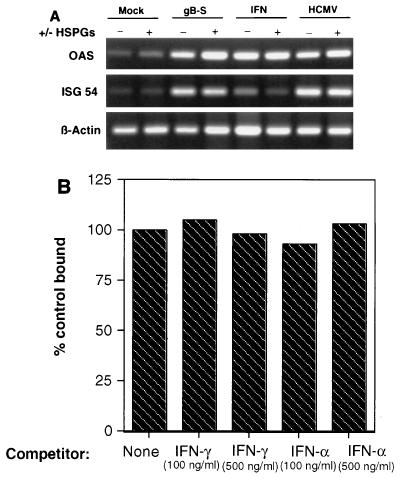
HSPGs serve as the initial interaction between virus and host cell (23, 52). Typical of the functional redundancy observed with herpesvirus envelope glycoproteins, two HCMV envelope glycoprotein complexes, glycoprotein complex II and gB, have heparin binding ability (11, 23, 38). To investigate the role of HSPGs in gB gene induction, fibroblasts were treated with heparinase at a concentration known to render the cell void of cell surface heparin molecules (11, 23, 38). As shown in Fig. 5A, gB induced OAS and ISG54 mRNA expression to similar levels in the presence or absence of enzyme. Comparably, loss of HSPGs did not result in a significant reduction in HCMV- or IFN-mediated signaling. Parallel virus entry assays were performed under identical conditions, and virus infection of heparinase-treated fibroblasts was reduced by 95% compared to untreated controls (data not shown). These data suggest that interaction of HCMV via gB to cell surface heparin did not mediate the observed signaling, but rather that subsequent binding to the as yet unidentified gB receptor is the crucial signaling interaction. To determine whether gB engaged either of the IFN receptors, ligand competition experiments were performed. Neither IFN-γ or IFN-α competed with gB binding to cells (Fig. 5B). We have also observed that gB does not activate the Tyk2 Jak kinase, which is associated with the IFN-α receptor (19), nor does expression of the IFN-γ receptor in HCMV-entry defective L cells confer virus entry (data not shown). These data suggest that the gB-induced signaling is mediated through its non-heparin receptor which is not either of the known IFN receptors.
FIG. 5.
(A) HSPGs are not required for gB-mediated signaling. Fibroblasts were either mock treated (serum-free medium) or incubated with heparinase (2 U/ml) for 60 min at 37°C. The cells were washed, incubated with the signaling ligand for 60 min at 37°C, washed, and harvested for total RNA 6 h poststimulation. (B) Fibroblasts were incubated with IFN-α or IFN-γ for 30 min at 4°C prior to the addition of 35S-labeled gB. Homologous cold competition blocked gB binding by greater than 95% (not shown).
Involvement of protein kinases in gB signal transduction.
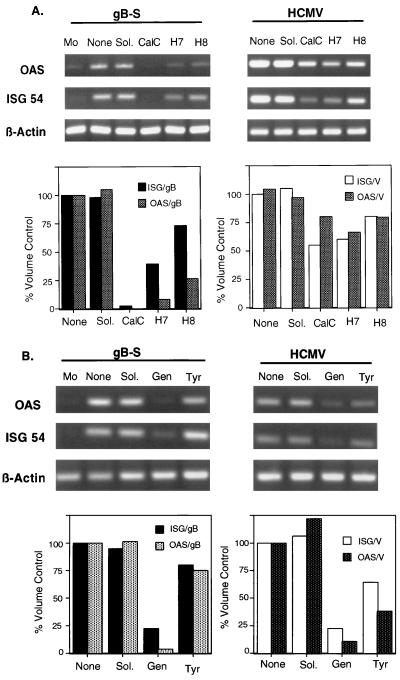
Protein phosphorylation represents one of the most important molecular mechanisms by which extracellular signals produce their biological responses in cells, while protein kinase stimulation is considered to be the most common activation mechanism in signal transduction systems. Although the protein kinases can be divided into two subsets, based on phosphate acceptors (serine/threonine or tyrosine residues), many substrates are known to undergo phosphorylation by multiple protein kinases. Fortunately, both activators and inhibitors of various branches of the protein kinase activation pathways have been identified, and their substrate specificities and mechanisms of action have been delineated. To assess the role of PKC activation, cells were treated before and during stimulation with one of three PKC inhibitors, calphostin C, H7, or H8. As shown in Fig. 6A, cells stimulated with gB in the presence of calphostin C, a highly specific and irreversible inhibitor of all PKC isoforms, no longer exhibited OAS and ISG54 gene induction. Incubation with H7, a broad-based serine/threonine kinase inhibitor, also reduced the ability of gB to elevate OAS (90% reduction) and ISG54 (70% reduction) mRNA levels, whereas H8 (inhibitor of cyclic AMP-dependent protein kinases) had differential effects on OAS and ISG54 mRNA expression. HCMV-stimulated cells retained the capacity to upregulate OAS mRNA in the presence of all three PKC inhibitors, albeit less effectively. ISG54 mRNA upregulation by HCMV involved the activation of serine/threonine protein kinases since calphostin C and H7 treatment lessened the level of gene induction, while H8-treated cells continued to elicit cell signaling. Therefore, gB-mediated signaling is dependent on a functional PKC pathway whereas HCMV was able to retain most signaling in the presence of the inhibitors, perhaps due to titration of the inhibitor by synthesis of viral transcriptional transactivators. PTK involvement was also investigated by treating cells with genistein or tyrphostin A25 before and during ligand stimulation (Fig. 6B). gB failed to induce OAS and ISG54 mRNA levels in cells treated with genistein, whereas tyrphostin A25 had no inhibitory effect on mRNA upregulation. Likewise, stimulation by intact virus demonstrated a similar pattern of sensitivity to the PTK inhibitors, suggesting that a conserved tyrosine kinase cascade(s), likely at the receptor level, may be utilized by gB and virions to stimulate ISGs.
FIG. 6.
Protein kinase activation is involved in gB signaling. (A) OAS and ISG54 mRNA levels were assessed in fibroblasts that were treated 2 h before and during stimulation with medium alone (Mo), ligand in the absence of inhibitor (None), solvent (Sol.), 50 nM calphostin C (CalC), 27.5 μM H7, or 29.5 μM H8. Quantitation of the relative yields of PCR product was determined with a Gel Documentation device (Bio-Rad). Extrapolation of the mean pixel density in each band to the relative percent volume allowed comparisons within an individual PCR. Each value was graphed as percent volume control relative to the untreated PCR product (None). (B) Fibroblasts were incubated with medium alone (Mo), ligand in absence of inhibitor (None), solvent (Sol.), 185 μM genistein (Gen), or 49.4 μM tyrphostin A25 (Tyr) 2 h before and during stimulation. RT-PCR analysis was performed to assess the effects of these inhibitors on cellular gene expression. Quantitation of the relative yields of the PCR products was determined as described for panel A. V, HCMV.
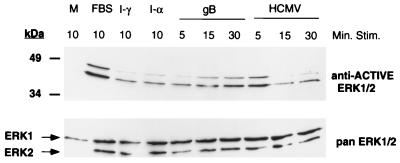
Finally, we examined potential activation of the MAPKs. The MAPKs comprise a group of protein serine/threonine kinases which are activated in response to extracellular stimuli through dual phosphorylation at conserved threonine and tyrosine residues (18, 59). This prominent pathway is utilized by a diverse array of biological ligands to regulate and modify the intracellular environment, including IFN-α/β (25). To address if HCMV and/or gB could activate the MAPK cascade, cytoplasmic fractions were isolated from ligand-stimulated cells and tested in immunoblotting experiments using polyclonal antibodies specific for the dually phosphorylated form of the terminal phosphate acceptor, ERK1/2. As shown in Fig. 7, the MAPK pathway is functional in HF cells, as demonstrated by the FBS-induced dual phosphorylation of ERK1/2 when probed with the anti-ACTIVE ERK1/2 antibodies. ERK1/2 phosphorylation in response to FBS stimulation was comparable to that observed with epidermal growth factor stimulation (data not shown). Similarly, incubation of fibroblasts with gB or HCMV resulted in the activation of the MAPKinase pathway. The accumulation of active ERK1/2 was a function of time. gB-treated cells exhibited maximum activation after 15 min of stimulation, whereas the virus activated ERK1/2 with faster kinetics. We also observed that both IFNs activated ERK1/2.
FIG. 7.
Incubation with gB stimulated the MAPK pathway. Fibroblasts were serum starved for 18 h prior to ligand stimulation. Cells were incubated in medium alone (M) or stimulated with FBS, IFN-γ (I-γ) or IFN-α (I-α) for 10 min. Cells were stimulated (Stim.) with gB or HCMV for 5-, 15-, or 30-min intervals. The cytoplasmic fraction was isolated and subjected to SDS-PAGE and immunoblotting. Reciprocol blots were probed with the anti-ACTIVE ERK1/2 polyclonal or pan ERK1/2 polyclonal antibody and detected with a secondary goat anti-rabbit HRP-conjugated antibody in conjunction with a chemiluminescent substrate.
DISCUSSION
gB of HCMV is the viral structural component responsible for intracellular signaling and gene induction.
Signal transduction is a common process used by an extensive array of biological ligands to modulate various host cell processes such as growth, differentiation, and proliferation. It is well documented that cells respond to cytomegalovirus by invoking a cascade of biological and physiological responses resulting in signal transduction and upregulation of cellular gene expression, including induction of genes in the IFN-responsive family. The majority of cellular activation is an early-phase response that does not require viral gene expression, leading to the conclusion that a viral structural element is responsible for these effects. Using purified, recombinant viral ligand, we observed efficient and robust activation of immediate-response genes such as c-fos and c-jun as well as IFN-responsive genes OAS and ISG54. Gene induction was independent of cellular protein synthesis but required active transcription. Native ligand structure and the input dose were also influential in signaling activity. Thus, we reason that a primary mechanism by which HCMV initiates intracellular signaling is via an interaction of its principal ligand, gB, with a cellular receptor.
Epstein-Barr virus (EBV), a B-cell-tropic herpesvirus, also initiates intracellular signaling via an engagement of its ligand, gp350/220, with its cellular receptor, CR2 (45, 56, 66). Protein phosphorylation in response to the EBV ligand is required for early events in internalization and early viral gene expression (16, 56). The human immunodeficiency virus (HIV) envelope protein, gp120/41, initiates signaling via an interaction between its primary receptor CD4 and its fusion coreceptor (7, 12), but signal transduction was not required for fusion and entry (73). HCMV, like HIV, penetrates via a direct fusion event at the cell surface, whereas EBV undergoes receptor-mediated endocytosis (22, 51, 64). It will be of great interest to determine the consequences of lack of signaling activity in HCMV infection. Specifically, a future goal of our research will be to determine if gB-induced signaling is an essential component of virus entry and the initiation of productive infection.
The gB signaling receptor is likely a novel cellular protein that interfaces with the IFN-responsive pathways.
HSPGs, broadly distributed cell surface molecules, are the initial attachment site for HCMV (23, 52). Virus attachment absolutely requires this initial interaction with HSPGs, but the binding step is transient and rapidly converts to stable adherence. Recombinant, soluble gB also has a biphasic Scatchard plot and can bind to HSPGs; however, the interaction of gB with its second binding site is independent and does not require the HSPG binding step (11). This is in contrast to many growth factors, such as basic fibroblast growth factor, which sequentially interacts with HSPGs and a high-affinity protein receptor, but similar to other herpesviruses which are known to encode functional redundancy for the heparin binding step (54, 63). Cells lacking HSPGs were equally responsive to gB and HCMV in intracellular signaling (Fig. 5A), suggesting that it is the nonheparin receptor mediating the response. HCMV has a broad cellular tropism in vivo and is capable of infecting cells of distinct and divergent developmental lineages. Both the type 1 and type 2 IFN receptors are distributed on cell types infected by the virus. Several lines of evidence argue against utilization of either of these receptors by HCMV via gB. First, cells treated with relatively high concentrations of IFN-α or IFN-γ exhibited wild-type levels of gB binding (Fig. 5B), and monoclonal antibodies to the IFN-γ receptor also had no gB or HCMV blocking activity (data not shown). Similarly, entry-defective L cells stably transfected with the IFN-γ receptor components remained refractory to HCMV entry (data not shown). HCMV is known to disrupt, not activate, the Jak1 kinase (47), and we determined that the Tyk2 Jak kinase associated with the IFN-α/β receptor was not activated by the gB ligand (18a). Confirming the original report by Zhu et al. (78), it was recently reported that HCMV induces expression of ISG54 (50). These authors found that HCMV infection did not result in the assembly of STAT1, STAT2, and p48 into the IFN-α-inducible ISGF3 transcription factor. Instead HCMV virions induced the formation of a novel transcription complex composed of, in part, the recently identified interferon-regulatory factor of unknown cellular function (IRF3 [5]) and CRE/B binding protein but not STAT1 or STAT2 (50). Taken together, the data suggest that the gB signaling receptor is a novel cellular receptor that activates IFN gene induction through a unique transcriptional activation complex.
Why would a virus activate an antiviral response: clever ploy or fatal flow?
IFNs, which were named for their ability to interfere with virus replication, are considered the major contributors to the first line of antiviral defense. OAS produced in response to IFN and HCMV is an integral component of the classical antiviral pathway that targets double-stranded RNAs produced in infection by RNA viruses (6, 17, 55). For DNA viruses, however, the antiviral activity of IFNs is less clear. Both IFN-γ and combinations of IFNs in the presence or absence of other cytokines such as tumor necrosis factor alpha are reported to have anti-HCMV activity (26, 70, 74). Yet, HCMV is an ancient, ubiquitous virus that persists in its human host for life. A majority of HCMV-associated disease is the result of spread of reactivated latent virus during conditions of immunosuppression (4, 13). Perhaps not the exclusive site of latency, but certainly a predominant cell type harboring latent HCMV, is monocytes (48, 68, 69). Reactivation and HCMV replication in latently infected monocytes requires differentiation and activation of the cells into macrophages, and administration of IFN-γ to latently infected monocytes is sufficient to activate cellular differentiation and HCMV replication (37, 41, 60–62). The IE proteins of HCMV are promiscuous transcriptional transactivators of viral and cellular genes, and they play a critical role in the progression of the virus replication cycle (reviewed in references 46 and 65). Recently it was discovered the IE proteins undergo phosphorylation by the ERK2 MAPK, and the phosphorylated IE protein had increased transcriptional activity (36). We found that ERK2 was activated as a consequence of the gB-receptor interaction (Fig. 7). Thus, it appears that HCMV has uniquely adapted a cellular signaling and gene activation pathway for its own benefit, ensuring its survival in the human host.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grant RO1 AI-34998 and a Basic Research grant from the March of Dimes Birth Defects Foundation.
We thank Donna Paulnock and Mary Lokuta for the reagents and expertise regarding RT-PCR analysis, and we thank Paul Bertics and Jon Houtman for the anti-ACTIVE ERK antibodies.
REFERENCES
- 1.Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- 2.Albrecht T, Boldogh I, Fons M, AbuBakar S, Deng C Z. Cell activation signals and the pathogenesis of human cytomegalovirus. Intervirology. 1990;31:68–75. doi: 10.1159/000150140. [DOI] [PubMed] [Google Scholar]
- 3.Albrecht T, Boldogh I, Fons M, Lee C H, AbuBakar S, Russell J M, Au W W. Cell-activation responses to cytomegalovirus infection. Relationship to the phasing of CMV replication and to the induction of cellular damage. Subcell Biochem. 1989;15:157–202. [PubMed] [Google Scholar]
- 4.Alford C A, Britt W J. Cytomegalovirus. In: Roizman B, Whitley R J, Lopex C, editors. Human herpesviruses. New York, N.Y: Raven Press; 1993. pp. 227–255. [Google Scholar]
- 5.Au W C, Moore P A, Lowther W, Juang Y T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baglioni C, Minks M A, Maroney P A. Interferon action may be mediated by activation of a nuclease by pppA2′p5′A2′p5′A. Nature. 1978;273:684–687. doi: 10.1038/273684a0. [DOI] [PubMed] [Google Scholar]
- 7.Baldari C T, Milia E, Di Somma M M, Baldoni F, Valitutti S, Telford J L. Distinct signaling properties identify functionally different CD4 epitopes. Eur J Immunol. 1995;25:1843–1850. doi: 10.1002/eji.1830250708. [DOI] [PubMed] [Google Scholar]
- 8.Boldogh I, AbuBakar S, Albrecht T. Activation of proto-oncogenes: an immediate early event in human cytomegalovirus infection. Science. 1990;247:561–564. doi: 10.1126/science.1689075. [DOI] [PubMed] [Google Scholar]
- 9.Boldogh I, Fons M P, Albrecht T. Increased levels of sequence-specific DNA-binding proteins in human cytomegalovirus infected cells. Biochem Biophys Res Commun. 1993;197:1505–1510. doi: 10.1006/bbrc.1993.2647. [DOI] [PubMed] [Google Scholar]
- 10.Boulton T G, Gregory J S, Cobb M H. Purification and properties of extracellular signal-regulated kinase 1, and insulin-stimulated microtubule-associated protein 2 kinase. Biochemistry. 1991;30:278–286. doi: 10.1021/bi00215a038. [DOI] [PubMed] [Google Scholar]
- 11.Boyle K A, Compton T. Receptor binding properties of a soluble form of human cytomegalovirus glycoprotein B. J Virol. 1998;72:1826–1833. doi: 10.1128/jvi.72.3.1826-1833.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Boyle, K. A., and T. Compton. Unpublished results.
- 12.Briant L, Signoret N, Gaubin M, Robert-Hebmann V, Zhang X, Murali R, Greene M I, Piatier-Tonneau D, Devaux C. Transduction of activation signal that follows HIV-1 binding to CD4 and CD4 dimerization involves the immunoglobulin CDR3-like region in domain 1 of CD4. J Biol Chem. 1997;272:19441–19450. doi: 10.1074/jbc.272.31.19441. [DOI] [PubMed] [Google Scholar]
- 13.Britt W J, Alford C A. Cytomegalovirus: In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 14.Britt W J, Mach M. Human cytomegalovirus glycoproteins. Intervirology. 1996;39:401–412. doi: 10.1159/000150510. [DOI] [PubMed] [Google Scholar]
- 15.Carlson C, Britt W J, Compton T. Expression, purification and characterization of a soluble form of human cytomegalovirus glycoprotein B. Virology. 1997;239:168–205. doi: 10.1006/viro.1997.8892. [DOI] [PubMed] [Google Scholar]
- 16.Cirone M, Angeloni A, Barile G, Zompetta C, Venanzoni M, Torrisi M R, Frati L, Faggioni A. Epstein Barr virus internalization and infectivity are blocked by selective protein kinase C inhibitors. Int J Cancer. 1990;45:490–493. doi: 10.1002/ijc.2910450320. [DOI] [PubMed] [Google Scholar]
- 17.Clemens M J, Williams B R G. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon treated cell extracts. Cell. 1976;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- 18.Cobb M H, Goldsmith E J. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 18a.Colamonici, O. R., and T. Compton. Unpublished results.
- 19.Colamonici O R, Uyttendaele H, Domanski P, Yan H, Krolewski J J. p135tyk2, an interferon-alpha-activated tyrosine kinase, is physically associated with an interferon-alpha receptor. J Biol Chem. 1994;269:3518–3522. [PubMed] [Google Scholar]
- 20.Compton T. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J Virol. 1993;67:3644–3648. doi: 10.1128/jvi.67.6.3644-3648.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Compton T. Towards a definition of the HCMV entry pathway. Scand J Infect Dis. 1995;99:30–32. [PubMed] [Google Scholar]
- 22.Compton T, Nepomuceno R R, Nowlin D M. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology. 1992;191:387–395. doi: 10.1016/0042-6822(92)90200-9. [DOI] [PubMed] [Google Scholar]
- 23.Compton T, Nowlin D M, Cooper N R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology. 1993;193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 24.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 25.David M, Petricoin III E, Benjamin C, Pine R, Weber M J, Larner A C. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 26.Davignon J L, Castanie P, Yorke J A, Gautier N, Clement D, Davrinche C. Anti-human cytomegalovirus activity of cytokines produced by CD4+ T-cell clones specifically activated by IE1 peptides in vitro. J Virol. 1996;70:2162–2169. doi: 10.1128/jvi.70.4.2162-2169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domanski P, Colamonici O R. The type-I interferon receptor. The long and short of it. Cytokine Growth Factor Rev. 1996;7:143–151. doi: 10.1016/1359-6101(96)00017-2. [DOI] [PubMed] [Google Scholar]
- 28.Farrar G H, Greenaway P J. Characterization of glycoprotein complexes present in human cytomegalovirus envelopes. J Gen Virol. 1986;67:1469–1473. doi: 10.1099/0022-1317-67-7-1469. [DOI] [PubMed] [Google Scholar]
- 29.Farrar M A, Schreiber R D. The molecular cell biology of interferon-gamma and its receptor. Annu Rev Immunol. 1993;11:571–611. doi: 10.1146/annurev.iy.11.040193.003035. [DOI] [PubMed] [Google Scholar]
- 30.Fu X-Y, Schindler C, Improta T, Aebersold R, Darnell J E. The proteins of ISGF3, the interferon a-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gazit A, Yaish P, Gilon C, Levitzki A. Tyrphostins. I. Synthesis and biological activity of protein tyrosine kinase inhibitors. J Med Chem. 1989;32:2344–2352. doi: 10.1021/jm00130a020. [DOI] [PubMed] [Google Scholar]
- 32.Gilmore K C, Reich N C. Signal transduction and activation of gene transcription by interferons. Gene Exp. 1995;5:1–18. [PMC free article] [PubMed] [Google Scholar]
- 33.Gopalakrishna R, Chen Z H, Gundimeda U. Irreversible oxidative inactivation of protein kinase C by photosensitive inhibitor calphostin C. FEBS Lett. 1992;314:149–154. doi: 10.1016/0014-5793(92)80962-g. [DOI] [PubMed] [Google Scholar]
- 34.Greenberg M E, Hermanowski A L, Ziff E B. Effect of protein synthesis inhibitors on growth factor activation of c-fos, c-myc, and actin gene transcription. Mol Cell Biol. 1986;6:1050–1057. doi: 10.1128/mcb.6.4.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gretch D R, Gehrz R C, Stinski M F. Characterization of a human cytomegalovirus glycoprotein complex (gcI) J Gen Virol. 1988;69:1205–1215. doi: 10.1099/0022-1317-69-6-1205. [DOI] [PubMed] [Google Scholar]
- 36.Harel N Y, Alwine J C. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J Virol. 1998;72:5481–5492. doi: 10.1128/jvi.72.7.5481-5492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibanez C E, Schrier R, Ghazal P, Wiley C, Nelson J A. Human cytomegalovirus productively infects primary differentiated macrophages. J Virol. 1991;65:6581–6588. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kari B, Gehrz R. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J Virol. 1992;66:1761–1764. doi: 10.1128/jvi.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi E, Nakano H, Morimoto M, Tamaoki T. Calphostin C (UCN-1028C) a novel microbial compound, is a highly potent and specific inhibitor of protein kinase C. Biochem Biophys Res Commun. 1989;159:548–553. doi: 10.1016/0006-291x(89)90028-4. [DOI] [PubMed] [Google Scholar]
- 40.Kowalik T F, Wing B, Haskill J S, Azizkhan J C, Baldwin A S, Huang E. Multiple mechanisms are implicated in the regulation of NF-κB activity during human cytomegalovirus infection. Proc Natl Acad Sci USA. 1993;90:1107–1111. doi: 10.1073/pnas.90.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lathey J L, Spector S A. Unrestricted replication of human cytomegalovirus in hydrocortisone-treated macrophages. J Virol. 1991;65:6371–6375. doi: 10.1128/jvi.65.11.6371-6375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levy D E, Kessler D S, Pine R, Darnell J E., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 43.Levy D E, Lew D J, Kessler D S, Darnell J E. Synergistic interaction between interferon-α and interferon-γ through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Stinski M F. Human cytomegalovirus contains a tegument protein that enhances transcription from promoters with upstream ATF and AP-1 cis-acting elements. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luxembourg A T, Cooper N R. Modulation of signaling via the B cell antigen receptor by CD21, the receptor for C3dg and EBV. J Immunol. 1994;153:4448–4457. [PubMed] [Google Scholar]
- 46.Meier J L, Stinski M F. Regulation of human cytomegalovirus immediate-early gene expression. Intervirology. 1996;39:331–42. doi: 10.1159/000150504. [DOI] [PubMed] [Google Scholar]
- 47.Miller D M, Rahill B M, Boss J M, Lairmore M D, Durbin J E, Waldman J W, Sedmak D D. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Minton E J, Tysoe C, Sinclair J H, Sissons J G. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarro D, Paz P, Tugizov S, Topp K, La Vail J, Pereira L. Glycoprotein B of human cytomegalovirus promotes virion penetration into cells, transmission of infection from cell to cell, and fusion of infected cells. Virology. 1993;197:143–158. doi: 10.1006/viro.1993.1575. [DOI] [PubMed] [Google Scholar]
- 50.Navarro L, Mowen K, Rodems S, Weaver B, Reich N, Spector D, David M. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol Cell Biol. 1998;18:3796–3802. doi: 10.1128/mcb.18.7.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nemerow G R, Cooper N R. Early events in the infection of human B lymphocytes by Epstein-Barr virus: the internalization process. Virology. 1984;132:186–198. doi: 10.1016/0042-6822(84)90102-8. [DOI] [PubMed] [Google Scholar]
- 52.Neyts J, Shoeck R, Schols D, Balzarini J, Esko J D, Van S A, De C E. Sulfated polymers inhibit the interaction of human cytomegalovirus with cell surface heparan sulfate. Virology. 1992;189:48–58. doi: 10.1016/0042-6822(92)90680-n. [DOI] [PubMed] [Google Scholar]
- 53.Pereira L. Function of glycoprotein B homologues of the family Herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 54.Rapraeger A C, Krufka A, Olwin B B. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- 55.Ratner L, Wiegand R C, Farrell P J, Sen G C, Lengyel P. Interferon, double stranded RNA and RNA degradation. Fractionation of the endonuclease INT system into two macromolecular components: role of a small molecule in nuclease activation. Biochem Biophys Res Commun. 1978;81:947–954. doi: 10.1016/0006-291x(78)91443-2. [DOI] [PubMed] [Google Scholar]
- 56.Roberts M L, Luxembourg A T, Cooper N R. Epstein-Barr virus binding to CD21, the virus receptor, activates resting B cells via an intracellular pathway that is linked to B cell infection. J Gen Virol. 1996;77(Pt. 12):3077–3085. doi: 10.1099/0022-1317-77-12-3077. [DOI] [PubMed] [Google Scholar]
- 57.Rossi D, Del Giacco L, Doneda L, Nicolini U, Acaia B, Brioschi D, Larizza L. Expression pattern of c-sis, c-fos, and c-jun in human placenta and embryofetal organs. Gynecol Obstetr Investig. 1996;42:1–7. doi: 10.1159/000291878. [DOI] [PubMed] [Google Scholar]
- 58.Sambucetti L C, Cherrington J M, Wilkenson G W G, Mocarski E S. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segar R, Krebs E G. The MAPK signaling cascade. FASEB J. 1995;9:726–735. [PubMed] [Google Scholar]
- 60.Sinclair J H, Baillie J, Bryant L A, Taylor-Wiedeman J A, Sissons J G. Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J Gen Virol. 1992;73(Pt. 2):433–435. doi: 10.1099/0022-1317-73-2-433. [DOI] [PubMed] [Google Scholar]
- 61.Soderberg-Naucler C, Fish K N, Nelson J A. Interferon-gamma and tumor necrosis factor-alpha specifically induce formation of cytomegalovirus-permissive monocyte-derived macrophages that are refractory to the antiviral activity of these cytokines. J Clin Investig. 1997;100:3154–3163. doi: 10.1172/JCI119871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 63.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 64.Stein B S, Gowda S D, Lifson J D, Penhallow R C, Bensch K G, Engleman E G. pH-independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 65.Stenberg R M, Kerry J A. Cytomegalovirus genes: their structure and function. Scand J Infect Dis Suppl. 1995;99:3–6. [PubMed] [Google Scholar]
- 66.Sugano N, Chen W, Roberts M L, Cooper N R. Epstein-Barr virus binding to CD21 activates the initial viral promoter via NF-kappaB induction. J Exp Med. 1997;186:731–737. doi: 10.1084/jem.186.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi I, Kobayashi E, Nakano H, Murakata C, Saitoh H, Suzuki K, Tamaoki T. Potent selective inhibition of 7-O-methyl UCN-01 against protein kinase C. J Pharmacol Exp Ther. 1990;255:1218–1221. [PubMed] [Google Scholar]
- 68.Taylor-Wiedeman J, Sissons J G, Borysiewicz L K, Sinclair J H. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72(Pt. 9):2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 69.Taylor-Wiedeman J, Sissons P, Sinclair J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Torigoe S, Campbell D E, Starr S E. Cytokines released by human peripheral blood mononuclear cells inhibit the production of early and late cytomegalovirus proteins. Microbiol Immunol. 1997;41:403–413. doi: 10.1111/j.1348-0421.1997.tb01871.x. [DOI] [PubMed] [Google Scholar]
- 71.Tugizov S, Navarro D, Paz P, Wang Y, Qadri I, Pereira L. Function of human cytomegalovirus glycoprotein B: syncytium formation in cells constitutively expressing gB is blocked by virus-neutralizing antibodies. Virology. 1994;201:263–276. doi: 10.1006/viro.1994.1291. [DOI] [PubMed] [Google Scholar]
- 72.Valy-Nagy T, Bandi Z, Boldogh I, Albrecht T. Hydrolysis of inositol lipids: an early signal of human cytomegalovirus infection. Arch Virol. 1988;101:199–207. doi: 10.1007/BF01311001. [DOI] [PubMed] [Google Scholar]
- 73.Weissman D, Rabin R L, Arthos J, Rubbert A, Dybul M, Swofford R, Venkatesan S, Farber J M, Fauci A S. Macrophage-tropic HIV and SIV envelope proteins induce a signal through the CCR5 chemokine receptor. Nature. 1997;389:981–985. doi: 10.1038/40173. [DOI] [PubMed] [Google Scholar]
- 74.Yamamoto N, Shimokata K, Maeno K, Nishiyama Y. Effect of recombinant human interferon-gamma against human cytomegalovirus. Arch Virol. 1987;94:323–329. doi: 10.1007/BF01310726. [DOI] [PubMed] [Google Scholar]
- 75.Yurochko A D, Hwang E, Rasmussen L, Keay S, Pereira L, Huang E. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J Virol. 1997;71:5051–5059. doi: 10.1128/jvi.71.7.5051-5059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yurochko A D, Kowalik T F, Huong S, Huang E. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J Virol. 1995;69:5391–5400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yurochko A D, Mayo M W, Poma E E, Baldwin A S, Huang E. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J Virol. 1997;71:4638–4648. doi: 10.1128/jvi.71.6.4638-4648.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu H, Cong J, Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: Induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94:13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]